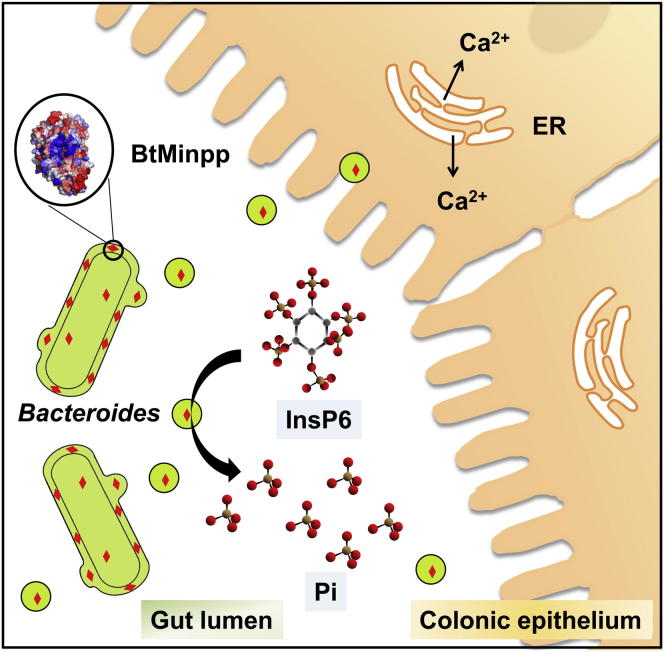
Summary
Dietary InsP6 can modulate eukaryotic cell proliferation and has complex nutritive consequences, but its metabolism in the mammalian gastrointestinal tract is poorly understood. Therefore, we performed phylogenetic analyses of the gastrointestinal microbiome in order to search for candidate InsP6 phosphatases. We determined that prominent gut bacteria express homologs of the mammalian InsP6 phosphatase (MINPP) and characterized the enzyme from Bacteroides thetaiotaomicron (BtMinpp). We show that BtMinpp has exceptionally high catalytic activity, which we rationalize on the basis of mutagenesis studies and by determining its crystal structure at 1.9 Å resolution. We demonstrate that BtMinpp is packaged inside outer membrane vesicles (OMVs) protecting the enzyme from degradation by gastrointestinal proteases. Moreover, we uncover an example of cross-kingdom cell-to-cell signaling, showing that the BtMinpp-OMVs interact with intestinal epithelial cells to promote intracellular Ca2+ signaling. Our characterization of BtMinpp offers several directions for understanding how the microbiome serves human gastrointestinal physiology.
Graphical Abstract
Highlights
-
•
Bacteroides thetaiotaomicron (Bt) secretes a cell-signaling InsP6 phosphatase MINPP
-
•
BtMinpp is exceptionally active and rationalized from its crystal structure
-
•
BtMinpp is secreted in outermembrane vesicles
-
•
BtMinpp/OMVs promote Ca2+ signaling in intestinal epithelial cells
Brearley, Carding, and colleagues now find that the prevalent gut bacterium Bacteroides thetaiotaomicron (Bt) contributes to human physiology, metabolism, and homeostasis through gastrointestinal InsP6 metabolism and secretion of a mammalian cell-signaling InsP6 phosphatase, MINPP. BtMinpp catalytic activity was exceptionally high and rationalized from its crystal structure. BtMinpp is packaged inside outermembrane vesicles (OMVs) protecting against intestinal proteases. BtMinpp and OMVs also promoted Ca2+ signaling in intestinal epithelial cells, an example of cross-kingdom dialog.
Introduction
The adult human gastrointestinal (GI) tract accommodates a bacterial community that comprises trillions of cells. This microbiota has many essential roles in human health (Tremaroli and Bäckhed, 2012): it suppresses proliferation of pathogenic microbes and has important nutritional consequences, including vitamin synthesis and fermentation of complex dietary carbohydrates. Microbial metabolites also regulate the signaling activities of the host’s intestinal epithelium, which, for example, aids the development and maintenance of local immune responses. Thus, considerable efforts are now being made to determine the precise nature of the dialog between gut bacteria and the human host. Much of the previous work in this area has focused on the roles of diffusible, small-molecule hormones and nutrients. We now describe a vehicle produced by a prevalent gut bacterium, Bacteroides thetaiotaomicron (Bt): a vesicle-enclosed homolog of a mammalian cell-signaling InsP6 phosphatase, MINPP.
Enzymatic homeostasis of InsP6 levels in the gut can have far-reaching consequences for human health. Considerable quantities of InsP6 are ingested daily as it is the primary storage form of phosphorus in cereals and legumes (Kumar et al., 2010). InsP6 is therefore a source of inositol and phosphate, two vital nutrients. However, InsP6 is also considered to have antinutritive properties, given that it inhibits polysaccharide digestibility and chelates divalent cations, thereby limiting their bioavailability in the GI tract (Kumar et al., 2010). More recently, with the emergence of cell-signaling activities for InsP6 and other members of the inositol phosphate family, dietary InsP6 has received separate attention for its anticarcinogenic properties (Fox and Eberl, 2002, Vucenik and Shamsuddin, 2003), at least when present at high concentrations. Conversely, lower concentrations of InsP6 may stimulate tumor cell proliferation (Windhorst et al., 2013). In any case, exogenous InsP6 can enter cells and be metabolized (Windhorst et al., 2013), thereby potentially contributing to intracellular signaling processes in intestinal epithelial and immune cells. For example, InsP6 is the precursor for inositol pyrophosphates, which have pleiotropic functions including the regulation of energy metabolism (Szijgyarto et al., 2011), insulin sensitivity (Chakraborty et al., 2010), and bactericidal activities of immune cells (Prasad et al., 2011).
Humans and other monogastric animals lack enzymes capable of participating in dietary InsP6 homeostasis, relying instead on exogenous phosphatases provided by resident microbes (Haros et al., 2007, Steer et al., 2004). Surprisingly, the nature of the genes that encode these phosphatases is unknown for virtually the entire microbiome of the human GI tract, although they are generally assumed to be “phytases” (Haefner et al., 2005, Steer et al., 2004, Tamayo-Ramos et al., 2012). Such enzymes occur in a number of environmental niches in which they scavenge inorganic phosphate from InsP6, but they have no signaling function. Moreover, if phytases were to be secreted into the GI tract, it is unclear how they might access dietary InsP6 while avoiding digestive proteases. In the current study, our phylogenetic, biochemical, and structural characterization of BtMinpp characterizes an unexpected repertoire of widely distributed and highly active class of InsP6 phosphatases in the GI tract.
Results
B. thetaiotaomicron Express a Homolog of a Mammalian Inositol Phosphate Signaling Phosphatase
The Bacteroidetes are a dominant constituent of the mammalian GI tract microbiota. The genome of B. thetaiotaomicron (Xu et al., 2003) contains a gene, BT_4744, which has a primary annotation (Hidden Markov Models-based annotation) as encoding a putative multiple inositol-polyphosphate phosphatase (MINPP, EC 3.1.3.62). This is a notable observation because bacteria have not previously been suggested to utilize the inositol phosphate signaling cascade (Michell, 2008). Indeed, MINPPs have only previously been considered to function inside animal cells, mainly by regulating levels of InsP5 and InsP6 (Chi et al., 2000, Romano et al., 1998). Strikingly, BT_4744 shares no significant sequence similarity with the bacterial or fungal phytases described in the literature (see Table S1). We shall refer to the BT_4744-encoded enzyme as BtMinpp, and below we further justify this characterization.
