Abstract
Zinc is an essential metal for cellular homeostasis and function in both eukaryotes and prokaryotes. To acquire this essential nutrient, bacteria employ transporters characterized by different affinity for the metal. Several studies have investigated the role of the high affinity transporter ZnuABC in the bacterial response to zinc shortage, showing that this transporter has a key role in adapting bacteria to zinc starvation. In contrast, the role of the low affinity zinc importer ZupT has been the object of limited investigations. Here we show that a Salmonella strain lacking ZupT is impaired in its ability to grow in metal devoid environments and that a znuABC zupT strain exhibits a severe growth defect in zinc devoid media, is hypersensitive to oxidative stress and contains reduced level of intracellular free zinc. Moreover, we show that ZupT plays a role also in the ability of S. Typhimurim to colonize the host tissues. During systemic infections, the single zupT mutant strain was attenuated only in Nramp1+/+ mice, but competition experiments between znuABC and znuABC zupT mutants revealed that ZupT contributes to metal uptake in vivo independently from the presence a functional Nramp1 transporter. Altogether, the here reported results show that ZupT plays an important role in Salmonella zinc homeostasis, being involved in metal import both in vitro and in infected animals.
Introduction
Metals play essential roles in all organisms as structural or catalytic cofactors in a wide number of proteins. This concept has been highlighted by a recent survey of proteins with known 3D structure, which has revealed that almost half of all enzymes bind metals, with approximately 40% containing metals at their catalytic centers.1 Among metals, zinc plays particularly important roles in cellular homeostasis and function. It has been estimated that zinc can selectively associate with about 5% of all bacterial proteins, including enzymes of all six functional classes.2,3 Zinc is often the cofactor of choice for enzymatic reactions because of its ability to stabilize negative charges and to activate substrates due to its strong Lewis acid properties.3 The binding of zinc to proteins is facilitated by its highly versatile coordination chemistry, as this metal can be ligated by nitrogen, oxygen and sulphur atoms and can assume different coordination numbers. This property, however, is potentially detrimental to cells because it favors the unspecific binding of zinc to polypeptides and free thiols. Therefore, the levels of intracellular zinc are tightly regulated and the pool of unbound metal is kept to a minimum.1,4
Because zinc is critical for many cellular functions, bacteria have evolved several mechanisms to respond to zinc deficiency, and acquire this metal in the host. In Gram-negative bacteria, the transcriptional regulator Zur controls the expression of a small number of genes required to adapt the cell to conditions of severe zinc paucity.5 Under zinc-replete conditions, the zinc-containing form of Zur tightly binds to the promoter region of said genes, preventing their expression.6 Conversely, when the intracellular zinc concentration falls below a critical threshold, the zinc-devoid form of Zur no longer represses transcription.
The number of Zur–regulated genes varies between different bacteria, but in all species they include the genes encoding for the different subunits of a high affinity zinc importer (ZnuABC in Gram-negative bacteria), as well as the genes encoding for one or more paralogs of zinc-containing ribosomal proteins.7,8,9,10 ZnuABC significantly enhances the bacterial ability to recruit zinc under different environmental conditions characterized by low availability of this metal. At the same time, the substitution of zinc-containing ribosomal proteins with metal-independent homologues significantly reduces the zinc requirements of bacterial cells. In line with this change in ribosomal structure organization, bacteria growing in zinc-depleted media contain much less zinc than bacteria growing in zinc-replete conditions.11
Several studies have investigated the consequences of the inactivation of znuABC in different bacteria. Besides showing that this transporter is necessary to ensure bacterial growth in vitro in zinc-limiting environments, ZnuABC was found to play a critical role in bacterial pathogenesis, thus implying that zinc availability is limited within infected hosts.12,13,14,15,16,17,18,19,20,21,22
