Abstract
Epidermal growth factor receptor (EGFR) is a crucial protein that plays an important role in the maintenance and development of glioblastomas. The silencing or knockdown of EGFR is possible by administering a small interfering ribonucleic acid (siRNA). Lipid nanocapsules (LNCs) covered by chitosan were developed in our laboratory by a transacylation process. The resulting nanocapsules have a positive zeta potential that enables electrostatic interactions with the negatively-charged siRNA. Prior to transfection, the cytotoxicity of the nanocapsules by (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) (MTS) test was performed on the U87MG cell line to determine non-toxic levels of the LNCs to avoid cell mortality. Treatment of the U87MG cells with the chitosan-transacylated LNCs/anti-EGFR siRNA complex resulted in a reduction of EGFR expression by 51.95%±6.03% (P≤0.05) after 96 hours of incubation. It also increased the cellular sensitivity to temozolomide in comparison to untreated cells with siRNA. The largest increase in mortality was 62.55%±3.55% (P<0.05). This successful knockdown provides proof for the concept of surface grafting of siRNA onto LNCs to modify cell sensitivity to temozolomide. The method could be implemented in future clinical models regarding the experimental treatment of glioblastoma cancer.
Keywords: EGFR, glioblastoma, siRNA, lipid nanocapsules, chitosan, temozolomide
Introduction
Glioblastoma is the most common primary brain tumor in adults.1,2 Median survival is 14 months and the percentage of patients living for 5 years or more is lower than 10%.3 Despite the advances made during the last few years in brain tumor therapy, the prognosis of this disease has not improved. Current therapy includes surgical intervention, radiotherapy, and chemotherapy with temozolomide.3
Glioblastoma cells overexpress many kinds of proteins, growth factors, and receptors.4–6 Epidermal growth factor receptor (EGFR) is one of the receptors that play an important role in tumor development by stimulating cell proliferation and cell resistance to apoptosis and autophagy and hence contributes to a malignant phenotype.7 It also plays an important role in anticancer drug resistance by stimulation of the Ras/Raf/ERK8 and PI3K/Akt/m-TOR9 signaling pathways implicated in the inhibition of cell autophagy and apoptosis.10 Some studies have shown that the inhibition of EGFR may have clinical importance for patients with glioblastoma.11
In recent years, the strategy of ribonucleic acid interference (RNAi), whose principle process is to reduce the expression of target messenger RNA (mRNA), was discovered, opening up new perspectives in different areas of medicine. RNAi is an intrinsically cellular pathway discovered in 1998.12–14 This pathway can be initiated by introducing small interfering RNA (siRNA) consisting of double-stranded RNA of 21–23 nucleotides. In the cytoplasm, these siRNA molecules are incorporated into a nuclease-containing multi-protein complex called RNA-induced silencing complex (RISC). Following that, the antisense strand remains in the RISC complex and guides the RNAi enzymatic machinery while the sense strand of the RISC complex is eliminated. Degradation of the complementary mRNA is provided by nuclease argonaute-2.
The use of therapeutic siRNAs alone is not possible for several reasons, including their negative charge; this charge prevents them from crossing the plasma membrane to reach their target in the cytoplasm.15–17 On the other hand, the presence of nucleases in plasma and the cytoplasm are likely to deteriorate siRNAs rapidly after their injection into the blood; additionally, the immune response that can be triggered by their presence in the blood may also lead to their degradation.18 These reasons emphasize the need for a suitable vector able to deliver the siRNA into the desired cells.
The vectors used thus far include viral vectors19 (adenovirus, adeno-associated virus [AAV], retroviral vectors derived from murine leukemia virus [MLV], or lentiviruses) and non-viral vectors (eg, liposomes, cationic polymers, polymeric micelles).20 Despite their efficiency, viral vectors have some limitations, including their residual pathogenic effect, which represents a potential risk to patients. Moreover, a mutagenesis effect was observed in some clinical trials using these vectors.21 Regarding non-viral vectors, the drawbacks are related to the high toxicity of their compounds, especially organic solvents and some cationic polymers.22
Nanoparticles can be loaded by various therapeutic agents that can protect against degradation and ensure their controlled release.23 In addition, their surface may be functionalized with specific ligands which can increase penetration into the tumor. However, poor intratumoral distribution and failure to target a disseminated tumor mass are still a great challenge for clinical application.24
In our laboratory, we prepared lipid nanocapsules (LNCs) by using a phase-inversion process.25 This has the advantage of using no organic solvents and only GRAS (generally recognized as safe) excipients and GMO (genetically modified organisms)-free products.26 The obtained nanocapsules have a structure consisting of an oily core of Labrafac® WL 1349 (Gattefossé Group, Saint-Priest, France), and a crown of surfectants (Solutol® HS 15 [BASF, Ludwigshafen, Germany] and Lipoïd® S75-3 [Lipoïd GmbH, Ludwigshafen, Germany]). The oily core could be used to encapsulate lipophilic drugs such as paclitaxel, a drug that is widely used in oncology.27
The siRNAs are insoluble in Labrafac®, a quality that prevents their encapsulation in nanocapsules. For this reason, we chose to attach them to the surface of nanocapsules by electrostatic attraction. However, due to the negative charge on the surface of blank LNCs (zeta potential =−6.4±0.4 mV), the setting of siRNA (also negatively charged) can only be obtained after modifying the surface of the LNCs by grafting positively-charged polymers (chitosan). Grafting chitosan is carried out by a transacylation reaction. This polymer was chosen as a transacylation polymer because of its interesting properties, including its low toxicity and its biocompatibility.28
The objectives of this work were therefore to graft an anti-EGFR siRNA to nanocapsules, then to study the knockdown of EGFR and to verify if this modification of protein production induced an increased sensitivity to temozolomide in U87MG glioblastoma cells.
Material and methods
Materials
Solutol® HS 15 (BASF, Ludwigshafen, Germany) is a mixture of free polyethylene glycol 660 and polyethylene glycol 660 hydroxystearate. Labrafac® WL 1349 (Gattefossé Group, Saint-Priest, France) is a mixture of capric and caprylic acid triglycerides. NaCl was purchased from Prolabo (Fontenay-sous-Bois, France). Lipoïd® S75-3 (Lipoïd GmbH, Ludwigshafen, Germany) is a soybean lecithin made of 69% phosphatidylcholine, 10% phosphatidylethanolamine, and other phospholipids. Milli-Q water was obtained from a Milli-Q-plus® system (Merck Millipore, Billerica, MA, USA). Chitosan oligosaccharide lactate 5,000 Da with a degree of deacetylation ≥75% was purchased from Sigma Aldrich (St Louis, MO, USA). The dialysis membrane was purchased from Spectrum Laboratories (Rancho Dominguez, USA) and had a molecular weight cut-off point equal to 50,000 Da. O-Phthalaldehyde (OPA) was purchased from Thermo Fisher Scientific (Waltham, MA, USA). MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) and MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assays (Cell Titer 96® kit) were purchased from Promega Corporation (Fitchburg, WI, USA).