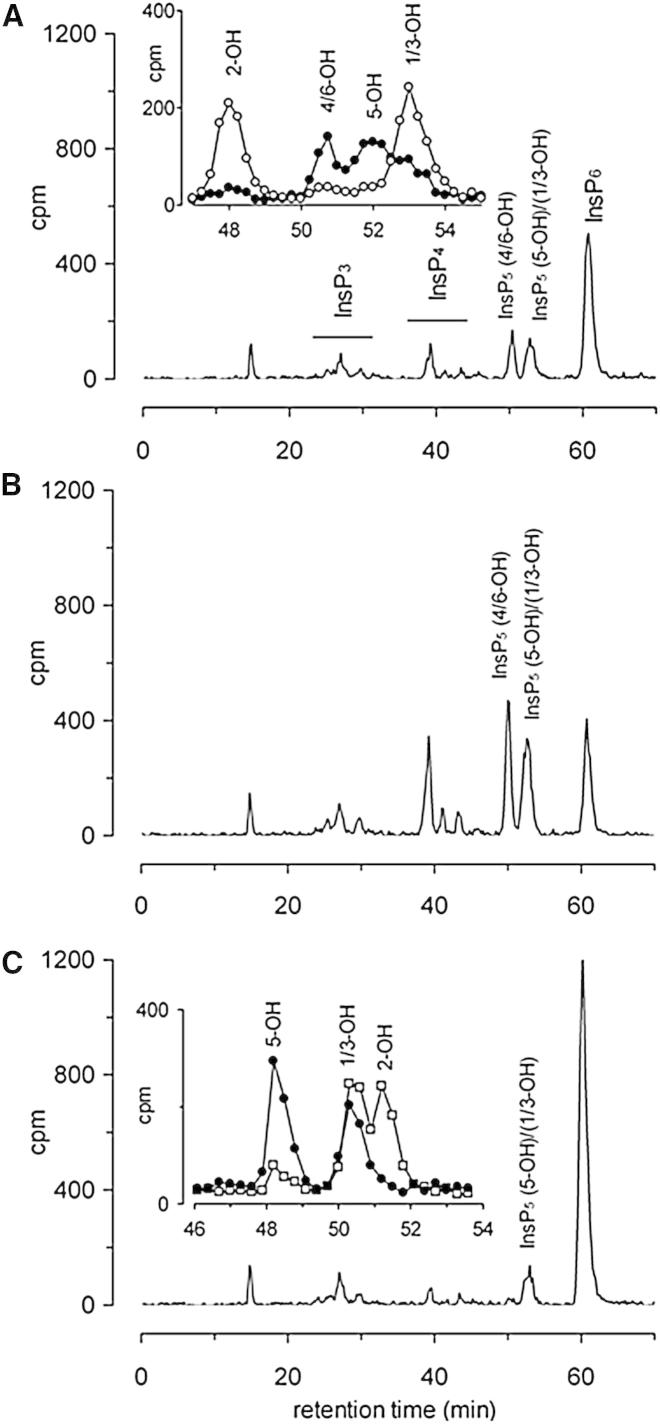
High-performance liquid chromatography (HPLC) analysis of the catalytic activity of a His-tagged recombinant form of BtMinpp confirmed that this protein has InsP6 phosphatase activity (Figure 1A). The Vmax value for InsP6 (178 μmol/mg/min; Table S2) is the highest yet reported for any member of the MINPP family, being several orders of magnitude greater than that for the human (0.006 μmol/mg/min) and avian (0.7 μmol/mg/min) representatives (Ali et al., 1995, Cho et al., 2006). We incubated BtMinpp with Ins(1,[32P]2,3,4,5,6)P6 substrate and identified the InsP5 products from their HPLC elution positions relative to myo-[2-3H]InsP5 standards (Figure 1A, inset). Although Ins(1,3,4,5,6)P5 (InsP5 [2-OH]) cannot be detected using Ins(1,[32P]2,3,4,5,6)P6 as substrate, we can be confident that InsP5 [2-OH] is not a major product because of the retention of radiolabel in progressively less-phosphorylated products, InsP4, InsP3, and InsP2 (Figure S1C). In any case, the use of Ins(1,[32P]2,3,4,5,6)P6 as substrate revealed that multiple InsP5 products were formed by BtMinpp, including D-and/or L-Ins(1,2,3,4,5)P5 (InsP5 [4/6-OH]), Ins(1,2,3,4,6)P5 (InsP5 [5-OH]), and D-and/or L-Ins(1,2,4,5,6)P5 (InsP5 [1/3-OH]) (Figure S2). Such a lack of specificity toward the site of initial attack on InsP6 is a defining characteristic of MINPPs of both plants (Dionisio et al., 2007) and animals (Figure S1A; Ali et al., 1995, Craxton et al., 1997). This catalytic flexibility contrasts with the more precise positional specificity of phytases. In fact, the distinct specificities of different types of phytases are sufficient to serve as a classification system: there are 3-phytases (E.C. 3.1.3.8) (see also Figure S1B), 6-phytases (E.C. 3.1.3.26), and 5-phytases (E.C. 3.1.3.72).
Figure 1.
Mutation of Active Site Residues Alters the Specificity of Initial Attack on InsP6 Substrate
(A–C) Products of reaction of native BtMinpp- (A), A31Y- (B), and R183D- (C) substituted enzyme with myo-inositol(1,[32P]2,3,4,5,6)P6 were resolved by Partisphere SAX HPLC. Mutated proteins were incubated at a concentration of 2.5 μg/ml and native protein at 0.25 μg/ml, with 1 mM InsP6. The regions of the chromatogram in which InsP3, InsP4, and specific InsP5 isomers elute are indicated in (A). For native enzyme (inset in A), reaction products were mixed with standards of myo-[2-3H]inositol (1,3,4,5,6)P5 (InsP5 [2-OH]), D/L- myo-[2-3H]inositol (1,2,4,5,6)P5 (InsP5 [1/3-OH]), and D/L- myo-[2-3H]inositol (1,2,3,4,5)P5 (InsP5 [4/6-OH]). Fractions, 0.25 min, were collected and radioactivity was estimated by scintillation counting; 3H, open circles; 32P, filled circles. For R183D-substituted enzyme (inset in C), the reaction products were additionally mixed with standards of myo-[14C]InsP5 [2-OH] and D/L- myo-[14C]InsP5 [1/3-OH] and resolved on a Adsorbosphere SAX HPLC column. This column separates InsP5 [2-OH] from InsP5 [1/3-OH]. Fractions, 0.25 min, were collected and radioactivity estimated by scintillation counting: 14C, open squares; 32P, filled circles.
Although mammalian MINPPs exhibit positional promiscuity toward InsP6 (Craxton et al., 1997), these enzymes are highly specific toward certain other inositol phosphate substrates. For example, they primarily remove the 3-phosphate from Ins(1,3,4,5,6)P5 and Ins(1,3,4,5)P4 (Craxton et al., 1997) and display a considerably higher Vmax toward Ins(1,3,4,5)P4 substrate than to InsP6 (Nogimori et al., 1991). The Bacteroides enzyme shares with mammalian MINPPs increased catalytic activity toward Ins(1,3,4,5)P4 substrate (Figure S2A), yielding a Vmax value 20- to 36-fold higher than that obtained with InsP6 (Table S2). In contrast, incubation of BtMinpp with Ins(1,3,4,5)P4 (Figure S2A) yielded two InsP3 peaks that coeluted with Ins(1,4,5)P3 and Ins(1,3,4)P3 standards. The pH optima of 2.5, 4.0, and 7.5 (Figure S3) overlaps with the pH profile of the human GI tract (Fallingborg, 1999), consistent with the successful adaptation of BtMinpp to this environment.
The Crystal Structure of BtMinpp in Complex with a Substrate Analog
There is no published description of the structures of any members of the MINPP family. The crystal structure (diffraction data were collected on beamlines I02 and I24 at the Diamond Light Source, UK) of BtMinpp expressed in Escherichia coli as an N-terminal 6 × His fusion protein was solved at pH 5.0 by SAD using phases from a selenomethionyl derivative. Refinement against native data at 1.93 Å resolution gave a final structural model with an Rcryst of 16.6% (Rfree 21.3%) and excellent geometry. A single inorganic phosphate group is bound at the active sites of both molecular copies of BtMinpp in the crystallographic asymmetric unit, presumably scavenged from the phosphate buffer used during purification of the enzyme (Table 1; Figure S4). A second structure was solved by difference Fourier analysis at 2.42 Å resolution in which the inorganic phosphate ions are displaced by the nonhydrolysable InsP6 analog, myo-inositol hexakissulfate (InsS6), again bound in the active sites of both molecules (Rcryst 15.6%; Rfree 21.7%) (Figure 2; Table 1). Comparison of the structures of analog- and phosphate-bound forms revealed an rmsd of 0.20 Å for Cα atoms suggesting limited conformational changes occur between the two states.
Table 1.
