Highlights
-
•
Type IV secretion systems are nanomachines that transport substrates through bacterial membranes.
-
•
Structures of components obtained by crystallography are presented.
-
•
Higher resolution core complex structures revealed localisations of protein components.
-
•
Docking of known and modelled atomic structures uncovers interactions between components.
Abstract
Type IV secretion (T4S) systems are large dynamic nanomachines that transport DNAs and/or proteins through the membranes of bacteria. Because of their complexity and multi-protein organisation, T4S systems have been extremely challenging to study structurally. However in the past five years significant milestones have been achieved by X-ray crystallography and cryo-electron microscopy. This review describes some of the more recent advances: the structures of some of the protein components of the T4S systems and the complete core complex structure that was determined using electron microscopy.
Current Opinion in Microbiology 2014, 17:24–31
This review comes from a themed issue on Host–microbe interactions: bacteria
Edited by Olivia Steele-Mortimer and Agathe Subtil
For a complete overview see the Issue and the Editorial
Available online 5th December 2013
1369-5274/$ – see front matter, © 2013 The Authors. Published by Elsevier Ltd. All rights reserved.
Introduction
Nearly all types of bacteria (Gram-negative, Gram-positive, cell wall-less bacteria) and some archaea have evolved secretion systems essential for both their survival and virulence [1•]. Secretion systems transport DNA and effector molecules such as enzymes or toxins from the bacterial interior to its exterior. Understanding the principles of action of these nanomachines has broad clinical significance not only due to delivery of bacterial toxins or effector proteins straight into targeted host cells, but also for the direct involvement in the rapid horizontal spread of antibiotic resistance genes among the microbial population.
Secretion systems found in bacteria are classified currently into seven major classes, among which type IV secretion (T4S) systems form the most functionally versatile class [2–7]. The variety of substrates and their nature (single proteins, protein complexes, DNA and nucleoprotein complexes) secreted by the T4S systems indeed single out this class of secretion systems from the others. The T4S systems are classified into three functional groups. The first group mediates the transfer of DNA from one bacterial cell to another in a process called conjugation that plays an important role in bacterial genome plasticity and diversity. Another group of T4S systems mediates the translocation of proteins, ranging from small protein effectors to large protein complexes. Pathogenic Gram-negative bacteria such as Helicobacter pylori, Brucella suis and Legionella pneumophila use the T4S system to inject virulence proteins into mammalian host cells [8–10] and Bordetella pertussis use the T4S systems to secrete pertussis toxin into the extracellular milieu [11]. The third group mediates DNA release and uptake. H. pylori and Neisseria gonorrhoeae typify bacteria with this type of T4S systems [13]. T4S systems of the first class represent an enormous public-health problem as they are involved in the rapid dissemination of antibiotic-resistance genes and other virulence traits among pathogens. While the fact that DNA can move from one cell to another has been established a long time ago [12,15], the mechanism of secretion was poorly understood since no structural information was available until very recently.
The T4S systems are evolutionarily related: all nucleoprotein and protein complexes are secreted by the T4S systems in an ATP-dependent process using a specific channel through a cell envelope [14]. The T4S systems share a number of main components and it seems that the secretion process of all types of substrates has common features [16]. In this review we describe the latest progress in structural studies of the T4S system components and their major complexes.
Overall organisation of T4S systems
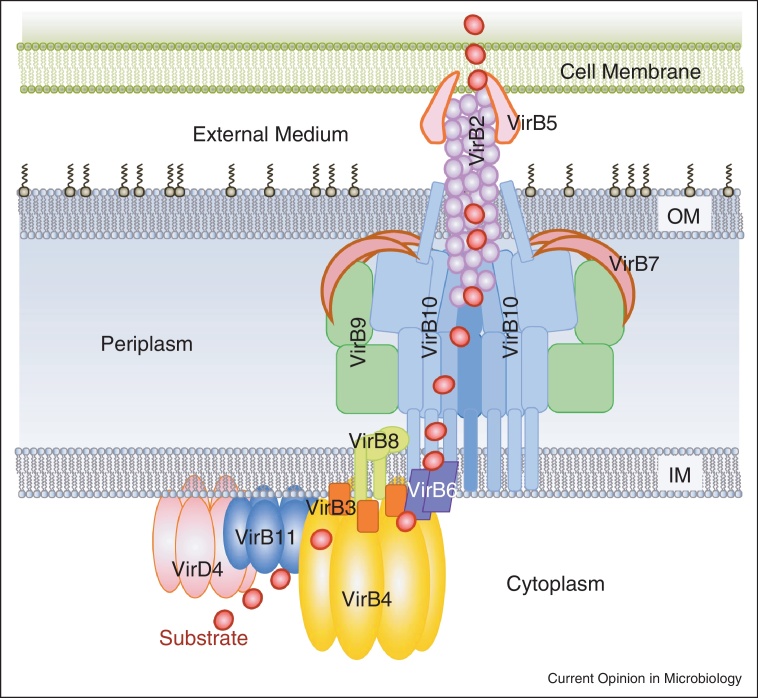
It has been shown that the T4S systems span the entire bacterial cell envelope, creating a translocation channel for various substrates [1•]. Almost all T4S systems of Gram-negative bacteria minimally consist of 12 proteins named VirB1 to VirB11 and VirD4 (based on the nomenclature used for Agrobacterium tumefaciens T4S system) that form a multi-protein envelope-spanning transport apparatus (Figure 1). Each protein is present in multiple copies. The twelve components are organised in three major subcomplexes. A cytoplasmic-inner membrane (IM) subcomplex is composed of three ATPases (VirB4, VirB11 and VirD4) and the VirB3, VirB6 and parts of the VirB8 and VirB10 proteins. The ATPases power system assembly and substrate translocation [17–21]. VirB3, VirB6, and VirB8 are likely to anchor VirB4 (and also perhaps VirB11) to the IM, participate in the IM channel, and provide a link to the core complex (CC). The CC is composed of three proteins: VirB7, VirB9 and VirB10, and is a large central structure of the T4S system, that serves as scaffolding for the rest of the T4S system components. This complex forms a trans membrane spanning pore inserted both in the outer membrane (OM) and IM of Gram-negative bacteria and participates actively in T4S substrate transfer through the bacterial envelope [1•,22•,23,24]. The third subcomplex is formed by the VirB2 and VirB5 proteins that make the extracellular pilus that is important for direct contact with the recipient cell surface and may act as a conduit delivering the substrate to the recipient cell (Figure 1). While there are some deviations in the composition of these subcomplexes between different types of T4S systems they are likely conserved in their overall organisation [1•].
