Abstract
Background
High risk human papillomavirus (HR-HPV) infection is common and only a small minority of infections become persistent and lead to cervical cancers. Women positive for HR-HPV usually require a second test to avoid unnecessary colposcopies and over treatment. Elevated DNA methylation of HR-HPV L1 and L2 genes in high grade disease has emerged as a promising molecular triage tool.
Objectives
Our aim was to accurately measure methylation levels at selected CpG positions in the HPV18, HPV31 and HPV33 genomes. We focused on the L2, L1, URR and E6 regions because these were previously shown to be interesting areas for study.
Study design
Pyrosequencing was used to measure methylation in 208 HPV18, 207 HPV31, and 126 HPV33 positive women selected from a London colposcopy referral population.
Results
After adjustment for multiple testing, at FDR 5%, elevated methylation was significantly associated with cervical intraepithelial neoplasia grades 2 or worse (CIN2+) in all investigated CpGs in HPV18 L2 and L1. Two of 6 L2 and 12 of 15 L1 sites in HPV31 and 6 of 8 L2 and 3 of 13 L1 sites in HPV33 showed significantly elevated methylation in CIN2+. Methylation of CpG sites in the URR and E6 region of the HPV types was low and most differences were not significant.
Conclusion
Elevated methylation of CpG sites in the L1 and L2 regions of HPV18, HPV31 and HPV33 is associated with CIN2+ and a panel test may be useful for triage of women with HR-HPV infections.
Abbreviations: HR-HPV, high risk human papilloma virus; CIN, cervical intraepithelial neoplasia; CI, confidence interval; FDR, false discovery rate; P1, predictors 1; P2, predictors 2; ROC, receiver operator characteristics; AUC, area under the curve; SCC, squamous cell carcinoma
Keywords: High risk human papillomavirus, Methylation, L1, L2, Pyrosequencing
1. Background
The main causative agent of cervical cancer is infection by approximately fifteen high risk human papillomavirus types (HR-HPV) [1]. Although common, with an estimated worldwide prevalence of 11–12% [2], a majority of the infections clear within 6 months to 2 years [3]. Consequently, a considerable number of women will acquire HR-HPV infection but very few will progress to invasive disease. The persisting infection fuels a decades long, stepwise progression to cancer; recognition of which initiated the use of cytological screening for the precursor cervical intraepithelial neoplasia (CIN) as a common preventative practice [4]. In the past decade, HR-HPV testing has been increasingly adapted as a secondary triage tool in women 30 years or older [5,6]. Due to its cost-effectiveness, approachability and high sensitivity, HR-HPV testing is of interest as a primary preventative tool. However, current HR-HPV tests cannot distinguish transient from persistent infections, resulting in low specificity for detection of high grade 2 or 3 CIN lesions (CIN2/3). To compensate for the limitation, different complementary methods have been suggested such as cytology, p16 immunochemistry, genotyping and DNA methylation [7]. While p16 immunochemistry and cytology rely on adequate cellular material, PCR based techniques have fewer requirements for preservation of the sample and can be applied to clinician or self-collected specimens in liquid or dry form [8], making them attractive for effective screening programme implementation in less developed regions of the world.
Carefully orchestrated, DNA methylation plays a crucial role for activating and silencing genes during normal development, however its disruption contributes to development of disease [9]. Intensive research in a variety of cancers shows great promise in quantification of DNA methylation as diagnostic and prognostic biomarkers. In cervical cancer many studies have shown that elevated methylation of CpG sites in the HPV16 L1 and L2 genes are associated with CIN2/3 and cancers [10–14]. We recently reported that a classifier score S1 developed in a study of Central American women based on HPV16 methylation of 7 CpG sites in L1 and L2 had similar differentiating potential to identify CIN2/3 in HPV16 infected European women, with 92% sensitivity and 40% specificity [13]. Methylation of other high risk types such as HPV18 [15–17] and HPV31 [17] have so far been investigated to a lesser extent than HPV16 and there are no methylation data on many HR-HPV types such as HPV33. Considering that HPV16 and 18 contribute to ∼70% of cervical cancers [2,18] and that HPV31 and HPV33 are among the next most prevalent HR-HPV types causing ∼8% of the cancers [19], we believe that extending methylation studies to these HR-HPV types may lead to development of a more comprehensive methylation test for triage of women to colposcopy.
2. Objective
The main aim of our study was to investigate DNA methylation levels in different regions of the HPV18, HPV31 and HPV33 genomes and to test for associations between methylation and presence of CIN2+ in a London colposcopy referral population. A secondary aim was to compare the methylation of HPV18, 31 and 33 in CIN2+ with single versus multiple HR-HPV infection.