In previous studies, we investigated the role of ZnuABC during infection of mice with the Gram-negative pathogen Salmonella enterica serovar Typhimurium (S. Typhimurium). We found that S. Typhimurium strains lacking the znuA gene are significantly attenuated in mice in both systemic and gastrointestinal infections.17,23,24 Deletion of the znuA gene, which encodes for the periplasmic component ZnuA, is sufficient to disrupt the function of the ZnuABC transporter, as no further attenuation is observed in mutant strains where the entire znuABC operon is deleted.23,25 By producing the ZnuABC transporter, S. Typhimurium evades the antimicrobial activity of calprotectin, a zinc-sequestering protein released by neutrophils during infection in the intestinal lumen.26 In this host environment, a strain lacking a functional ZnuABC transporter cannot compete with the microbiota and is killed by the inflammatory response.26 As a matter of fact, the absence of znuABC also causes significant proteomic and ionomic alterations11 and changes in the expression of putative virulence factors.27
While ZnuABC is exclusively produced under zinc starvation, in environments where zinc in more abundant metal uptake is ensured by other transporters with lower affinity for zinc and broader metal specificity.28 The most important permease allowing entrance of zinc is thought to be ZupT, a membrane protein belonging to the ZIP (ZRT-, IRT-like Protein) protein family.29 Studies on Escherichia coli ZupT have established that this transporter mediates the import of different metals, although a preference for zinc over manganese, copper and iron was shown.30,31,32 In E. coli, the activity of ZupT is constitutive and it is not regulated in response to changes in metal availability.31 Even so, ZupT activity is inhibited by ionophores, suggesting that the proton motive force drives ZupT-mediated metal-uptake.32 This hypothesis was also supported by the observation that the phage shock protein (Psp) system, which is involved in the maintenance of the proton motive force, facilitates metal uptake though different importers, including ZupT.33
The respective roles of ZupT and ZnuABC were also analyzed in an E. coli uropathogenic (UPEC) strain.19 This study confirmed that ZupT mediates zinc uptake in vitro and showed that ZnuABC contributes to UPEC colonization of kidneys and bladders in CBA/J mice. Nonetheless, the znuABC zupT and the znuABC mutants showed a very similar ability to colonize host tissues, thereby suggesting that ZupT only modestly contributes to zinc uptake during UPEC infections. In contrast, the ZupT transporter is required for S. Typhimurium virulence in a typhoid model of infection in mice expressing the natural resistance-associated macrophage protein 1 (Nramp1) metal transporters.33 However, it is not known whether the contribution of ZupT to Salmonella pathogenicity is related to the import of zinc and/or other metals. Here we set out to further investigate the role of ZupT in zinc homeostasis by analyzing its regulation during in vitro growth, as well as its contribution to pathogenesis in typhoid and gastrointestinal models of S. Typhimurium infection.
Experimental
Salmonella strains and growth conditions
The S. Typhimurium strains used in this work are listed in Table 1. Cultures were routinely grown aerobically in liquid Luria-Bertani broth (LB) or on LB agar plates at 37°C. Agarose plates were prepared using molecular biology grade agarose (Vivantis) at 15 g/l in LB medium. Growth under zinc limiting conditions was achieved using either the Vogel-Bonner minimal medium E (VB-MM) (anhydrous MgSO4 [0.04 g/liter], citric acid [2 g/liter], anhydrous K2HPO4 [10 g/liter], NaH4PO4 [3.5 g/liter], glucose [2 g/liter]), or the M9 minimal medium (M9-MM) (Na2HPO4 [7.52 g/liter], KH2PO4 [3 g/liter], NH4Cl [1 g/liter], NaCl [5 g/liter], MgSO4·7H2O [1.23 g/liter], CaCl2·2H2O [0.007 g/liter], glucose [0.2%]). To avoid zinc contaminations from glassware, minimal media were prepared in disposable plastic containers and sterilized by filtration in 500 ml Vacuum Filter/Storage Bottle Systems, 0.22μm (Corning). The quality of each minimal medium batch was verified by monitoring the accumulation of ZnuA in strain SA140 (which is abolished by zinc at concentrations below 1 μM).17 Antibiotics were used at the following concentration: ampicillin 100 mg/l, kanamycin 50 mg/l and chloramphenicol 30 mg/l.
Table 1.