X-Ray Data Collection and Structure Refinement Statistics
| SAD Data | Phosphatea | InsS6a | |
|---|---|---|---|
| Data Collection | |||
| Wavelength (Å) | 0.9799 | 0.9778 | 0.9778 |
| Space group | P 21 | P 21 | P 21 |
| Cell parameters | |||
| a, b, c (Å) | 53.1, 121.0, 76.0 | 52.7, 120.6, 76.1 | 52.2, 117.3, 75.5 |
| β (°) | 107.8 | 107.9 | 107.46 |
| Resolution limits (Å) (high-resolution bin)b | 50.00–2.50 (2.64–2.50) | 62.07–1.93 (1.98–1.93) | 61.44–2.42 (2.48–2.42) |
| Rsymc | 0.098 (0.265) | 0.048 (0.473) | 0.069 (0.529) |
| <(I)/sd(I) > | 16.4 (7.3) | 12.6 (2.1) | 10.5 (2.9) |
| Completeness (%) | 100.0 (100.0) | 97.1 (97.0) | 99.2 (99.2) |
| Multiplicity | 7.4 (7.5) | 2.3 (2.4) | 3.3 (3.2) |
| Anomalous completeness (%) | 99.9 (100.0) | — | — |
| Anomalous multiplicity | 3.8 (3.8) | — | — |
| Overall temperature factor (Å2) | 26.0 | 48.7 | |
| Refinement | |||
| Protein monomers per asymmetric unit | 2 | 2 | |
| Total nonhydrogen atoms | 7194 | 6747 | |
| Water molecules | 451 | 127 | |
| Rcrystd (%) | 16.6 (23.5) | 15.6 (19.7) | |
| Rfreee (%) | 21.3 (28.2) | 21.7 (27.0) | |
| Ramachandran analysis (%) | |||
| Most favored | 97.58 | 97.37 | |
| Outliers | 0.25 | 0.25 | |
| Rmsds | |||
| Bonds, Å | 0.007 | 0.008 | |
| Angles, ° | 0.976 | 1.17 | |
| Planes, Å | 0.005 | 0.006 | |
| Mean atomic b value (Å2) | 24.2 | 39.8 | |
Phosphate and IHS refer to the complexes of BtMinpp with inorganic phosphate and myo-inositol hexakis sulfate, respectively.
Figures in brackets refer to the high resolution data bin as indicated.
Rsym = Σ∣Ii - 〈I〉∣/ Σ Ii where 〈I〉 is the average of symmetry equivalent reflections and the summation extends over all observations for all unique reflections.
Rcryst = Σ∣∣Fo∣-∣Fc∣∣/ Σ∣Fo∣ where Fo and Fc are the measured and calculated structure factors, respectively
For Rfree the summations extends over a randomly selected subset (5%) of reflections excluded from all stages of refinement
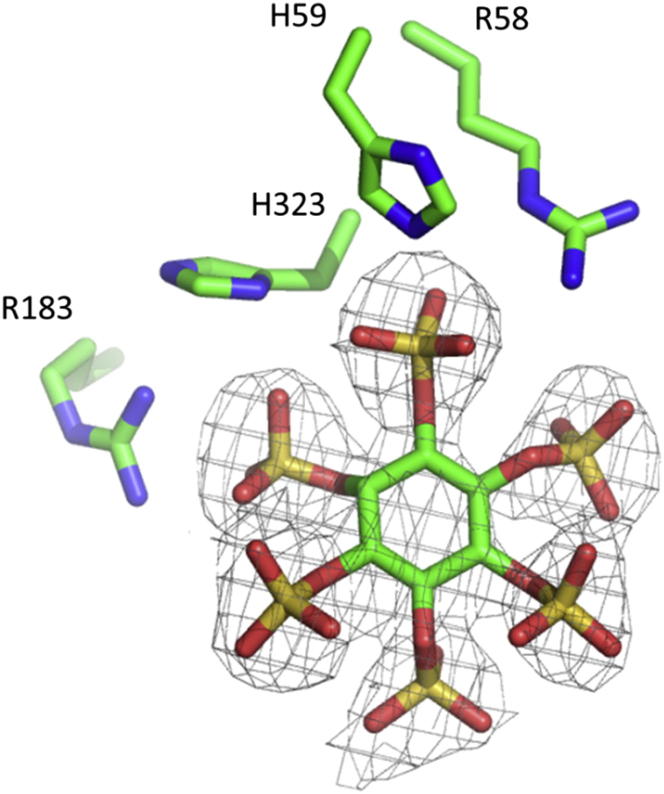
Figure 2.
Simulated Annealing Omit Map Revealing the Location of InsS6 Bound to the Active Site of BtMinpp
A region of the simulated annealing omit electron density map (gray lines) (Brünger and Rice, 1997) calculated with data to a resolution of 2.42 Å and contoured at 1.8 σ. Selected active site residues are shown in stick representation and labeled. The location of the inhibitor in the final refined structure of the complex is shown superimposed on the omit map electron density.
BtMinpp folds into two domains, an α/β domain and an α domain. Despite their low amino acid sequence homology, and functional diversity, all available crystal structures of members of branch 2 of the histidine phosphatase superfamily possess this general domain arrangement with a structurally conserved α/β domain (the “core” domain) and a more variable α-domain (the “cap” domain) (Rigden, 2008). The cleft between the two domains always contains the active site, which is highly specialized in each of the distinct branch two subfamilies, allowing them to perform different functions. The active site in BtMinpp is lined with predominantly basic amino acids, thereby rationalizing how the enzyme can bind unusually highly phosphorylated substrates such as InsP6 (Figure 3A). InsS6 is bound in the active site with its 3-sulfate group occupying the position taken by inorganic phosphate in the enzyme-phosphate complex and presumably that of the hydrolyzed phosphate group in substrates. This conclusion is consistent with the sulfur atom of this group lying 3.1 Å from the imidazole Nε2 atom of the side chain of H59. By analogy with other members of the superfamily, we predict that H59 of BtMinpp acts as a nucleophile during catalysis to generate a phosphohistidine intermediate. In phytases, the aspartate residue from a proximal HD sequence motif takes the role of proton donor for the leaving group (Kostrewa et al., 1997). However, a distinguishing feature of mammalian and plant MINPPs is the substitution of this motif with an HAE tripeptide in which the glutamic acid residue (E325 in BtMinpp) is the candidate proton donor. Indeed, in our crystal complex with InsS6 one of the E325 side-chain carboxyl oxygen atoms is only 3.3 Å from the ether oxygen bridging the 3-sulfate to the inositol ring, the analog to the phosphate-leaving group in InsP6.
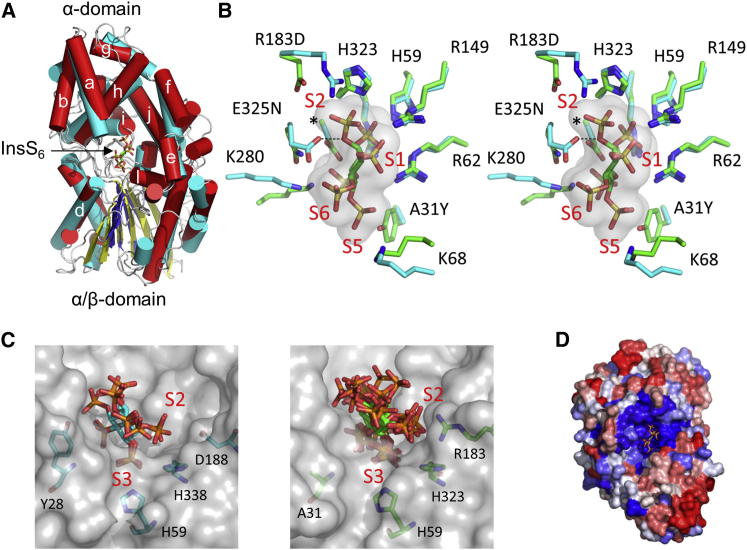
Figure 3.
X-Ray Crystal Structure of the BtMinpp
(A) Cylinder and ribbon diagram showing a superposition of the crystal structures of BtMinpp (α helices in red, β sheet in yellow) and the Aspergillus niger phytase PhyA (Oakley, 2010) (α helices in cyan, β sheet in blue) both taken from their complexes with the InsP6 analog, InsS6. The core α+β and capping α-domains are indicated. The naming of α helices in the capping domain of BtMinpp (indicated by a lowercase letter) follows that suggested for PhyA (Oakley, 2010). The atoms of the bound substrate analog are shown as sticks.
(B) A stereoview of the superposition of the side chains of InsS6-binding residues (shown as sticks) of BtMinpp (carbon atoms colored cyan) and Aspergillus niger PhyA (carbon atoms colored green). The residues selected for display form the primary sites of interaction with InsS6 in both complexes. The van der Waals surface of InsS6 is shown in gray. Residue labels follow the numbering for the BtMinpp structure. The leading character identifies the residue in BtMinpp, whereas the trailing identifies the corresponding variant residue found in PhyA. An asterisk (∗) indicates the position of the A324D substitution. Selected binding pockets (S1, S2, S5, and S6) are indicated, numbered according to corresponding sulfate group on the bound ligand. Pocket S4 is obscured in this orientation by the 6-sulfate group of the ligand. The interaction distance between a carboxyl group oxygen of residue E325 and the bridging ether oxygen of the S3 sulfate group of InsS6 is 3.3 Å and is indicated by a dashed line.
