Summary
Background
In Drosophila, male courtship behavior is regulated in large part by the gene fruitless (fru). fru encodes a set of putative transcription factors that promote male sexual behavior by controlling the development of sexually dimorphic neuronal circuitry. Little is known about how Fru proteins function at the level of transcriptional regulation or the role that isoform diversity plays in the formation of a male-specific nervous system.
Results
To characterize the roles of sex-specific Fru isoforms in specifying male behavior, we generated novel isoform-specific mutants and used a genomic approach to identify direct Fru isoform targets during development. We demonstrate that all Fru isoforms directly target genes involved in the development of the nervous system, with individual isoforms exhibiting unique binding specificities. We observe that fru behavioral phenotypes are specified by either a single isoform or a combination of isoforms. Finally, we illustrate the utility of these data for the identification of novel sexually dimorphic genomic enhancers and novel downstream regulators of male sexual behavior.
Conclusions
These findings suggest that Fru isoform diversity facilitates both redundancy and specificity in gene expression, and that the regulation of neuronal developmental genes may be the most ancient and conserved role of fru in the specification of a male-specific nervous system.
Highlights
-
•
Isoform-specific fru mutants reveal both functional redundancy and specificity
-
•
Fru isoform-specific genomic occupancy is characterized in the Drosophila nervous system
-
•
All Fru isoforms directly target neuronal morphogenesis genes
-
•
Isoform-specific motifs are associated with specific Fru isoform occupancy
Neville et al. characterize the roles of sex-specific Fruitless isoforms in specifying male behavior in Drosophila by generating novel isoform-specific mutants, along with using a genomic approach to identify direct Fruitless isoform targets during development.
Introduction
How the development and physiology of neuronal networks shapes innate and species-specific behaviors remains largely unknown. Building these networks requires making the appropriate cell types in the right places at the correct time and wiring these cells together to produce functional circuits. Deciphering the complex gene regulatory networks that act during neuronal development is essential to our understanding of the relationship between genes, the brain, and behavior.
Drosophila male courtship behavior is an excellent paradigm for exploring the genetic, developmental, and neural logic underlying complex behaviors. Much of what is known about the neuronal basis of male behavior has come from studies of the genes fruitless (fru) and doublesex (dsx) [1]. Both dsx and fru lie at the bottom of the sex-determination hierarchy and in males act in concert to specify sex-specific neural circuitry and physiology [2]. fru is a pleiotropic gene with at least two major functions, one that controls male sexual behavior and another that is essential for viability in both sexes (Figure 1A) [1]. All Fru proteins are putative transcription factors containing a common BTB (protein-protein interaction) N-terminal domain and, through alternative splicing, one of four C-terminal zinc-finger (Zn-finger) DNA-binding domains [3–5]. Many BTB-Zn-finger proteins are known to be sequence-specific transcriptional regulators that often play key roles in development [6]. Male-specific Fru proteins (FruM) are produced from fru transcripts whose expression initiates from the most distal promoter (P1), the only fru promoter controlled by the sex-determination hierarchy. FruM proteins contain a 101 aa male-specific sequence and one of three alternative C2H2 Zn-finger domains (A, B, or C) [7]. FruM proteins are first detected in the nervous system at the beginning of metamorphosis, when the CNS is remodeled from the larval to adult form [8] and neural substrates governing sex-specific behaviors are specified [9]. Although FruM is clearly critical for male sexual behavior, the genes it controls to specify male sexual behavior have remained elusive. Recently, FruM has been shown to act in concert with key chromatin regulators to establish male-specific neurite projections and dendritic branching [10]. Through the formation of antagonistic Fru-containing chromatin-regulating complexes, Fru can act to either masculinize or demasculinize specific neuronal subtypes, suggesting that it can act as both a transcriptional activator and repressor.
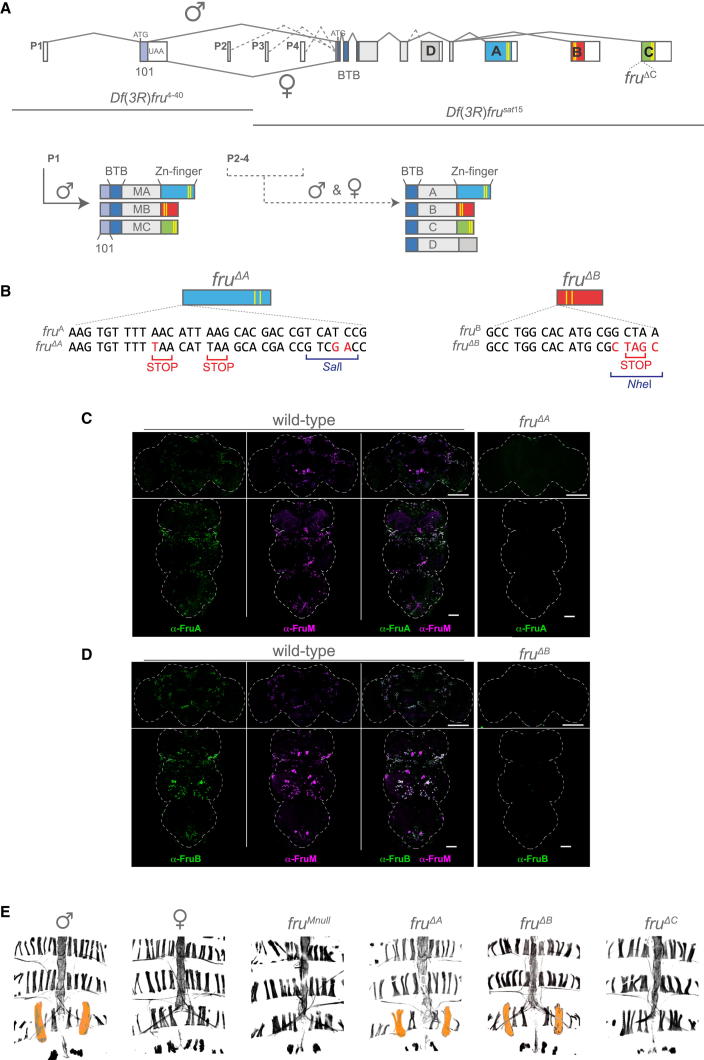
Figure 1.
Generation of fruΔA and fruΔB Isoform-Specific Mutants
(A) Organization of the fru locus. fru contains at least four independent promoters, P1–P4. mRNA transcripts generated from the most distal P1 promoter undergo alternative splicing under the control of the sex-determination hierarchy, thereby producing three protein isoforms in males (FruMA, FruMB, and FruMC). Females produce no functional protein. Protein products from promoters P2–P4 are not sex-specifically regulated and produce four alternative protein products in males and females (FruA, FruB, FruC, and FruD). Alternative 3′ exons are shown in blue (A), red (B), green (C), and gray (D). C2H2 DNA-binding domains are represented by yellow lines. Deficiencies utilized to genetically isolate the function of specific fru promoters are shown: Df(3R)fru4-40 and Df(3R)frusat15. The position of the extant fru C null mutation, fruΔC, is indicated.
(B) Ends-in homologous recombination was used to insert premature stop codons (shown in red) into individual Zn-finger-encoding 3′ exons of the fru locus, resulting in the generation of the fruΔA and fruΔB mutants. The restriction enzyme recognition sequences SalI and NheI were also introduced into the A and B exons, respectively. See also Figures S1A and S1B.
(C and D) Five-day-old adult male CNSs stained with anti-FruM and either anti-FruA or anti-FruB. Brain scale bars represent 100 μm; ventral nerve cord (VNC) scale bars represent 50 μm.
(C) FruMA isoform expression in the adult CNS. The FruMA isoform shows expression in a subset of FruM-expressing neurons (anti-FruA/-FruM merged). FruMA expression is absent in males heterozygous for fruΔA and a deficiency of the fru locus Df(3R)fru4-40 in the brain, and in the VNC of fruΔA homozygous males.
(D) FruMB isoform expression in the adult CNS. The FruMB isoform shows expression in most FruM-expressing neurons (anti-FruB/-FruM merged). FruMB expression is absent in the CNS of males heterozygous for fruΔB and a deficiency of the fru locus Df(3R)fru4-40.
(E) The formation of the muscle of Lawrence (MOL) depends solely on the FruMC isoform. Dorsal abdominal musculature is revealed by phalloidin staining (black) of adults. The MOL is pseudocolored orange for visualization purposes.