Figure 1.
Overall organisation of the T4S system. VirD4 (in pink), VirB11 (in blue), VirB4 (in gold) ATPases, polytopic VirB6 (in purple), bitopic VirB8 (in light green) and VirB3 (in orange) form the cytoplasmic IM part of the complex. VirB7 (in brown), VirB9 (in green), and VirB10 (in blue) compose the periplasmic part of the secretion system. VirB2 and VirB5 constitute the outer part of the secretion system. Red dot indicates the path of the substrate through the machinery as established by Cascales and Christie [22•]. The stoichiometry of the various components in a native, fully assembled, T4S system is unknown.
Advances in the structural analysis of T4S systems
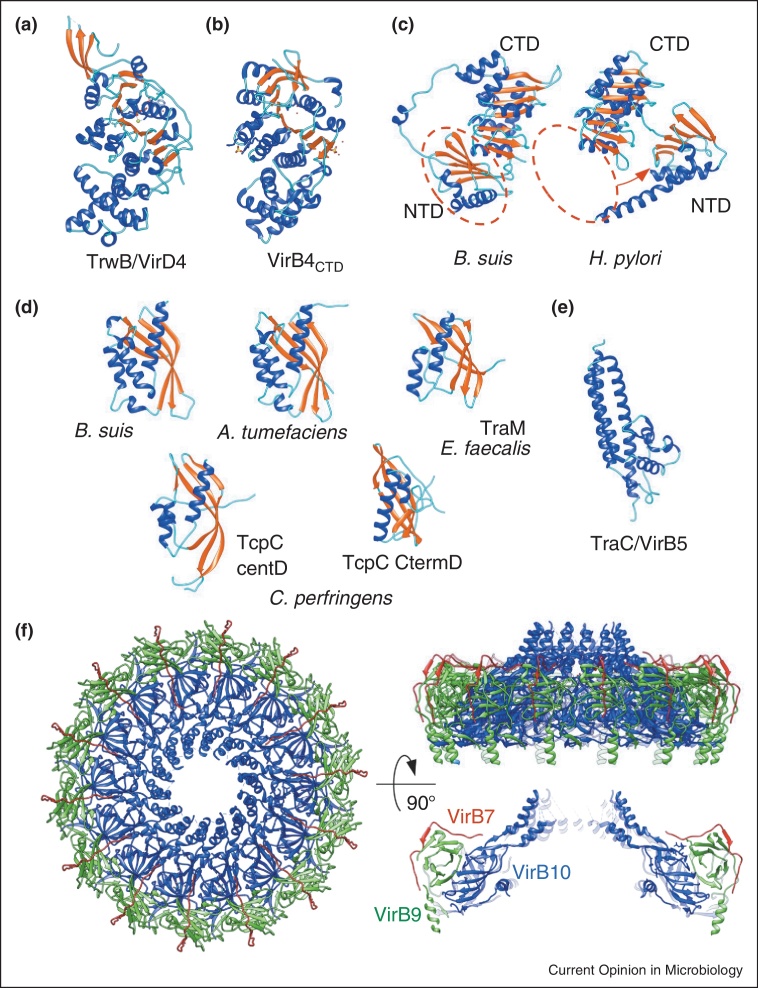
The complexity and dynamics of the T4S systems and their multiprotein organisation represent serious difficulties for the structural analysis of T4S systems or even their components. Only during the last decade structural information has emerged for the VirB/D system from A. tumefaciens and the closely related systems from Escherichia coli encoded by the conjugative plasmids F, R388, pKM101 and RP4 [25•,26]. X-ray crystallographic studies of the T4S system have been performed on soluble protein components (VirB11, the cytoplasmic regions of VirD4 and VirB4, the periplasmic domain of VirB8, and VirB5) and on the OM-embedded region of the CC (Figure 2).