Preparation and characterization of chitosan-transacylated LNCs
Formulation of LNCs
In this study, LNCs with a diameter of 50 nm were prepared according to a phase-inversion process described by Heurtault et al.25
In brief, Solutol® HS15, Lipoïd® S75-3, NaCl, Labrafac®, and water (0.846 g, 0.075 g, 0.089 g, 1.028 g, 2.962 g, respectively) were mixed and warmed up to 85°C under magnetic stirring. This was followed by cooling down the formulation to 60°C; this cycle was repeated three times. During the last cycle, 12.5 mL of water at 4°C was added to the formulation. With this phase-inversion method, LNCs with an oily core consisting of triglycerides (Labrafac®) and a surface consisting of Solutol® HS15 and Lipoïd® S75-3 were obtained.
Surface modification of LNCs by a transacylation reaction
A transacylation reaction was obtained between the pegylated hydroxystearate and a functional amino group of a polymer (Figure 1). To perform a transacylation reaction, 20 mL of the previously-made LNCs were used. This quantity was mixed with 1 mL of NaOH 10 M and different amounts of chitosan. The reaction took place at 25°C in a water bath for 15 minutes and afterwards the reaction was stopped by adding 20 mL of a glycine buffer. The pH was adjusted to around 5.5±0.5 to ensure a correct ionization of the chitosan before adding siRNA. Finally purification was performed by dialysis with membranes having a molecular weight cut-off of 50 K Daltons. This allowed transacylated nanoparticles to stay inside the dialysis membrane while the free chitosan diffused in the dialysis water. Dialysis was performed for 24 hours with Milli-Q water under magnetic stirring.
Figure 1.
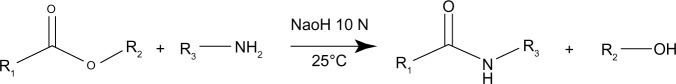
Transacylation reaction between the pegylated hydroxystearate contained in the Solutol® HS 15 and a polyamine.
Notes: R1, 12-hydroxystearic acid; R2, polyethylene glycol; R3, chitosan. Solutol® HS 15 (BASF, Ludwigshafen, Germany).
Determination of sizes and zeta potentials of chitosan-transacylated LNCs
The average hydrodynamic diameter, polydispersity index and zeta potential of all particles were measured by dynamic light scattering (DLS) using NanoZS® (Malvern Instruments, Malvern, UK). Measurements were performed at a 90° angle after dispersion of 50 μL of the LNC suspension in 2.95 mL of Milli-Q water. Measurements were performed in triplicate at 25°C with comparable conductivity values for zeta potentials determination.
Quantification of bound chitosan on the LNC surface
To quantify the amount of bound chitosan on the surface of the transacylated particles, the o-Phthalaldehyde (OPA) quantification method was used. Dialyzed transacylated LNCs (20 μL) were placed in a black, 96-well plate. OPA (280 μL) was added to each sample and the fluorescence was recorded with a Fluoroskan (Thermo Fisher Scientific) with an excitation wavelength of 355 nm and an emission wavelength of 460 nm.29 A calibration curve ranging from 0.013–0.83 mg/mL of chitosan was obtained.
Characterization of grafted siRNA onto chitosan-transacylated LNCs
The characterization of grafted siRNA serves two purposes: first to determine the ability of the LNCs to graft siRNA; second, to correlate the amount of grafted siRNA onto the LNCs to the knockdown of the target gene in vitro.
In order to facilitate the quantification of the siRNA fixed to the surface of chitosan-transacylated nanocapsules, the fluorescence quenching of a suspension of fluorescein-labeled siRNA was used. A suspension containing an anti-EGFR siRNA labeled with fluorescein isothiocyanate (FITC) at a concentration of 19.2 μM was prepared. Five microliters of this suspension was added to volumes of LNCs from 0 μL to 50 μL and supplemented with RNase free water up to one final volume of 300 μL. The fluorescence was measured immediately by Fluoroskan at wavelengths of 485 nm excitation and 520 nm emission.
Stability of chitosan-transacylated LNCs
The stability of nanocapsules transacylated with 1.2 mg/mL of chitosan was evaluated at 4°C and 25°C for 30 days. For this purpose, measurements of hydrodynamic diameter, polydispersity index, and zeta potential of nanocapsules transacylated with 1.2 mg/mL of chitosan were made by NanoZS®. Moreover, the visual aspect of the formulation was analyzed at both temperatures.
Sizes and zeta potentials of chitosan-transacylated LNCs/anti-EGFR siRNA complexes
The average hydrodynamic diameter, polydispersity index, and zeta potential of LNCs transacylated with 1.2 mg/mL of chitosan/anti-EGFR siRNA complexes were measured by DLS using NanoZS®. The mixture of transacylated LNCs and anti-EGFR siRNA was performed so as to have a final concentration of LNCs of about 100 μg/mL and concentrations of anti-EGFR siRNA of 10, 20, 30, and 40 nM. Measurements were performed in RNase-free water. Measurements were performed in triplicate at 25°C with comparable conductivity values for zeta potentials determination.
MTS cytotoxicity test on U87MG cells
An MTS cytotoxicity test was performed to determine the maximal nontoxic concentration of transacylated LNCs. The MTS assay depends on the reduction of tetrazolium salt into formazan by the mitochondrial enzyme succinate dehydrogenase. The reaction takes place only in living cells and the quantity of produced formazan is proportional to cell viability. Fluorescence was measured by Multiskan Ascent® (Labsystems, Haverhill, MA, USA) at a wavelength of 492 nm. The effect of LNCs transacylated with 1.2 mg/mL of chitosan on the U87MG (American Type Culture Collection) cell line was investigated. In a 96-well plate, 5,000 cells were incubated in each well for 48 hours in 100 μL of culture medium (Dulbecco’s Modified Eagle Medium [DMEM] with 1 g/L of glucose and supplemented with 10% of fetal calf serum [FCS] and 1% of antibiotics). After 48 hours, 20 μL of sample was added. The final concentrations of the LNCs in the wells were 15, 50, 75, 100, 150, and 200 μg/mL. After 24 hours of incubation, the medium was removed and the cells were washed with 120 μL of phosphate-buffered saline (PBS). MTS reagent was added to each well and then stored in the dark for 2 hours at 37°C. Finally, a reading was carried out at 492 nm.
Calculating the percentage of living cells by MTS test was done by the following report:
| [1] |
siRNA transfection and determination of EGFR expression by fluorescence-activated cell sorting
Twenty-five thousand cells were incubated in 500 μL of DMEM supplemented with 10% fetal bovine serum and 1% antibiotics. The U87MG cells were treated with five concentrations of anti-EGFR siRNA from 10–50 nM. The anti-EGFR siRNA was carried by LNCs transacylated with 1.2 mg/mL of chitosan at a concentration of 100 μg/mL.