3. Study design
We followed the REMARK guidelines for evaluation of diagnostic tests [20]. Methylation of 29 sites in HPV18, HPV31 and 27 sites in HPV33 (Supplemental Fig. 1) was measured by pyrosequencing in 208 HPV18, 207 HPV31 and 126 HPV33 positive patients.
3.1. Population
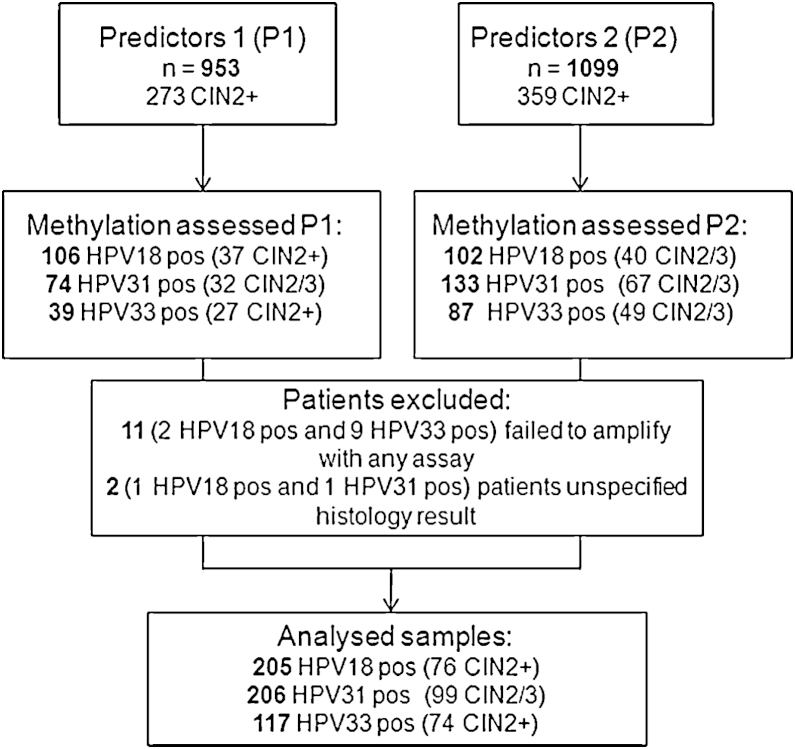
Patients from two cohorts, Predictors 1 (P1) [21] and Predictors 2 (P2) [22] were pooled to obtain a large number of HPV18, HPV31 and HPV33 positive samples (Fig. 1). The cohorts were identical in all parameters except the time of sample collection – P1 2005–2007 and P2 2007–2009. In summary, the two cohorts comprised 2052 women attending colposcopy at Hammersmith or St Mary Hospital in London after a “mild dyskaryosis or worse” cytology finding. Women were eligible if they had been referred as a result of one or more abnormal cervical smears, were not pregnant, had not been treated previously for CIN, nor had a hysterectomy. Prior to colposcopy, cervical specimens were obtained and stored in PreservCyt transport medium. Initially, the samples were analysed by a number of commercial HR-HPV test kits [21,22] and aliquots stored in −70 °C until used for the current study. To identify HPV18, HPV31 and HPV33 positive samples, Linear Array (Roche Molecular Systems, Inc., Pleasanton, CA, USA) data was used for P1 and BD HPV test (BD Diagnostics, Burlington, NC, USA) for P2. In total, 208 HPV18, 207 HPV31 and 126 HPV33 positive patients were identified. All analyses are based on a centrally reviewed histopathology and take the highest grade of abnormality seen in the biopsy or treatment specimen within 9 months of the initial base-line visit as the final diagnosis. If no abnormal areas were visible at colposcopy, biopsy was not taken and these women were classified in the <CIN2 group.
Fig. 1.
Consort diagram of the studied patients.
All women received a patient information sheet explaining the study and provided written consent. Approvals were obtained from the relevant local research ethics committee.
3.2. DNA isolation and bisulfite conversion
300 μl of PreservCyt was centrifuged at 13,200 rpm for 2 min and the pellet was resuspended in 200 μl PBS. The genomic DNA was extracted with QIAamp DNA Mini Kit (Qiagen Inc., Hilden, Germany) following the spin protocol recommended by the manufacturer except that DNA was eluted in 60 μl AE buffer. 250 ng of DNA was used in the bisulfite conversion reactions where unmethylated cytosines were converted to uracil with the EZ DNA methylation kit (Zymo research, Irvine, CA) according to manufacturer's instructions.