Figure 2.
Structures of T4S system components or domains. (a) VirD4: structure of the soluble domain of TrwB. One subunit of the hexamer is shown (coils are shown in cyan, helices in blue, and strands in red). (b) VirB4: C-terminal domain of Thermoanaerobacter pseudethanolicus VirB4; (c) VirB11: crystal structures of B. suis VirB11 and H. pylori HP5025, CTD-C terminal domain, NTD-N terminal domain (shown in red dashed oval on B. suis VirB11). Red arrow indicates the shift of the NTD in H. pylori Vir11 compared to B. suis VirB11. (d) VirB8: crystal structure of the periplasmic domain (C-terminal domain) of VirB8 from B. suis (left panel), A. tumefaciens (middle panel), TraM214–322 protein (right panel); the central and C-terminal domains of the TcpC99–359 structure are shown in the bottom panel. (e) VirB5: crystal structure of TraC encoded by the E. coli conjugative plasmid pKM101. (f) Crystal structure of the CC's O-layer composed of VirB7 (in dark red), VirB9CT (in green) and VirB10CT (blue). All structures are shown in the ribbon representation.
Structures of ATPases
VirD4 homologues are essential T4S ATPases that act as substrate receptors at the entry of the T4S apparatus and might power substrate translocation through the IM [21]. Because of their role in delivering the substrate to the translocation channel, VirD4 proteins are termed ‘coupling proteins’. The crystal structure of the soluble ∼50 kDa cytoplasmic domain of TrwB (VirD4 homologue) from the R388 conjugative plasmid (PDB 1GKI, Figure 2A) revealed a homohexamer in which each subunit has two distinct domains: an all-alpha domain facing the cytoplasm and an ATP-binding domain linked to the IM by the N-terminal membrane anchor [17]. The structure implies that VirD4 could function as a rotary motor that assists ssDNA and/or protein translocation through the IM. Another recent evidence of involvement of coupling proteins in engagement of substrates and activation of the secretion system was demonstrated for DotL, an IM component of the Dot/Icm secretion system in L. pneumophila. DotL acts as an IM receptor for substrates and requires adaptor proteins for the secretion of a major class of substrates [27]. The DotL protein is analogous to VirD4 in A. tumefaciens, and TraB in E. coli pKM101 plasmid [28]. As in all members of the VirD4 family, DotL also contains a Walker A motif for ATP hydrolysis and is believed to provide energy to actively drive the export of substrates across the inner and outer membranes of the bacterial cell wall [27].
The VirB4 ATPase of the T4S system is also essential for the assembly of the system and substrate transfer. Recently the crystal structure of the C-terminal domain of Thermoanaerobacter pseudethanolicus VirB4 (PDBs 4AG6 and 4AG5) has been obtained [20]. This structure is similar to VirD4, although it shares only 12% sequence identity. The VirB4 domain crystallizes as a monomer, but the full-length protein is observed in monomer–dimer equilibrium, even in the presence of nucleotides and/or DNAs. The VirB4 proteins share four common sequence motifs including the consensus Walker A and B nucleoside triphosphate-binding motifs (Figure 2B). A VirB4 topology model suggests possible periplasmic loops, one near the N terminus and a second on the N-terminal side of the Walker A motif [20]. The cellular localization, topology and oligomerization state of VirB4 remain unclear: different studies have reported dimeric and higher oligomeric (tetrameric and hexameric) forms of the protein [29–32]. It was shown that VirB4 can be bound through its N-terminal domain to the core's VirB9 protein [20]. VirB4 is involved in earlier steps of substrate engagement, and makes a number of additional interactions, notably with VirB3 and VirB8 [33–35].
VirB11 is a peripheral membrane protein and belongs to a family of ATPases termed ‘traffic ATPases’ [19,18,36]. Recent studies have shown that conformation of VirB11(TrwD) can be regulated by magnesium, that at physiological concentrations induces a more rigid conformation of the enzyme hence slowing down its activity [37]. The crystal structure of the ADP-bound H. pylori VirB11 homologue (PDB 2PT7) revealed that each monomer consists of two domains formed by the N-terminal and C-terminal halves of the protein [19]. The nucleotide-binding site is at the interface between the two domains. In the hexamer, the N-terminal domain and C-terminal domain form two separate rings defining a chamber of ∼50 Å. Comparison of the structures of B. suis (2GZA) and H. pylori VirB11 (PDB 2PT7) showed that the B. suis VirB11 monomer differs significantly from that of H. pylori by a N-terminal domain swap that greatly modifies the nucleotide-binding site and the interface between subunits (Figure 2C) [18].
