Abstract
Orsay virus is the first identified virus that is capable of naturally infecting Caenorhabditis elegans. Although it is most closely related to nodaviruses, Orsay virus differs from nodaviruses in its genome organization. In particular, the Orsay virus RNA2 segment encodes a putative novel protein of unknown function, termed delta, which is absent from all known nodaviruses. Here we present evidence that Orsay virus utilizes a ribosomal frameshifting strategy to express a novel fusion protein from the viral capsid (alpha) and delta ORFs. Moreover, the fusion protein was detected in purified virus fractions, demonstrating that it is most likely incorporated into Orsay virions. Furthermore, N-terminal sequencing of both the fusion protein and the capsid protein demonstrated that these proteins must be translated from a non-canonical initiation site. While the function of the alpha–delta fusion remains cryptic, these studies provide novel insights into the fundamental properties of this new clade of viruses.
Keywords: Orsay virus, Ribosomal frameshifting, Mass spectrometry, Caenorhabditis elegans
Highlights
-
•
Orsay virus encodes a novel fusion protein by a ribosomal frameshifting mechanism.
-
•
Orsay capsid and fusion protein is translated from a non-canonical initiation site.
-
•
The fusion protein is likely incorporated into Orsay virions.
Introduction
Orsay virus, Santeuil virus and Le Blanc virus are three recently identified viruses that are capable of infecting Caenorhabditis nematodes. Phylogenetic analysis indicates that these three nematode viruses form a distinct clade of viruses that are distantly, but most closely related to viruses in the family Nodaviridae. The discovery of these viruses and their ability to infect the genetically tractable model organisms Caenorhabditis elegans and Caenorhabditis briggsae provides a new opportunity for the study of host–virus interactions. While the ability of these viruses to infect nematodes has been established, many fundamental aspects of the molecular virology of these viruses, including the complement of proteins expressed by these viruses, remain unknown.
Nodaviruses are non-enveloped icosahedral viruses with a bipartite positive sense RNA genome (Ball and Johnson, 1999, Odegard et al., 2010, Venter and Schneemann, 2008). The nodavirus genome consists of two 5' capped and 3' non-polyadenylated single stranded positive sense RNA segments. The RNA1 segment, typically ~3 kb to 3.4 kb, encodes an RNA dependent RNA polymerase (RdRP) that is essential for viral transcription and replication. In addition, there is a subgenomic transcript derived from the RNA1 segment, which encodes both the B1 protein within the same frame and the B2 protein in the +1 frame relative to the polymerase. The nodavirus B2 protein is a functional viral suppressor of RNA silencing (Chao et al., 2005), while the function of the B1 protein remains unknown. The RNA2, ~1.4 kb, encodes the viral capsid protein (alpha) which is subsequently cleaved into beta and gamma peptides during virus maturation in at least a subset of the nodaviruses. The most widely studied nodavirus is Flock house virus (FHV), which can infect a wide range of host cells, including mammalian, insect, plant, and yeast cells (Johnson and Ball, 1997, Lu et al., 2005, Price et al., 1996, Selling et al., 1990, Venter and Schneemann, 2008).
Orsay, Le Blanc and Santeuil viruses differ from nodaviruses in several distinct ways. For example, there is currently no experimental evidence that any of these three viruses generates a subgenomic transcript from the RNA1 genome segment, and thus they appear to lack B1/B2 proteins. Most strikingly, these three viruses have much larger RNA2 segments than the nodaviruses. In all three viruses, a novel ORF delta whose theoretical product has no homology to any known protein in GenBank occupies the 3' half of the RNA2 segment. Currently, the function of delta is unknown.
A first step towards evaluating the potential function of the delta ORF would be to establish whether it is in fact expressed in vivo. In the absence of a subgenomic transcript, non-5'-proximal ORFs in RNA viruses are commonly expressed via ribosomal frameshifting, a mechanism whereby specific signals – usually in the mRNA – stimulate a proportion of ribosomes to change reading frame, most commonly in the −1 direction, and continue translating an alternative ORF to produce a ‘transframe’ fusion protein (Firth and Brierley, 2012). Ribosomal −1 frameshifting was first identified in Rous sarcoma virus (Jacks et al., 1988), and later found to be utilized by many viruses – including retroviruses, astroviruses and coronaviruses – for polymerase expression (Brierley, 1995). Moreover, a few viruses use ribosomal frameshifting to append an extension domain onto a proportion of their capsid proteins (van der Wilk et al., 1997). The eukaryotic −1 ribosomal frameshift site typically consists of a ‘slippery’ heptanucleotide sequence fitting the consensus motif X_XXY_YYZ, where XXX normally represents any three identical nucleotides (though certain exceptions have been found, such as GUU and GGA); YYY represents AAA or UUU; Z represents A, C or U; and underscores separate codons in the original reading frame. This consensus motif is generally followed by a stimulatory element comprising a stable RNA secondary structure such as a pseudo-knot or stem-loop, beginning 5–9 nt downstream of the shift site.
In this study, we determined that the Orsay virus delta ORF is primarily expressed as a fusion protein with the alpha ORF via a ribosomal frameshifting mechanism. This fusion protein co-purified with the major capsid protein and viral RNA after gradient ultracentrifugation, suggesting that it is incorporated into Orsay virus particles. In addition, we defined multiple physical properties of Orsay virus such as the virion density and diameter. Collectively these data provide new insights into the fundamental biology of this clade of viruses and provide a basic foundation for future exploitation of the virus-nematode infection system.