3.3. The methylation assays
Primers for 8 PCRs in L2, L1, URR and E6 covering 29 CpG positions in HPV18 (Supplemental Table 1), 7 PCRs in L2, L1 and URR covering 29 CpG positions in HPV31 (Supplemental Table 2) and 7 PCRs in L2, L1 and URR covering 27 CpG positions in HPV33 (Supplemental Table 3) were obtained using PyroMark Assay Design software version 2.0.1.15 (Qiagen). All CpGs within L2, L1 and URR were mapped (Supplemental Fig. 1) and the primers were designed with intent to cover the densest CpG areas in a single amplicon of less than 300 bp. CpG positions in 3′ end of HPV18 E6 gene were investigated as a surrogate for the 5′ end of URR for which we could not obtain a functional assay. Overlapping of CG dyads was avoided to prevent amplification biases. To provide the internal control for total bisulfite conversion, a non-CpG cytosine in the region for pyrosequencing was included where possible. PCRs were performed using a converted DNA equivalent of 1600 cells employing the PyroMark PCR kit (Qiagen) assuming 6.6 pg DNA per diploid cell for calculations. Briefly, 12.5 μl PCR master mix, 2.5 μl Coral red, 1 μl primer-mix, 2 μl DNA and optimised amount of MgCl2 were adjusted with water to give a final 25 μl reaction. The final concentration of each primer for HPV31 and HPV33 assays was 0.2 μM, while the concentration differed for HPV18 assays (Supplemental Table 1). All the assays were run with thermal cycling conditions: 95 °C for 15 min, optimised number of cycles: 30 s at 94 °C; 30 s at the optimised annealing temperature; 30 s at 72 °C and a final extension varied between 5 and 10 min at 72 °C (Supplemental Tables 1–3). In each run, a non-template negative control was run in addition to a standard curve consisting of 1 pg/ml of 0, 50 and 100% methylated HPV plasmid in a background of 10 ng/ml human DNA. The amplified DNA was confirmed on QIAxel capillary electrophoresis instrument (Qiagen). 10 μl of PCR product was pyrosequenced using a PyroMark™Q96 ID (Qiagen) instrument as previously described [23].
3.4. Statistical analyses
The non-parametric Wilcoxon test was used to determine whether the proportion of methylation at each individual CpG site was associated with HPV infection outcome. Heatmaps were constructed according to Spearman rho correlation values for each CpG site. To account for the high number of CpG positions tested on the same data, the Benjamin and Hochberg step-up procedure for controlling false discovery rate (FDR) was applied with FDR of 5% [24]. The classification ability of each CpG position was evaluated by receiver operating characteristics (ROCs), area under the ROC curve (AUC). For the analysis of methylation in CIN2+ with multiple infections, the 15 types considered as HR-HPV were 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73, 82 [1]. STATA v12 and GraphPad Prism v5.03 were used for statistical analysis and illustrations.
4. Results
4.1. Methylation of HPV18
The methylation of 29 sites was measured in 208 HPV18 positive patients. Three patients were excluded from the analysis – 2 patients failed to amplify for all assays and 1 patient had unspecified histology. The CIN2+ group consisted of 74 CIN2/3, 1 invasive squamous cell carcinoma (SCC) and one adenocarcinoma. 129 patients had diagnosis <CIN2. The median age in all HPV18 positive samples was 29 years with interquartile range (IQR) 6. Per diagnostic group, median age was 28 in CIN2+ and 29 in <CIN2. Age was not correlated to the methylation of any sites as the Spearman rho ranged from −0.08 to 0.18.
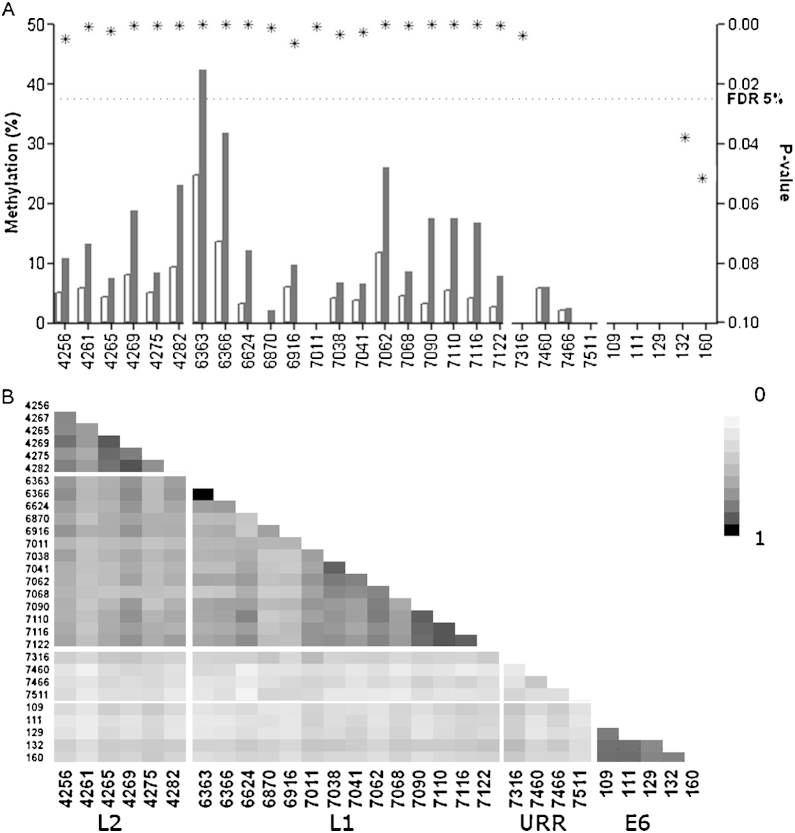
Overall median methylation measured higher in the CIN2+ in L2 and L1 genes (Fig. 2). All interrogated sites in HPV18 L2 and L1 sites were significantly higher in CIN2+ after adjustment for multiple comparisons. Low methylation was observed in URR and E6 CpG sites with only site 7316 showing a significant difference between the two diagnostic groups, this site had an AUC of 0.59 [95CI 0.53–0.66] p = 0.0037. High correlation was observed between methylation of CpG positions within the genes (Fig. 2b). The AUC and p-values for each site in L1 and L2 are presented in Supplemental Table 4.