Structures of VirB5 and VirB8 proteins
VirB8 homologues are essential for substrate transfer through the IM. It has been proposed that they are part of the inner-membrane pore and interact with VirB3, VirB4, VirB5 and VirB6 [35,38,39]. The N-terminal extremity of the structure is connected to a transmembrane segment inserted in the IM. A structure of the periplasmic domain (C-terminal domain) of VirB8 has been obtained for B. suis and A. tumefaciens (PDBs 2BHM and 2CC3) [38–40]. Structural comparison of B. suis and A. tumefaciens VirB8 confirms that the monomers have a similar fold (Figure 2D). It comprises an extended β-sheet flanked with α-helices. Both proteins exist as homodimers, and amino acids at the dimer interface are critical for protein function. Smith et al. have found that E115 and K182 interact with inhibitors which apparently induce conformational changes that prevents VirB8-VirB8 and VirB8-VirB10 interactions [41•]. Recently it was shown that the C-terminal component of the transfer TraM protein (214–322 aa, PDB 4EC6) from the Enteroccocus faecalis conjugative plasmid pIP501 and the conjugation protein TcpC99–359 (PDB 3UB1) from Clostridium perfringens (Gram-positive) plasmid pCW3 are structurally homologous to the periplasmic region of VirB8 [42••,43••]. TraM and TcpC are found localized to the cell membrane. Despite the absence of sequence-based similarity, the crystal structures of both proteins display folds similar to the T4S system VirB8 proteins from B. suis and A. tumefaciens. Surprisingly, the TcpC99–359 structure comprises two VirB8-like domains separated by a linker. The C-terminal domain is critical for interactions with other conjugation proteins (Figure 2D). The structure, molecular dynamics, and cross-linking studies indicate that TraM is active as a trimer. The oligomerization state of TcpC is not known.
The crystal structure of TraC, a VirB5 homologue encoded by the E. coli conjugative plasmid pKM101 (PDB 1R8I, Figure 2E), has been obtained at 3 Å resolution [44]. The VirB5 structure comprises a 3-helix bundle flanked by a smaller globular domain. Together with the main pilin VirB2, VirB5 homologues form the T4S pilus, possibly acting as adhesins.
Structures of the CCs
The most advanced structural work described to date is the X-ray crystal structure for the part of the CC that locates in and near the OM. This part, termed the ‘O-layer’, forms the OM pore of the T4S system (PDB 3JQO) [45••]. It is composed of three proteins/domains: the TraF/VirB10 C-terminal domain (TraF/VirB10CT), the TraO/VirB9 C-terminal domain (TraO/VirB9CT), and TraN/VirB7, each in 14 copies. The structure has unveiled a surprisingly intricate network of interactions between these three major components of the CC (Figure 2F). VirB7 seems to work as a stapler that fastens VirB9 and VirB10 together; the inner surface of the OM channel is lined by VirB10. A diameter of the α-helical pore in the crystal structure is ∼32 Å that could accommodate the passage of DNA and unfolded protein substrates.
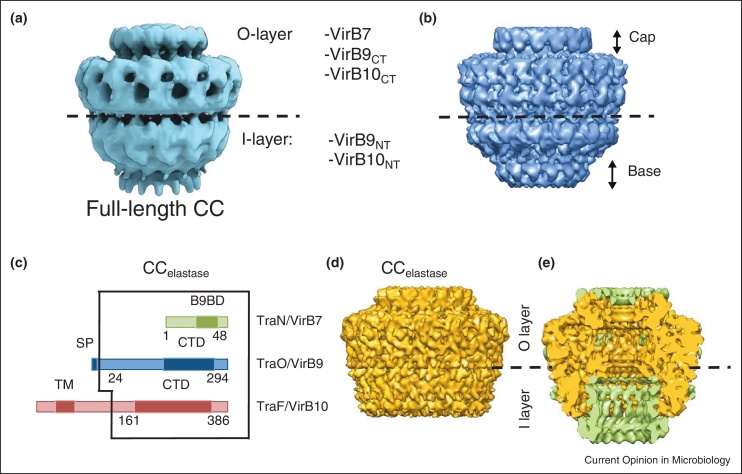
Experiments combining cryo-EM structure determination of full-length CC and partly deleted CCs, docking of X-ray crystallographic and pseudo atomic models, and immuno-labelling have resulted in the elucidation of the structure of the entire CC (Figure 3). The first structure of the complete CC (Figure 3A) was solved by cryo-electron microscopy (cryo-EM) at 15 Å resolution using images of frozen samples [25•]. The overall dimensions of the complex are 185 Å in both height and diameter. It consists of two layers, termed O and I. The O-layer has an inner chamber that is open to the extracellular media through an opening of 10–20 Å in the cap. The complex has an inner channel that spans the structure from the OM to the IM. Later on, the CC structure was refined to a resolution of 12.4 Å (Figure 3B) which revealed more details of the inner central channel [46••]. In this higher resolution structure, the I-layer has a diameter of ∼50–55 Å and its inner wall is formed by 14 columns of density with a diameter of ∼8 Å. The columns project from the O-layer of the CC to the bottom of the I-layer where they are connected to the base of the complex defining an opening of ∼55 Å in diameter on the cytoplasmic side of the complex.
Figure 3.
Electron microscopy of the T4S system. (a) Structure of the CC complex at 15 Å; (b) structure of the CC complex at 12 Å; (c) schematic illustration of the regions of TraN/VirB7, TraO/VirB9, and TraF/VirB10 present in the CCelastase complex. Domains corresponding to the VirB9 binding domain (B9BD), the signal peptide (SP), the N-terminal trans-membrane (TM) helix and the C-terminal domains (CTD) are shown in darker colours; (d) Cryo-EM structure of the CCelastase complex. (e) Cutaway view of the superposition of the difference map (in green) between the full length CC and CCelastase and the cryo-EM structure of the CCelastase complex (in gold).