Strains used in this work
| Strain | Relevant genotype or description | Source or reference |
|---|---|---|
| DH5α | E. coli F- ( 80d lacZ M15) (lacZYA-argF)U169 hsdR17(r - m +) recA1 endA1 relA1 deoR | lab collection |
| MA6926 | Wild type | lab collection |
| MA6926 (pKD46) | Wild type harboring plasmid pKD46 | lab collection |
| SA212 | ilvI::Tn10dTac-cat::3xFLAG-SCAR | lab collection |
| SA140 | znuA::3xFLAG-kan ilvI::Tn10dTac-cat::3xFLAG-kan | 17 |
| SA186 | znuABC::scar | 25 |
| SA321 | zupT::kan | this work |
| SA327 | znuABC::scar zupT::kan | this work |
| SA336 | sitABCD::kan mntH::scar | lab collection |
| SA337 | feoB::scar fepA/entF::kan | lab collection |
| SA340 | sitABCD::scar mntH::scar zupt::kan | lab collection |
| IR715 | ATCC 14028, NalR derivative | 49 |
| JZL3 | znuA::cm | 26 |
| AJP3 | zupT::kan | this work |
| AJP4 | znuA::cm zupT::kan | this work |
| PP127 | zur::kan | 23 |
| PP134 | zinT::3X Flag-kan | 23 |
| PP137 | zinT::3X Flag-kan znuA::cam | 23 |
| MC113 | zinT::3X Flag-kan znuABC::scar zupT::scar | this work |
| PP150 | Wild type harboring plasmid pPzupT-lacZ | this work |
| PP152 | znuABC::scar harboring plasmid pPzupT-lacZ | this work |
| PP153 | zur::kan harboring plasmid pPzupT-lacZ | this work |
Mutants construction
S. Typhimurium ATCC14028 zupT mutants (SA321 and AJP3, obtained respectively in A.B. and M.R. laboratories) were achieved following the one-step inactivation protocol,34 using plasmid pKD4 as the kanamycin resistance cassette template. The insertions were confirmed by PCR with oligonucleotides annealing upstream the mutated allele and into the inserted antibiotic resistance cassette. The mutated alleles were then transduced by generalized transduction with phage P22 HT 105/1 int-20 35 into wild type and znuABC::scar (SA186) or znuA::cam (JZL3) strains to obtain respectively the zupT (SA321) and the znuABC zupT (SA327) or znuA zupT (AJP4) mutant strains.
Strain MC113 was obtained transducing the zinT::3X Flag-kan allele23 in the znuABC::scar zupT::scar mutant, previously obtained electroporating plasmid pCP20, harboring the FLP recombinase function,34 in SA327 strain. The mutant was selected for kanamycin resistance.
Growth analyses
Overnight cultures grown in LB were washed with PBS and normalized to an optical density at 600 nm of 1.0. Bacterial suspensions were serially diluted 1:10 and 5 μl of each dilution were plated onto appropriate LB agarose plates. Growth was imaged after an incubation of 24 h at 37°C.
To analyse bacterial growth in liquid media, strains were grown overnight in VB-MM at 37°C and then diluted 1:500 in the same medium supplemented or not with the appropriate concentration of metals. Aliquots of 300 μl of these dilutions were inoculated in 96-well plates (Becton-Dickinson) and incubated at 37°C with shaking. Bacterial growth was monitored at 595 nm every hour for 15 hours using a microtiter-plate reader (Sunrise Tm, Tecan). Assays were performed in triplicate and each strain was tested in three independent experiments.
Western blot analyses
ZinT accumulation was analyzed in Salmonella strains expressing an epitope-tagged protein, containing a 3X FLAG epitope at its C-terminus, as previously described23. An amount of bacteria corresponding to 0.25 optical density at 600 nm was loaded in each lane. Strain SA212, constitutively expressing a 3xFLAG epitope-tagged chloramphenicol acetyl transferase protein (CAT), was used as a control of protein loading. All the strains used for these experiments exhibited an identical correspondence between optical density at 600 nm and number of colony forming units in VB-MM (with or without zinc supplementation) as well as an identical content of soluble proteins (as evaluated by quantifying proteins with the method of Lowry), thus suggesting that the mutations have no evident effects on cell size and intracellular protein content. Proteins tagged with the 3XFLAG epitope were revealed by incubation of nitrocellulose filters with a mouse anti-FLAG antibody (anti-FLAG M2, Sigma) and an anti-mouse horseradish peroxidase-conjugated antibody (Bio-Rad), followed by the enhanced chemiluminescence reaction (ECL, Amersham).