In this study, we investigated the role that FruM isoforms play, individually and collectively, in the establishment of a male-specific nervous system. We generated novel isoform-specific mutants and characterized their individual roles in male courtship behavior. We show that although fru isoform-specific mutants impair the male’s ability to perform wild-type levels of courtship behavior, the loss of individual isoforms does not lead to a complete loss of a male’s ability to court. We established genome-wide binding profiles of all male-specific Fru isoforms throughout development in the nervous system using a DNA adenine methyltransferase identification (DamID) approach [11]. FruM interacts with genes in key nervous system developmental pathways, most notably those involved in neuronal morphogenesis. We identified putative Fru-DNA binding motifs and found that genomic regions containing this motif exhibit sexually dimorphic expression in fru neurons. Elucidating the FruM transcriptional network is an essential step forward in understanding the molecular mechanisms that underlie the complex behavioral phenotypes associated with the fru gene.
Results
Generation of fru Isoform-Specific Mutant Flies
To understand the contribution of each male-specific isoform to the formation of the male’s diverse behavioral repertoire, we constructed flies that carry new isoform-specific knockouts of the fru A and B exons. We previously isolated an isoform-specific mutant in the fru C exon, containing a premature stop codon ensuring that no functional Fru C-containing proteins are produced (fruΔC; Figure 1A) [7]. We generated novel isoform-specific null mutants in fru exons A and B by targeted mutagenesis using ends-in homologous recombination (see Figure S1A available online) [12]. Premature stop codons were introduced close to the splice junctions of either fru exon A or B, in addition to restriction enzyme recognition sequences for use as markers of the mutagenesis (Figures 1B and S1B). The resulting mutants fruΔA and fruΔB are thus unable to produce full-length Fru exon A- or B-containing proteins, respectively.
We previously established that the male-specific FruMA and FruMC isoforms have different patterns of expression in the male CNS [7]. FruMC is broadly expressed in most neurons labeled by an antibody against the male-specific 101 aa domain (FruM neurons). In contrast, FruMA expression is more restricted to a subset of FruM neurons. We generated a new exon-specific antibody, anti-FruB, to localize FruB-containing isoforms. The FruMB isoform, like FruMC, shows broad expression in FruM-expressing neurons in the adult male CNS. In parallel, we confirmed that FruMA is expressed in a restricted subset of FruM neurons in the adult CNS. We used these antibodies to demonstrate the absence of FruMA and FruMB expression in our fruΔA and fruΔB isoform-specific mutant flies (Figures 1C and 1D), while confirming that FruM expression is still detected (Figure S1C). FruM isoform expression analysis revealed considerable overlap in cell-specific expression, while also showing clear differential regulation of alternative splicing. The FruMB and FruMC isoforms are largely coexpressed, whereas FruMA is expressed in a subset of the cells expressing the other isoforms.
The formation of the male-specific muscle of Lawrence (MOL) is controlled by the fruM-expressing MIND motor neuron that innervates it [13, 14]. We previously demonstrated that FruMC is necessary and sufficient for the formation of the MOL [7]. By examining the abdominal musculature of all fru isoform mutants, we can now confirm that the formation of the MOL depends solely on the presence of the FruMC isoform (Figures 1E and S2B).
Courtship Analysis of fru Isoform-Specific Mutant Flies
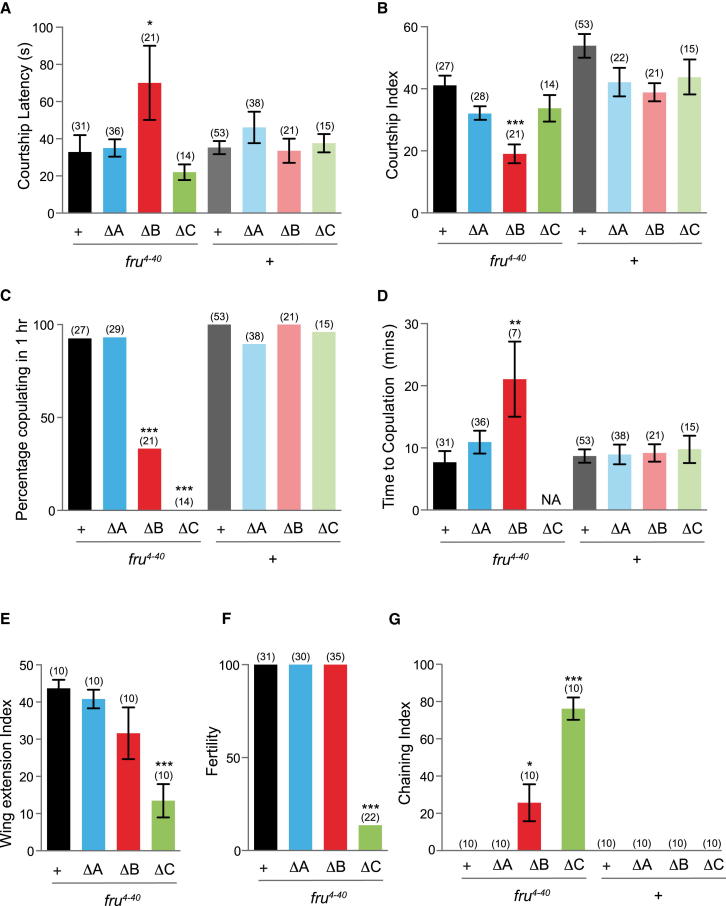
In the absence of all FruM isoforms, males show no measureable levels of courtship toward females [15]. We used our full complement of fru isoform-specific mutants to investigate their individual contribution to male courtship behavior (Figure 2). Our fru mutant alleles disrupt both the male-specific isoforms and those common to both sexes (Figure 1A). We used an extant fru deficiency (Df(3R)fru4-40) that selectively removes FruM expression when combined with each mutant, to assay distinct behavioral phenotypes resulting from the absence of individual FruM isoforms. Males lacking FruMB showed a significant delay in latency to courtship (Figure 2A), and their overall levels of courtship toward females were greatly reduced (Figure 2B). FruMB mutant males also showed low levels of copulation. In addition, males that did mate were significantly delayed in time taken to successfully copulate (Figures 2C and 2D). In contrast, mutant males lacking either FruMA or FruMC did not show any significant delay in courtship latency or overall levels of courtship (Figures 2A and 2B). However, mutants lacking FruMC were markedly unsuccessful at achieving copulation, with none of the males examined managing to copulate within the 1 hr observation period (Figure 2C). Fertility analysis of the mutants showed that only males lacking FruMC showed dramatically low levels of fertility over a one-week observation period (Figure 2F). One of the most conspicuous phenotypes observed in previously characterized fruM mutant males is the formation of male-male courtship chains, observed when mutant males are grouped together. Vigorous chaining behavior was observed among mutants lacking FruMC, whereas mutant males lacking FruMB exhibited chaining behavior at reduced levels (Figure 2G). Strikingly, mutant males lacking FruMA did not exhibit any significant behavioral defects in male behavior (Figure 2), suggesting that in the context of single-pair-based mating assays, the FruMA isoform does not appear to be necessary for a male to perform robust levels of courtship. This contrasts to the FruMB and FruMC isoforms, which both appear to be necessary for the male to exhibit wild-type levels of the courtship behavioral repertoire. All isoform mutants were able to perform some courtship; therefore, no individual isoform is essential for the overall performance of this behavior.
Figure 2.
FruM Isoform-Specific Analysis of Male Courtship Behavior
All genotypes indicated are males; target females are wild-type Canton-S. NA indicates measurements that were not applicable. For statistical analysis, comparisons were made against the control group fru4-40/+ for all mutants in combination with Df(3R)fru4-40, whereas comparisons for all mutants in combination with Canton-S were +/+. Error bars represent ± SEM. n values are shown in parentheses.
(A) Courtship latencies. ∗p < 0.05 (Kruskal-Wallis ANOVA test).
(B) Courtship indices. ∗∗∗p < 0.001 (Kruskal-Wallis ANOVA test).
(C) Percentage of males mating within 1 hr. ∗∗∗p < 0.001 (Fisher’s exact test).
(D) Time to copulation. ∗∗p < 0.001 (Kruskal-Wallis ANOVA test).
(E) Wing extension indices. ∗∗∗p < 0.001 (Kruskal-Wallis ANOVA test).
(F) Male fertility. ∗∗∗p < 0.001 (Fisher’s exact test).
(G) Chaining indices. ∗p < 0.05; ∗∗∗p < 0.001 (Kruskal-Wallis ANOVA test).