Fig. 2.
(A) The median methylation presented for each of the investigated CpG positions in HPV18. The methylation was overall higher in CIN2+ (black bar) comparing to <CIN2 (white bar) in the capsid coding genes. The p-values (*) were plotted on the right Y-axis and the 5% FDR is marked by a dashed line. (B) The heat map showing Spearman r correlation between all investigated sites.
4.2. Methylation of HPV31
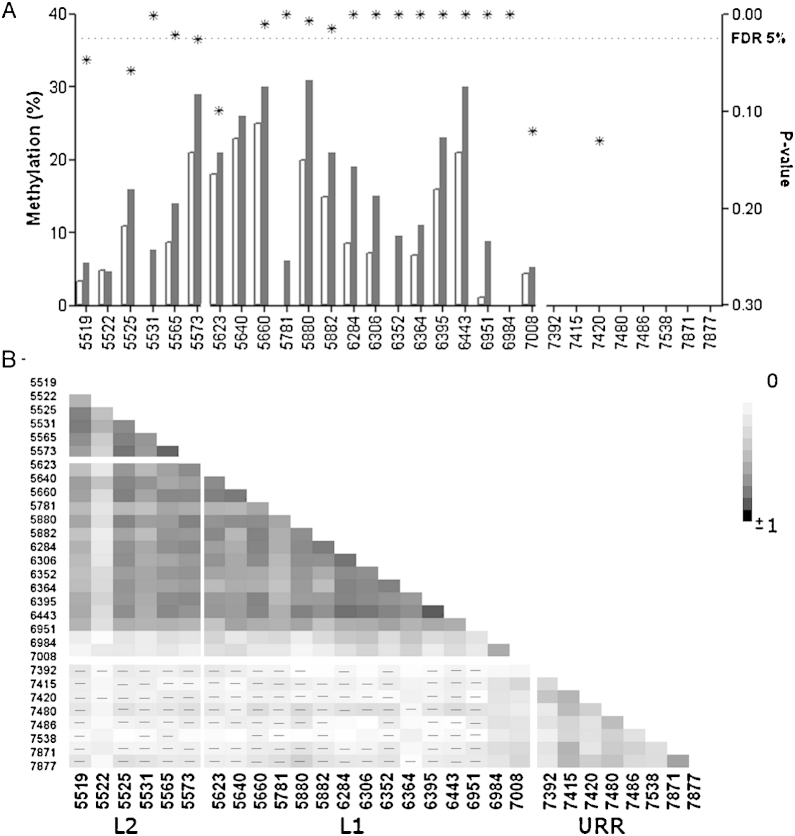
The methylation of 29 sites was measured in 207 HPV31 positive patients. One patient was excluded due to an unspecified histology. In the case group, 99 patients were diagnosed with CIN2/3 and none with cancer. 107 patients comprised the control group (<CIN2). The median age was 28 years in each group as well as in all HPV31 positive women with interquartile range (IQR) 6. Age was not correlated to the methylation of HPV31 sites with Spearman rho ranging from −0.10 to 0.13. Median methylation in the investigated sites in HPV31 L2 and L1 measured higher in CIN2+ compared to <CIN2 (Fig. 3a). Two of 6 L2 and 12 of 15 L1 sites showed significantly higher methylation after adjustment for multiple comparisons. Highest correlation was observed between the sites within the L1 and L2 gene (Fig. 3b). The AUC and p-values for each site in L1 and L2 is presented in Supplemental Table 5.
Fig. 3.