Mouse infections and competition assays
Experiments with mice either performed at the Italian Istituto Superiore di Sanità (ISS) or at the University of California, were carried out according to the respective national regulations. All experiments were previously approved by the ISS Ethical Committee and the Institutional Animal Care and Use Committee at the University of California, Irvine and carried out under the supervision of certified veterinarians. We used C57BL/6J (Taconic farms) and BALB/c and DBA/2 (Charles River, Calco, Italy) female mice of 8–12 weeks of age. Mice were fed a commercial diet and water was provided ad libitum. All mice were acclimatized for a minimum of 1 week prior to experimentation.
Intraperitoneal mouse infections and competition assays were performed as previously described.17 Bacteria recovered from spleens were plated for single colonies, and then 200 colonies were picked on selective plates. The competitive index (CI) was calculated by the formula CI = output (strain A/strain B)/input (strain A/strain B). Statistical differences between outputs and inputs were determined by Student t test. Oral infections were carried out in streptomycin-pretreated animals, as previously described.26
Cloning the zupT promoter region and β-galactosidase activity assay
A 218 bp fragment located upstream the zupT coding sequence, including the GTG start codon, was amplified by PCR from S. Typhimurium chromosomal DNA (extracted with the ZR fungal/bacterial DNA Kit™, Zymo Research) using oligonucleotides PzupTpMC For (CAGAATTCATAATCGTTATCGTCCAGCA) and PzupTpMC Rev (GCGGATCCATCCATTACTCCTTATCAAT). The purified fragment (DNA Clean and Concentration ™, Zymo Research) was then digested with the restriction enzymes EcoRI and BamHI (New England Biolabs) and cloned into plasmid pMC1403,36 obtaining plasmid pMCPzupT-lacZ. The nucleotide sequence of the DNA insert was verified by sequencing (Genechron). Subsequently, the plasmid was extracted from E. coli (HiYeld RBC ™ Plasmid Mini Kit) and electroporated into S. Typhimurium strains MA6926 (wild type), SA186 (znuABC::scar) and PP127 (zur::kan) obtaining respectively strains PP150, PP152 and PP153 (see Table 1). β-galactosidase activity of the resulting strains was measured as previously described.37
H2O2 sensitivity assay
S. Typhimurium strains were grown for 6 hours in LB medium and then diluted 1:1000 in M9-MM with or without the addition of zinc. After growing the strains over-night (at 37°C with aeration) cultures were diluted to 106 bacteria/ml in sterile PBS (time 0) and H2O2 0.5 mM was added. After 1 hour incubation at 37°C with aeration, 1000 units of catalase (Boehringer) was added to each sample. At time 0 and 1 hour bacteria were plated on agar plates for counting, after suitable dilutions. Percentage of viable bacteria was calculated by assuming bacterial viability at time 0 as 100% survival. The assay was performed in duplicate and the reported results are the average of three independent experiments.
Results
zupT expression is increased under zinc limiting conditions
Whereas it has been reported that in plants a ZIP transporter may be induced in roots in response to zinc or iron deficiency,38 zupT is constitutively expressed in E. coli.31 To determine whether zinc deficiency influences the expression of zupT in S. Typhimurium, we analyzed zupT expression in wild type S. Typhimurium (PP150) and in a znuABC mutant strain (PP152), either in zinc-limiting conditions or in zinc-replete media, using a transcriptional fusion between the zupT promoter and the lacZ reporter gene. As shown in Fig. 1, basal levels of zupT expression can be measured in both the wild type (black bars) and znuABC (grey bars) strains cultured in rich medium (LB). Transcription from the zupT promoter was not affected by the addition of 1 mM zinc per se. However, zupT expression increased upon treatment with the metal chelator EDTA, and it was restored to basal levels by the addition of zinc to the medium. Notably, EDTA-mediated induction of zupT was more pronounced in the znuABC background, supporting the hypothesis that zupT expression may be modulated by zinc availability. Moreover, a slight decrease in zupT transcriptional activity was observed in bacteria grown in a defined medium (VB-MM) supplemented with zinc (Fig. 1), both in the wild type and in the znuABC background.
Fig. 1.