These cells were incubated for 72 hours and 96 hours, then analyzed by fluorescence-activated cell sorting (FACS). The anti-EGFR siRNA sequence used was: sens, 5′CACAGUGGAGCGAAUUCCUdTdT 3′; anti-sens, 3′dTdTGUGUCACCUCGCUUAAGGA 5′.
The siRNA negative control sequence used was a scrambled sequence carried by chitosan-transacylated LNCs.
After the incubation period, U87MG cells were detached from the bottom of the plate by Versene® (EDTA) 0.02% (Lonza, Verviers, Belgium) and then recovered in PBS/FCS/azide (0.02%). The cells were then placed in a 96-well plate and incubated with the anti-EGFR and isotype (IgG2bk) antibodies (BD Biosciences, San Diego, CA, USA). After 1 hour of incubation at 4°C, washes with PBS/FCS/azide (0.02%) were made to remove unbound antibodies. Following this, an incubation with a secondary antibody labeled with FITC (polyclonal goat anti-mouse Ig/FITC Goat F(ab′)2) (Dako Denmark A/S, Glostrup, Denmark) was then performed for 30 minutes at 4°C in the shelter of light. After the incubation period with the secondary antibody, washes with PBS/FCS/azide (0.02%) were made to remove unbound secondary antibody. A 50 μL of PBS/azide (0.02%) solution was added to each cell pellet. Each cell suspension was then placed in a volume of 250 mL PBS/azide (0.02%)/formaldehyde (1%). The analyses were performed by FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA).
The calculation of the percentage of EGFR expression in each sample was performed by comparing the geometric mean of the samples and the control cells according to the formula:
| [2] |
Evaluation of chitosan transacylated LNCs/anti-EGFR siRNA effect on proliferation of U87MG cells
The objective of this part was to evaluate the effect on the proliferation of U87MG cells after treatment with LNCs transacylated with 1.2 mg/mL of chitosan/anti-EGFR siRNA; in a 24-well plate, 25,000 cells were incubated with 500 μL of medium culture in each well, for 48 hours. After that, these cells were treated for 72 hours with complex LNCs transacylated with 1.2 mg/mL of chitosan/anti-EGFR siRNA. The final concentration of anti-EGFR siRNA tested was 40 nM and the concentration of chitosan-transacylated LNCs was 100 μg/mL. Cell proliferation was then assessed by MTT assay whose principle is based on the reduction of a tetrazolium salt (bromide 3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium) to formazan by succinate dehydrogenase, a mitochondrial enzyme only active in living cells. The amount of precipitate formed is proportional to the percentage of living cells.
Calculating the percentage of living cells by MTT test was done by the following report:
| [3] |
Evaluation of the increased sensitivity of U87MG cells to temozolomide after siRNA treatment by MTT assay
In a 24-well plate, 25,000 cells were incubated with 500 μL of culture medium in each well, for 48 hours. The cells were then treated and incubated for 72 hours with the anti-EGFR siRNA at a concentration of 40 nM. At the end of the incubation period, the culture medium was changed and the cells were treated with increasing concentrations of temozolomide (0.06, 0.125, 0.25, and 0.5 mg/mL) for 24 hours. At the end of the incubation period with temozolomide, the culture medium was changed in each well and a volume of 40 μL of the MTT reagent was added and incubated for 4 hours in the dark at 37°C. The supernatant was then aspirated and a volume of 400 μL of acidic isopropanol was added to each well to dissolve the previously-obtained formazan crystals. The plate was left under mechanical stirring and the contents of each well were transferred to a 96-well plate to perform the Multiskan Ascent® reading at 580 nm.
Statistical analysis
Results were expressed as mean values ± standard error of the mean (SEM). Mann-Whitney U test was used for statistical comparison between controls and other groups.
Results
Characterization of chitosan-transacylated LNCs
Size and zeta potential of chitosan-transacylated LNCs
LNCs were obtained and chitosan was successfully grafted to their surfaces. The sizes and zeta potentials of blank nanocapsules and four formulations of LNCs transacylated with different amounts of chitosan are reported in Table 1. These results show an increase of about 19 nm in the size of transacylated LNCs compared to the blank nanocapsules. On the other hand, the zeta potential of these formulations became positive and increased with the amount of chitosan.
Table 1.
Sizes and zeta potentials of different LNC formulations
| Formulation | Size (nm) | Polydispersity index | Zeta potential (mV) |
|---|---|---|---|
| Blank LNCs | 52.6±0.6 | 0.02 | −6.4±0.4 |
| LNC 50 nm + 0.6 mg/mL chitosan | 70.6±1.5 | 0.23 | 17.4±0.5 |
| LNC 50 nm + 1.2 mg/mL chitosan | 71.6±1.1 | 0.22 | 24.7±2.1 |
| LNC 50 nm + 1.8 mg/mL chitosan | 70.6±0.7 | 0.24 | 31.4±0.6 |
| LNC 50 nm + 2.4 mg/mL chitosan | 72.2±0.6 | 0.25 | 36.0±0.7 |
Note:
Data are shown as mean ± standard error of the mean (n=3).
Abbreviation: LNC, lipid nanocapsules.
Quantification of bound chitosan onto the surface of the LNCs
The evolution of yield of transacylation depending on the amount of chitosan is shown in Figure 2. Indeed, the graph shows a decrease in the yield of transacylation of 20.11% to 6.5% after addition of increasing amounts of chitosan.
Figure 2.
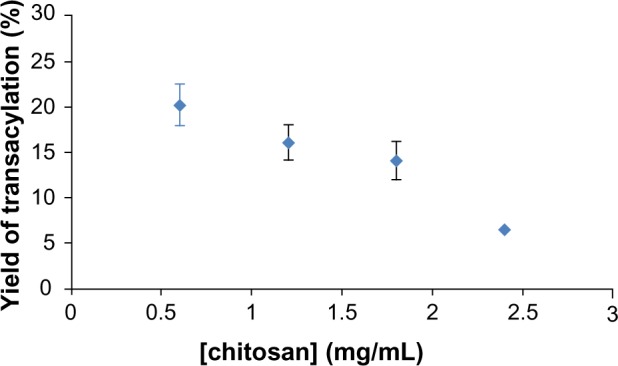
Yield of transacylation measured by OPA reaction related to amounts of chitosan used in the reaction.
Note: Data are shown as mean ± standard error of the mean (n=3).
Abbreviation: OPA, o-Phthalaldehyde.