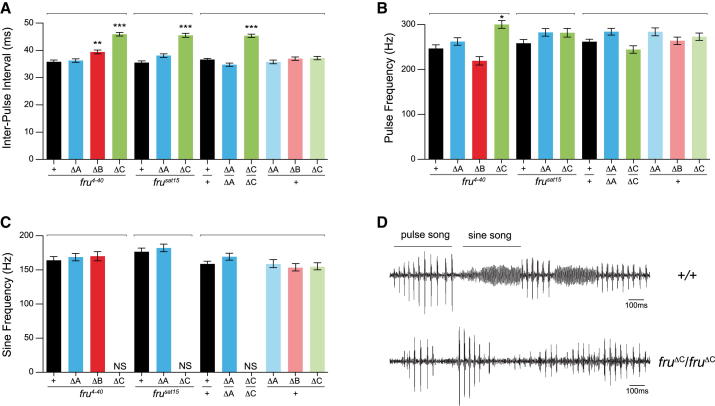
Examination of mutant males for deficits in unilateral wing extension, associated with males’ production of courtship song, revealed that only males lacking FruMC showed a significant deficit (Figure 2E). This led us to examine the attributes of song production in the fru isoform-specific mutants (Figures 3 and S3). Courtship song consists of alternating continuous oscillations (sine song) and trains of pulses (pulse song). Temporal variation in the time between pulses or interpulse intervals (IPIs) [16] and frequency components of song [17] influence species-specific female preferences, though differences in functions of pulse versus sine song are poorly understood [18]. Previous analysis of song production in a variety of fru mutant genotypes showed a range of phenotypes, from significantly longer IPIs to a total lack of song production [15]. Our analysis revealed the ability to produce pulse song in all mutants; however, a significant lengthening of IPI was found in mutants lacking both the FruMB and FruMC isoforms (Figure 3A). This was especially dramatic in the fruΔC mutant and was consistent across the genetic backgrounds examined; indeed, the 45 ms IPI observed exceeds the natural range typically seen within D. melanogaster and is more like the IPI of D. simulans [19, 20]. The pulse frequency appeared normal in all fru mutants, apart from the fruΔMC mutant, in which a small but significant increase in frequency was detected; however, this was not seen consistently across fruΔC mutant backgrounds (Figure 3B). The most dramatic phenotype observed was the consistent and complete absence of sine song in the fruΔC mutant (Figures 3C and 3D), whereas the fruΔA and fruΔB mutants did not show any significant changes to sine song frequency. These analyses suggest a role for both the FruMB and FruMC isoforms in the production of a species-specific IPI, while the ability to produce sine song appears to depend solely on the FruMC isoform.
Figure 3.
FruM Isoform-Specific Analysis of Male Courtship Song
All genotypes indicated are males; target females are wild-type Canton-S. Bars above represent groupings for statistical comparisons where the controls are fru4-40/+ for all mutants in combination with Df(3R)fru4-40, frusat15/+ for all mutants in combination with Df(3R)frusat15, and +/+ for all mutants in combination with Canton-S. Error bars represent 95% confidence intervals. Numerical values associated with each measurement are shown in Figure S3 along with n values for each genotype.
(A) Interpulse interval in ms. ∗∗p < 0.001; ∗∗∗p < 0.001 (Kruskal-Wallis ANOVA test).
(B) Pulse frequency in Hz. ∗p < 0.01 (Kruskal-Wallis ANOVA test).
(C) Sine frequency in Hz (Kruskal-Wallis ANOVA test). SA, sine song absent.
(D) Song recording from wild-type (+/+) and homozygous fruΔC mutant males. Examples of pulse and sine song are highlighted.
The fruΔA and fruΔB mutant alleles also disrupt the expression of the non-sex-specific (common) FruA and FruB isoforms. Since the loss of all fru expression is lethal [21], the isoform-specific mutants enable an analysis of the roles of fruA and fruB in fly development and viability. Homozygous fruΔB mutant male and female flies died as pupae, whereas fruΔA mutant males showed no decrease in viability (Figure S2A). Since fruΔC mutant males have reduced viability ([7]; Figure S2A), we conclude that only common FruB and FruC isoforms play essential developmental roles in both sexes. This result is consistent with the behavioral analysis described above: exon B- and C-containing Fru isoforms play overt roles in courtship and development, and exon A-containing isoforms likely play more subtle roles.
FruM Isoform-Specific Genomic Occupancy in the Drosophila Nervous System
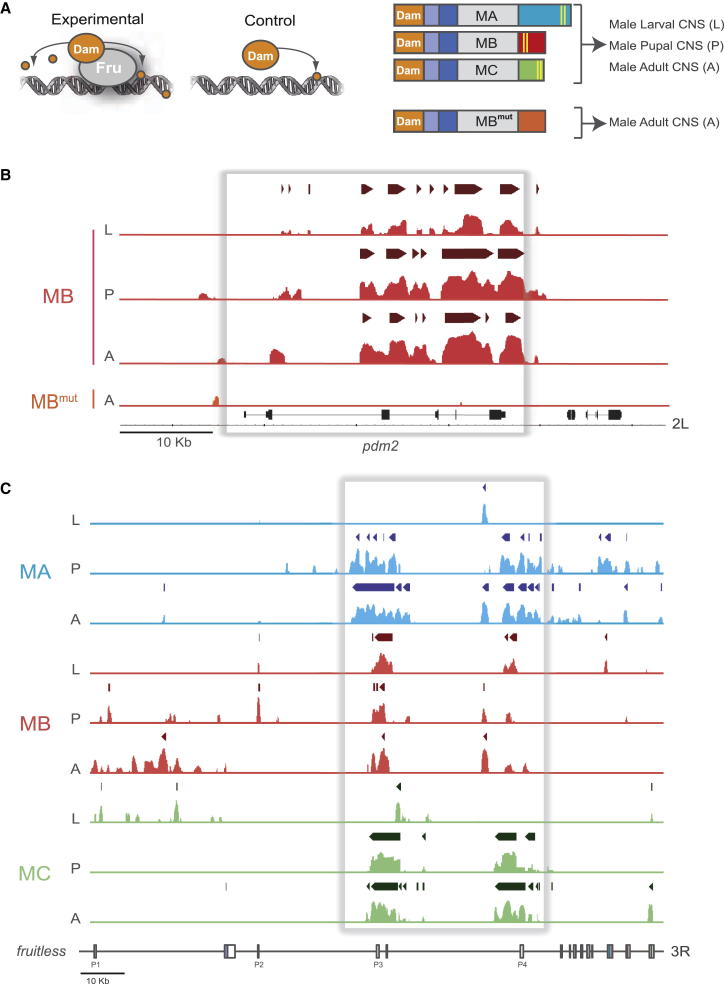
As isoform-specific FruM mutants provide insight into the behavioral roles of the individual FruM isoforms, we sought to connect these behavioral outputs with an understanding of genes regulated by each isoform. We used DamID [11], a technique enabling genome-wide analysis of DNA-protein interaction sites in vivo, to determine FruM-DNA associations throughout the genome (Figure 4A). We generated functional Dam-Fru fusion constructs for all three fru male-specific isoforms (Dam-FruMA, Dam-FruMB, and Dam-FruMC) driven by the uninduced hsp70 promoter in the pUAST vector, ensuring very low levels of Dam-fusion protein expression and avoiding overexpression artifacts (Figure 4A). In addition, to control for the DNA-binding specificity of our DamID analysis, we generated a DNA-binding-defective Dam-FruMBmut, which contains multiple mutations in key cysteine residues of the FruMB C2H2 Zn-finger domain. As a control for nonspecific activity of the methylase, we expressed a Dam-only construct in parallel.
Figure 4.
Developmental and Isoform-Specific Analysis of FruM Genomic Occupancy
(A) FruM-DamID experimental design. Each experimental replicate compares the Dam methylation footprint of tissue expressing Dam alone (control) against tissue expressing a Dam-FruM fusion protein (experimental). The schematic shows Dam-FruM isoform fusion proteins along with the developmental stages and tissues examined. Fru isoforms are abbreviated as follows: FruMA (MA), FruMB (MB), FruMC (MC), and FruMB DNA-binding mutant (MBmut).
(B and C) Binding profiles were generated using Integrated Genomic Browser (IGB) software as described in Supplemental Experimental Procedures. Darker bars above binding profiles represent identified binding intervals (1% false discovery rate, apart from FruMA larvae and pupae, which are 11% and 13%, respectively); the direction indicated by bars is 5′–3′ relative to the annotated Drosophila genome.