(A) The median methylation presented for each of the investigated CpG positions in HPV31. The methylation was overall higher in CIN2+ (black bar) comparing to <CIN2 (white bar) in the capsid coding genes. The p-values (*) were plotted on the right Y-axis and the FDR 5% is shown. (B) The heat map showing Spearman r correlation between all investigated sites. Negative correlations are marked by (−).
4.3. Methylation of HPV33
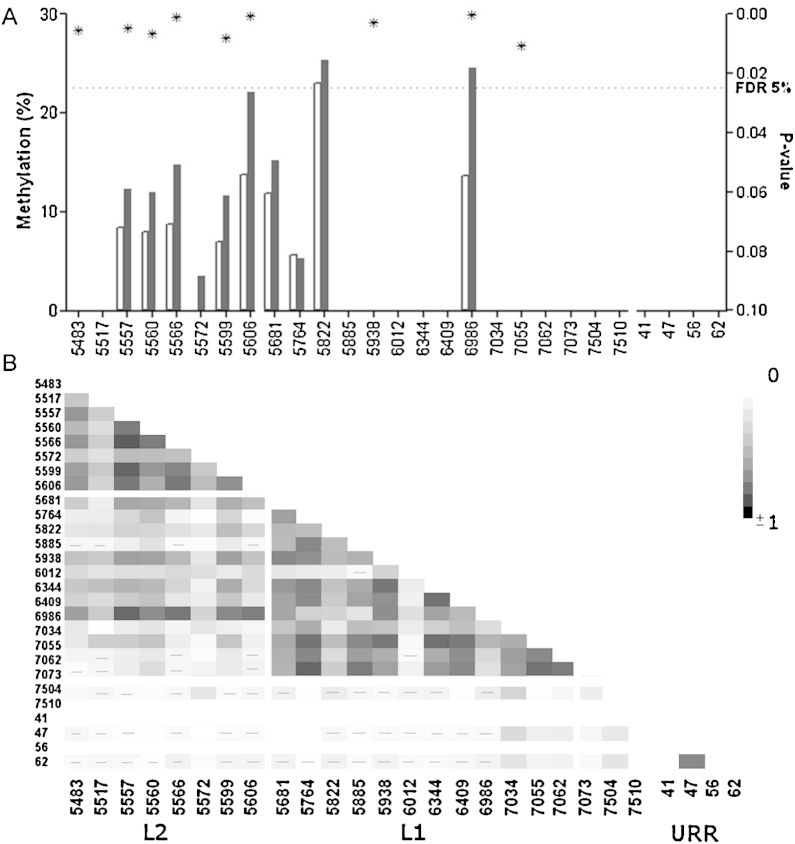
The methylation of 27 sites was measured in 126 HPV33 positive patients. Nine patients (2 cases and 7 controls) failed to amplify with all assays and were therefore excluded from the analysis. Out of remaining 117 patients, there were 74 cases (72 CIN2/3 and 2 SCC) and 43 controls (<CIN2). Median age was 29 each group, and 28 in all analysed samples with interquartile range (IQR) 4. Age was not correlated to the methylation of HPV33 sites as the Spearman rho ranged from −0.06 to 0.10.
Median methylation measured higher in the CIN2+ group in 6 out of 8 sites in L2 and 3 out of 13 sites investigated in HPV33 L1 (Fig. 4a). The AUC and p-values for each site in L1 and L2 is presented in Supplemental Table 6. The correlation of methylation was highest within L2 CpG sites and L1 CpG sites (Fig. 4b).
Fig. 4.
(A) The median methylation presented for each of the investigated CpG positions in HPV33. The methylation was higher in CIN2+ (black bar) comparing to <CIN2 (white bar) in 6 L2 and 3 L1 sites. The p-values (*) were plotted on the right Y-axis and the FDR 5% is shown. (B) The heat map showing Spearman's r correlation between all investigated sites. Negative correlations are marked by (−).