The yield of transacylation is the percentage of polymer (chitosan) remaining on the surface of nanocapsules after purification. It is determined by the following formula:
| [4] |
where IF460 sample is the fluorescence intensity of the sample at 460 nm; IF460 blank is the fluorescence intensity of reference sample at 460 nm; [chitosan] theoretical is the concentration of chitosan in mg/mL inside the LNC samples before purification; F is the dilution factor used for the preparation of samples; D is the coefficient of calibration curve; and Y is the yield of transacylation.
Determination of quantity of siRNA grafted onto chitosan-transacylated LNCs
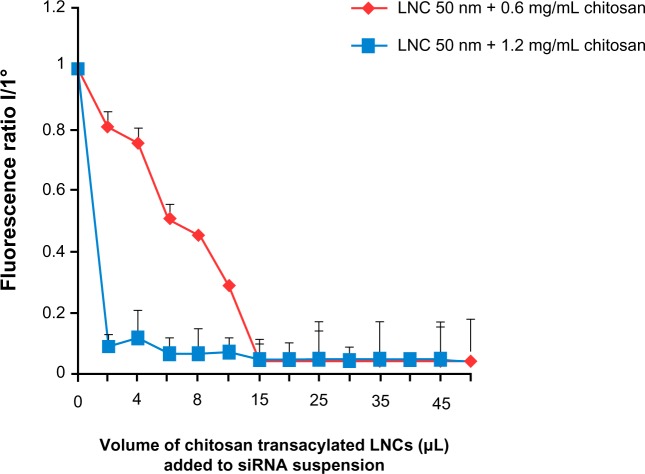
In this part of the study, two formulations of transacylated LNCs (with 0.6 mg/mL and 1.2 mg/mL of chitosan) were studied. The results showed a decrease in fluorescence intensity of anti-EGFR siRNA suspensions following the addition of chitosan-transacylated LNCs in the medium (Figure 3). This indicates that these siRNAs were grafted by electrostatic attraction to the surface of the nanocapsules. In addition, differences in the binding capacity of the siRNA appear between the two formulations. Indeed, the ratio of 95.43%±0.05% fixation of siRNA was obtained with a volume of 15 μL of LNCs transacylated with 0.6 mg/mL of chitosan, whereas 93.12%±0.25% fixation was obtained with 6 μL of LNCs transacylated with 1.2 mg/mL of chitosan.
Figure 3.
Amount of siRNA grafted to the surface of chitosan-transacylated LNCs determined by fluorescence quenching.
Notes: Fluorescence ratio I/I° corresponds to fluorescence of transacylated LNCs/siRNA over the fluorescence of siRNA suspension I° without transacylated LNCs. Data are shown as mean ± standard error of the mean (n=3).
Abbreviations: LNC, lipid nanocapsules; siRNA, small interfering ribonucleic acid.
Determination of sizes and zeta potentials of chitosan-transacylated LNCs/anti-EGFR siRNA complexes
Table 2 indicates the sizes and zeta potentials of complexes formed by the association of LNCs transacylated with 1.2 mg/mL of chitosan and anti-EGFR siRNA. These results indicate an average 24 nM size increase of these complexes compared to LNCs transacylated with 1.2 mg/mL of chitosan. The polydispersity index remained constant compared to that of LNCs transacylated with 1.2 mg/mL of chitosan. On the other hand, the zeta potential of the LNCs transacylated with 1.2 mg/mL of chitosan decreased from 24.7 mV to 15.2 mV with the addition of 40 nM of anti-EGFR siRNA.
Table 2.
Sizes and zeta potentials of LNCs transacylated with 1.2 mg/mL of chitosan/anti-EGFR siRNA complexes
| Formulation | Size (nm) | Polydispersity index | Zeta potential (mv) |
|---|---|---|---|
| LNC 50 nm + 1.2 mg/mL chitosan + 10 nM anti-EGFR siRNA | 94.9±3.9 | 0.25 | 19.2±0.6 |
| LNC 50 nm + 1.2 mg/mL chitosan + 20 nM anti-EGFR siRNA | 93.7±0.8 | 0.23 | 18.9± 1.1 |
| LNC 50 nm + 1.2 mg/mL chitosan + 30 nM anti-EGFR siRNA | 95.5±4.1 | 0.24 | 17.5±0.4 |
| LNC 50 nm + 1.2 mg/mL chitosan + 40 nM anti-EGFR siRNA | 96.4±1.0 | 0.23 | 15.2±0.9 |
Note:
Data are shown as mean ± standard error of the mean (n=3).
Abbreviations: LNC, lipid nanocapsules; anti-EGFR, anti-epidermal growth factor receptor; siRNA, small interfering ribonucleic acid.
Stability of LNCs transacylated with 1.2 mg/mL of chitosan
The results showed that there was no significant variation of the hydrodynamic diameter, polydispersity index, and zeta potential of the formulations tested until day 30 at 4°C and 25°C (Table 3). The visual appearance of the dispersion at 4°C was identical to that of the dispersion left to 25°C.
Table 3.
Stability at 4°C and 25°C of LNC 50 nm + 1.2 mg/mL of chitosan
| Day | Size (nm) | Polydispersity index | Zeta potential (mV) | |
|---|---|---|---|---|
| 4°C | 1 | 71.3±1.5 | 0.21 | 24.8±0.4 |
| 30 | 70.9±0.7 | 0.22 | 23.5±0.7 | |
| 25°C | 1 | 71.3±1.5 | 0.21 | 24.8±0.4 |
| 30 | 74.1±0.7 | 0.23 | 22.3±0.9 |
Note:
Data are shown as mean ± standard error of the mean (n=3).
Abbreviation: LNC, lipid nanocapsules.
MTS cytotoxicity test on U87MG cells
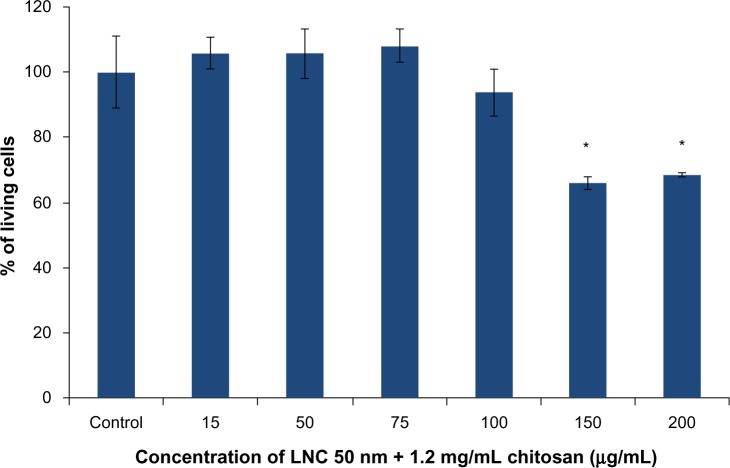
The results obtained with the LNCs transacylated with 1.2 mg/mL of chitosan showed cell death at concentrations of 150 and 200 μg/mL (Figure 4). A concentration of 100 μg/mL of chitosan-transacylated LNCs was chosen as a safe concentration to avoid toxicity and cell death.