(B) DNA-binding specificity of Dam-FruMB at the pdm2 locus. DamID binding profiles comparing chromatin profiles of Dam-FruMB and the DNA-binding mutant Dam-FruMBmut across the POU-domain protein 2 (pdm2) locus (shown 3′–5′).
(C) DamID binding profiles of all FruM isoforms at the fru locus showing potential self-regulation at promoters P3 and P4 (shown 5′–3′).
The temporal expression patterns of FruM in the male CNS are well documented. Expression begins at the third-instar larval stage, peaks in 48 hr pupae, and continues at a low level into adulthood [8]. We analyzed nervous system tissue from wandering third-instar larval male CNS, 46 hr male pupal CNS, and 24 hr adult male CNS. CNS tissue (brain and ventral nerve cord) was manually dissected from flies expressing either a Dam-FruM isoform fusion (experimental) or Dam alone (control), and genomic DNA was isolated (Figure 4A). Samples were pooled and processed as described in Experimental Procedures, prior to being hybridized to Drosophila genome-wide tiling arrays. DamID chromatin profiles were generated and biological replicates subjected to a series of analyses to establish statistically significant data sets for each individual experiment [22]. For each data set, we identified bound regions (peaks) according to a false discovery rate (FDR) model using Ringo software [23]. The numbers of genes identified for each data set are shown in Figure S4A. Examples of FruM DamID chromatin profiles, including peaks that were found to be significantly enriched, are shown in Figures 4B and 4C. Comparison of the DamID chromatin profiles between the FruMB and FruMBmut data sets (Figure 4B) reveals that binding at the key neuronal homeobox gene POU domain protein 2 (pdm2) is dependent on functional C2H2 Zn fingers. We examined FruM isoform-specific binding at the fru locus itself (Figure 4C). Interestingly FruM isoforms show potential autoregulation at fru promoters P3 and P4.
FruM Isoforms Bind a Largely Overlapping Set of Genes Involved in Nervous System Development
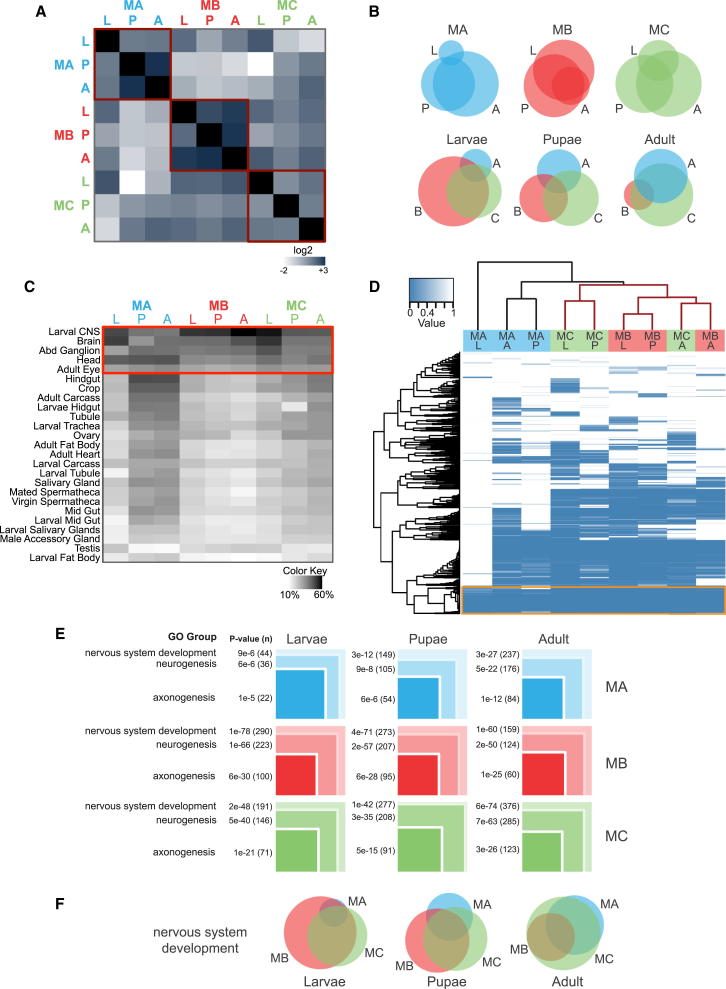
We analyzed developmental profiles of the genomic regions bound by each FruM isoform (Figure 5A) and relationships to neighboring genes (Figure 5B). FruMA and FruMC isoform occupancy was significantly overrepresented in intronic regions and at transcriptional start sites (TSSs), whereas FruMB associated preferentially to intergenic regions (Figure S4B). Developmental profiles revealed that FruM isoforms target many of the same genes throughout development, with the greatest extent of overlap between the pupal and adult stages. FruMB isoform occupancy is the most constant across the three developmental stages (Figures 5A and 5B). Interisoform comparisons revealed that FruM isoforms share many genomic targets throughout development in addition to targeting distinct sets of genes. Most FruM target genes are highly expressed in the nervous system; the heatmap in Figure 5C shows that the largest percentage of FruM target genes are expressed in tissues consisting of largely neuronal cell types (red box).
Figure 5.
FruM Isoforms Show Both Overlap and Specificity in Their Genomic Occupancy
Developmental times and isoforms are abbreviated as follows: L, larvae; P, pupae; A, adult; MA, FruMA; MB, FruMB; MC, FruMC. See also Figure S4.
(A) Heatmap showing pairwise comparisons between FruM data sets (see also Table S1). Within-isoform developmental comparisons are boxed in red.
(B) Venn diagrams comparing target genes between different isoforms at specific developmental times, as well as individual FruM isoforms throughout development. The full lists of FruM-associated genes are shown in Table S2.
(C) Expression of FruM targets throughout the fly. The percentage of genes upregulated in specific tissues of the fly is shown based on FlyAtlas [24]. The red box highlights nervous-system-enriched tissues. Enrichment is shown on a scale between 10% (white) and 60% (black).
(D) Gene ontology and clustering heatmap of FruM data sets. The dendrogram at the top shows the relationship between the data sets; the red group highlights the relationship between the FruMB and FruMC data sets. The dendrogram on the left represents the groupings of the ontologies; the orange box highlights the grouping of ontologies shared between all data sets (group 1). The significance of the enrichments (p values) are shown on a scale from dark blue (most significant) to white (not significant). The full GO analysis for each data set is shown in Table S3.
(E) Graphical representation of nested gene ontologies using the broadest ontology represented in group 1, “nervous system development” (GO:0007399). The number of genes (n) associated with each ontology (see Table S3) is shown in parentheses; p values represent the significance of enrichment. The sizes of the boxes in each set represent the proportion of genes represented in each ontology relative to each other.
(F) Venn diagram highlighting the high degree of overlap in genes associated with the “nervous system development” ontology between FruM isoforms throughout development.
To investigate the functional relationship between putative FruM target genes, we performed a global gene ontological (GO) enrichment analysis of biological functions (Figure 5D; Table S3). Statistically significant (p < 0.05) ontology terms associated with all data sets were hierarchically clustered. Similarities between the different FruM data sets are represented by clustering in the top dendrogram, the red portion of which highlights a relationship between the FruMB and FruMC data sets.
Since FruM functions in the development of a sexually dimorphic nervous system, we expected that Fru targets would be enriched in genes with known roles in neural development. Indeed, “nervous system development” (NSD) is the most significantly enriched GO term for all three isoforms (>level 4); the associated p value for this enrichment ranged from 9 × 10−6 in the FruMA larval data set to 10−78 in the FruMB larval data set (Figure 5E). Many of the other most highly enriched ontologies in this cluster are descendent groups of the NSD ontology, such as “neurogenesis” and “axonogenesis” (Figure 5E). A further comparison between the FruM isoforms and the genes associated with the NSD ontology (Figure 5F) revealed a high degree of overlap, especially in the adult.