(C) Predicted binding modes within 1 kcal mol-1 of the minimum binding energy resulting from molecular docking experiments for InsP6 with PhyA (left, three modes) and BtMinpp (right, seven modes). Selected active site residues are shown as sticks and labeled as are binding pockets S2 and S3. The enzyme molecular surfaces are shown in gray.
(D) View of a ConSurf (Landau et al., 2005) color-coded surface representation of BtMinpp. The normalized conservation scores calculated by ConSurf are a relative measure of evolutionary conservation at each residue position based on the alignment of 23 MINPP sequences. The highest scores (8 and 9 on the ConSurf scale) represent the most conserved residue positions and are shown colored blue. The residues with the lowest scores (i.e., the most variable) are colored red. The bound InsS6 ligand is shown in stick format.
The structure of the InsS6-protein crystal complex rationalizes the ability of BtMinpp to remove the 3-phosphate from its natural substrate, InsP6. However, BtMinpp also removes alternative phosphates from InsP6 (Figure 1A), indicating that the enzyme’s active site can bind substrate in alternate orientations. To analyze which features of the catalytic core of BtMinpp permit this positional promiscuity, we compared its structure with that of A. niger PhyA phytase (Oakley, 2010), a member of a different subfamily of clade 2 histidine acid phosphatases, that hydrolyzes InsP6 by specific removal of its 3-phosphate (as does the closely related A. ficuum PhyA; Figure S2B). A comparison of this PhyA with BtMinpp yielded an rmsd of 2.5 Å, albeit with low sequence identity (16% over 360 structurally equivalent residues; Table S3). The overall topologies of these two enzymes are superficially similar, which is why we have labeled those secondary structural elements in BtMinpp that correspond to those found in PhyA (Figures 3A and S5).
Accordingly, we performed an overlay of the InsS6-bound structures of BtMinpp and A. niger phytase based on the coordinates of the backbone atoms of the active site fingerprint sequence (RHGXRXP; residues 58–64 in BtMinpp) found in the A-a loop and those of the short region containing the presumed proton donor, E325 (HAE; residues 323–325) from the N-terminal region of helix l. The rmsd was 0.30 Å for these 30 atoms. The conformations of InsS6 were very similar (rmsd 0.50 Å for the six inositol ring carbon atoms) in these overlayed structures. Nevertheless, this superposition (Figure 3B) revealed some significant differences in the nature of the ligand-binding residues in the two proteins. Binding pocket S2 shows the most extensive differences. Here, R183, conserved in MINPP enzymes and forming an ion pair with the 2-sulfate of the ligand, replaces the Asp in PhyA. Additionally, the HAE tripeptide of BtMinpp replaces the characteristic HD sequence motif in PhyA. Both substitutions substantially alter the volume and character of S2. The substitution of the phytase proton donor (Asp) with Ala324 in BtMinpp increases volume and decreases polarity of both S2 and the adjacent S4 pockets. Finally, a tyrosine residue that contributes to pockets S4 and S5 is replaced by Ala31in BtMinpp. These substitutions at positions 31 and 324 make the S4 and S5 pockets of BtMinpp significantly larger than those in PhyA. The significance of the larger BtMinpp ligand-binding site is that it may permit alternate substrate binding modes. Molecular docking experiments further support the concept of a reduced discrimination against alternative InsP6-binding modes in BtMinpp compared to PhyA (Figure 3C; Table S4): PhyA showed an exclusive preference for a limited set of highly similar InsP6 binding modes all presenting the D-3 phosphate in binding pocket S3. Termed the obverse binding mode, this places the D-2 axial phosphate in the S2 binding pocket. BtMinpp was more permissive, allowing binding of not only the D-3 phosphate in pocket S3 but also the D-5 and D-6 phosphates. These additional binding modes present equatorial phosphates of InsP6 in the S2 pocket and are distributed between obverse and reverse presentations (the latter having the inositol ring flipped over to present its opposite face to the enzyme). Given that the A31Y and R183D substitutions in phyA relative to BtMinpp were identified by our structural studies as making particularly significant contributions to the volume and polarity of the active site, we tested that hypothesis with a mutagenic approach.
Impact of Mutating the Active Site of BtMinpp on the Specificity of Initial Attack on InsP6 Substrate
When incubated with InsP6, the A31Y mutant of BtMinpp yielded an HPLC profile of InsP5 products (Figure 1B) very similar to that observed for wild-type enzyme (Figure 1A). Thus, the positional specificity toward InsP6 was not affected by this mutation. Nevertheless, it should be noted that the HPLC profile of the wild-type enzyme was obtained with one-tenth the protein concentration of that of the A31Y mutant (we estimated that the A31Y mutant was approximately 2.5-fold less active). Elevating the wild-type enzyme concentration 10-fold resulted in complete dephosphorylation of InsP5 and InsP6 (Figure S2B). Remarkably, the R183D substitution abolished the production of InsP5 [4/6-OH], but retained a peak with the retention time of InsP5 [5-OH]/ InsP5 [1/3-OH] (Figure 1C). Using an Adsorbosphere SAX column to resolve InsP5 [5-OH] from InsP5 [1/3-OH] and InsP5 [2-OH], we confirmed that the R183D mutant had lost the ability to attack the 4/6-position (compare Figures 1A and 1C), but retained the ability to attack the 5- and 1/3-positions (Figure 1C, inset). Thus, our mutagenic data confirm that the positional flexibility of BtMinpp toward InsP6 reflects a larger and less discriminating active site compared to phyA. The R183D mutant was also about 10-fold less active than wild-type enzyme (compare Figures 1A and 1C), suggesting that increased catalytic activity may be an evolutionary advantage of BtMinpp’s more flexible active site.
Given that human MINPP1 and BtMinpp share a common ability to attack InsP6 substrate at different positions and remove a 3-phosphate from Ins(1,3,4,5)P4 (see above), we considered whether our crystallographic analysis of BtMinpp offers an explanation of the substrate specificity of mammalian MINPPs. Analysis of the conservation of residues that are predicted to contribute atoms to the molecular surface of 23 representative MINPP enzymes from different kingdoms of life reveals highest sequence identity in a region of the active site centered on H59 (Figure 3D). There is 52% sequence identity between human MINPP1 and BtMinpp for those active site residues that have an atom within 8 Å of the InsS6 bound to the bacterial enzyme. Interestingly, residue A31, forming part of binding pocket S5 in BtMinpp, is commonly replaced with a lysine in other MINPP enzymes (Figure S5). Introduction of a lysine residue may allow additional polar contacts with highly negatively charged substrates. Its size may also serve to make binding pocket S5 smaller. This substitution may therefore be of relevance in explaining the differential activity of Bacteroides and eukaryotic enzymes toward lower inositol phosphate substrates by enforcing more positional specificity on the mammalian enzyme.
The MINPP Protein Family Includes Microbial Members
Having defined Minpp as a bacterial product, we next performed a phylogenetic analysis to determine how widespread is the distribution of minpp homologs in microbial organisms. A BLASTP search of BtMinpp encoded protein sequence against the nonredundant protein database identified 326 sequences with significant alignments (e value ≤ 1 × 10−4 and a minimal number of 60 identical amino acids over the entire sequence length). These included proteins of bacterial, animal, and plant origin. Whereas most bacterial phyla were represented, no representative of the Firmicutes, a major phylum of the human GI tract microbiota, was included. Nevertheless, a significant number of the other dominant GI-resident organisms (Bacteroides, Bifidobacterium, Prevotella, and Alistipes) encode proteins belonging to the MINPP family. In contrast, a search for microbial genes predicted to encode microbial phytase enzymes among 341 genomes of the human GI tract (Human Microbiome Project at http://commonfund.nih.gov/hmp/index) revealed that microbial phytases are scarce in this environment and are only present in the genome of subdominant bacterial species (Table S5). This analysis leads us to conclude that bacterial Minpps are the major representative of InsP6-degrading enzymes within the GI tract of humans and are encoded by dominant bacterial species that are niche-adapted human symbionts.