Figure 4.
MTS cytotoxicity test on U87MG cells performed with LNC 50 nm + 1.2 mg/mL of chitosan.
Notes: Data are shown as mean ± standard error of the mean (n=3). *Statistical difference compared to control was set at P<0.05 by Mann-Whitney U test.
Abbreviations: MTS, (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium); LNC, lipid nanocapsules.
Determination of EGFR expression on U87MG cells after anti-EGFR siRNA treatment by FACS
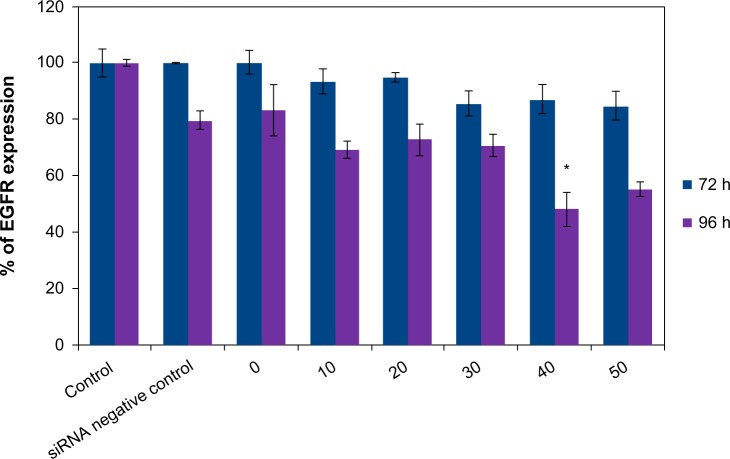
The treatment of U87MG cells by the vector alone at the concentration of 100 μg/mL did not cause a significant decrease in production of EGFR after the two incubation periods. The same finding was observed with the complex formed by the association of LNCs transacylated with 1.2 mg/mL of chitosan/siRNA negative control. Treatment of U87MG cells by LNCs transacylated with 1.2 mg/mL of chitosan/anti-EGFR siRNA for 72 hours did not significantly reduce the rate of EGFR. The same treatment for 96 hours resulted in a significant decrease in the production of EGFR, with a maximum of 51.95%±6.03% obtained with 40 nM of anti-EGFR siRNA (Figure 5). A dose effect was observed between 20 and 40 nM of anti-EGFR siRNA.
Figure 5.
Determination of EGFR expression on U87MG cells by FACS after treatment by the anti-EGFR siRNA for 72 hours and 96 hours.
Notes: Data are shown as mean ± standard error of the mean (n=3). *Statistical difference compared to control was set at P<0.05 by Mann-Whitney U test. siRNA negative control, LNC 50 nm + 1.2 mg/mL chitosan + siRNA negative control; 0, LNC 50 nm + 1.2 mg/mL chitosan; 10, LNC 50 nm + 1.2 mg/mL chitosan + 10 nM anti-EGFR siRNA; 20, LNC 50 nm + 1.2 mg/mL chitosan + 20 nM anti-EGFR siRNA; 30, LNC 50 nm + 1.2 mg/mL chitosan + 30 nM anti-EGFR siRNA; 40, LNC 50 nm + 1.2 mg/mL chitosan + 40 nM anti-EGFR siRNA; 50, LNC 50 nm + 1.2 mg/mL chitosan + 50 nM anti-EGFR siRNA.
Abbreviations: LNC, lipid nanocapsules; EGFR, epidermal growth factor receptor; siRNA, small interfering ribonucleic acid; FACS, fluorescence-activated cell sorting; h, hours.
Evaluation of chitosan-transacylated LNCs/anti-EGFR siRNA effect on proliferation of U87MG cells
The results obtained showed that the LNCs transacylated with 1.2 mg/mL chitosan at a concentration of 100 μg/mL had no effect on proliferation of U87MG cells. The same result was obtained with the association of LNCs transacylated with 1.2 mg/mL chitosan/siRNA negative control. Regarding the treatment of the cells with anti-EGFR siRNA at the concentration of 40 nM carried by LNCs transacylated with 1.2 mg/mL chitosan, the result showed a 26.7%±3.81% (P<0.05) decrease in cell proliferation compared to untreated cells (Figure 6).
Figure 6.
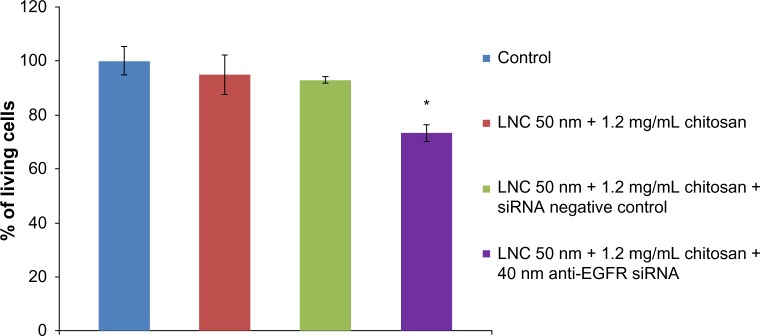
Evaluation of anti-EGFR siRNA/transacylated LNCs effect on proliferation of U87MG after 72 hours of incubation.
Notes: Data are shown as mean ± standard error of the mean (n=3). *Statistical difference compared to control was set at P<0.05 by Mann–Whitney U test.
Abbreviations: LNC, lipid nanocapsules; EGFR, epidermal growth factor receptor; siRNA, small interfering ribonucleic acid.
Evaluation of the increased sensitivity in U87MG cells to temozolomide after siRNA treatment
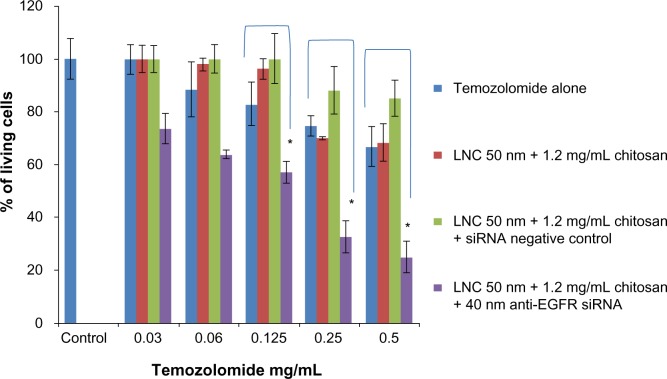
The MTT assay performed in this section was used to assess the increased mortality of U87MG cells pretreated with anti-EGFR siRNA followed by treatment with temozolomide compared to cells treated with temozolomide alone.
The U87MG cells treatment by different concentrations of temozolomide alone showed significant resistance and an absence of dose effect. The same results were obtained after treatment of cells with LNCs transacylated with 1.2 mg/mL of chitosan/siRNA negative control and vector alone at the concentration of 100 μg/mL.