Results
Complete genome of Orsay virus
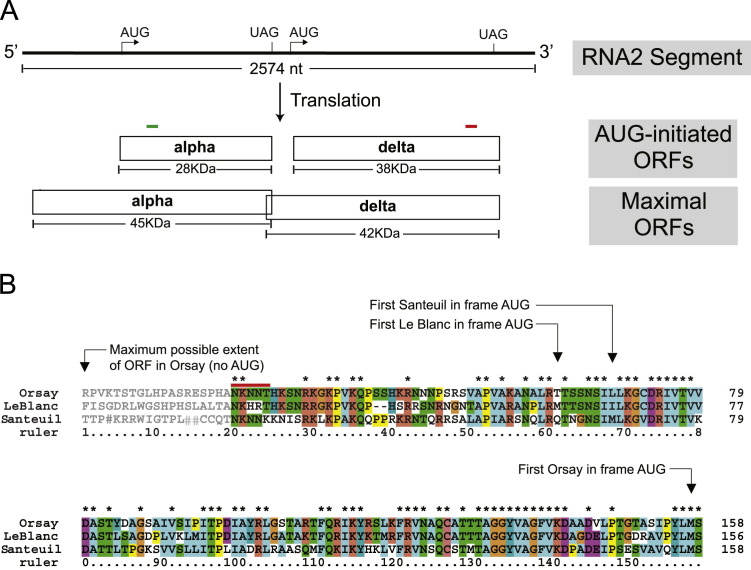
In the initial publication describing the discovery of Orsay and Santeuil viruses, we reported partial sequences of the RNA1 and RNA2 segments of Orsay virus (GenBank: HM030970.1, HM030971.1) (Felix et al., 2011). Here, we sequenced the complete genome of Orsay virus using a combination of 5' and 3' RACE to define the segment termini. The complete RNA1 segment was 3421 nt while the RNA2 segment was 2574 nt. Standard gene prediction using AUG as a start codon suggested that the RNA2 encodes a putative capsid protein (nt 500 to 1261; ~28 kDa) and the previously described delta ORF (nt 1345–2385;~38 kDa). Curiously, the Orsay capsid annotation yielded a protein much smaller than that predicted for Santeuil and Le Blanc viruses, and resulted in a very long predicted 5' UTR. To confirm that there were no sequencing errors leading to misannotation, we repeated 10 independent RT-PCR reactions of the Orsay RNA2 5' end, all of which were consistent with the initial sequence. While there were no other AUG codons in frame with the capsid ORF present in the 5' end of RNA2, the capsid ORF could be extended up to 156 codons in the 5' direction if initiation were to occur at a non-AUG codon (Firth and Brierley, 2012). Such a non-AUG initiated ORF could encode a protein with a molecular size of up to ~45 kDa. Moreover, alignment of the N-terminally extended Orsay virus capsid sequence with the corresponding sequences of Le Blanc and Santeuil viruses demonstrated extensive sequence conservation in the region 5' to the first in-frame AUG codon in all three viruses ( Fig. 1B), raising the possibility that all three viruses may utilize non-AUG initiation to express their capsid proteins.
Fig. 1.
Annotation of Orsay virus RNA2 and capsid sequence alignment for three nematode viruses. (A) A schematic model of Orsay virus RNA2 organization and capsid translation initiation. Arrows indicate the first in-frame AUG codon in each ORF. Vertical lines indicate the stop codons. The largest possible ORF that could be generated by a non-AUG start in RNA2 is shown. Green and red bars indicate where the peptide sequences were used to generate alpha and delta peptide derived antibodies, respectively. (B) Alignment of the 5'-proximal regions of RNA2 of all three nematode viruses conceptually translated in the capsid reading frame (‘♯’ in sequence represents a stop codon). Arrows indicate the first methionine in each sequence. The red bar shows the Orsay virus capsid N-terminal sequence confirmed by sequencing. Sequence in grey shows the conceptual N-terminal translation back to the first in-frame stop codon for Orsay virus. For LeBlanc virus and Santeuil virus, ORF products were also conceptually translated without requiring AUG for initiation. For clarity, the LeBlanc reading frame was truncated to match the length of the maximal Orsay ORF.
Western blotting of Orsay virus infected C. elegans
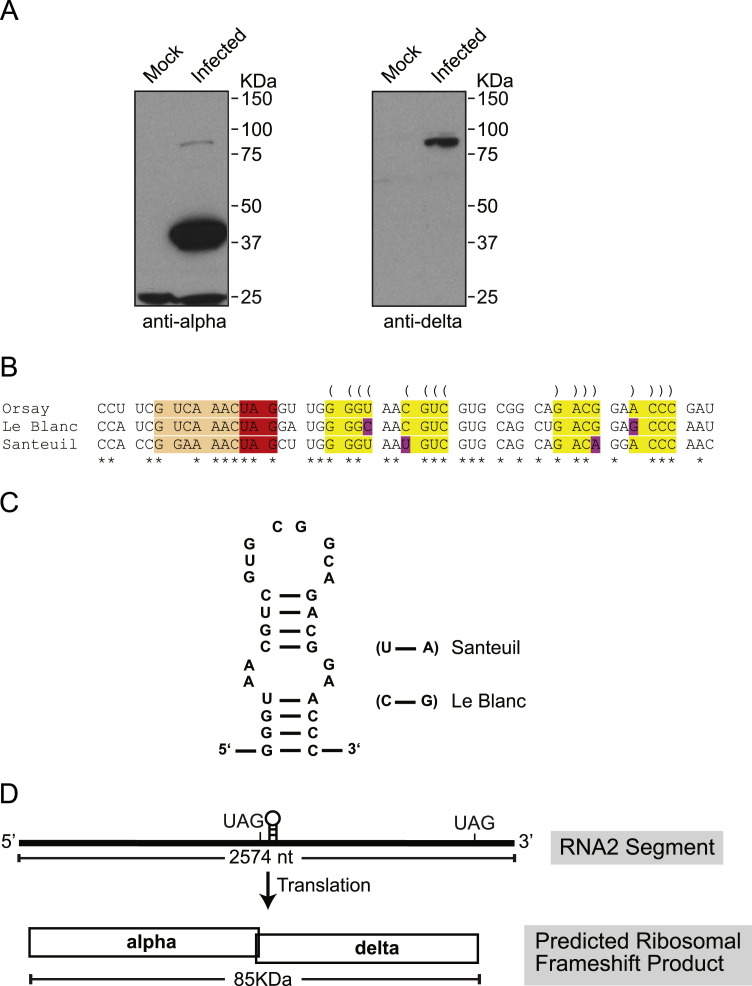
In order to detect the viral proteins translated from the RNA2 segment, we generated polyclonal antibodies raised against synthesized peptides from the capsid and the putative delta protein (Fig. 1A). Both capsid and delta peptide derived antibodies specifically bound proteins in Orsay virus infected C. elegans as compared to mock infected C. elegans ( Fig. 2A). The anti-capsid antibody detected a band with a molecular size higher than 37 kDa compared to the protein molecular size marker. This band migrated more slowly than expected for the capsid protein based on its theoretical size, ~28 kDa, if translation initiated at the first in-frame AUG codon, but was potentially consistent with translation initiation at an upstream (i.e. non-AUG) codon (up to 45 kDa). In addition, a second band migrating between 75 kDa and 100 kDa was detected in the infected C. elegans with capsid peptide derived antibodies, but not in the mock infected C. elegans. With the anti-delta derived antibody, no protein band that matched the theoretical translational size of the delta ORF (~38 kDa if AUG-initiated) was detected. Instead, a single band migrating between 75 kDa and 100 kDa was clearly detected in the infected C. elegans lysates. These observations raised the possibility that the two antibodies may be detecting a common protein derived from the RNA2 segment. One plausible mechanism that could generate such a protein is ribosomal frameshifting. Analysis of the Orsay virus RNA2 segment demonstrated the presence of a putative heptameric slippery sequence, G_UCA_AAC (underscores separate alpha-frame codons), immediately before the alpha ORF stop codon (Fig. 2B). Although G_UCA_AAC is not normally regarded as a canonical shift site for −1 ribosomal frameshifting, it has in fact been tested in reporter constructs and found to be an efficient shift site (Brierley et al., 1992). Further, we identified by RNA secondary structure modeling with Mfold a putative RNA stem loop structure beginning eight nucleotides downstream of the slippery sequence, consistent with a canonical eukaryotic −1 frameshift cassette (Fig. 2C). Nucleotide sequence alignment of all three nematode viruses demonstrated conservation of the slippery sequence and the stem loop structure (Fig. 2B). Notably, we found that Le Blanc virus has the same slippery sequence as that of Orsay virus, while Santeuil virus has a different shift-prone sequence G_GAA_AAC (Brierley et al., 1992). Although the predicted RNA stem-loop structure has a 2-nt internal symmetric bulge (Fig. 2C), its base is stabilized by three consecutive G–C base-pairings, and its functional significance is supported by compensatory substitutions (i.e. paired substitutions that preserve the predicted base-pairings), U–A to C–G (lower part of stem, Le Blanc virus) and C–G to U–A (upper part of stem, Santeuil virus). While in some cases of −1 ribosomal frameshifting the stimulatory element comprises a simple 3'-adjacent RNA stem-loop structure, in many other cases a pseudoknot is involved. In this case, however, analysis with pknotsRG (Reeder et al., 2007) yielded no evidence for a pseudoknot to form in the appropriate location. Collectively, these data suggested that the nematode viruses may utilize a ribosomal frameshifting mechanism to generate a fusion protein of the alpha and delta ORFs with an estimated molecular size of ~85 kDa (Fig. 2D).