4.4. Methylation in single vs. multiple infections
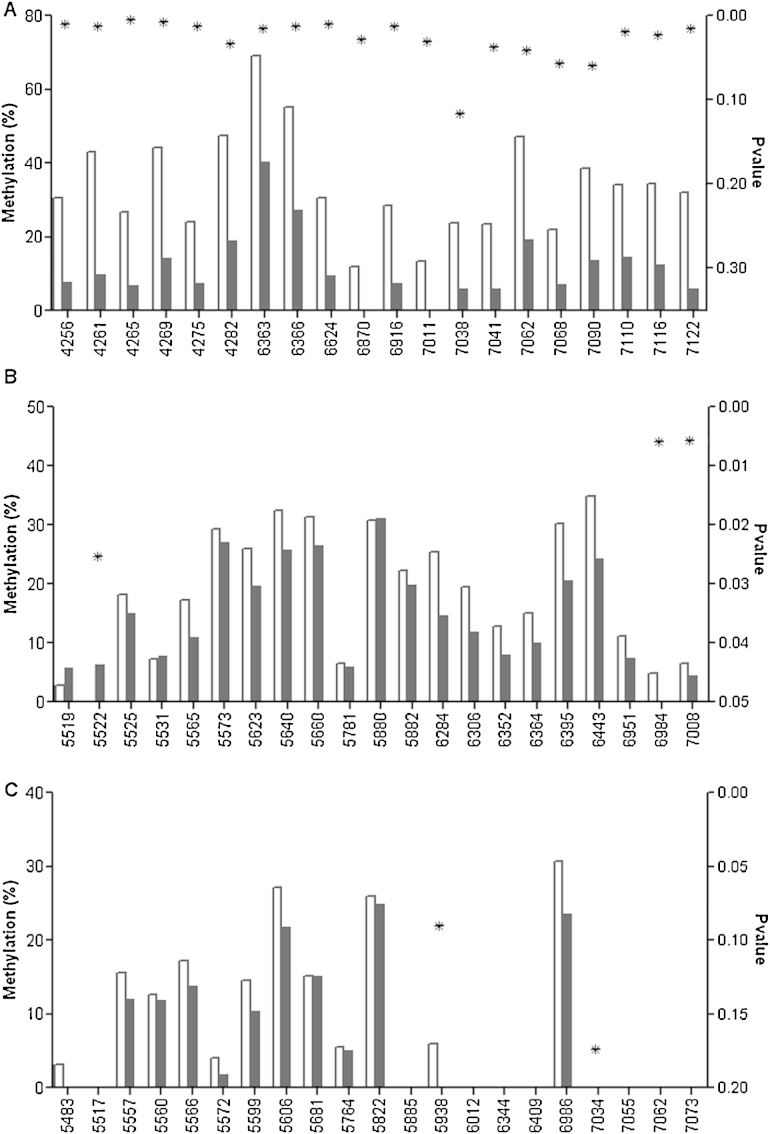
Methylation was compared between single and multiple infections in CIN2+ (Fig. 5). A multiple infection was defined as CIN2+ infected with at least two of the 15 HR-HPV types. For HPV18, median methylation was significantly higher in single infections in all L2 sites and 11 of 14 L1 sites (Fig. 5a). Median methylation was significantly higher in HPV31 L1 sites 6984 and 7008 in single infections and in L2 site 5522 in multiple infections (Fig. 5b). For HPV33, although methylation appeared higher in several sites in L2 and 6986 in L1, none were significant at α = 0.05 (Fig. 5c). The p-values in Fig. 5 were not corrected for multiple testing.
Fig. 5.
Median methylation of (a) HPV18, (b) HPV31 and (c) HPV33 L2 and L1 in CIN2+ group in single infections (white bar) vs. multiple infections (black bar). A patient was considered to have multiple infection if positive for any of the 15 HR-HPV types. Out of 77 HPV18 positive CIN2+, 14 were single infections and 63 multiple. For HPV31, there were 37 single and 62 multiple infections. HPV33 infected CIN2+ consisted of 21 single and 53 multiple infections. Unadjusted p-values (*) were plotted on the right Y-axis.
5. Discussion
The methylation of many CpG sites in L2 of HPV18, HPV31 and HPV33 were significantly higher in CIN2+ compared to <CIN2 (Figs. 2–4) while the methylation of CpGs in the URR and E6 gene were overall low and did not differ between the diagnostic groups. For the L1 gene, methylation was significantly higher in all investigated sites in HPV18, a majority of HPV31 sites and in only 3 out of 13 sites in HPV33. This may reflect that we had the smallest group of patients positive for HPV33, warranting a larger study with HPV33. Elevated methylation in CpG positions in capsid genes of HPV18 and HPV31 is in line with previous reports [15–17] and confirm that assessment of methylation in these HPV types may be a good approach for development of an effective triage tool to complement HPV16, which has been previously validated as a promising triage biomarker [13]. The highest AUC reached was 0.69 for HPV18 sites 6363, 6366 and 7090 (Supplemental Table 4) and site 6951 in HPV31 (Supplemental Table 5). A previous study reported AUC values for the corresponding sites of approximately 0.8 [17]. This observed difference may be due to differences in the populations under study as ours was a colposcopy referral group and also we included CIN1 in the control group. For HPV33, the highest AUC of 0.70 was reached for site 6986 in spite of the smaller patient group and presence of CIN1 in the control group. We report a significant and fair separation by at least one site for each of the investigated HPV CpGs and suggest that an expansion of S1 classifier [13] to methylation of additional HPV is warranted and likely to increase its effectiveness.