After 3 days of incubation with anti-EGFR siRNA at a concentration of 40 nM carried by LNCs transacylated with 1.2 mg/mL chitosan, treated U87MG cells showed increased sensitivity to temozolomide at all concentrations tested compared to cells not treated by siRNA. In addition, a dose effect was observed. The maximum increase in sensitivity was 62.55%±3.55% (P<0.05) and was obtained with a concentration of 0.5 mg/mL of temozolomide (Figure 7).
Figure 7.
U87MG survival cells in function of temozolomide concentration after anti-EGFR siRNA treatment for 72 hours.
Notes: Data are shown as mean ± standard error of the mean (n=3). *Statistical difference compared to control was set at P<0.05 by Mann–Whitney U test.
Abbreviations: LNC, lipid nanocapsules; EGFR, epidermal growth factor receptor; siRNA, small interfering ribonucleic acid.
Temozolomide concentrations tested were chosen taking into account the sensitivity of U87MG cells and trying to get closer to the plasma concentrations described in the literature. The plasma concentration of temozolomide was determined for patients with high grades gliomas. The pharmacokinetics of temozolomide appeared linear, with the area under the curve (AUC) increasing proportionally to the dose after intravenous administration. For a dose of 250 mg/m2, the Cmax (maximum concentration) was 10 μg/mL. Measured in the cerebrospinal fluid, concentration was about 29% of the plasma concentration.30
Discussion
Glioblastoma cells are resistant to apoptosis and autophagy due to increased activation of signaling pathways related to cell death, these being controlled by proteins such as EGFR. Reducing the ability of tumor cells to proliferate and to restoring their capacity to die is one of the therapeutic strategies developed against cancer. To achieve this goal, targeting EGFR by RNA interference strategy has been sought to reduce EGFR expression and thus control its effects on tumor development. Indeed, in one study, anti-EGFR siRNA and a wild phosphatase and tensin homolog (PTEN) gene were introduced in U251 cells with a plasmid in order to assess the effect on the inhibition of cell growth. Suppression of cell proliferation, cell cycle arrest, a reduced cell invasion, and promotion of cell apoptosis have all been observed in vitro and in vivo.31
In our study, we investigated the effect of EGFR knockdown on the sensitivity of the U87MG cell line to temozolomide by a using a specific siRNA carried by chitosan-transacylated LNCs. Chitosan was used for the transacylation reaction due to its high transfection efficiency and low toxicity32 compared to other polymers such as polyethyleneimine (PEI); despite its potential for transfection of nucleic acids, PEI, particularly the branched form, has a high cell toxicity which limits its clinical use.33,34 Different chitosan nanoparticles are described in the literature.35–37 The high molecular weight and a high degree of deacetylation of the chitosan molecules are essential characteristics that allow for the obtaining of nanoparticles of small size with a large capacity to interact with siRNA.37 However, due to its low solubility in water at physiological pH and low buffering capacity, the chitosan nanoparticles can dissociate and lose their positive charge which causes the premature release of siRNA and reduces its cellular effect. In this study, the chitosan used had a low molecular weight, allowing it to obtain nanoparticles with small size which form complexes with siRNA and dissociate at the cellular level to allow the action of siRNA. The chitosans with high molecular weights have the drawback of forming a very stable complex with siRNA that reduces the release of siRNA and causes a smaller effect on the target mRNA.38
Transacylation reaction was carried out using small amounts of chitosan, which avoided the problems of solubility of the polymer. Increasing the size of the nanoparticles observed after this reaction confirms the grafting of chitosan to the surface of the LNCs. The zeta potential became positive and increased with the amount of chitosan used in the reaction until a plateau, probably due to the saturation of the surface of the LNCs. In addition, a decreasing yield of transacylation was observed and probably related to the same effect.
The determination of the quantity of siRNA grafted onto the surface of LNCs showed differences in the binding capacity of the siRNA between the two formulations of transacylated LNCs tested. This effect was caused by the difference of zeta potentials between the formulations. Indeed, small quantities of LNCs with the highest zeta potential fixed the maximum content of siRNA. This indicates that the LNCs transacylated with 1.2 mg/mL of chitosan were the most effective at fixing siRNA.
Regarding the hydrodynamic diameter of LNCs transacylated with 1.2 mg/mL of chitosan/anti-EGFR siRNA complexes, the results showed an increase compared to chitosan-transacylated LNCs, which indicates the fixation of siRNA on the surface of the transacylated LNCs. Given the small size of siRNA ~ 7 nM, it is conceivable that a multilayer of siRNA can be produced on the surface of LNCs. However, the grafting of siRNA to the transacylated LNCs is ensured by electrostatic attraction which excludes this hypothesis. The increase of 25 nM in the hydrodynamic diameter is related to siRNA and water molecules associated with siRNA that give a hydration layer detected by DLS.
The decrease in zeta potential of chitosan-transacylated LNCs/anti-EGFR siRNA complexes was the result of neutralization of positive charges of transacylated LNCs by the negatively charged siRNA. However, this charge neutralization was not complete with the amounts of siRNA tested, which allowed it to keep a positive zeta potential for better interaction with the plasma membrane.
Concerning the toxicity of LNCs transacylated with 1.2 mg/mL of chitosan on U87MG cells, cell mortality was observed with 150 μg/mL and 200 μg/mL of LNCs. This effect is due to the characteristics of cationic chitosan that allow binding to negatively-charged cellular components, thereby causing damage to the membrane and disrupting organelles and ultimately leading to cell death.39
Treatment of the cells with anti-EGFR siRNA carried by LNCs transacylated with 1.2 mg/mL of chitosan for 3 days slowed cell proliferation, which confirms the involvement of the EGFR in the tumor cell proliferation.
The present study showed that with this strategy, it was possible to increase the sensitivity of glioma cells to temozolomide using small quantities of anti-EGFR siRNA. Similar results have been found with anti-Galectin-1 siRNA and temozolomide.40 However, these results had been obtained with ProFection® Mammalian Transfection Systems (Promega) which is not suitable for human injection.
Other nanocarriers were used for siRNA-targeting EGFR, including the dendriworms developed by Agrawal et al,41 which come from the combination of PAMAM dendrimers (polyamidoamine) and magnetic nanoworms; dendriworms have demonstrated their efficiency in transfection of siRNA in glioblastoma cells. However, the production of this nanocarrier is achieved by a long process using many reagents and requires multiple purifications to obtain a good final quality. In comparison, our vector is produced by a simple process without solvents and is simply purified by dialysis. Moreover, LNCs have proved their very good toxicity profile in mice.27 It is also interesting to note that the maximum effect of dendriworms was obtained with 100 nM of anti-EGFR siRNA, whereas in our study, we obtained a reduction of 51.95% with 2.5 times less anti-EGFR siRNA.