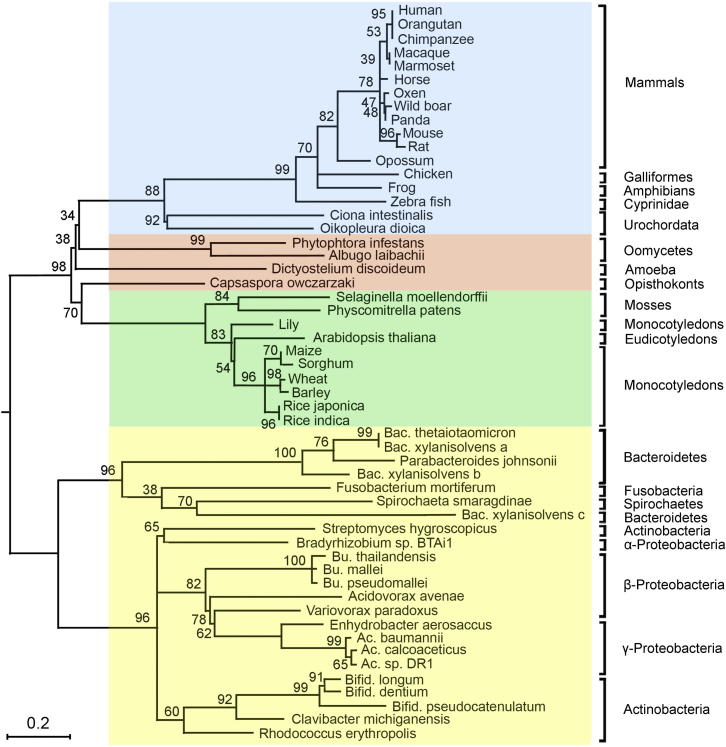
A phylogenetic tree (Figure 4) was constructed from the alignment of 54 representatives (Table S6) of different kingdoms of life. In general, MINPP-related sequences are found within the bacteria, plant, and animal kingdoms with the sequences in plants and animals being evolutionarily conserved within their respective clade. However, in the bacterial group, the Bacteroidetes phylum does not group with the Actinobacteria phylum as previously reported (Ciccarelli et al., 2006). The different Minpps from the bacterial species displayed in the tree all appear equally related to their eukaryotic homologs (Figure 4) with the eukaryotic and bacterial proteins each forming monophyletic groups, suggesting an ancient origin for the MINPP family of proteins.
Figure 4.
The MINPP Family Phylogenetic Tree
A phylogenetic tree derived from the alignment of 54 MINPP protein representatives from different kingdoms of life was constructed using the Maximum Likelihood method. The different kingdoms have been assigned the following background colors: animals, blue; plants, green; protists, salmon-pink; bacteria, yellow. Ac, Acinetobacter; Bac, Bacteroides; Bu, Burkholderia; Bifid, Bifidobacterium. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site.
Among the 2,536 completed bacterial genomes listed in EMBL-EBI (http://www.ebi.ac.uk/genomes/bacteria.html), only 55 contained one or more copies of a gene homologous to BtMinpp, accounting for 2.2% of the completed genomes. In contrast, greater than 50% of animal and plant completed genomes contained at least one copy of a gene predicted to encode a MINPP representative. Our phylogenetic analyses strengthen the conclusion from our biochemical and structural studies (see above) that the bacterial Minpp-related protein sequences constitute a different family, distinct from phytases, but nevertheless a subset of clade 2 of the histidine phosphatase superfamily (HAP; IPR000560) (Rigden, 2008).
InsP6 Metabolism by BtMinpp Secreted from B. thetaiotaomicron in Outer Membrane Vesicles
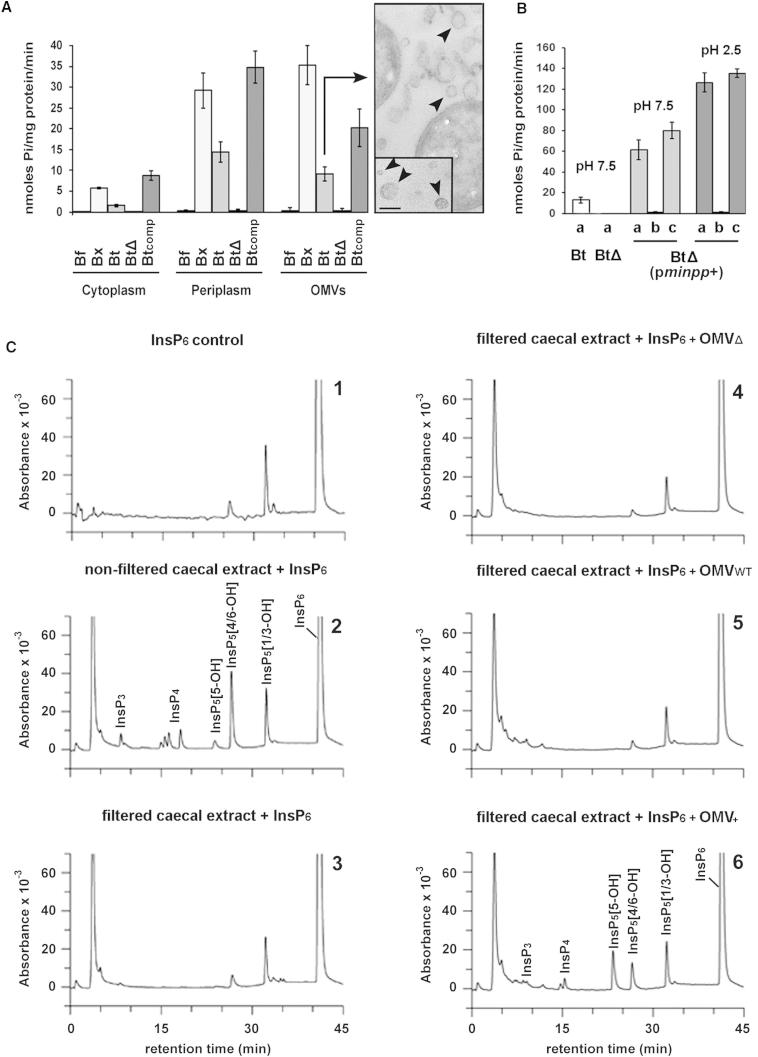
The key question of how BtMinpp in the gut survives a hostile protease-containing environment yet accesses extracellular InsP6 was addressed. BtMinpp is predicted to contain an N-terminal signal peptide (http://www.cbs.dtu.dk/services/SignalP/), so we hypothesized that the protein would be secreted into the periplasmic space. Indeed, the periplasmic fraction of B. thetaiotaomicron showed substantial Minpp activity (Figure 5A). This observation led us to investigate if BtMinpp might also be packaged into outer membrane vesicles (OMVs). Although OMV blebbing from the cell surface has been observed in different Bacteroides species, B. fragilis is the only example for which various enzymatic activities in OMVs have previously been assayed (Patrick et al., 1996). Transmission electron microscopy confirmed the presence of membrane blebs and the release of intact OMVs by B. thetaiotaomicron (Figure 5A). Substantial InsP6 phosphatase activity was detected in OMV protein extracts (Figure 5A).
Figure 5.
Intact OMVs Produced by B. thetaiotaomicron Degrade Exogenous InsP6
(A) Minpp activity was assessed in different fractions of B. thetaiotaomicron and other Bacteroides spp. using InsP6 as a substrate and a molybdate/malachite green-based enzyme assay to measure released phosphate. The different fractions were extracted from B. fragilis (Bf), B. xylanisolvens (Bx), B. thetaiotaomicron (Bt), Bt minpp-deleted mutant strain GH59 (BtΔ), and the same strain containing the pGH38 plasmid (GH120) expressing BtMinpp (Btcomp). The values shown represent the mean ± SEM values obtained from five to six independent experiments. The TEM images show Bt cells and associated OMVs (arrowed) with the inset showing images of isolated OMVs. Scale bar, 100 nm.
(B) (a) Degradation of InsP6 by intact OMVs produced by B. thetaiotaomicron wild-type strain (Bt), the minpp-deleted mutant (BtΔ), and the minpp-deleted mutant containing the pGH037 plasmid overexpressing BtMinpp (BtΔ (pminpp+). OMVs were isolated, concentrated, and resuspended in a Tris buffer (pH 7.5) and for BtΔ (pminpp+) in a glycine-HCl buffer (pH 2.5) to which InsP6 substrate was added and the suspension was incubated for 1 hr at 37°C. (b) BtMinpp activity was measured in the buffer fraction of BtΔ (pminpp+) OMV suspensions after incubation for 1 hr at 37°C and removal of OMVs. (c) Enzyme activity was measured in sonicated extracts of OMVs recovered from the mixtures described in (b). The values shown represent the mean ± SEM values obtained from at least five independent experiments.
(C) Cecal contents were obtained from C57BL/6 mice as described in the Experimental Procedures. Each HPLC chromatogram panel shown is representative of samples from four different mice. InsP6 was added to all samples. (C1) InsP6 control, showing contaminating InsP5s of this commercial InsP6 sample; (C2) nonfiltered cecal extract; (C3) cecal extracts filtered (100 kDa molecular weight cutoff membrane); (C4–6) cecal extracts filtered (100 kDa molecular weight cutoff membrane) and supplemented with OMVs from either a BtMinpp-deleted mutant (C4), the WT strain (C5), or a strain overexpressing Minpp (C6).