Motif Enrichment Associated with FruM Genomic Occupancy
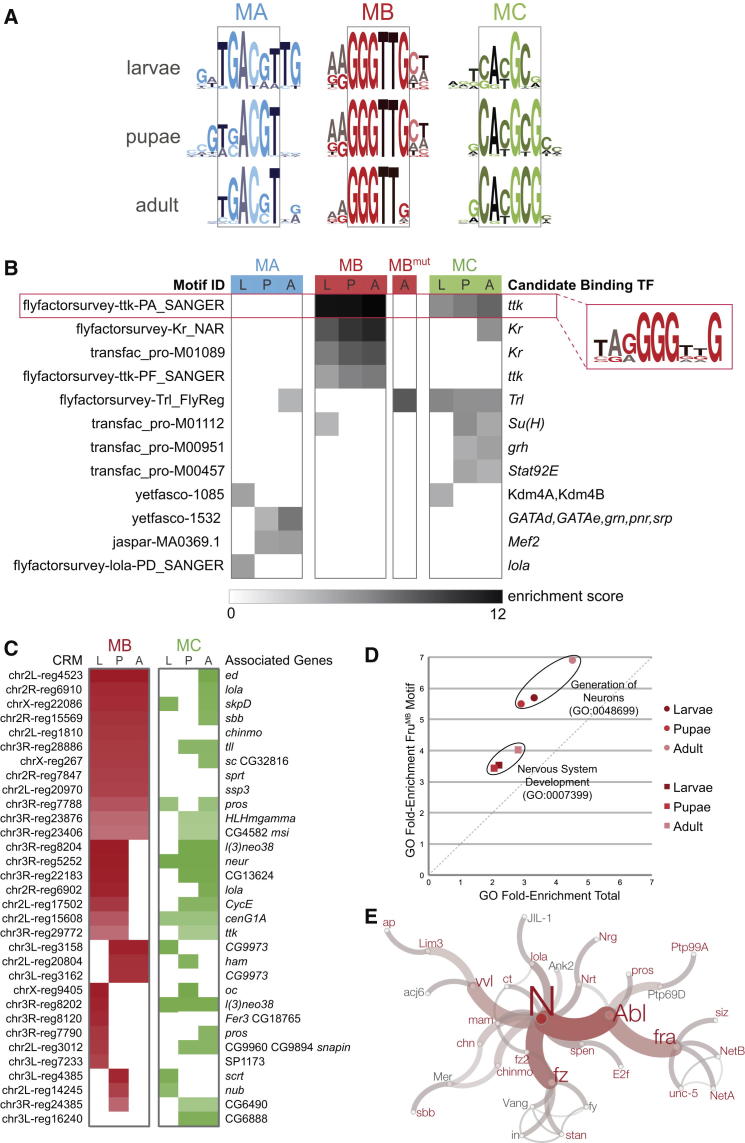
To investigate DNA-binding specificities of individual FruM isoforms, we identified cis-regulatory motifs enriched in our DamID-FruM genomic occupancy data. We utilized motif identification using conservation and relative abundance (MICRA), a motif discovery tool designed to analyze the low-resolution data produced using the DamID technique [25]. MICRA extracts 1 kb of sequence from each binding site, filters it for conserved sequences, and calculates enrichment of the binding site compared to background frequency. We generated position weight matrices using the sequences from the top ten enriched motifs (Figure 6A; Table S4). The top motif identified for each data set shows that each FruM isoform has unique sequence specificity. Importantly, the same core sequence was identified for each isoform across development, suggesting that our motif identification technique is robust. The FruMB isoform exhibited the most consistent DNA-binding specificity throughout development.
Figure 6.
Motif Analysis of FruM Data Sets
(A) MICRA-based motif analysis of FruM data sets. The top motif identified for each data set is shown. The representative core motif identified with each isoform is boxed. The full results of the MICRA analysis can be seen in Table S4.
(B) i-cisTarget motif analysis of FruM data sets. A representative heatmap is shown (full analysis in Table S5). Motif IDs are shown on the left; the associated candidate binding transcription factors are shown on the right. The grayscale represents the enrichment score associated with each motif. The most significantly enriched motif is boxed in red; its sequence is shown boxed on the right.
(C) Top 5% of cis-regulatory modules (CRMs) associated with the FruMB motif in the FruMB and FruMC data sets. Heatmaps represent the presence and ranking of the CRMs in the data sets: darker red and green colors signify the ranking of CRMs (1–100) in the FruMB and FruMC data sets, respectively. Genes associated with the identified CRMs are shown on the right.
(D) Nervous system ontology enrichments associated with the FruMB motif. Fold enrichments are graphed comparing those identified using the total FruMB data sets against those identified using the FruMB motif-associated data sets.
(E) Network analysis of the top 5% of FruMB motif-associated genes. Genes associated with the top 5% CRMs (shown in Figure 6C) were analyzed using Cytoscape [26]. The largest connected network is shown. Nodes labeled in red are genes associated with the FruMB motif, whereas those labeled in gray were identified as part of the network but are not associated with identified CRMs. The size of nodes and thickness and color of edges represent betweenness centrality and edge betweenness, respectively (described in Supplemental Experimental Procedures).
See also Table S6.
We independently searched for binding motifs using i-cisTarget, a method that identifies cis-regulatory modules (CRMs) [27] by ranking conserved regions in the Drosophila genome. We identified significant motifs and then determined the optimal subset of genomic regions that are predicted as direct targets in all FruM data sets. The top-ranked motifs identified in all of the data sets are shown in Figure 6B (Table S5 contains complete results). The most significantly enriched motif throughout the FruMB and FruMC data sets (enrichment score of 11.1–12.3 in FruMB and 4.7–6.5 in FruMC) was one previously identified as a binding site for isoform A of the Tramtrack (TtkA) protein (flyfactorsurvey-ttk-PA_SANGER). This is striking because ttk is the gene most closely related to fru in the Drosophila genome (Figure S5) [28, 29]. Importantly, the FruMBmut analysis did not reveal significant motif enrichment, other than a motif associated with the Trithorax-like (Trl) transcription factor. This motif was also enriched in the FruMC data sets throughout development and in the FruMA adult data set. However, this may not represent a direct binding motif, because Trl, like Fru and Ttk, is a BTB-Zn-finger transcriptional regulator that has been shown to interact with other BTB-containing proteins [30]. It is therefore possible that the genomic regions associated with this motif represent associations mediated through the BTB domain of FruM, rather than direct DNA binding.
The highly significant flyfactorsurvey-ttk-PA motif has a stretch that is identical to the core of the FruMB motif identified in the MICRA analysis (GGGTTG). We therefore designated this sequence as the putative FruMB DNA-binding motif for further analysis. We determined the genes in the FruMB data set associated with the top-ranked CRMs containing this motif (see Table S6 for complete results). As i-cisTarget also identified the putative FruMB motif in FruMC data sets, we included a parallel analysis of the FruMC data using this motif, although MICRA suggests that FruMC preferentially binds to a different site. The top 5% of CRMs identified in FruMB and FruMC data sets are shown with their presence and CRM rank in red or green, respectively (Figure 6C). The FruMB data sets show a consistent association with this candidate motif throughout development, whereas FruMC appears to preferentially associate with these CRMs later in development. Many of the target genes are common between the isoforms in one or more developmental stages.
Comparison of the genes associated with the putative FruMB DNA-binding motif versus all FruMB target genes again revealed the most significantly enriched GO term to be of neural origin, “generation of neurons” (GO:0048699; Figure 6D). A network analysis of the genes associated with the “nervous system development” GO term (GO:0007399) produced a connected network involving 32 genes (Figure 6E). Of the 32 genes, 26 are putative direct FruMB target genes (labeled in red), 23 of which have established roles in neuronal projection morphogenesis. The most connected node in the network is Notch (N), which plays key roles in neuronal development, neural integration, and neural plasticity [31]. Our motif analysis provides further insight into the relationship between FruM isoforms and strengthens the functional connection between FruMB target genes and neuronal development.