In another study, Michiue et al42 associated a peptide transduction domain delivery (PTD) (TAT peptide) with a dsRNA binding domain (DRBD) used for its high affinity to siRNA, in order to obtain a PTD-DRBD vector. This team associated anti-EGFR and anti-Akt siRNAs to PTD-DRBD. Transfections were performed on U87MG cells that were injected into the brains of mice that were then sacrificed 48 hours after injection. Although the authors obtained good results, the proposed vector was made by genetic engineering of Escherichia coli, which is a cost in terms of production and purification. In our case, chitosan-transacylated nanocapsules are produced by a low cost process which is a considerable advantage for a possible large-scale production.
Targeting EGFR is a promising strategy to reduce the resistance of malignant gliomas to anticancer drugs. However, the amplification of EGFR affects 40%43 of patients, implying that targeting EGFR alone cannot solve the problem of resistance to temozolomide. Therefore, simultaneous targeting of other proteins involved in tumor resistance (taking into account the genetic profile of each patient) could improve the treatment of the glioblastoma.
Our vector has been designed for intracerebral injection, a strategy which offers two advantages. First, it limits the interaction with plasma proteins observed with the intravenous route. This is why a noncovalent link between siRNA and LNCs seems adequate for transfection efficiency. Second, this approach solves the issue of crossing the blood–brain barrier. In vivo, the formulation is better adapted to injection during tumor resection or can be injected in the tumor by stereotactic procedure.
Conclusion
Chitosan-transacylated nanocapsules have shown both their ability to bind siRNA by electrostatic attraction and their capacity to deliver it at the cellular level; in this way they have a biological effect and are able to increase the sensitivity of glioma cells to anticancer drugs such as temozolomide. The main advantages of these new carriers are their low toxicity and injectability. Moreover, other siRNAs can be fixed in the same way in similar experimental conditions due to the homogeneous structure of these biomolecules. This will allow us to consider the use of two messenger RNA targets, which will in turn increase the possibilities of enhancing the sensitivity of cancer cells to chemotherapy. The effectiveness of these siRNA carriers remains to be studied in animal models in combination with chemotherapy.
Acknowledgments
The authors are very grateful to the “Comité interrégional de la Ligue contre le Cancer, région Maine et Loire [InterRegional Committee, League against Cancer, Pays de la Loire region]” for providing financial support for this work. The authors also wish to thank Thomas Perrier for his help with the chemistry in this study.
Footnotes
Disclosure
Khaled Messaoudi is supported by the Centre Hospitalier Universitaire d’Angers. The other authors report no conflicts of interest in this work.
Author contributions
Substantial contributions to the conception or design of the work or the acquisition, analysis, or interpretation of data for the work: KM, KB, FL, PS, JPB. Drafting the work or revising it critically for important intellectual content: KM, KB, FL, PS, JPB. Final approval of the version to be published: KM, KB, FL, PS, JPB. Agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved: KM, KB, FL, PS, JPB.
References
- 1.Lefranc F, Sadeghi N, Camby I, Metens T, Dewitte O, Kiss R. Present and potential future issues in glioblastoma treatment. Expert Rev Anticancer Ther. 2006;6(5):719–732. doi: 10.1586/14737140.6.5.719. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, et al. European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Hegi ME, Mason WP, et al. European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups; National Cancer Institute of Canada Clinical Trials Group. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 4.Le Mercier M, Fortin S, Mathieu V, Kiss R, Lefranc F. Galectins and gliomas. Brain Pathol. 2010;20(1):17–27. doi: 10.1111/j.1750-3639.2009.00270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao XG, Yan M, Xue XY, et al. Overexpression of ZNF217 in glioblastoma contributes to the maintenance of glioma stem cells regulated by hypoxia-inducible factors. Lab Invest. 2011;91(7):1068–1078. doi: 10.1038/labinvest.2011.56. [DOI] [PubMed] [Google Scholar]
- 6.Zhu D, Hunter SB, Vertino PM, Van Meir EG. Overexpression of MBD2 in glioblastoma maintains epigenetic silencing and inhibits the antiangiogenic function of the tumor suppressor gene BAI1. Cancer Res. 2011;71(17):5859–5870. doi: 10.1158/0008-5472.CAN-11-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang PH, Xu AM, White FM. Oncogenic EGFR signaling networks in glioma. Sci Signal. 2009;2(87):re6. doi: 10.1126/scisignal.287re6. [DOI] [PubMed] [Google Scholar]
- 8.Guha A, Feldkamp MM, Lau N, Boss G, Pawson A. Proliferation of human malignant astrocytomas is dependent on Ras activation. Oncogene. 1997;15(23):2755–2765. doi: 10.1038/sj.onc.1201455. [DOI] [PubMed] [Google Scholar]
- 9.Narita Y, Nagane M, Mishima K, Huang HJ, Furnari FB, Cavenee WK. Mutant epidermal growth factor receptor signaling down-regulates p27 through activation of the phosphatidylinositol 3-kinase/Akt pathway in glioblastomas. Cancer Res. 2002;62(22):6764–6769. [PubMed] [Google Scholar]
- 10.Furuta S, Hidaka E, Ogata A, Yokota S, Kamata T. Ras is involved in the negative control of autophagy through the class I PI3-kinase. Oncogene. 2004;23(22):3898–3904. doi: 10.1038/sj.onc.1207539. [DOI] [PubMed] [Google Scholar]
- 11.Haas-Kogan DA, Prados MD, Tihan T, et al. Epidermal growth factor receptor, protein kinase B/Akt, and glioma response to erlotinib. J Natl Cancer Inst. 2005;97(12):880–887. doi: 10.1093/jnci/dji161. [DOI] [PubMed] [Google Scholar]
- 12.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 13.McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nat Rev Genet. 2002;3(10):737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- 14.Ngo H, Tschudi C, Gull K, Ullu E. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc Natl Acad Sci U S A. 1998;95(25):14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aagaard L, Rossi JJ. RNAi therapeutics: principles, prospects and challenges. Adv Drug Deliv Rev. 2007;59(2–3):75–86. doi: 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8(3):173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 17.Reischl D, Zimmer A. Drug delivery of siRNA therapeutics: potentials and limits of nanosystems. Nanomedicine. 2009;5(1):8–20. doi: 10.1016/j.nano.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Volkov AA, Kruglova NS, Meschaninova MI, et al. Selective protection of nuclease-sensitive sites in siRNA prolongs silencing effect. Oligonucleotides. 2009;19(2):191–202. doi: 10.1089/oli.2008.0162. [DOI] [PubMed] [Google Scholar]