We prepared the protein fraction from the cytoplasm, periplasm and OMVs of a strain of B. thetaiotaomicron from which we had deleted the minpp gene. All of these fractions had negligible InsP6 phosphatase activity (Figure 5A), indicating that BtMinpp accounts for the InsP6 phosphatase activity observed in the corresponding fractions prepared from the wild-type strain (Figure 5A). Expression of minpp in trans in the mutant increased the activity to levels higher than in the wild-type parental strain in each of the subcellular fractions (Figure 5A). It is noteworthy that no InsP6 hydrolysis activity was detected in another human gut Bacteroides species, B. fragilis, that does not contain a gene related to minpp. In contrast, high levels of InsP6 phosphatase activity were detected in the periplasm and OMVs of B. xylanisolvens (Figure 5A), which contains three copies of the minpp gene.
We next investigated the capacity of intact vesicles containing BtMinpp to degrade InsP6 in the external milieu. InsP6 was hydrolyzed by OMVs produced and isolated from the wild-type strain, whereas no phosphate release from InsP6 was detected by OMVs produced by the Minpp1-deficient strain (Figure 5B). We also engineered a B. thetaiotaomicron strain that overexpresses BtMinpp and recorded between 6 and 12 times more InsP6 degradation for its isolated OMVs compared to those from wild-type B. thetaiotaomicron (Figure 5B). The conditioned media from which OMVs had been removed showed no enzyme activity; sonication of the OMVs was required for enzyme activity to be released (Figure 5B). These findings are consistent with OMVs being intact and that the enzyme is retained in the vesicles and is not released due to leakage.
The functional significance of this packaging of BtMinpp in OMVs was investigated in physiological experiments using the cecal contents of mice to which InsP6 substrate was added and using HPLC to detect InsP6 metabolites. Note that in these experiments the commercial InsP6 sample was contaminated with small peaks of InsP5 (Figure 5C1). Control cecal extracts contained InsP6-phosphatase activity (Figure 5C2) that was no longer detected after filtration using a 100 kDa cutoff membrane (Figure 5C3), capable of removing large protein complexes or OMVs produced by resident bacteria. In further experiments, no significant InsP6-phosphatase activity was observed in filtered cecal extracts supplemented with OMVs prepared from Minpp-deficient bacteria (Figure 5C4). The assay was also unable to detect InsP6 phosphatase activity in OMVs produced by wild-type B. thetaiotaomicron (Figure 5C5), probably due to limitations in its sensitivity (see Figure 5B). However, when filtered cecal contents were supplemented with OMVs produced by B. thetaiotaomicron that overexpress Minpp, considerable InsP6-phosphatase activity was detected (Figure 5C6). Collectively, these findings demonstrate that BtMinpp is retained inside the OMVs, which InsP6 must enter in order to access the BtMinpp.
Minpp1-Loaded Vesicles Induce the Release of Intracellular Calcium in Colonic Epithelial Cells
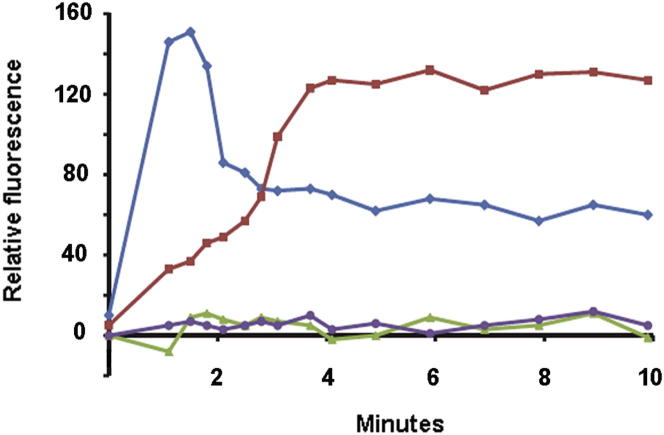
It is well established that OMVs produced by bacterial pathogens such as Helicobacter pylori, Legionella pneumophila, or E. coli are capable of interacting with host cells via a membrane fusion event or via adhesin-receptor-mediated attachment to deliver virulence factors such as proteases and toxins (Ellis and Kuehn, 2010). We hypothesized therefore that either mechanism might be used by B. thetaiotaomicron OMVs to deliver BtMinpp into host intestinal epithelial cells, with the potential consequence that BtMinpp could interact with the inositol polyphosphate signaling pathways of host cells. Indeed, Yu et al. (2003) have shown that in mammalian cells expression of an engineered truncated cytosolic form of the endoplasmic reticulum-confined MINPP1 leads to a significant enhancement of Ins(1,4,5)P3 concentration, triggering the release of calcium from intracellular stores via the InsP3 receptor/Ca2+ channel. To test these ideas, BtMinpp-containing OMVs were added to HT29 human colonic epithelial cells in the absence of extracellular calcium. Immediately after addition of the OMVs, a gradual increase in intracellular calcium concentration was observed that plateaued within approximately 4 min (Figure 6). By contrast, no calcium response was observed when HT29 cells were incubated with OMVs that did not contain any BtMinpp. This result suggests that OMVs interact with epithelial cells leading to the release of BtMinpp and generation of InsP3 products. Additionally, it is possible that attachment of OMVs to target cells may locally increase the concentration of an InsP6 metabolite that stimulates Ca2+ mobilization. Nevertheless, irrespective of their mechanism of action, OMVs containing BtMinpp are able, in vitro, to trigger the release of calcium from intracellular stores to the cytosol of colonic epithelial cells.
Figure 6.
Minpp1-Loaded Vesicles Modify Colonic Epithelial Cell Signaling
Fluorescence intensity of Fluo-8 AM-loaded HT29 cells in response to treatment with ionomycin (blue), OMVs from a BtMinpp-deleted mutant (purple), OMVs containing the overexpressed BtMinpp (red), or PBS alone (green). The data shown are representative of three experiments, each with three replicates.
Discussion
The main impact of our study centers on the identification of a homolog of a eukaryotic inositolphosphate phosphatase, MINPP, in major species of human gut bacterial genomes. Detailed biomolecular and phylogenetic analyses of Minpp from B. thetaiotaomicron (BtMinpp) validated it as a member of the MINPP family with an exceptionally high catalytic activity that makes it exquisitely suited for facilitating InsP6 homeostasis in the mammalian GI tract. Moreover, we rationalize the catalytic activity and evolutionary conservation of BtMinpp by mutagenic studies and by an atomic-level description of the structure of this enzyme at 1.9 Å resolution. We further demonstrate that BtMinpp is packaged inside OMVs, thereby protecting the phosphatase activity from degradation by gastrointestinal proteases, and also facilitating an example of cross-kingdom, long-range, cell-to-cell signaling; we show that the OMVs that are released by B. thetaiotaomicron are able to deliver BtMinpp to intestinal epithelial cells, promoting intracellular Ca2+ signaling.
The physiological significance of InsP6 phosphatase activity in the human gut is likely complex and multifactorial. First, there is nutritional benefit to both the host and the bacterial community from the inorganic phosphate and the inositol moiety that are both released. Additionally, the hydrolysis of InsP6 eliminates its antinutritive properties, such as divalent ion chelation and inhibition of polysaccharide digestibility. High concentrations of InsP6 have been considered to have anticarcinogenic properties in the human colon (Fox and Eberl, 2002, Vucenik and Shamsuddin, 2003). However, a recent paper indicates that lower and more physiologically relevant levels of InsP6 in the diet might promote tumor development (Windhorst et al., 2013). This last finding raises the possibility of further human health impacts of bacterial Minpp in the GI tract.
B. thetaiotaomicron is a dominant human GI symbiont. Moreover, our phylogenetic studies indicate that the majority of other bacteria known to harbor a minpp gene are also members of the human intestinal microbiota. These 149 representatives are from the Bacteroides, Bifidobacterium, Prevotella, and Alistipes communities. Indeed, our analysis of InsP6 hydrolysis ex vivo from endogenous cecal contents indicates that the endogenous enzymatic activity is positionally promiscuous toward its substrates, and contained in OMVs. These are both properties of BtMinpp making it likely that bacterial Minpps are a widespread yet hitherto unappreciated source of highly active InsP6 phosphatase activity in the GI tract.