Fig. 2.
Western blot of Orsay infected Caenorhabditis elegans suggests possibility of ribosomal frameshifting. (A) Detection of Orsay virus capsid and delta protein in virus infected C. elegans by Western blot using peptide derived antibodies. (B) Alignment of all three nematode virus alpha delta junction regions showed conservation of slippery sequence and stem loop sequence. Spaces separate alpha ORF codons upstream of the shift site (orange) and delta ORF codons downstream of the shift site. The predicted RNA stemloop structure is highlighted with yellow and predicted base-pairings are indicated with parentheses. Paired substitutions that preseve the predicted base-pairings are highlighted with pink. Asterisks indicate conserved positions. (C) The presence of a stem loop structure was modeled by M-fold. Compensatory substitutions are shown for Santeuil and Le Blanc viruses. (D) Predicted model of Orsay virus RNA2 alpha–delta fusion protein translation.
Orsay virus CsCl ultracentrifugation based purification
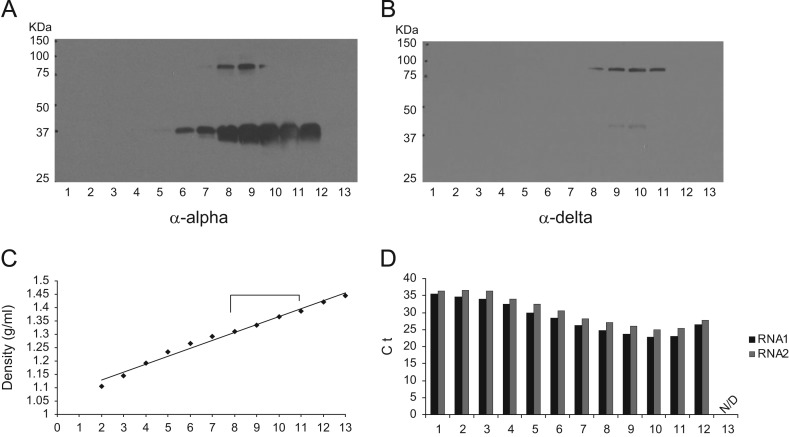
In order to obtain sufficient quantities of purified viral proteins for biochemical protein analyses, we used CsCl density gradient ultracentrifugation to purify Orsay viral particles from large-scale infected C. elegans liquid culture. Western blotting with the anti-capsid antibody yielded a major band slightly above 37 kDa and a minor band between 75 kDa and 100 kDa as seen in the infected lysates ( Fig. 3A). The anti-delta antibody also detected two bands—a major band between 75 kDa and 100 kDa, and a very faint band at~42 kDa (Fig. 3B). This lower band was not detected in the Western blots of infected C. elegans lysates (Fig. 2A). It is possible that this band represents a degradation product of the larger alpha–delta fusion, or possibly some form of the delta ORF alone. Its abundance is much lower than that of the fusion band, which may explain why it is not detected in the infected C. elegans lysates (Fig. 2A). For both antibodies, the peak fractions of Orsay virus protein (fractions 8–11) were found in a density range of 1.311 g/ml to 1.387 g/ml (Fig. 3C). For comparison, FHV virions have an average density of 1.351 g/ml (Scotti et al., 1983).
Fig. 3.
CsCl density gradient characterization of Orsay virus. Orsay virus from large scale C. elegans liquid culture was resolved by 20–40% CsCl density gradient. Fractions were numbered 1 to 13 from the top to the bottom of the gradient. (A) Western blot using capsid peptide derived antibody. (B) Western blot using delta peptide derived antibodies. (C) Orsay virus fraction densities. (D) Real time qRT-PCR detection of Orsay virus RNA1 and RNA2. The Y-axis shows the Ct (cycle threshold) value of each fraction. N/D: Ct values were not detected.
To evaluate whether the peak protein fractions also contained Orsay virus RNA, we performed real time qRT-PCR for both the RNA1 and RNA2 segments. The highest level of both viral RNAs was detected in fractions 8–11, the same fractions as the peak protein levels (Fig. 3D).
N-terminal sequencing of Orsay capsid protein
Orsay virus CsCl density gradient fractions 8 and 9 were pooled and further purified by a second CsCl density gradient. The capsid band from fraction 9 of the second gradient was used for Edman degradation sequencing which demonstrated that the N-terminal sequence was NKNNT ( Table 1). The asparagine (Asn) corresponds to nucleotides 89–91, an AAC codon, which is not traditionally thought to be a suitable translation initiation site. In the complete RNA2 segment, there was no AUG codon 5' to the codon corresponding to the N-terminal Asn amino acid. The predicted size of the alpha-ORF-encoded protein beginning with this Asn is 42.9 kDa, consistent with its migration in SDS-PAGE. We were unable to obtain sufficient quantities of the alpha–delta fusion from the CsCl gradient purification to perform N-terminal sequencing.
Table 1.