Digestion of the CC with elastase produced a stable truncated complex, termed the ‘CCelastase’ complex. The sequencing of the bands in SDS-PAGE gels confirms that the complex is made of full-length TraN/VirB7 and TraO/VirB9, and TraF/VirB10CT (Figure 3C). A structure of CCelastase was obtained at a resolution of 8.5 Å using cryo-EM and single particle analysis (Figure 3D) [46••]. Comparison of full length CC and CCelastase demonstrated considerable differences in the I-layer region. In the full-length CC, the I-layer comprises the N-terminal domains of VirB9 (TraF/VirB9NT) and VirB10 (TraF/VirB10NT) and is inserted in the IM via the VirB10 N-terminal trans-membrane segment. The removal of TraF/VirB10NT by elastase results in an increase of the opening on the cytoplasmic side (65 Å compared to ∼55 Å in the CC) and a shortening of the I-layer (Figure 3D).
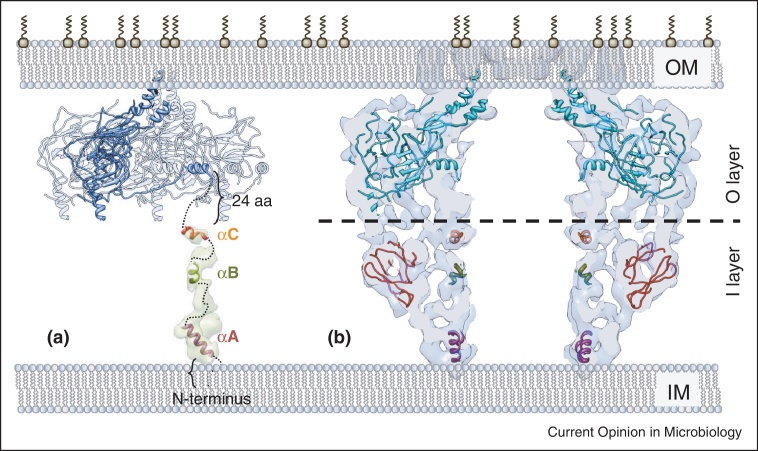
In the CCelastase complex, the columns within the central channel are absent (Figure 3E). Therefore the inner columns of the CC I-layer were assigned to TraF/VirB10NT (Figure 4A). The major part of TraF/VirB10NT was shown to be unstructured; the only secondary structural elements that were reliably identified were three short helical elements: the helix corresponding to the inner-membrane spanning region (αA: K40AF-LVF53) and two other α-helical regions (αB: A107RA-QAA113; αC: P138EE-QRR146). Their length is compatible with the diameter and the length of the observed columns in the CC cryo-EM map ([46••], and references herein]). At the very bottom of the complex these stretches join each other to form a ring at the base of the CC. This ring might correspond to the 14 trans-membrane segments (helix αA) associated with detergent micelles. VirB10 is linked to the IM by the VirB10 N-terminal trans-membrane helix (30–50 aa) characterised with two types of putative dimerization motifs: a GxxxA (GA4) motif and two leucine (Leu1, Leu2) zippers. While mutations in the Leu1 motif disrupt T-pilus biogenesis, this and other mutations in the GA4 or Leu2 motif do not abolish substrate transfer [47].
Figure 4.
Schematic model for the full length organisation of the core complex. (a) Four TraF/VirB10CT subunits of the 14-mer present in the O-layer atomic structure are shown. One subunit is highlighted in blue. The density of one subunit column in the difference map (in light green) is shown with the tentative docking of the three TraF/VirB10NT α-helical regions. This subunit is located immediately below TraF/VirB10CT shown in dark blue. The connections are shown in dashed lines. (b) Central slice of the full length CC with fitted O-layer atomic structure (in cyan) and atomic models obtained for TraO/VirB9NT (in dark red) and the α-helices predicted in TraF/VirB10NT (shown as in (a)).
The modelling of the atomic structure of TraO/VirB9NT (residues 24–135) and the examination of its possible docking into EM maps helped locate this domain within the structure. The results suggest that TraO/VirB9NT adopts a beta-sandwich fold (Figure 4B). Fitting of models for TraO/VirB9NT, and labelling experiments with nanobodies targeted against TraO/VirB9NT are all pointing towards the conclusion that TraO/VirB9NT is located at the outside surface of the I-layer.
Conclusions
During the last decade structural analysis has made a great impact on our understanding of how T4S systems are organised and function.