Our characterization of the crystal structure of BtMinpp provides a description of members of the MINPP family at the atomic level. In particular, the structure of the protein in complex with InsS6, a substrate analog, gave insight into the reaction mechanism. We propose that H59 acts as a nucleophile for cleavage of the 3-phosphate from InsP6, with E325 in an HAE tripeptide being the proton donor for the leaving group. Our structural work also illuminates the positional promiscuity of the MINPP superfamily, which distinguishes it from the various classes of phytases in the HAP family that each exhibits a distinct preference for a particular phosphate group (e.g., 3-phytases, 4/6-phytases, and 5-phytases; Konietzny and Greiner, 2002, Kumar et al., 2010). Our overlay of the structure of the active sites of A. niger 3-phytase and BtMinpp revealed that the latter has a larger and less polar ligand-binding pocket that, according to our molecular models, accepts InsP6 in several different orientations (Figure 3C). Furthermore, in a synthetic biology approach, we mutated R183 in BtMinpp to the corresponding Arg in the 3-phytase in A. niger. This resulted in BtMinpp losing the ability to remove the 4/6-phosphate from InsP6 (Figure 1). That is, the R183D mutation caused the positional preference of the enzyme to become more phytase-like in nature.
Interestingly, A31 in BtMinpp is commonly replaced with Lys in other MINPPs (Figure S5), and so this residue may be of relevance in explaining the differential positional specificities of Bacteroides and eukaryotic enzymes toward lower inositol phosphate substrates. For example, the mammalian MINPPs only removes the 3-phosphate from Ins(1,3,4,5)P4 (Caffrey et al., 1999), whereas the bacterial enzyme is less selective. It is an intriguing possibility that a lack of positional selectivity may be associated with elevated catalytic activity of BtMinpp against InsP6. Nevertheless, we found a 52% sequence identity between human MINPP1 and BtMinpp for those active site residues that have an atom within 8 Å of the InsS6 that cocrystallized in the active site. Such a high degree of evolutionary conservation would suggest that bacterial and mammalian Minpps have originated from a common primordial ancestor. It is alternately possible that bacteria acquired Minpps from eukaryotes by horizontal gene transfer (HGT), although known cases of this phenomenon are rare (Keeling and Palmer, 2008). Our phylogenetic analyses were unable to distinguish between these two alternatives.
Our characterization of the properties of a vesicularized bacterial homolog of a mammalian signaling enzyme challenges established orthodoxies concerning our understanding of the mechanisms by which symbiotic bacteria in the mammalian GI tract interact with their host. First, it has been thought that OMVs primarily mediate pathogenic processes (Ellis and Kuehn, 2010). Our data reveal that commensal gut bacteria also utilize OMVs in a manner that is beneficial to the host, by contributing to InsP6 homeostasis. The ability of BtMinpp-containing OMVs to stimulate intracellular Ca2+ release in human colonic epithelial cells suggests a further biological significance to bacterial Minpps, namely, a role in interkingdom communication pathways (Figure 6). Another example of this phenomenon is the OMV-mediated delivery of immunoregulatory capsular polysaccharide A from B. fragilis to host dendritic cells to effect disease protection (Shen et al., 2012). Nevertheless, the application of an enzyme to mediate dialog between gut bacteria and the human host is an addition to a field of research that has previously focused on the roles of diffusible, small-molecule hormones and nutrients. Further research into variations in the expression and secretion of Minpps by the different species of GI bacteria could increase our insight into the complex interrelationships between the microbiome and the human host.
Experimental Procedures
Bacterial Strains and Growth Conditions
All E. coli and Bacteroides strains used in this study are listed in Table S7. The bacterial growth conditions are described in the Supplemental Information.
HPLC
Inositol phosphate products of assays using 32P-labeled substrate were resolved by HPLC, details of which are provided in the Supplemental Information.
Crystal Structure Determination
A single crystal of selenomethionyl-derivitized BtMinpp was used to collect a SAD data set, which was processed using the CCP4 package to obtain a crystal structure as described in the Supplemental Information.
Phylogenetic Analysis
The evolutionary history of the MINPP protein was inferred using the Maximum Likelihood method in the MEGA5 software tool (Tamura et al., 2011). Amino acid sequences were aligned with PRANK and highly variable regions removed from the data set. The Whelan and Goldman (2001) model of evolution was used, and initial tree(s) for the heuristic search was obtained automatically as follows. When the number of common sites was <100 or less than one-fourth of the total number of sites, the maximum parsimony method was used; otherwise, BIONJ method with MCL distance matrix was used. To provide statistical support for each node on the tree, a consensus tree was generated from 1,000 bootstrap data sets.
Periplasmic and OMV Protein Extraction and BtMinpp Phosphatase Activity Measurement
The method of Osborn et al. (1972) was used to obtain periplasmic fractions of B. thetaiotaomicron and OMVs were harvested from bacterial cultures by ultracentrifugation and assayed for phosphatase activity using the PiColorLock Gold Phosphate Detection System as described in the Supplemental Information.
Intracellular Calcium Measurement
HT-29 cells were propagated in Sarstedt culture flasks (25 cm2) in a 5% CO2 humidified atmosphere at 37°C. Cells were fed with DMEM (Lonza) supplemented with 10% heat-inactivated FBS (Biosera) and 2 mM L-glutamine (Lonza). For the intracellular calcium measurement assay, cells were seeded in 24-well microplates (Corning CellBind Surface) at a density of 105 cells/well. After 16 hr, the culture medium was replaced with fresh medium containing 5 μM of the calcium indicator Fluo-8 AM (AAT Bioquest), 4 mM of Probenecid (Sigma-Aldrich), and 0.025% (w/v) Pluronic acid F-127 (Invitrogen) according to a method adapted from Abrahamse and Rechkemmer, 2001, and cells were incubated for 30 min at 37°C, 5% CO2. The medium was removed, and calcium-free PBS was added before the cells were incubated at 37°C for another 30 min. The cell monolayer was washed twice with calcium-free PBS and PBS was added. Fluo-8 fluorescence was measured immediately after addition of Ionomycin (Sigma-Aldrich) 1 μg/ml, PBS, or purified OMVs (corresponding to 10 μg of soluble protein) with a FLUOstar Optima fluorescence plate reader (BMG Labtechnologies) fitted with custom excitation/emission filters (485/538 nm). OMVs were purified from 500 ml culture supernatants, washed with calcium-free PBS, and concentrated to a 2 ml suspension.
Author Contributions
R.S. and S.R.C. conceived the study and R.S., S.R.C., C.A.B., S.B.S., and A.M.H. wrote the paper. R.S., S.R.C., C.A.B., A.M.H., P.B., and S.B.S. designed experiments and analyzed data. R.S., S.O., N.H., A.W.H.L., C.A.B., I.H., R.B., M.R., and S.B.S. performed the experiments. S.R.C. and S.B.S. obtained funding to support the study.
Acknowledgments
We would like to thank Kathryn Cross for help with electron microscopy, Dr. Ida Porcelli for providing advice on periplasmic extraction, and Dr. Nathalie Juge for scientific discussions. This work was supported by institutional grants from Biotechnology and Biological Sciences Research Council (BBSRC; BB/J004529/1, S.R.C.), by a BBSRC postgraduate studentship (BB/F016816/1, S.O.), and by the Intramural Research Program of the National Institutes of Health and National Institute of Environmental Health Sciences (S.B.S.).
Published: February 13, 2014
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information includes Supplemental Experimental Procedures, five figures, and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.01.021.
Contributor Information
Charles A. Brearley, Email: c.brearley@uea.ac.uk.
Simon R. Carding, Email: simon.carding@ifr.ac.uk.
Accession Numbers
The Protein Data Bank ID codes for the atomic coordinates and structure factors for the phosphate- and InsS6-bound complexes of BtMinpp reported in this paper are 4FDT and 4FDU, respectively.