Sexually Dimorphic Expression of FruMB Motif-Containing Genomic Enhancers in fru Neurons
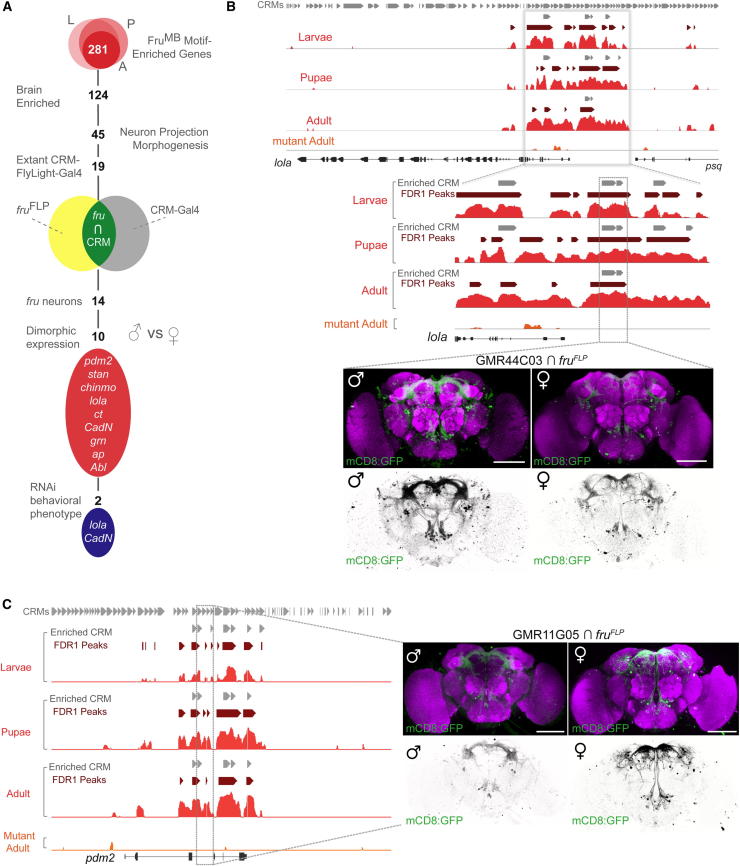
As FruM is male specific, we tested whether our identified genomic enhancers containing the FruMB motif exhibit sexually dimorphic expression in fru-positive neurons (Figure 7). We took an intersectional approach using extant FlyLight transgenic fly lines that express GAL4 under the control of defined genomic enhancers [32]. From the complete list of FruMB motif-enriched genes, we selected brain-enriched genes and those that clustered into the “neuron projection morphogenesis” GO term (Figure 7A). We used this subset of genes to select 19 FlyLight-GAL4 lines driven by CRMs associated with 15 FruMB motif-enriched genes (CRM-GAL4) for neuroanatomical analysis (Table S7). To assess whether each CRM-GAL4 is expressed in fru-positive neurons, we intersected them with fruFLP [33] and a UAS>stop>mCD8::GFP reporter transgene (where “>stop>” represents a FLP recombinase target sequence-flanked transcriptional termination cassette). fruFLP expresses FLP recombinase in FruM-expressing neurons in males, as well as the homologous set of neurons in females. Therefore, if a particular CRM-GAL4 drives expression in fru-positive neurons, the neurons will be labeled by membrane-bound GFP (Figure 7A). We screened for sexually dimorphic expression in the male and female CNS, where the female is “equivalent” to a fru null (i.e., there is no FruM protein produced in females). Strikingly, we observed expression in fru neurons in 14 of the 19 assayed CRM-GAL4 lines. Furthermore, 10 of the 14 lines (>70%) exhibited overt sexually dimorphic expression patterns, (Figures 7A and S6; Table S7).
Figure 7.
Sexually Dimorphic Expression of FruMB Motif-Containing Genomic Enhancers in fru Neurons
(A) Strategy for analyzing genomic regions containing FruMB motif-enriched CRMs in fru neurons.
(B) Dimorphic expression of lola FruMB-CRM-GAL4 in fru neurons in the CNS. Top: DamID binding profiles of the FruMB isoform through development at the lola locus, as well as FruMBmut in the adult (lola is shown oriented 3′–5′). All CRMs in i-cisTarget are shown above (gray). Statistically significant peaks (FDR1 peaks) are shown in dark red; enriched CRMs are shown in gray. Bottom: images of male and female brains from the fruFLP intersected FlyLight-GAL4 line GMR44C03 (the relevant CRM region is boxed in gray above).
(C) Dimorphic expression of pdm2 FruMB-CRM-GAL4s in fru neurons in the CNS. Top: DamID binding profiles of the FruMB isoform through development at the pdm2 locus. Bottom: images of male and female brains from the fruFLP intersected FlyLight-GAL4 line GMR11G05 (CRM region is boxed in gray).
In (B) and (C), brains are stained with anti-GFP (green) and the nc82 general neuropil reference (magenta). Inverted views of the GFP signal are also shown in black and white below. Brain scale bars represent 100 μm; VNC scale bars represent 50 μm. See also Figure S6 and Table S7.
We identified binding of the FruMB isoform throughout development at the lola genomic locus, which encodes BTB-Zn-finger proteins (Figure 7B). Binding is dependent on the DNA-binding domain, as FruMBmut showed no significant binding in this region. We observed marked sexually dimorphic expression of the lola CRM-GAL4 GMR44C03 in fru neurons in the male and female CNS. The sexual dimorphism is evident in the number of neurons labeled in the brain, whereas the gross morphology of the neuronal arbors appears to be similar between male and female brains. The most overt difference was the increased density of neural arbors in males compared with females. This is especially evident in the projections of neurons in the dorsal superior protocerebral bridge [33] and within the fru-mcAL cluster [8] localized to the region just ventromedial to the antennal lobe. In another example, we observed sexually dimorphic expression patterns in a FruMB motif-enriched CRM-GAL4 line associated with the pdm2 locus (Figure 7C). In this case, however, the expression of the pdm2 CRM-GAL4 line (GMR11G05) was far more intense in females, especially in areas specific to the medial and lateral superior protocerebrum. There is also an overt increase in expression in projections to or from the periesophageal neuropil. Additional dimorphic expression patterns of CRM-GAL4 in fru neurons are shown in Figure S6.
As a preliminary screen, we independently targeted eight of the identified genes by expressing gene-specific RNAi transgenes under the control of a fruGAL4 driver [34] and examined male courtship behavior (Figure S7). Both lola- and CadN-disrupted flies showed dramatic decrements in courtship. No appreciable courtship behavior was detected in fru-GAL4/CadNRNAi males during a 1 hr observation period (Figure S7B). In comparison, only 50% of fru-GAL4/lolaRNAi males initiated courtship within the observation period, and those that did court showed a significant delay (Figure S7A). Defects in courtship behavior were not due to overt defects in locomotion (Figure S7D), and additional RNAi lines targeting lola and CadN were shown to significantly disrupt courtship behavior (Figure S7E). Although our analysis has revealed the functional importance of CadN and lola in fru neurons, future developmental studies will be needed to refine these relationships.
Discussion
Our study of fru isoform function exemplifies how complex behaviors involved in courtship can be controlled by a single locus. Differential expression of multiple isoforms with different binding specificities produces a “neural code” of downstream gene expression, in which phenotypes can be specified by either a single isoform or a combination of isoforms. The DamID approach allowed us to identify the association of FruM proteins to specific regions of the genome and relate this binding with downstream target genes. Genes known to play a role in the development of the nervous system are significantly overrepresented within these identified FruM target genes. This is certainly consistent with the established role that fru plays in the development of a number of neuronal structures [2]. However, until this study, the identity of fru-regulated genes had not been determined. The identification of putative FruM binding motifs, our strategy for identifying and characterizing Fru-regulated genomic enhancers, and the production of a comprehensive set of fruM isoform-specific mutant flies facilitates an unprecedented leap forward in our ability to study FruM transcriptional regulation.
For a more in-depth analysis, we concentrated on a subset of putative FruMB target genes. The identification of a putative FruMB DNA-binding motif allowed us to show that the majority of genomic enhancers containing this motif exhibit sexually dimorphic expression in fru neurons. Among the genes associated with these enhancers are the related BTB-Zn-finger genes lola and chinmo, both key neuronal morphogenesis genes [35, 36]. In addition, decreased expression of lola, specifically in fru neurons, led to dramatically reduced levels of male sexual behavior, establishing the necessity of this protein in fru neurons. Since Fru targets other BTB-Zn-finger genes, we speculate that regulatory diversity of these transcription factors contributes to a neuron-specific transcriptional code leading to specific developmental outcomes. Future examination of these and other FruM target genes will allow us to decipher this code and connect specific dimorphic neural cell fates with behavioral outputs.
FruM Isoform Function: Cooperativity, Specificity, and Redundancy
We determined that there is a great deal of overlap in the genomic loci targeted by all of the FruM isoforms when it comes to genes involved in the development of the nervous system. Since each Fru isoform appears to have unique binding specificity, it follows that FruM isoforms could act independently on the same genes, either cooperatively or redundantly. FruMB and FruMC isoforms can associate with the same genomic regions containing the putative FruMB motif (Figure 6B). FruMB exhibits the most consistent binding specificity, which we determined is dependent on amino acid residues that are required for DNA binding. In contrast, although the FruMC isoform is enriched for the FruMB motif, it appears to have a unique DNA binding specificity (Figure 6A). Our previous analysis of serotonergic neurons in the abdominal ganglion that innervate the male reproductive organs showed evidence of cooperative function between the FruMB and FruMC isoforms, as both were required for the development of these neurons [7]. However, other functions appeared to be isoform specific: for example, only FruMC controls the innervation and formation of the male-specific muscle of Lawrence (Figure 1E) [7]. Although the elimination of individual FruM isoforms generated overt behavioral deficits, they were not sufficient to abrogate courtship behaviors completely, suggesting some degree of redundancy in the determination of the neural networks directing these behaviors.