The EM structures have greatly altered our conception of the entire T4S system architecture and its possible mechanism of action. It was found that the CC is an essential structural portion of T4S systems. It is self-assembling and therefore is likely to be assembled first. The CC forms the IM channel and is a major component of the OM pore. It provides the central scaffold for assembly of all other components. Structural comparison of the complete and truncated CCs demonstrate that TraF/VirB10 spans the entire cell envelope. This observation together with fitting of crystal and modelled structures into EM maps of large complexes is of particular interest since it provides the explanation how VirB10 is able to sense conformational changes induced by ATP binding to or ATP hydrolysis by the T4S system ATPases situated at the T4S channel's entrance on its cytoplasmic side [23]. VirB10 responds to these changes by undergoing its own structural modifications that are required for substrate transfer. Cascales et al. have demonstrated that DNA binding to VirD4 and VirB11 stimulates ATP binding/hydrolysis, which in turn activates VirB10 through a structural transition [48•]. Segure and co-authors have shown that the transmembrane domain of TrwB/VirD4 is needed for the interaction with the transmembrane domain of TrwB/VirB10 and for R388 conjugative transfer. The removal of first 64 amino acids in TrwB/VirB10 reduced conjugation by six orders of magnitude [47]. Thus proteins analogues to VirB10 may serve as a signal transmitter throughout the secretion complex [1•,21,23,25•,47,48•,49•].
The next mission in T4S studies is to establish the organisation and structure of even larger complexes of T4S system components. For example, it is important to complete our understanding of the IM channel by determining the structure of VirB6 and VirB8 bound to the CC. Ultimately, the structure of the entire translocation machine will need to be determined. A related question is how substrate transfer takes place. This task represents a tremendous challenge that can only be met by implementing novel biochemical and genetic approaches resulting in trapping T4S systems at different stages of substrate transfer. The elucidation of T4S mechanisms will thus require joint efforts from biochemistry and structural biology. The derived knowledge might be used to find ways of inhibiting these systems to combat infections and control the spread of antibiotic resistance genes.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
of special interest
of outstanding interest
Acknowledgements
The work on the CC and the “CCelastase” complex of the T4S system was funded by grant 082227 from the Wellcome Trust to GW and K012401 from MRC to EVO and GW. The authors would also like to thank the Wellcome Trust for the EM equipment.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Contributor Information
Gabriel Waksman, Email: g.waksman@mail.cryst.bbk.ac.uk.
Elena V Orlova, Email: e.orlova@mail.cryst.bbk.ac.uk.
References
- Alvarez-Martinez C.E., Christie P.J. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the most complete review on Prokaryotic Type IV Secretion systems.
- 2.Omori K., Idei A. Gram-negative bacterial ATP-binding cassette protein exporter family and diverse secretory proteins. J Biosci Bioeng. 2003;95:1–12. doi: 10.1016/S1389-1723(03)80141-X. [DOI] [PubMed] [Google Scholar]
- 3.Filloux A. The underlying mechanisms of type II protein secretion. Biochim Biophys Acta. 2004;1694:163–179. doi: 10.1016/j.bbamcr.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Cornelis G.R., Van Gijsegem F. Assembly and function of type III secretory systems. Annu Rev Microbiol. 2000;54:735–774. doi: 10.1146/annurev.micro.54.1.735. [DOI] [PubMed] [Google Scholar]
- 5.Henderson I.R., Navarro-Garcia F., Desvaux M., Fernandez R.C., Ala’Aldeen D. Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev. 2004;68:692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cascales E. The type VI secretion toolkit. EMBO Rep. 2008;9:735–741. doi: 10.1038/embor.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdallah A.M., Gey van Pittius N.C., Champion P.A., Cox J., Luirink J., Vandenbroucke-Grauls C.M., Appelmelk B.J., Bitter W. Type VII secretion — mycobacteria show the way. Nat Rev Microbiol. 2007;5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- 8.Backert S., Meyer T.F. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol. 2006;9:207–217. doi: 10.1016/j.mib.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 9.de Jong M.F., Sun Y.H., den Hartigh A.B., van Dijl J.M., Tsolis R.M. Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Mol Microbiol. 2008;70:1378–1396. doi: 10.1111/j.1365-2958.2008.06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ninio S., Roy C.R. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 2007;15:372–380. doi: 10.1016/j.tim.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 11.O’Callaghan D. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol Microbiol. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- 12.Hofreuter D., Karnholz A., Haas R. Topology and membrane interaction of Helicobacter pylori ComB proteins involved in natural transformation competence. Int. J. Med. Microbiol. 2003;293:153–165. doi: 10.1078/1438-4221-00258. [DOI] [PubMed] [Google Scholar]
- 13.Lederberg J., Tatum E.L. Sex in bacteria; genetic studies 1945-1952. Science. 1953;118:169–175. doi: 10.1126/science.118.3059.169. [DOI] [PubMed] [Google Scholar]
- 14.Lessl M., Lanka E. Common mechanisms in bacterial conjugation and Timediated T-DNA transfer to plant cells. Cell. 1994;77:321–324. doi: 10.1016/0092-8674(94)90146-5. [DOI] [PubMed] [Google Scholar]
- 15.Pansegrau W., Lanka E. Enzymology of DNA transfer by conjugative mechanisms. Prog. Nucleic Acid Res. Mol. Biol. 1996;54:197–251. doi: 10.1016/s0079-6603(08)60364-5. [DOI] [PubMed] [Google Scholar]
- 16.Christie P.J., Cascales E. Structural and dynamic properties of bacterial type IV secretion systems. Mol Membr Biol. 2005;22:51–61. doi: 10.1080/09687860500063316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomis-Rüth F.X., Moncalián G., Pérez-Luque R., González A., Cabezón E., de la Cruz F., Coll M. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature. 2001;409:637–641. doi: 10.1038/35054586. [DOI] [PubMed] [Google Scholar]
- 18.Hare S., Bayliss R., Baron C., Waksman G. A large domain swap in the VirB11 ATPase of Brucella suis leaves the hexameric assembly intact. J Mol Biol. 2006;360:56–66. doi: 10.1016/j.jmb.2006.04.060. [DOI] [PubMed] [Google Scholar]
- 19.Savvides S.N., Yeo H.J., Beck M.R., Blaesing F., Lurz R., Lanka E., Buhrdorf R., Fischer W., Haas R., Waksman G. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 2003;22:1969–1980. doi: 10.1093/emboj/cdg223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walldén K., Williams R., Yan J., Lian P.W., Wang L., Thalassinos K., Orlova E.V., Waksman G. Structure of the VirB4 ATPase, alone and bound to the core complex of a type IV secretion system. Proc Natl Acad Sci U S A. 2012;109:11348–11353. doi: 10.1073/pnas.1201428109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atmakuri K., Cascales E., Christie P.J. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol Microbiol. 2004;54:1199–11211. doi: 10.1111/j.1365-2958.2004.04345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E., Christie P.J. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004;304:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is an essential review that describes an event sequence during secretion in Type IV Secretion systems.