Supplemental Information
References
- Abrahamse S.L., Rechkemmer G. Identification of an organic anion transport system in the human colon carcinoma cell line HT29 clone 19A. Pflugers Arch. 2001;441:529–537. doi: 10.1007/s004240000437. [DOI] [PubMed] [Google Scholar]
- Ali N., Craxton A., Sumner M., Shears S.B. Effects of aluminium on the hepatic inositol polyphosphate phosphatase. Biochem. J. 1995;305:557–561. doi: 10.1042/bj3050557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T., Rice L.M. Crystallographic refinement by simulated annealing: methods and applications. Methods Enzymol. 1997;277:243–269. doi: 10.1016/s0076-6879(97)77015-0. [DOI] [PubMed] [Google Scholar]
- Caffrey J.J., Hidaka K., Matsuda M., Hirata M., Shears S.B. The human and rat forms of multiple inositol polyphosphate phosphatase: functional homology with a histidine acid phosphatase up-regulated during endochondral ossification. FEBS Lett. 1999;442:99–104. doi: 10.1016/s0014-5793(98)01636-6. [DOI] [PubMed] [Google Scholar]
- Chakraborty A., Koldobskiy M.A., Bello N.T., Maxwell M., Potter J.J., Juluri K.R., Maag D., Kim S., Huang A.S., Dailey M.J. Inositol pyrophosphates inhibit Akt signaling, thereby regulating insulin sensitivity and weight gain. Cell. 2010;143:897–910. doi: 10.1016/j.cell.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi H.B., Yang X.N., Kingsley P.D., O’Keefe R.J., Puzas J.E., Rosier R.N., Shears S.B., Reynolds P.R. Targeted deletion of Minpp1 provides new insight into the activity of multiple inositol polyphosphate phosphatase in vivo. Mol. Cell. Biol. 2000;20:6496–6507. doi: 10.1128/mcb.20.17.6496-6507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J., Choi K., Darden T., Reynolds P.R., Petitte J.N., Shears S.B. Avian multiple inositol polyphosphate phosphatase is an active phytase that can be engineered to help ameliorate the planet’s “phosphate crisis”. J. Biotechnol. 2006;126:248–259. doi: 10.1016/j.jbiotec.2006.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli F.D., Doerks T., von Mering C., Creevey C.J., Snel B., Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Craxton A., Caffrey J.J., Burkhart W., Safrany S.T., Shears S.B. Molecular cloning and expression of a rat hepatic multiple inositol polyphosphate phosphatase. Biochem. J. 1997;328:75–81. doi: 10.1042/bj3280075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionisio G., Holm P.B., Brinch-Pedersen H. Wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.) multiple inositol polyphosphate phosphatases (MINPPs) are phytases expressed during grain filling and germination. Plant Biotechnol. J. 2007;5:325–338. doi: 10.1111/j.1467-7652.2007.00244.x. [DOI] [PubMed] [Google Scholar]
- Ellis T.N., Kuehn M.J. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev. 2010;74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallingborg J. Intraluminal pH of the human gastrointestinal tract. Dan. Med. Bull. 1999;46:183–196. [PubMed] [Google Scholar]
- Fox C.H., Eberl M. Phytic acid (IP6), novel broad spectrum anti-neoplastic agent: a systematic review. Complement. Ther. Med. 2002;10:229–234. doi: 10.1016/s0965-2299(02)00092-4. [DOI] [PubMed] [Google Scholar]
- Haefner S., Knietsch A., Scholten E., Braun J., Lohscheidt M., Zelder O. Biotechnological production and applications of phytases. Appl. Microbiol. Biotechnol. 2005;68:588–597. doi: 10.1007/s00253-005-0005-y. [DOI] [PubMed] [Google Scholar]
- Haros M., Bielecka M., Honke J., Sanz Y. Myo-inositol hexakisphosphate degradation by Bifidobacterium infantis ATCC 15697. Int. J. Food Microbiol. 2007;117:76–84. doi: 10.1016/j.ijfoodmicro.2007.02.021. [DOI] [PubMed] [Google Scholar]
- Keeling P.J., Palmer J.D. Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- Konietzny U., Greiner R. Molecular and catalytic properties of phytate-degrading enzymes (phytases) Int. J. Food Sci. 2002;37:791–812. [Google Scholar]
- Kostrewa D., Grüninger-Leitch F., D’Arcy A., Broger C., Mitchell D., van Loon A.P. Crystal structure of phytase from Aspergillus ficuum at 2.5 A resolution. Nat. Struct. Biol. 1997;4:185–190. doi: 10.1038/nsb0397-185. [DOI] [PubMed] [Google Scholar]
- Kumar V., Sinha A.K., Makkar H.P.S., Becker K. Dietary roles of phytate and phytase in human nutrition: A review. Food Chem. 2010;120:945–959. [Google Scholar]
- Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33(Web Server issue):W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michell R.H. Inositol derivatives: evolution and functions. Nat. Rev. Mol. Cell Biol. 2008;9:151–161. doi: 10.1038/nrm2334. [DOI] [PubMed] [Google Scholar]
- Nogimori K., Hughes P.J., Glennon M.C., Hodgson M.E., Putney J.W., Jr., Shears S.B. Purification of an inositol (1,3,4,5)-tetrakisphosphate 3-phosphatase activity from rat liver and the evaluation of its substrate specificity. J. Biol. Chem. 1991;266:16499–16506. [PubMed] [Google Scholar]
- Oakley A.J. The structure of Aspergillus niger phytase PhyA in complex with a phytate mimetic. Biochem. Biophys. Res. Commun. 2010;397:745–749. doi: 10.1016/j.bbrc.2010.06.024. [DOI] [PubMed] [Google Scholar]
- Osborn M.J., Gander J.E., Parisi E., Carson J. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J. Biol. Chem. 1972;247:3962–3972. [PubMed] [Google Scholar]
- Patrick S., McKenna J.P., O’Hagan S., Dermott E. A comparison of the haemagglutinating and enzymic activities of Bacteroides fragilis whole cells and outer membrane vesicles. Microb. Pathog. 1996;20:191–202. doi: 10.1006/mpat.1996.0018. [DOI] [PubMed] [Google Scholar]
- Prasad A., Jia Y.H., Chakraborty A., Li Y.T., Jain S.K., Zhong J., Roy S.G., Loison F., Mondal S., Sakai J. Inositol hexakisphosphate kinase 1 regulates neutrophil function in innate immunity by inhibiting phosphatidylinositol-(3,4,5)-trisphosphate signaling. Nat. Immunol. 2011;12:752–760. doi: 10.1038/ni.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigden D.J. The histidine phosphatase superfamily: structure and function. Biochem. J. 2008;409:333–348. doi: 10.1042/BJ20071097. [DOI] [PubMed] [Google Scholar]
- Romano P.R., Wang J., O’Keefe R.J., Puzas J.E., Rosier R.N., Reynolds P.R. HiPER1, a phosphatase of the endoplasmic reticulum with a role in chondrocyte maturation. J. Cell Sci. 1998;111:803–813. doi: 10.1242/jcs.111.6.803. [DOI] [PubMed] [Google Scholar]
- Shen Y., Giardino Torchia M.L., Lawson G.W., Karp C.L., Ashwell J.D., Mazmanian S.K. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe. 2012;12:509–520. doi: 10.1016/j.chom.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steer T.E., Gee J.N., Johnson I.T., Gibson G.R. Biodiversity of human faecal bacteria isolated from phytic acid enriched chemostat fermenters. Curr. Issues Intest. Microbiol. 2004;5:23–39. [PubMed] [Google Scholar]
- Szijgyarto Z., Garedew A., Azevedo C., Saiardi A. Influence of inositol pyrophosphates on cellular energy dynamics. Science. 2011;334:802–805. doi: 10.1126/science.1211908. [DOI] [PubMed] [Google Scholar]
- Tamayo-Ramos J.A., Sanz-Penella J.M., Yebra M.J., Monedero V., Haros M. Novel phytases from Bifidobacterium pseudocatenulatum ATCC 27919 and Bifidobacterium longum subsp. infantis ATCC 15697. Appl. Environ. Microbiol. 2012;78:5013–5015. doi: 10.1128/AEM.00782-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremaroli V., Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- Vucenik I., Shamsuddin A.M. Cancer inhibition by inositol hexaphosphate (IP6) and inositol: from laboratory to clinic. J. Nutr. 2003;133(Suppl 1):3778S–3784S. doi: 10.1093/jn/133.11.3778S. [DOI] [PubMed] [Google Scholar]
- Whelan S., Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Windhorst S., Lin H.Y., Blechner C., Fanick W., Brandt L., Brehm M.A., Mayr G.W. Tumour cells can employ extracellular Ins(1,2,3,4,5,6)P(6) and multiple inositol-polyphosphate phosphatase 1 (MINPP1) dephosphorylation to improve their proliferation. Biochem. J. 2013;450:115–125. doi: 10.1042/BJ20121524. [DOI] [PubMed] [Google Scholar]
- Xu J., Bjursell M.K., Himrod J., Deng S., Carmichael L.K., Chiang H.C., Hooper L.V., Gordon J.I. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- Yu J., Leibiger B., Yang S.N., Caffery J.J., Shears S.B., Leibiger I.B., Barker C.J., Berggren P.O. Cytosolic multiple inositol polyphosphate phosphatase in the regulation of cytoplasmic free Ca2+ concentration. J. Biol. Chem. 2003;278:46210–46218. doi: 10.1074/jbc.M303743200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.