A limitation of our study is that we used archived specimens from a set of women referred to colposcopy due to one or more abnormal cytology tests. Although this allowed us to investigate methylation in a large group of CIN2+, our results need to be validated in women who were identified as HR-HPV positive during routine screening in the general population. Furthermore, pyrosequencing is a highly efficient and accurate quantification method but is limited to relatively short reads. Our study did not include an assessment of all CpG sites in each HPV types; however, a previous report has shown high correlation between CpG sites within the HPV18 and 31 genes [17] and therefore we believe that we investigated a sufficient number of sites to allow the development of new triage classifiers for HPV18, HPV31 and HPV33.
Interestingly, similarly to previous findings [17], the methylation of HPV18 and HPV31 capsid genes appeared to be higher in CIN2+ with a single infection (Fig. 5), the mechanism behind which is unknown at the present time. A plausible explanation is that the HPV type driving the carcinogenesis is subjected to more methylation as a cellular defence. In specimens that contain DNAs of multiple HPVs it is often not clear which HPV type is the driver; if methylation pressure is actually exerted preferentially on the driver it may allow a differentiation between driver and passenger infections.
In summary, we confirm that methylation of the capsid genes of HPV18 and HPV31 is elevated in high grade disease. We report for the first time the methylation of 27 sites in L2 and L1 HPV33 showing that 8 sites in L2 and 3 sites in L1 were elevated in high grade disease. A next step is development of a comprehensive classifier where methylation of HR-HPV types 16, 18, 31, 33 and 45 is combined to identify high grade disease. We believe that an expanded classifier may serve as an effective triage tool for HR-HPV positive women and improve cervical cancer prevention.
Author contributions
NV made substantial contribution to acquisition, analysis and interpretation of the data and drafting of the manuscript. DSB made substantial contribution to acquisition of data and revising of the manuscript. AB made substantial contribution to analysis and interpretation of data as well as revising of the manuscript. JC and ATL contributed to the original concept, design of the study, data interpretation, and manuscript revisions. All authors read and approved the final manuscript.
Funding
This work was funded by Cancer Research UK [Grant number C569/A10404].
Competing interest
None to declare.
Ethical approval
Hammersmith and Queen Charlottes & Chelsea Research Ethics Committee REC no 05/Q0406/57.
Acknowledgments
The authors thank all the participating patients. We thank Louise Cadman, Janet Austin and Anne Szarewski for providing us with the samples and collecting, updating and maintaining the P1 and P2 cohorts. Dr Anne Szarewski passed away during the final stages of this study. We are very grateful to her for the wonderful enthusiasm and the great efforts she made to ensure that studies were done to the highest level.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Munoz N., Bosch F.X., de Sanjose S., Herrero R., Castellsague X., Shah K.V. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 2.Bruni L., Diaz M., Castellsague X., Ferrer E., Bosch F.X., de Sanjose S. Cervical human papillomavirus prevalence in 5 continents: meta-analysis of 1 million women with normal cytological findings. J Infect Dis. 2012;202:1789–1799. doi: 10.1086/657321. [DOI] [PubMed] [Google Scholar]
- 3.Ho G.Y., Bierman R., Beardsley L., Chang C.J., Burk R.D. Natural history of cervicovaginal papillomavirus infection in young women. N Engl J Med. 1998;338:423–428. doi: 10.1056/NEJM199802123380703. [DOI] [PubMed] [Google Scholar]
- 4.Cervix Cancer Screening . IARC Press; Lyon: 2005. IARC handbooks of cancer prevention. [Google Scholar]
- 5.Saslow D., Solomon D., Lawson H.W., Killackey M., Kulasingam S.L., Cain J. American Cancer Society, American Society for Colposcopy and Cervical Pathology, and American Society for Clinical Pathology screening guidelines for the prevention and early detection of cervical cancer. Am J Clin Pathol. 2012;137:516–542. doi: 10.1309/AJCPTGD94EVRSJCG. [DOI] [PubMed] [Google Scholar]
- 6.Castle P.E., de Sanjose S., Qiao Y.L., Belinson J.L., Lazcano-Ponce E., Kinney W. Introduction of human papillomavirus DNA screening in the world: 15 years of experience. Vaccine. 2012;30(Suppl. 5):F117–F122. doi: 10.1016/j.vaccine.2012.05.071. [DOI] [PubMed] [Google Scholar]