- 23.Jakubowski S.J., Kerr J.E., Garza I., Krishnamoorthy V., Bayliss R., Waksman G., Christie P.J. Agrobacterium VirB10 domain requirements for type IV secretion and T pilus biogenesis. Mol Microbiol. 2009;71:779–794. doi: 10.1111/j.1365-2958.2008.06565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Llosa M., Zunzunegui S., de la Cruz F. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc Natl Acad Sci U S A. 2003;100:10465–10470. doi: 10.1073/pnas.1830264100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fronzes R., Schäfer E., Wang L., Saibil H.R., Orlova E.V., Waksman G. Structure of a type IV secretion system core complex. Science. 2009;323:266–268. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes the structure of first largest complex from the type IV secretion system obtained by cryo electron microscopy.
- 26.Waksman G., Fronzes R. Molecular architecture of bacterial type IV secretion systems. Trends Biochem Sci. 2010;35:691–698. doi: 10.1016/j.tibs.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 27.Sutherland M.C., Nguyen T.L., Tseng V., Vogel J.P. The Legionella IcmSW complex directly interacts with DotL to mediate translocation of adaptor-dependent substrates. Plos Pathogens. 2012;8:e1002910. doi: 10.1371/journal.ppat.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buscher B.A., Conover G.M., Miller J.L., Vogel S.A., Meyers S.N., Isberg R.R., Vogel J.P. The DotL protein, a member of the TraG-coupling protein family, is essential for viability of Legionella pneumophila strain Lp02. J Bacteriol. 2005;187:2927–2938. doi: 10.1128/JB.187.9.2927-2938.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Durand E., Waksman G., Receveur-Brechot V. Structural insights into the membrane-extracted dimeric form of the ATPase TraB from the Escherichia coli pKM101 conjugation system. BMC Struct Biol. 2011;11:1–13. doi: 10.1186/1472-6807-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arechaga I., Peña A., Zunzunegui S., del Carmen Fernández-Alonso M., Rivas G., de la Cruz F. ATPase activity and oligomeric state of TrwK, the VirB4 homologue of the plasmid R388 type IV secretion system. J Bacteriol. 2008;190:5472–5479. doi: 10.1128/JB.00321-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dang T.A., Zhou X.R., Graf B., Christie P.J. Dimerization of the Agrobacterium tumefaciens VirB4 ATPase and the effect of ATP-binding cassette mutations on the assembly and function of the T-DNA transporter. Mol Microbiol. 1999;32:1239–1253. doi: 10.1046/j.1365-2958.1999.01436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peña A., Matilla I., Martín-Benito J., Valpuesta J.M., Carrascosa J.L., de la Cruz F., Cabezón E., Arechaga I. The hexameric structure of a conjugative VirB4 protein ATPase provides new insights for a functional and phylogenetic relationship with DNA translocases. J Biol Chem. 2012;287:39925–39932. doi: 10.1074/jbc.M112.413849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li F., Alvarez-Martinez C., Chen Y.Q., Choi K.J., Yeo H.J., Christie P.J. Enterococcus faecalis PrgJ, a VirB4-Like ATPase, mediates pCF10 conjugative transfer through substrate binding. J Bacteriol. 2012;194:4041–4051. doi: 10.1128/JB.00648-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones A.L., Shirasu K., Kado C.I. The product of the virB4 gene of Agrobacterium tumefaciens promotes accumulation of VirB3 protein. J Bacteriol. 1994;176:5255–5261. doi: 10.1128/jb.176.17.5255-5261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mossey P., Hudacek A., Das A. Agrobacterium tumefaciens type IV secretion protein VirB3 is an inner membrane protein and requires VirB4, VirB7, and VirB8 for stabilization. J Bacteriol. 2010;192:2830–2838. doi: 10.1128/JB.01331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hare S., Fischer W., Williams R., Terradot L., Bayliss R., Haas R., Waksman G. Identification, structure and mode of action of a new regulator of the Helicobacter pylori HP0525 ATPase. EMBO J. 2007;26:4926–4934. doi: 10.1038/sj.emboj.7601904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ripoll-Rozada J., Pena A., Rivas S., Moro F., de la Cruz F., Cabezon E., Arechaga I. Regulation of the type IV secretion ATPase TrwD by magnesium implications for catalytic mechanism of the secretion ATPase superfamily. J Biol Chem. 2012;287:17408–17414. doi: 10.1074/jbc.M112.357905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terradot L., Bayliss R., Oomen C., Leonard G.A., Baron C., Waksman G. Structures of two core subunits of the bacterial type IV secretion system, VirB8 from Brucella suis and ComB10 from Helicobacter pylori. Proc Natl Acad Sci U S A. 2005;102:4596–4601. doi: 10.1073/pnas.0408927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giraldo A.M.V., Sivanesan D., Carle A., Paschos A., Smith M.A., Plesa M., Coulton J., Baron C. Type IV secretion system core component VirB8 from brucella binds to the globular domain of VirB5 and to a periplasmic domain of VirB6. Biochemistry. 2012;51:3881–3890. doi: 10.1021/bi300298v. [DOI] [PubMed] [Google Scholar]