Orsay virus capsid protein N-terminal sequence.
| Cycle | Residue | Amino acid |
|---|---|---|
| 1 | Blank | − |
| 2 | Standard | |
| 3 | 1 | N |
| 4 | 2 | K |
| 5 | 3 | N |
| 6 | 4 | D/(N) |
| 7 | 5 | T |
Evidence of Orsay virus ribosomal frameshifting by protein mass spectrometry
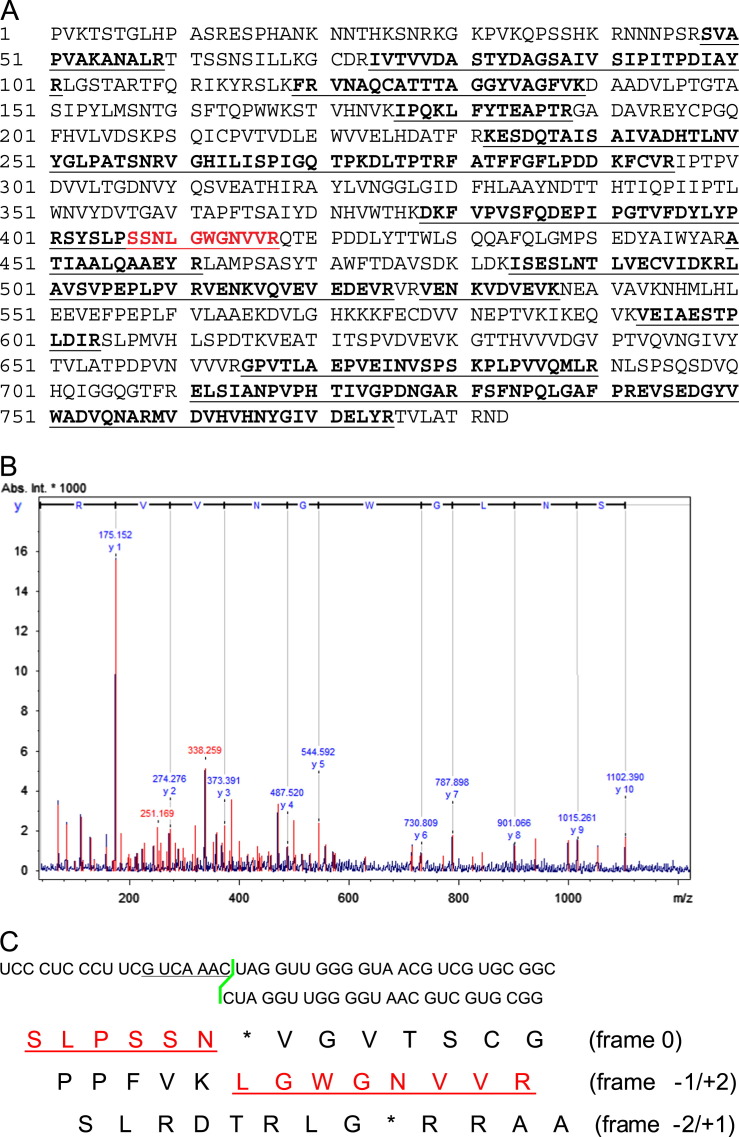
To investigate whether Orsay virus uses ribosomal frameshifting, we analyzed the high molecular weight band that migrates between 75 kDa and 100 kDa from fraction 9 of the CsCl gradient by mass spectrometry. In the mass spectrometry peptide mass fingerprinting (PMF) analysis, 44% of the sequence of the theoretical alpha–delta fusion protein was detected by MASCOT searching against the custom Orsay virus genome derived protein database. Critically, we detected the peptide SYSLPSSNLGWGNVVR that could only be generated through ribosomal frameshifting on the G_UCA_AAC motif as it included amino acids encoded by the 3' end of the alpha ORF linked to amino acids encoded by the sequence downstream of the alpha ORF but in the delta ORF reading frame (i.e. the −1 reading frame relative to the alpha ORF) ( Fig. 4A and C). In the MALDI–MS/MS peptide sequencing analysis, seven of the 15 sequenced peptides mapped to the theoretical alpha–delta fusion protein and six of the seven peptides showed a high ion score (Table 3). The MS/MS peptide sequencing spectrum definitively identified the peptide SNLGWGNVVR (Fig. 4B right to left).
Fig. 4.
Mass spectrometry analysis of putative fusion protein. (A) Peptides detected by PMF analysis are highlighted in bold with underline. The peptide sequence spanning the frameshift site is indicated in red. (B) Fragmented peptide ions detected from the peptide spanning the frameshift site in MS/MS analysis. Each charged peptide fragment m/z is shown on the peak. (C) Conceptual translation of the nucleotide sequence spannning the frameshift region in all three reading frames. The red underlined sequence indicates −1 frameshifting product.
Table 3.
Orsay virus alpha–delta fusion protein peptides analyzed by mass spectrometry.
| Peptide no | Start–end | Observed | Mr(expt) | Mr(calc) | Delta | Miss | Sequence | Ions score |
|---|---|---|---|---|---|---|---|---|
| 1 | 180–188 | 1097.59 | 1096.58 | 1096.56 | 25 | 0 | K.LFYTEAPTR.G | 44 |
| 2 | 260–273 | 1459.87 | 1458.86 | 1458.86 | 2 | 0 | R.VGHILISPIGQTPK.D | 70 |
| 3 | 402–417 | 1735.9 | 1734.9 | 1734.87 | 16 | 0 | R.SYSLPSSNLGWGNVVR.Q | 41 |
| 4 | 500–515 | 1747.04 | 1746.03 | 1746 | 15 | 1 | R.LAVSVPEPLPVRVENK.V | 7 |
| 5 | 711–730 | 2057.11 | 2056.11 | 2056.07 | 18 | 0 | R.ELSIANPVPHTIVGPDNGAR.F | 54 |
| 6 | 731–742 | 1380.73 | 1379.73 | 1379.7 | 20 | 0 | R.FSFNPQLGAFPR.E | 57 |
| 7 | 743–758 | 1837.87 | 1836.86 | 1836.83 | 17 | 0 | R.EVSEDGYVWADVQNAR.M | 56 |
Orsay virus alpha–delta fusion protein is present in highly purified virions
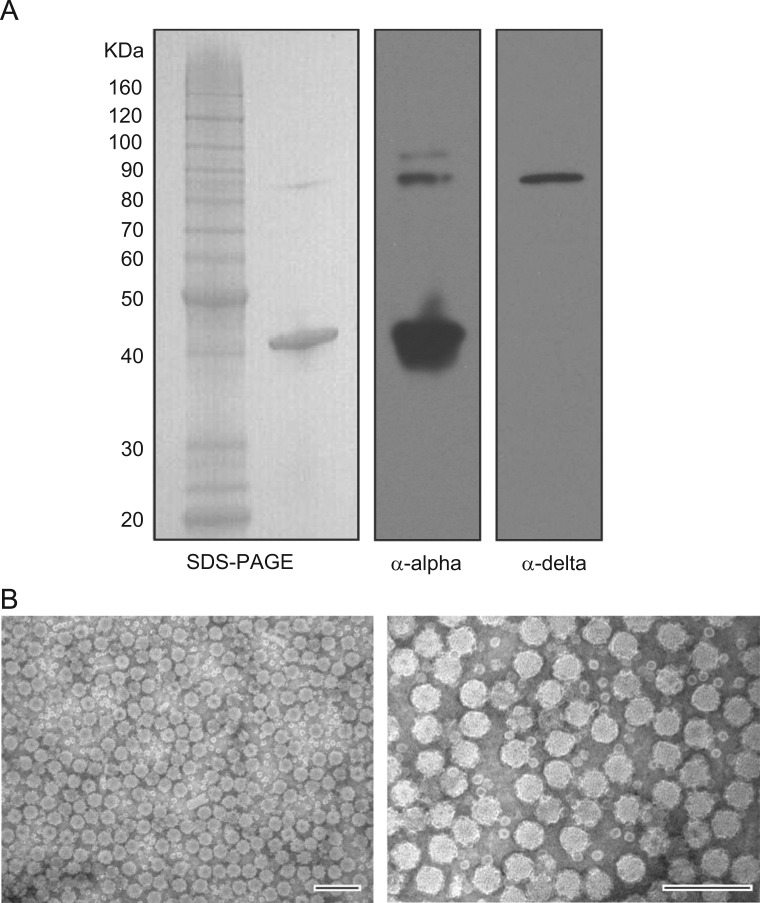
We used an orthogonal purification strategy to confirm that the presence of the alpha–delta fusion in the CsCl gradient fractions with peak capsid protein was not due to contamination or incidental co-purification. Three serial iodixanol gradients were used to purify the Orsay virions to >95% purity based on SDS-PAGE coomassie blue staining ( Fig. 5A). We could clearly detect the alpha–delta fusion protein band in the highly purified fraction by both Western blot and coomassie staining (Fig 5A). This observation, together with the CsCl density gradient results, demonstrated that the alpha–delta fusion protein is likely assembled into the Orsay virions. There was also an additional faint protein band above the fusion protein band recognized by the anti-capsid antibody, which might reflect post translational modification of the fusion protein. This band was not observed with the anti-capsid antibody in either the infected C. elegans lysates or in the CsCl gradient fractions nor was it detected by the anti-delta antibody in this preparation. Because of the high purity of the iodixanol purified virions, greater quantities of virus were loaded onto the analytical gels than in the previous experiments, which may explain why it was not detected in the other experiments. From this purification, we were able to obtain sufficient quantities of the alpha–delta fusion protein to perform N-terminal sequencing, which yielded the identical N-terminus, NKNNT, as with the capsid protein ( Table 2).
Fig. 5.