Evolutionary Considerations
Alternatively spliced isoforms, like gene duplications, enable a diversification of gene function, by allowing essential (often ancestral) functions to be maintained while others are able to diverge and take on new roles [37, 38]. Evolutionary analysis of the fru C2H2 Zn-finger domains in various insect species shows the appearance and disappearance of these domains throughout evolution. However, the high conservation between all fru C2H2 Zn fingers supports the idea that they all originated from one or a few ancestral sequences and retained a common function [29].
Our results support this scenario of evolution in fru, as we have found evidence for both conservation and divergence in function of the different isoforms. Loss of either FruMB or FruMC expression significantly disrupts the male’s ability to perform courtship behavior, whereas loss of the FruMA isoform has no obvious consequence. Our expression analysis of the individual isoforms in the CNS also mirrors these relationships. FruMB and FruMC isoforms are broadly expressed in most FruM-positive neurons, whereas FruMA expression is restricted to only a subset of FruM-positive neurons (Figure 1) [7]. The lack of an overt phenotype associated with the fruΔA mutant may be a reflection of the relative involvement of this subset of fru-positive neurons, or may indicate that the FruMA isoform fulfills more specialized nonessential functions. We recently found evidence of positive selection acting on fru exon A across Drosophila species, whereas exons B and C were found to be conserved, supporting the involvement of transcripts containing the A exon in nonessential functions, which may contribute to phenotypic differences between species [39].
The Fruitless/Ecdysone Relationship
Previous microarray experiments showed that genes regulated downstream of FruM (either directly or indirectly) appear to also be regulated by ecdysone. In addition, the ecdysone receptor was shown to act in fru neurons to mediate male courtship behavior [40]. More recently, it was shown that females depleted in ecdysone display male-like courtship behaviors [41], and it was proposed that distinct ecdysone peaks might regulate the formation of distinct FruM-containing chromatin regulatory complexes [42]. Although our developmental time course did not detect a dramatic shift in FruM DNA-binding specificity as a result of ecdysone pulses, there are more subtle dynamic shifts in binding throughout development that might result from these pulses. We found a small but significant enrichment of known ecdysone-responsive genes in all FruM data sets (40 of 61, p < 0.01, chi-square) [40]. Interestingly, this included the cell death gene reaper (rpr), which we identified as a putative target of all FruM isoforms throughout development. fru has been shown to be essential for the suppression of cell death in the male mAL neural cluster, potentially by downregulating key cell death genes [43]. Direct targeting of rpr by FruM isoforms would support this mechanism. We also identified the ecdysone-responsive transcription factor crooked legs (crol) as a putative target of both FruMB and FruMC isoforms in pupal and adult stages. A previous microarray analysis reported crol as being upregulated in the CNS of fruM mutant males [44]. Deficiency combinations resulting in complete loss of the fru locus (along with a small number of neighboring genes) result in early pupal developmental arrest, around the time of pupal ecdysis [21]. It was noted that the phenotype of fru-deficient flies was similar to that of flies mutant for the ecdysone receptor and the crol gene. Therefore, some of these targets may link the sex-specific and common isoform functions of Fru in response to ecdysone.
Fruitless and Doublesex: Regulator Partners?
Our identified FruM targets overlap with those of the other key sex-determination protein, Doublesex (Dsx) (Figure S3C). The male-specific form of dsx (dsxM) is expressed in far fewer cells in the adult CNS than fru, but almost all dsxM cells coexpress fru [45, 46]. Dsx and Fru are the only identified factors at the bottom of the sex-determination hierarchy, and both of these transcriptional regulators act in the same neurons to bring about male-specific neuronal wiring and male-specific behavioral patterns [45–47]. Given the overrepresentation of identified Dsx target genes in our FruM data sets, we speculate that FruM and DsxM act together, either in a physical complex or through coregulation of genomic targets, to determine the male-specific nervous system.
Acknowledgments
We thank TRiP at Harvard Medical School (NIH/NIGMS grant R01-GM084947) for RNAi stocks; Barry Dickson for fly stocks; Anthony Dornan, Carolina Rezaval, and Scott Waddell for comments on the manuscript; Andreas Beyer and members of the Goodwin lab for helpful discussions; and Bill Heitler for assistance with DataView. We are grateful to Anthony Dornan and J.C. Billeter for technical assistance. This work was supported by grants from the Wellcome Trust to S.F.G. (WT085521MA and WT082987MF) and the Natural Environment Research Council to S.F.G and M.G.R. (NE/J023647/1). J.W. and E.A were supported by Biotechnology and Biological Sciences Research Council Committee studentships.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Contributor Information
Megan C. Neville, Email: megan.goodwin@dpag.ox.ac.uk.
Stephen F. Goodwin, Email: stephen.goodwin@dpag.ox.ac.uk.
Accession Numbers
The NCBI GEO accession number for the DamID data reported in this paper is GSE52247.
Supplemental Information
References
- 1.Villella A., Hall J.C. Neurogenetics of courtship and mating in Drosophila. Adv. Genet. 2008;62:67–184. doi: 10.1016/S0065-2660(08)00603-2. [DOI] [PubMed] [Google Scholar]
- 2.Pavlou H.J., Goodwin S.F. Courtship behavior in Drosophila melanogaster: towards a ‘courtship connectome’. Curr. Opin. Neurobiol. 2013;23:76–83. doi: 10.1016/j.conb.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito H., Fujitani K., Usui K., Shimizu-Nishikawa K., Tanaka S., Yamamoto D. Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proc. Natl. Acad. Sci. USA. 1996;93:9687–9692. doi: 10.1073/pnas.93.18.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryner L.C., Goodwin S.F., Castrillon D.H., Anand A., Villella A., Baker B.S., Hall J.C., Taylor B.J., Wasserman S.A. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell. 1996;87:1079–1089. doi: 10.1016/s0092-8674(00)81802-4. [DOI] [PubMed] [Google Scholar]
- 5.Usui-Aoki K., Ito H., Ui-Tei K., Takahashi K., Lukacsovich T., Awano W., Nakata H., Piao Z.F., Nilsson E.E., Tomida J., Yamamoto D. Formation of the male-specific muscle in female Drosophila by ectopic fruitless expression. Nat. Cell Biol. 2000;2:500–506. doi: 10.1038/35019537. [DOI] [PubMed] [Google Scholar]
- 6.Siggs O.M., Beutler B. The BTB-ZF transcription factors. Cell Cycle. 2012;11:3358–3369. doi: 10.4161/cc.21277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Billeter J.C., Villella A., Allendorfer J.B., Dornan A.J., Richardson M., Gailey D.A., Goodwin S.F. Isoform-specific control of male neuronal differentiation and behavior in Drosophila by the fruitless gene. Curr. Biol. 2006;16:1063–1076. doi: 10.1016/j.cub.2006.04.039. [DOI] [PubMed] [Google Scholar]
- 8.Lee G., Foss M., Goodwin S.F., Carlo T., Taylor B.J., Hall J.C. Spatial, temporal, and sexually dimorphic expression patterns of the fruitless gene in the Drosophila central nervous system. J. Neurobiol. 2000;43:404–426. doi: 10.1002/1097-4695(20000615)43:4<404::aid-neu8>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 9.Arthur B.I., Jr., Jallon J.M., Caflisch B., Choffat Y., Nöthiger R. Sexual behaviour in Drosophila is irreversibly programmed during a critical period. Curr. Biol. 1998;8:1187–1190. doi: 10.1016/s0960-9822(07)00491-5. [DOI] [PubMed] [Google Scholar]
- 10.Ito H., Sato K., Koganezawa M., Ote M., Matsumoto K., Hama C., Yamamoto D. Fruitless recruits two antagonistic chromatin factors to establish single-neuron sexual dimorphism. Cell. 2012;149:1327–1338. doi: 10.1016/j.cell.2012.04.025. [DOI] [PubMed] [Google Scholar]