- 40.Bailey S., Ward D., Middleton R., Grossmann J.G., Zambryski P.C. Agrobacterium tumefaciens VirB8 structure reveals potential protein–protein interaction sites. Proc Natl Acad Sci U S A. 2006;103:2582–2587. doi: 10.1073/pnas.0511216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.A., Coincon M., Paschos A., Jolicoeur B., Lavallee P., Sygusch J., Baron C. Identification of the binding site of brucella VirB8 interaction inhibitors. Chem Biol. 2012;19:1041–1048. doi: 10.1016/j.chembiol.2012.07.007. [DOI] [PubMed] [Google Scholar]; This paper describes interactions of different inhibitors with VirB8.
- Goessweiner-Mohr N., Grumet L., Arends K., Pavkov-Keller T., Gruber C.C., Gruber K., Birner-Gruenberger R., Kropec-Huebner A., Huebner J., Grohmann E. The 2.5 angstrom structure of the enterococcus conjugation protein TraM resembles VirB8 type IV secretion proteins. J Biol Chem. 2013;288:2018–2028. doi: 10.1074/jbc.M112.428847. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes a structure of the transfer protein TraM that has the structural similarity to VirB8 but active as a trimer.
- Porter C.J., Bantwal R., Bannam T.L., Rosado C.J., Pearce M.C., Adams V., Lyras D., Whisstock J.C., Rood J.I. The conjugation protein TcpC from Clostridium perfringens is structurally related to the type IV secretion system protein VirB8 from Gram-negative bacteria. Mol Microbiol. 2012;83:275–288. doi: 10.1111/j.1365-2958.2011.07930.x. [DOI] [PubMed] [Google Scholar]; Here is presented the first structure of the TcpC conjugation protein from Gram positive bacteria that is surprisingly similar to VirB8.
- 44.Yeo H.-J., Yuan Q., Beck M.R., Baron C., Waksman G. Structural and functional characterization of the VirB5 protein from the type IV secretion system encoded by the conjugative plasmid pKM101. Proc Natl Acad Sci U S A. 2003;100:15947–15952. doi: 10.1073/pnas.2535211100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran V., Fronzes R., Duquerroy S., Cronin N., Navaza J., Waksman G. Structure of the outer membrane complex of a type IV secretion system. Nature. 2009;462:1011–1015. doi: 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is an impressive structural study of significant part of the core complex obtained by combination of electron microscopy and crystallography.
- Rivera-Calzada A., Fronzes R., Savva C.G., Chandran V., Lian P.W., Laeremans T., Pardon E., Steyaert J., Remaut H., Waksman G., Orlova E.V. Structure of a bacterial type IV secretion core complex at subnanometre resolution. EMBO J. 2013;32:1195–1204. doi: 10.1038/emboj.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reports comprehensive structural study of the core complex obtained by combination of electron microscopy and methods of structural modelling and docking into EM maps.
- 47.Garza I., Christie P.J. A Putative transmembrane leucine zipper of agrobacterium VirB10 is essential for T-Pilus biogenesis but not type IV secretion. J Bacteriol. 2013;195:3022–3034. doi: 10.1128/JB.00287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E., Atmakuri K., Sarkar M.K., Christie P.J. DNA substrate-induced activation of the agrobacterium VirB/VirD4 type IV secretion system. J Bacteriol. 2013;195:2691–2704. doi: 10.1128/JB.00114-13. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes a role of the DNA substrate in activation of VirD4 and in the function of the T4S system.
- Segura R.L., Aguila-Arcos S., Ugarte-Uribe B., Vecino A.J., de la Cruz F., Goni F.M., Alkorta I. The transmembrane domain of the T4SS coupling protein TrwB and its role in protein–protein interactions. Biochim Biophys Acta. 2013;1828:2015–2025. doi: 10.1016/j.bbamem.2013.05.022. [DOI] [PubMed] [Google Scholar]; This paper reports detailed study of interactions between TrwB/VirD4 and TrwE/VirB10, components of T4S system of the R388 plasmid.