Iodixanol gradient purification of Orsay virus and electron microscopy study. (A). Coomassie blue staining and Western blot analysis of iodixanol gradient purified Orsay virus. (B). Electron microscopy of iodixanol gradient purified Orsay virions. The structure of purified Orsay virus is shown by negative staining electron microscopy. The scale bar represents 100 nm.
Table 2.
Orsay virus alpha–delta fusion protein N-terminal sequence.
| Cycle | Residue | Amino acid |
|---|---|---|
| 1 | Blank | − |
| 2 | Standard | − |
| 3 | 1 | N/M/(G/R) |
| 4 | 2 | K |
| 5 | 3 | N |
| 6 | 4 | (D/N/L) |
| 7 | 5 | T/(E/V) |
EM study of the Orsay virion morphology
We performed negative staining with uranyl acetate on the purified Orsay virions and examined their morphology by electron microscopy. The virus particles have an icosahedral structure, with an average diameter of 34.2 nm (range from 29.4 nm to 39.9 nm; n=10 counted). For comparison, FHV virions average 29 nm in diameter Dearing et al. (1980). In contrast to FHV, clear protrusions on the Orsay virion surface were present (Fig. 5B) which are not present in either FHV assembled VLPs or FHV virions (Dearing et al., 1980, Schneemann et al., 1993). We also observed some much smaller particles which could be capsomeres from disrupted Orsay virions.
Discussion
Here we describe for the first time, multiple fundamental properties of Orsay virus. These include the density of the virus particles in CsCl, the first negative stain EM images, the size of the viral particles, evidence for a ribosomal frameshifting mechanism that leads to expression of a novel fusion protein of currently unknown function, and evidence for non-canonical translation initiation for the viral capsid protein.
One of the key differences between the three recently discovered nematode infecting viruses, Orsay, Le Blanc and Santeuil, and nodaviruses is the larger RNA2 segment size. In the three nematode viruses, they range from 2.5 kb to 3.0 kb while the nodaviruses have only ~1.4 kb RNA2 segments. In the original annotation of the RNA2 segment of the nematode infecting viruses, we identified an ORF encoding a product orthologous to the capsid proteins of nodaviruses in the 5' half of the RNA2 segment. In the 3' half, we identified an ORF of unknown function, the delta ORF. Here we present extensive data demonstrating that the delta ORF is primarily expressed as a fusion with the alpha capsid via ribosomal frameshifting. The alpha–delta fusion was unambiguously identified by mass spectrometry of purified protein through the detection of peptide fragments throughout the fusion protein and critically, the peptide encoded at the ribosomal frameshift site. In two distinct biochemical purification strategies, this fusion protein co-sedimented with the peak virion fractions, demonstrating that it is likely part of the viral particle. Thus, unlike the structures of the FHV virions, which are composed of the alpha capsid protein (cleaved into beta and gamma fragments) (Gallagher and Rueckert, 1988) alone, Orsay virus virions appear to contain a mixture of the traditional alpha capsid protein and the alpha–delta fusion protein. The function of the alpha–delta fusion protein remains unknown. It is possible that it plays a role in receptor binding. By analogy, a C-terminal domain appended to a small proportion of luteovirus capsid proteins via stop codon read-through plays a role in aphid transmission (Brault et al., 1995). The delta domain may also have additional non-structural roles, as have been described for many viral capsid proteins (Ni and Kao, 2013). One possibility is that this protein may play a role in antagonizing antiviral pathways in nematodes.
At this stage we have little evidence that the predicted delta ORF is expressed in vivo outside of the context of the fusion protein. Only when large quantities of highly purified virions were analyzed did we detect by Western blot a band that could be consistent with a delta ORF protein alone. However the observed band is more likely to be simply a degradation product of the alpha–delta fusion protein as there is no clear mechanism by which delta could be independently expressed. Furthermore, this band was not detected in the final highly purified iodixonal gradient fractions. A recent study that attempted to evaluate whether the delta ORF alone, when cloned into a heterologous expression vector, can act as a suppressor of RNA silencing, yielded a negative result (Guo and Lu, 2013). However, in light of our data demonstrating that the delta ORF is primarily expressed as a larger fusion with the capsid protein, the potential of the alpha–delta fusion to suppress RNA silencing should be evaluated.