- 7.Cuzick J., Bergeron C., Doeberitz M.K., Gravitt P.E., Jeronimo J., Lorincz A. New technologies and procedures for cervical cancer screening. Vaccine. 2012;30:F107–F116. doi: 10.1016/j.vaccine.2012.05.088. [DOI] [PubMed] [Google Scholar]
- 8.Lazcano-Ponce E., Lorincz A.T., Cruz-Valdez A., Salmeron J., Uribe P., Velasco-Mondragon E. Self-collection of vaginal specimens for human papillomavirus testing in cervical cancer prevention (MARCH): a community-based randomised controlled trial. Lancet. 2011;378:1868–1873. doi: 10.1016/S0140-6736(11)61522-5. [DOI] [PubMed] [Google Scholar]
- 9.Portela A., Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 10.Kalantari M., Calleja-Macias I.E., Tewari D., Hagmar B., Lie K., Barrera-Saldana H.A. Conserved methylation patterns of human papillomavirus type 16 DNA in asymptomatic infection and cervical neoplasia. J Virol. 2004;78:12762–12772. doi: 10.1128/JVI.78.23.12762-12772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun C., Reimers L.L., Burk R.D. Methylation of HPV16 genome CpG sites is associated with cervix precancer and cancer. Gynecol Oncol. 2011;121:59–63. doi: 10.1016/j.ygyno.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mirabello L., Schiffman M., Ghosh A., Rodriguez A.C., Vasiljevic N., Wentzensen N. Elevated methylation of HPV16 DNA is associated with the development of high grade cervical intraepithelial neoplasia. Int J Cancer. 2012;132:1412–1422. doi: 10.1002/ijc.27750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorincz A.T., Brentnall A.R., Vasiljevic N., Scibior-Bentkowska D., Castanon A., Fiander A. HPV16 L1 and L2 DNA methylation predicts high-grade cervical intraepithelial neoplasia in women with mildly abnormal cervical cytology. Int J Cancer. 2013;133(3):637–644. doi: 10.1002/ijc.28050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mirabello L., Sun C., Ghosh A., Rodriguez A.C., Schiffman M., Wentzensen N. Methylation of human papillomavirus type 16 genome and risk of cervical precancer in a Costa Rican population. J Natl Cancer Inst. 2012;104:1–10. doi: 10.1093/jnci/djs135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turan T., Kalantari M., Calleja-Macias I.E., Cubie H.A., Cuschieri K., Villa L.L. Methylation of the human papillomavirus-18 L1 gene: a biomarker of neoplastic progression. Virology. 2006;349:175–183. doi: 10.1016/j.virol.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 16.Turan T., Kalantari M., Cuschieri K., Cubie H.A., Skomedal H., Bernard H.U. High-throughput detection of human papillomavirus-18 L1 gene methylation, a candidate biomarker for the progression of cervical neoplasia. Virology. 2007;361:185–193. doi: 10.1016/j.virol.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wentzensen N., Sun C., Ghosh A., Kinney W.K., Mirabello L., Wacholder S. Methylation of HPV18, HPV31, and HPV45 genomes is associated with cervical intraepithelial neoplasia grade 3. J Natl Cancer Inst. 2012;104:1738–1749. doi: 10.1093/jnci/djs425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Sanjose S., Quint W.G., Alemany L., Geraets D.T., Klaustermeier J.E., Lloveras B. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. 2010;11:1048–1056. doi: 10.1016/S1470-2045(10)70230-8. [DOI] [PubMed] [Google Scholar]
- 19.Li N., Franceschi S., Howell-Jones R., Snijders P.J., Clifford G.M. Human papillomavirus type distribution in 30,848 invasive cervical cancers worldwide: Variation by geographical region, histological type and year of publication. Int J Cancer. 2010;128:927–935. doi: 10.1002/ijc.25396. [DOI] [PubMed] [Google Scholar]
- 20.McShane L.M., Altman D.G., Sauerbrei W., Taube S.E., Gion M., Clark G.M. REporting recommendations for tumor MARKer prognostic studies (REMARK) Breast Cancer Res Treat. 2006;100:229–235. doi: 10.1007/s10549-006-9242-8. [DOI] [PubMed] [Google Scholar]
- 21.Szarewski A., Ambroisine L., Cadman L., Austin J., Ho L., Terry G. Comparison of predictors for high-grade cervical intraepithelial neoplasia in women with abnormal smears. Cancer Epidemiol Biomarkers Prev. 2008;17:3033–3042. doi: 10.1158/1055-9965.EPI-08-0508. [DOI] [PubMed] [Google Scholar]
- 22.Szarewski A., Mesher D., Cadman L., Austin J., Ashdown-Barr L., Ho L. Comparison of seven tests for high-grade cervical intraepithelial neoplasia in women with abnormal smears: the predictors 2 study. J Clin Microbiol. 2012;50:1867–1873. doi: 10.1128/JCM.00181-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vasiljevic N., Wu K., Brentnall A.R., Kim D.C., Thorat M., Kudahetti S.C. Absolute quantitation of DNA methylation of 28 candidate genes in prostate cancer using pyrosequencing. Disease Markers. 2011;30:151–161. doi: 10.3233/DMA-2011-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.