- 19.Li CX, Parker A, Menocal E, Xiang S, Borodyansky L, Fruehauf JH. Delivery of RNA interference. Cell Cycle. 2006;5(18):2103–2109. doi: 10.4161/cc.5.18.3192. [DOI] [PubMed] [Google Scholar]
- 20.David S, Pitard B, Benoit JP, Passirani C. Non-viral nanosystems for systemic siRNA delivery. Pharmacol Res. 2010;62(2):100–114. doi: 10.1016/j.phrs.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 21.Hacein-Bey-Abina S, Garrigue A, Wang GP, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118(9):3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kakizawa Y, Furukawa S, Ishii A, Kataoka K. Organic-inorganic hybrid-nanocarrier of siRNA constructing through the self-assembly of calcium phosphate and PEG-based block aniomer. J Control Release. 2006;111(3):368–370. doi: 10.1016/j.jconrel.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Ambruosi A, Khalansky AS, Yamamoto H, Gelperina SE, Begley DJ, Kreuter J. Biodistribution of polysorbate 80-coated doxorubicin-loaded [14C]-poly(butyl cyanoacrylate) nanoparticles after intravenous administration to glioblastoma-bearing rats. Journal of Drug Targeting. 2006;14(2):97–105. doi: 10.1080/10611860600636135. [DOI] [PubMed] [Google Scholar]
- 24.Auffinger B, Morshed R, Tobias A, Cheng Y, Ahmed AU, Lesniak MS. Drug-loaded nanoparticle systems and adult stem cells: a potential marriage for the treatment of malignant glioma? Oncotarget. 2013;4(3):378–396. doi: 10.18632/oncotarget.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heurtault B, Saulnier P, Pech B, Proust JE, Benoit JP. A novel phase inversion-based process for the preparation of lipid nanocarriers. Pharm Res. 2002;19(6):875–880. doi: 10.1023/a:1016121319668. [DOI] [PubMed] [Google Scholar]
- 26.Hureaux J, Lagarce F, Gagnadoux F, et al. Lipid nanocapsules: ready-to-use nanovectors for the aerosol delivery of paclitaxel. Eur J Pharm Biopharm. 2009;73(2):239–246. doi: 10.1016/j.ejpb.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 27.Hureaux J, Lagarce F, Gagnadoux F, et al. Toxicological study and efficacy of blank and paclitaxel-loaded lipid nanocapsules after iv administration in mice. Pharm Res. 2010;27(3):421–430. doi: 10.1007/s11095-009-0024-y. [DOI] [PubMed] [Google Scholar]
- 28.Saranya N, Moorthi A, Saravanan S, Devi MP, Selvamurugan N. Chitosan and its derivatives for gene delivery. Int J Biol Macromol. 2011;48(2):234–238. doi: 10.1016/j.ijbiomac.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Perrier T, Fouchet F, Bastiat G, Saulnier P, Benoit JP. OPA quantification of amino groups at the surface of lipidic nanocapsules (LNCs) for ligand coupling improvement. Int J Pharm. 2011;419(1–2):266–270. doi: 10.1016/j.ijpharm.2011.07.028. [DOI] [PubMed] [Google Scholar]
- 30.Marzolini C, Decosterd LA, Shen F, et al. Pharmacokinetics of temozolomide in association with fotemustine in malignant melanoma and malignant glioma patients: comparison of oral, intravenous, and hepatic intra-arterial administration. Cancer Chemother Pharmacol. 1998;42(6):433–440. doi: 10.1007/s002800050842. [DOI] [PubMed] [Google Scholar]
- 31.Han L, Zhang AL, Xu P, et al. Combination gene therapy with PTEN and EGFR siRNA suppresses U251 malignant glioma cell growth in vitro and in vivo. Med Oncol. 2010;27(3):843–852. doi: 10.1007/s12032-009-9295-8. [DOI] [PubMed] [Google Scholar]
- 32.Rudzinski WE, Aminabhavi TM. Chitosan as a carrier for targeted delivery of small interfering RNA. Int J Pharm. 2010;399(1–2):1–11. doi: 10.1016/j.ijpharm.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 33.Fischer D, Bieber T, Li Y, Elsasser HP, Kissel T. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicity. Pharm Res. 1999;16(8):1273–1279. doi: 10.1023/a:1014861900478. [DOI] [PubMed] [Google Scholar]
- 34.Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J Control Release. 2006;114(1):100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 35.Katas H, Alpar HO. Development and characterisation of chitosan nanoparticles for siRNA delivery. J Control Release. 2006;115(2):216–225. doi: 10.1016/j.jconrel.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 36.Katas H, Chen S, Osamuyimen AA, Cevher E, Oya Alpar H. Effect of preparative variables on small interfering RNA loaded Poly(D,L-lactide-co-glycolide)-chitosan submicron particles prepared by emulsification diffusion method. J Microencapsul. 2008;25(8):541–548. doi: 10.1080/02652040802075567. [DOI] [PubMed] [Google Scholar]
- 37.Liu X, Howard KA, Dong M, et al. The influence of polymeric properties on chitosan/siRNA nanoparticle formulation and gene silencing. Biomaterials. 2007;28(6):1280–1288. doi: 10.1016/j.biomaterials.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 38.Techaarpornkul S, Wongkupasert S, Opanasopit P, Apirakaramwong A, Nunthanid J, Ruktanonchai U. Chitosan-mediated siRNA delivery in vitro: effect of polymer molecular weight, concentration and salt forms. AAPS PharmSciTech. 2010;11(1):64–72. doi: 10.1208/s12249-009-9355-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi LF, Xu ZR, Li Y, Jiang X, Han XY. In vitro effects of chitosan nanoparticles on proliferation of human gastric carcinoma cell line MGC803 cells. World J Gastroenterol. 2005;11(33):5136–5141. doi: 10.3748/wjg.v11.i33.5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Mercier M, Mathieu V, Haibe-Kains B, et al. Knocking down galectin 1 in human hs683 glioblastoma cells impairs both angiogenesis and endoplasmic reticulum stress responses. J Neuropathol Exp Neurol. 2008;67(5):456–469. doi: 10.1097/NEN.0b013e318170f892. [DOI] [PubMed] [Google Scholar]
- 41.Agrawal A, Min DH, Singh N, et al. Functional delivery of siRNA in mice using dendriworms. ACS Nano. 2009;3(9):2495–2504. doi: 10.1021/nn900201e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michiue H, Eguchi A, Scadeng M, Dowdy SF. Induction of in vivo synthetic lethal RNAi responses to treat glioblastoma. Cancer Biol Ther. 2009;8(23):2306–2313. doi: 10.4161/cbt.8.23.10271. [DOI] [PubMed] [Google Scholar]
- 43.Prados MD, Chang SM, Butowski N, et al. Phase II study of erlotinib plus temozolomide during and after radiation therapy in patients with newly diagnosed glioblastoma multiforme or gliosarcoma. J Clin Oncol. 2009;27(4):579–584. doi: 10.1200/JCO.2008.18.9639. [DOI] [PMC free article] [PubMed] [Google Scholar]