During the course of these studies, we noted that the standard AUG-initiation gene prediction models for Orsay virus yielded a capsid protein that is much smaller than expected compared to Le Blanc and Santeuil viruses, as well as when compared to nodaviruses. To address this issue, we experimentally defined the N-terminus of both the capsid protein and the alpha–delta fusion protein of Orsay virus in the context of purified virions. By N-terminal sequencing, NKNNT were defined as the N-terminal amino acids for both proteins. Based on sequence homology, it appears likely that this region is translated as part of the capsid protein in all three nematode viruses (Fig. 1B). Strikingly, such a protein cannot be derived from an AUG translation initiation site. A number of near-cognate non-AUG codons (e.g. CUG) can support some level of initiation, particularly if in a strong initiation context (Firth and Brierley, 2012). One possibility is that an upstream UUG triplet at nucleotide 56–58 is used for initiation in Orsay virus, and that a subsequent proteolytic cleavage generates the NKNNT termini found in these proteins in the virion. In the alphanodaviruses, the alpha protein is cleaved into beta and gamma proteins, providing an explicit precedent for such proteolytic cleavage. However the potential for upstream non-AUG initiation at one of the standard near-cognate non-AUG initiation codons (CUG, GUG, UUG, ACG, AUA, AUC, AUU) is not present in Santeuil virus due to an in-frame stop codon just five codons upstream of the sequence encoding NKNNK. Moreover, the UUG codon in Orsay virus lacks a strong initiation context (ggc_UUG_c) (Nakagawa et al., 2008). It is possible therefore that Orsay virus, and by homology Santeuil and Le Blanc viruses, can initiate capsid translation directly at the site of the NKNNT motif via some unknown mechanism, perhaps akin to the initiator-Met-tRNA-independent initiation utilized in the expression of the dicistrovirus capsid polyprotein (Jang and Jan, 2010). The precise mechanism and site of capsid protein initiation will be investigated in future work.
We have demonstrated that the delta protein is translated as a fusion protein with alpha and that the fusion protein is most likely incorporated into virions. Nevertheless many questions still remain such as: (1) How is the fusion protein structurally assembled into the virions? (2) What functional role does the fusion protein play in the context of the virion as well as intracellular? (3) Is the fusion protein enzymatically cleaved to release delta protein as many frameshift viral fusion proteins are during virus replication? We are currently pursuing additional biochemical, structural and genetic experiments to address these questions.
Material and methods
Orsay virus genome sequencing
5' RACE was performed by reverse transcription to synthesize viral cDNA. Next, viral cDNA was purified by a Zymo column to remove primers and free nucleotides and then poly(C) tailed by terminal transferase enzyme (Thermo). PCR was then performed by an abridged anchored primer (AAP) and a viral gene specific primer (GSP). The 3' RACE was performed by first poly(A) tailing of the viral RNA and then amplification by Qiagen one-step RT-PCR with a GSP primer and an oligo dT primer. PCR amplicons were cloned into a pCR4 TOPO TA vector and then sequenced by Sanger chemistry. The complete Orsay RNA1 and RNA2 segment sequences have been deposited in GenBank (HM030970, HM030971).
Genome annotation and sequence analyses
Genome annotation was performed using the ORF Finder (NCBI) [http://www.ncbi.nlm.nih. gov/gorf/gorf.html] to predict in-frame initiation and termination codons. ClustalX (version 1.83) was used for sequence alignments. RNA secondary structure was predicted with Mfold (Zuker, 2003).
Antibodies
Polyclonal antibodies were generated by immunizing rabbits with synthetic peptides (GenScript Inc.). The peptide sequence used for generating the Orsay virus alpha derived antibody was APTRGADAVREYCP. The peptide sequence used for generating the Orsay virus delta derived antibody was QLGAFPREVSEDGY. All antibodies were purified using a protein G affinity column.
Liquid culture of Orsay virus
Orsay virus filtrate was prepared as previously described (Felix et al., 2011). The C. elegans rde-1 strain (WM27) was infected by the Orsay virus filtrate and maintained by serial chunking in a 20 °C incubator. For liquid culture, a 1.5 l overnight culture of Escherichia coli OP50 was pelleted and resuspended in 280 ml S-medium (1 l S basal (5.85 g NaCl, 1 g K2HPO4, 6 g KH2PO4, 1 ml 5 mg/ml cholesterol in ethanol, H2O supplemented to 1 l), 10 ml 1 M potassium citrate pH 6.0, 10 ml Trace metals (6.3 mM EDTA, 2.4 mM FeSO4, 0.9 mM MnCl2, 1.2 mM ZnSO4, 0.1 mM CuSO4 ), 3 ml 1 M CaCl2, 3 ml 1 M MgSO4). Four 10 cm stably infected rde-1 plates grown to a mixed stage were washed off with 5 ml S-medium per plate. The 20 ml of resuspended rde-1 C. elegans were added to the liquid culture to make a final volume of 300 ml in a 2 l flask. The flask was maintained at 160 rpm in a 20 °C shaking incubator for seven days. The growth of the infected C. elegans was monitored by checking a drop of the culture mixture under a stereomicroscope.
CsCl density gradient assay of Orsay virus
The C. elegans liquid culture was chilled on ice for 30 min to allow the animals to settle. The culture supernatant was collected and cleared by centrifugation at 15,000 rpm for 30 min (JLA 16.250, Beckman coulter). Orsay virus in the supernatant was pelleted by centrifugation at 31,000 rpm for 2 h (SW32Ti rotor, Beckman coulter). The pellet was resuspended in 2 ml of CsCl gradient buffer (20 mM Tris–HCl pH7.8, 0.1% 2-Mercaptoethanol) at 4 °C overnight. The 2 ml of the virus suspension was applied to a 20–40% (weight/weight) CsCl linear gradient and centrifuged at 29,100 rpm (SW41Ti rotor, Beckman coulter) at 4 °C for 16 h. The 1 ml fractions were collected using a peristaltic fraction maker (Labconco Auto Dens-flow) linked to a Bio-rad fraction collector. The refractive index of each CsCl density fraction was measured and converted to the corresponding density (Fasman, 1976). To concentrate the virus, each fraction was mixed with 4 ml CsCl gradient buffer and centrifuged at 37,000 rpm (SW55Ti, Beckman coulter) for 2 h to pellet the virus. The pellet from each fraction was resuspended in 100 μl of CsCl gradient buffer.