- 11.van Steensel B., Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- 12.Rong Y.S., Golic K.G. Gene targeting by homologous recombination in Drosophila. Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- 13.Currie D.A., Bate M. Innervation is essential for the development and differentiation of a sex-specific adult muscle in Drosophila melanogaster. Development. 1995;121:2549–2557. doi: 10.1242/dev.121.8.2549. [DOI] [PubMed] [Google Scholar]
- 14.Nojima T., Kimura K., Koganezawa M., Yamamoto D. Neuronal synaptic outputs determine the sexual fate of postsynaptic targets. Curr. Biol. 2010;20:836–840. doi: 10.1016/j.cub.2010.02.064. [DOI] [PubMed] [Google Scholar]
- 15.Villella A., Gailey D.A., Berwald B., Ohshima S., Barnes P.T., Hall J.C. Extended reproductive roles of the fruitless gene in Drosophila melanogaster revealed by behavioral analysis of new fru mutants. Genetics. 1997;147:1107–1130. doi: 10.1093/genetics/147.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritchie M.G., Halsey E.J., Gleason J.M. Drosophila song as a species-specific mating signal and the behavioural importance of Kyriacou & Hall cycles in D. melanogaster song. Anim. Behav. 1999;58:649–657. doi: 10.1006/anbe.1999.1167. [DOI] [PubMed] [Google Scholar]
- 17.Riabinina O., Dai M., Duke T., Albert J.T. Active process mediates species-specific tuning of Drosophila ears. Curr. Biol. 2011;21:658–664. doi: 10.1016/j.cub.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Tauber E., Eberl D.F. Acoustic communication in Drosophila. Behav. Processes. 2003;64:197–210. [Google Scholar]
- 19.Colegrave N., Hollocher H., Hinton K., Ritchie M.G. The courtship song of African Drosophila melanogaster. J. Evol. Biol. 2000;13:143–150. [Google Scholar]
- 20.Ritchie M.G., Yate V.H., Kyriacou C.P. Genetic variability of the interpulse interval of courtship song among some European populations of Drosophila melanogaster. Heredity (Edinb) 1994;72:459–464. doi: 10.1038/hdy.1994.64. [DOI] [PubMed] [Google Scholar]
- 21.Anand A., Villella A., Ryner L.C., Carlo T., Goodwin S.F., Song H.J., Gailey D.A., Morales A., Hall J.C., Baker B.S., Taylor B.J. Molecular genetic dissection of the sex-specific and vital functions of the Drosophila melanogaster sex determination gene fruitless. Genetics. 2001;158:1569–1595. doi: 10.1093/genetics/158.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greil F., van der Kraan I., Delrow J., Smothers J.F., de Wit E., Bussemaker H.J., van Driel R., Henikoff S., van Steensel B. Distinct HP1 and Su(var)3-9 complexes bind to sets of developmentally coexpressed genes depending on chromosomal location. Genes Dev. 2003;17:2825–2838. doi: 10.1101/gad.281503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toedling J., Skylar O., Krueger T., Fischer J.J., Sperling S., Huber W. Ringo—an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics. 2007;8:221. doi: 10.1186/1471-2105-8-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chintapalli V.R., Wang J., Dow J.A. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 25.Southall T.D., Brand A.H. Neural stem cell transcriptional networks highlight genes essential for nervous system development. EMBO J. 2009;28:3799–3807. doi: 10.1038/emboj.2009.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smoot M.E., Ono K., Ruscheinski J., Wang P.L., Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herrmann C., Van de Sande B., Potier D., Aerts S. i-cisTarget: an integrative genomics method for the prediction of regulatory features and cis-regulatory modules. Nucleic Acids Res. 2012;40:e114. doi: 10.1093/nar/gks543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spokony R.F., Restifo L.L. Anciently duplicated Broad Complex exons have distinct temporal functions during tissue morphogenesis. Dev. Genes Evol. 2007;217:499–513. doi: 10.1007/s00427-007-0159-y. [DOI] [PubMed] [Google Scholar]
- 29.Bertossa R.C., van de Zande L., Beukeboom L.W. The Fruitless gene in Nasonia displays complex sex-specific splicing and contains new zinc finger domains. Mol. Biol. Evol. 2009;26:1557–1569. doi: 10.1093/molbev/msp067. [DOI] [PubMed] [Google Scholar]
- 30.Schwendemann A., Lehmann M. Pipsqueak and GAGA factor act in concert as partners at homeotic and many other loci. Proc. Natl. Acad. Sci. USA. 2002;99:12883–12888. doi: 10.1073/pnas.202341499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lieber T., Kidd S., Struhl G. DSL-Notch signaling in the Drosophila brain in response to olfactory stimulation. Neuron. 2011;69:468–481. doi: 10.1016/j.neuron.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenett A., Rubin G.M., Ngo T.T., Shepherd D., Murphy C., Dionne H., Pfeiffer B.D., Cavallaro A., Hall D., Jeter J. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu J.Y., Kanai M.I., Demir E., Jefferis G.S., Dickson B.J. Cellular organization of the neural circuit that drives Drosophila courtship behavior. Curr. Biol. 2010;20:1602–1614. doi: 10.1016/j.cub.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 34.Stockinger P., Kvitsiani D., Rotkopf S., Tirián L., Dickson B.J. Neural circuitry that governs Drosophila male courtship behavior. Cell. 2005;121:795–807. doi: 10.1016/j.cell.2005.04.026. [DOI] [PubMed] [Google Scholar]
- 35.Goeke S., Greene E.A., Grant P.K., Gates M.A., Crowner D., Aigaki T., Giniger E. Alternative splicing of lola generates 19 transcription factors controlling axon guidance in Drosophila. Nat. Neurosci. 2003;6:917–924. doi: 10.1038/nn1105. [DOI] [PubMed] [Google Scholar]
- 36.Zhu S., Lin S., Kao C.F., Awasaki T., Chiang A.S., Lee T. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell. 2006;127:409–422. doi: 10.1016/j.cell.2006.08.045. [DOI] [PubMed] [Google Scholar]
- 37.Chothia C., Gough J., Vogel C., Teichmann S.A. Evolution of the protein repertoire. Science. 2003;300:1701–1703. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- 38.Graveley B.R. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 39.Parker D.J., Gardiner A., Neville M.C., Ritchie M.G., Goodwin S.F. The evolution of novelty in conserved genes; evidence of positive selection in the Drosophila fruitless gene is localised to alternatively spliced exons. Heredity (Edinb.) 2013 doi: 10.1038/hdy.2013.106. Published online October 23, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dalton J.E., Lebo M.S., Sanders L.E., Sun F., Arbeitman M.N. Ecdysone receptor acts in fruitless-expressing neurons to mediate Drosophila courtship behaviors. Curr. Biol. 2009;19:1447–1452. doi: 10.1016/j.cub.2009.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ganter G.K., Desilets J.B., Davis-Knowlton J.A., Panaitiu A.E., Sweezy M., Sungail J., Tan L.C., Adams A.M., Fisher E.A., O’Brien J.R. Drosophila female precopulatory behavior is modulated by ecdysteroids. J. Insect Physiol. 2012;58:413–419. doi: 10.1016/j.jinsphys.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ito H., Sato K., Yamamoto D. Sex-switching of the Drosophila brain by two antagonistic chromatin factors. Fly (Austin) 2013;7:87–91. doi: 10.4161/fly.24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimura K., Ote M., Tazawa T., Yamamoto D. Fruitless specifies sexually dimorphic neural circuitry in the Drosophila brain. Nature. 2005;438:229–233. doi: 10.1038/nature04229. [DOI] [PubMed] [Google Scholar]
- 44.Goldman T.D., Arbeitman M.N. Genomic and functional studies of Drosophila sex hierarchy regulated gene expression in adult head and nervous system tissues. PLoS Genet. 2007;3:e216. doi: 10.1371/journal.pgen.0030216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rideout E.J., Billeter J.C., Goodwin S.F. The sex-determination genes fruitless and doublesex specify a neural substrate required for courtship song. Curr. Biol. 2007;17:1473–1478. doi: 10.1016/j.cub.2007.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rideout E.J., Dornan A.J., Neville M.C., Eadie S., Goodwin S.F. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat. Neurosci. 2010;13:458–466. doi: 10.1038/nn.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kimura K., Hachiya T., Koganezawa M., Tazawa T., Yamamoto D. fruitless and doublesex coordinate to generate male-specific neurons that can initiate courtship. Neuron. 2008;59:759–769. doi: 10.1016/j.neuron.2008.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.