Mass spectrometry and N-terminal sequencing
For both mass spectrometry and N-terminal peptide sequencing, 15 μl of purified virus were mixed with 3 μl of 6X protein loading buffer (350 mM Tris–HCl pH6.8, 30% glycerol, 10% SDS, 600 mM DTT, 0.012% bromophenol blue) and then boiled for 5 min. Protein lysates were loaded onto a 4–15% SDS-PAGE gel (Bio-rad) and separated by electrophoresis at 30 mA for 75 min. The SDS-PAGE gel was stained by fresh coomassie blue R250 for 5 min and then destained for 40 min with buffer containing 40% acetic acid and 10% methanol.
The protein band that corresponded to the suspected viral fusion protein as detected by Western blot with both anti-capsid and anti-delta polyclonal antibodies was excised and sent for mass spectrometry analysis (Alphalyse Inc.). In brief, the protein sample was reduced and alkylated with iodoacetamide and then trypsin digested. The digested peptides were spotted onto an anchorchip target for analysis on a Bruker Autoflex Speed MALDI TOF/TOF instrument. The accurate mass for the peptides was determined in positive reflector mode. Peptide fragmentation analysis was performed on 15 peptides by matrix assisted laser desorption/ionization mass spectrometry/mass spectrometry (MALDI MS/MS). All MS/MS spectra were recorded. The MS and MS/MS data were combined and searched against both the NCBI NR database and a custom Orsay virus genome derived database containing possible frameshifted sequences using MASCOT software (Matrix Science).
For the protein N-terminal sequence analysis, the protein on the gel was electro-transferred to a PVDF membrane (Invitrogen cat. No.LC2007) at 65 V for 90 min. The PVDF membrane was stained by coomassie blue R250 staining and the corresponding band was excised. Protein N-terminal sequencing was performed on an ABI Procise 494 sequencer by the Edman degradation chemistry (Alphalyse Inc.).
Iodixanol purification of Orsay virus
The alternative purification scheme using iodixanol (OptiPrepTM, Axis-Shield PoC AS) consisted of (1) a step gradient purification, (2) a density gradient purification and (3) a self-generated gradient purification. Briefly, 1.2 l of Orsay virus liquid culture supernatant was cleared and collected as described above. The culture supernatant was then treated with 1% triton X-100 (Santa Cruz Biotechnology) for 1 h at room temperature in a shaking incubator at 160 rpm. Viruses were then pelleted through a 10% iodixanol cushion (volume to volume) at 31,000 rpm (SW32Ti rotor, Beckman coulter) for 2 h. The virus pellet was then resuspended in iodixanol gradient buffer (20 mM Tris–HCl pH7.8, 0.1% 2-Mercaptoethanol, 100 mM NaCl, 1 mM EDTA, 0.1% triton X-100) overnight. Gradient 1. The resuspended virus was applied to a step iodixanol gradient. The step gradient consisted of 1 ml, 15% iodixanol in high salt buffer (20 mM Tris–HCl pH7.8, 0.1% 2-Mercaptoethanol, 1 M NaCl, 1 mM EDTA) in which high salt concentration can disrupt protein–protein and/or protein–vesicle charge interactions, and 3 ml each of 25%, 32.5% and 40% of iodixanol in low salt buffer (20 mM Tris–HCl pH7.8, 0.1% 2-Mercaptoethanol, 100 mM NaCl, 1 mM EDTA). Virus was centrifuged at 34,900 rpm for 3 h (SW41Ti rotor, Beckman coulter). The 1 ml fractions were collected and spun through a 30 kDa ultrafiltration column (Amicon Ultra, Millipore) to remove iodixanol. The ultrafiltered virus was then treated again with 1% detergent Triton X-100 for 1 h. Gradient 2. Fractions from gradient 1 that contained Orsay virus were pooled and then applied to a 25–45% iodixanol density gradient (20 mM Tris–HCl pH7.8, 0.1% 2-Mercaptoethanol, 100 mM NaCl, 1 mM EDTA) for purification by centrifugation at 34,200 rpm for 16 h (SW41Ti rotor, Beckman coulter). The gradient was fractionated and virus containing fractions were pooled and ultrafiltered as described above to remove the iodixanol. Gradient 3. Fractions from gradient 2 that contained Orsay virus were pooled and subjected to a self-generated iodixanol gradient purification (Dormond et al., 2010) (40% iodixanol and centrifuged at 61,300 rpm for 1.5 h (NVT 90 rotor, Beckman coulter)). The 0.5 ml fractions were collected by dripping from the bottom of the gradient. Virus containing fractions were pooled and iodixanol was removed by ultrafiltration. The purified virus was finally resuspended in buffer containing 20 mM Tris–HCl pH7.8, 100 mM NaCl, 0.1% 2-mercaptoethanol.
Negative staining and analysis by electron microscopy
A 15 µl droplet of 1.67 mg/ml virus was allowed to absorb onto formvar/carbon-coated copper grids (Ted Pella Inc., Redding, CA) for 10 min. Grids were then washed in distilled H2O and stained with 1% aqueous uranyl acetate (Ted Pella Inc.) for 1 min. Excess liquid was gently wicked off and grids were allowed to air dry. Samples were viewed on a JEOL 1200EX transmission electron microscope (JEOL USA, Peabody, MA) equipped with an AMT 8 megapixel digital camera (Advanced Microscopy Techniques, Woburn, MA).
Western blot
For infected sample preparation, stably infected C. elegans wild isolate JU1580 animals were washed off 10 cm nematode growth medium (NGM) plates. Infected C. elegans were collected by centrifugation at 2000 rpm (SORVALL, swing bucket rotor) for 2 min and then resuspended in 0.5 ml M9. The final C. elegans pellet was homogenized by using silicon beads (1.0 mm dia. ZIRCONIA/SILICA) in a tissue homogenizer (MagNA Lyzer, Roche) at 6000 rpm for 1 min, cooled for 2 min on ice, homogenized another 1 min and then pelleted. Each sample was mixed with 6X SDS sample buffer and then boiled for 5 min. Proteins were resolved on a 4–15% SDS-PAGE gel (Bio-Rad) and transferred to a PVDF membrane. The PVDF membrane was then blocked with 5% skim milk in PBST (PBS with 0.3% Tween 20) for 1 h shaking at room temperature. The membrane was blotted with specific viral peptide derived antibodies at a dilution of 1:2,000 (0.5 μg/ml) for 1 h at room temperature. Horseradish perioxidase (HRP) conjugated goat anti-rabbit IgG secondary antibody (Thermo Scientific) was used at a dilution of 1:10,000 (0.04 μg/ml). Protein bands were developed by using a chemiluminescent substrate (Thermo Scientific).
Real time qRT-PCR
Orsay viral RNAs were extracted from 50 μl of a CsCl gradient pellet by Trizol-LS (Invitrogen) according to the manufacturer's instruction, and the RNA was then ethanol precipitated in the presence of 25 μg of linear acrylamide (Ambion). The viral RNAs were finally resuspended in 20 μl of water and diluted to 1:100. Real time qRT-PCR was performed using Taqman one-step RT-PCR master mix reagents (Applied Biosystem) on a ViiaA7 real time PCR system (Applied Biosystem). Both Orsay virus RNA1 (primers GW314: 5' TGG ATC CAA CGC CGT TAA C and GW 315: 5' CGA TTT GCA GTG GCT TGC T, probe: 5' FAM TGG ACC ACT GAG CAA GTG ATC GAG AAC C) and RNA2 (primers GW303: 5' CCG GCG ACA ATG TGT ACC A and GW304: 5' CCA GCC CTC CGT TGA CAA, probe: 5' FAM CGA GGC CAC CCA TAT CAG GG CCT A) were detected and quantified by the amplification Ct value.
Acknowledgment
The authors thank Dr. Wandy Beatty for her help on EM analysis. DW holds an Investigator in the Pathogenesis of Infectious Diseases Award from the Burroughs -Wellcome Fund. AEF is supported by Wellcome Trust Grant 088789.
References
- Ball L.A., Johnson K.L. Reverse genetics of nodaviruses. Adv. Virus Res. 1999;53:229–244. doi: 10.1016/s0065-3527(08)60350-4. [DOI] [PubMed] [Google Scholar]
- Brault V., van den Heuvel J.F., Verbeek M., Ziegler-Graff V., Reutenauer A., Herrbach E., Garaud J.C., Guilley H., Richards K., Jonard G. Aphid transmission of beet western yellows luteovirus requires the minor capsid read-through protein P74. EMBO J. 1995;14:650–659. doi: 10.1002/j.1460-2075.1995.tb07043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I. Ribosomal frameshifting viral RNAs. J. Gen. Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- Brierley I., Jenner A.J., Inglis S.C. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao J.A., Lee J.H., Chapados B.R., Debler E.W., Schneemann A., Williamson J.R. Dual modes of RNA-silencing suppression by Flock house virus protein B2. Nat. Struct. Mol. Biol. 2005;12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- Dearing S.C., Scotti P.D., Wigley P.J., Dhana S.D. A small RNA virus isolated from the grass grub, Costelytra zealandica (Coleoptera: Scarabaeidae) N.Z. J. Zool. 1980;7:267–269. [Google Scholar]
- Dormond E., Chahal P., Bernier A., Tran R., Perrier M., Kamen A. An efficient process for the purification of helper-dependent adenoviral vector and removal of helper virus by iodixanol ultracentrifugation. J. Virol. Methods. 2010;165:83–89. doi: 10.1016/j.jviromet.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Fasman G.D. Vol. 1. 1976. Physical and chemical data; pp. 419–423. (Handbook of Biochemistry and Molecular Biology). (third ed.) [Google Scholar]
- Felix M.A., Ashe A., Piffaretti J., Wu G., Nuez I., Belicard T., Jiang Y., Zhao G., Franz C.J., Goldstein L.D., Sanroman M., Miska E.A., Wang D. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011;9:e1000586. doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth A.E., Brierley I. Non-canonical translation in RNA viruses. J. Gen. Virol. 2012;93:1385–1409. doi: 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T.M., Rueckert R.R. Assembly-dependent maturation cleavage in provirions of a small icosahedral insect ribovirus. J. Virol. 1988;62:3399–3406. doi: 10.1128/jvi.62.9.3399-3406.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Lu R. Characterization of virus-encoded RNA interference suppressors in Caenorhabditis elegans. J. Virol. 2013;87:5414–5423. doi: 10.1128/JVI.00148-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Madhani H.D., Masiarz F.R., Varmus H.E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang C.J., Jan E. Modular domains of the Dicistroviridae intergenic internal ribosome entry site. RNA. 2010;16:1182–1195. doi: 10.1261/rna.2044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K.L., Ball L.A. Replication of Flock house virus RNAs from primary transcripts made in cells by RNA polymerase II. J. Virol. 1997;71:3323–3327. doi: 10.1128/jvi.71.4.3323-3327.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Maduro M., Li F., Li H.W., Broitman-Maduro G., Li W.X., Ding S.W. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005;436:1040–1043. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Niimura Y., Gojobori T., Tanaka H., Miura K. Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res. 2008;36:861–871. doi: 10.1093/nar/gkm1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni P., Kao C.C. Non-encapsidation activities of the capsid proteins of positive-strand RNA viruses. Virology. 2013;446:123–132. doi: 10.1016/j.virol.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegard A., Banerjee M., Johnson J.E. Flock house virus: a model system for understanding non-enveloped virus entry and membrane penetration. Curr. Top. Microbiol. Immunol. 2010;343:1–22. doi: 10.1007/82_2010_35. [DOI] [PubMed] [Google Scholar]
- Price B.D., Rueckert R.R., Ahlquist P. Complete replication of an animal virus and maintenance of expression vectors derived from it in Saccharomyces cerevisiae. Proc. Nat. Acad. Sci. U.S.A. 1996;93:9465–9470. doi: 10.1073/pnas.93.18.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder J., Steffen P., Giegerich R. pknotsRG: RNA pseudoknot folding including near-optimal structures and sliding windows. Nucleic Acids Res. 2007;35:W320–324. doi: 10.1093/nar/gkm258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneemann A., Dasgupta R., Johnson J.E., Rueckert R.R. Use of recombinant baculoviruses in synthesis of morphologically distinct viruslike particles of Flock house virus, a nodavirus. J. Virol. 1993;67:2756–2763. doi: 10.1128/jvi.67.5.2756-2763.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti P.D., Dearing S., Mossop D.W. Flock house virus: a nodavirus isolated from Costelytra zealandica (White) (Coleoptera: Scarabaeidae) Arch. Virol. 1983;75:181–189. doi: 10.1007/BF01315272. [DOI] [PubMed] [Google Scholar]
- Selling B.H., Allison R.F., Kaesberg P. Genomic RNA of an insect virus directs synthesis of infectious virions in plants. Proc. Nat. Acad. Sci. U.S.A. 1990;87:434–438. doi: 10.1073/pnas.87.1.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wilk F., Dullemans A.M., Verbeek M., Van den Heuvel J.F. Nucleotide sequence and genomic organization of Acyrthosiphon pisum virus. Virology. 1997;238:353–362. doi: 10.1006/viro.1997.8835. [DOI] [PubMed] [Google Scholar]
- Venter P.A., Schneemann A. Recent insights into the biology and biomedical applications of Flock house virus. Cell. Mol. Life Sci. 2008;65:2675–2687. doi: 10.1007/s00018-008-8037-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]