Transcriptional analysis of S. Typhimurium zupT promoter in bacteria grown overnight in VB-MM or in LB, in presence or absence of 0.01mM ZnSO4 (VB-MM) or 1mM (LB) ZnSO4 or 1 mM EDTA or both. Black and grey bars represent the wild type strain (PP150) and znuABC strain (PP152) respectively. Each experiment was repeated at least twice and samples were always assayed in triplicate. P values: * p < 0,05; ** p < 0,01
As Zur is the main transcriptional regulator governing bacterial responses to zinc shortage, the observation that zupT expression is partly modulated by zinc prompted us to investigate whether Zur regulates the expression of zupT. A nucleotide sequence showing 62% homology to the Salmonella Zur-box upstream of the znuABC and zinT genes23 was also identified within the zupT promoter (about 100 bp upstream the start codon) (Fig. S1, ESI). In contrast, consistent with the hypothesis that zupT is constitutively expressed in E. coli, this putative consensus sequence was found to be absent in the E. coli zupT promoter.31
To test whether S. Typhimurium zupT is regulated by Zur, we electroporated plasmid pMCPzupT-lacZ in a strain lacking zur (PP127), obtaining the strain PP153 (pMCPzupT-lacZ zur). β-galactosidase assays were carried out on strains PP150, PP152 and PP153 grown under zinc limiting condition (VB-MM or LB supplemented with EDTA). As β-galactosidase activity was similar in PP150 and PP153 (data not shown), these results suggested that zupT expression is not regulated by Zur.
Contribution of ZupT to zinc uptake during growth of S. Typhimurium
To characterize the role of ZupT in the process of zinc acquisition, the growth of strains lacking zupT (SA321), znuABC (SA186) or both transporters (SA327) was compared to that of the wild type strain in zinc limiting conditions, both in semi-solid and in liquid media.
We have previously shown that the growth of S. Typhimurium strains lacking a functional ZnuABC transporter is inhibited in agar plates containing EDTA or TPEN.17,23 A potential problem with these experiments is that chelating agents have a broad metal specificity, thus some of the observed phenotypes could be only indirectly related to zinc uptake. We have more recently observed that the growth of strains lacking genes involved in zinc uptake is decreased in LB plates containing 1.5% pure agarose in place of agar, without the need of adding chelating agents. When we prepared LB plates with 1.5% pure agarose, we observed that this polysaccharide has an intrinsic ability to sequester zinc, but not other biologically relevant metals such as iron and manganese. Specifically, the growth of S. Typhimurium strains lacking znuABC was slightly inhibited in these plates (Figure 2). Although the zupT and the wild type strains displayed a comparable growth on agarose plates, the growth of the znuABC zupT double mutant was nearly completely abolished, suggesting that these two transporters have synergistic roles. Noticeably, the growth of the znuABC zupT strain was rescued to wild type levels by the addition of zinc, but not of manganese or iron, to LB-agarose plates. In contrast, the growth of a mntH sitABCD mutant strain lacking the two major manganese transporters and of a fepA/entF feoB mutant, deleted of the most important inorganic iron uptake systems, was not impaired in agarose plates, although these strains show clearly impaired growth in VB-MM (Fig. S2, ESI).
Fig. 2.
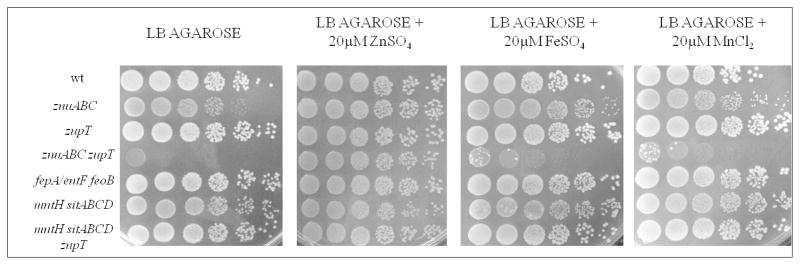
Growth of S. Typhimurium on LB-agarose plates. Plates were prepared with standard LB or with LB supplemtend with zinc, iron or manganese.
The role of ZupT in enhancing S. Typhimurium growth in zinc limiting conditions was also analyzed in liquid VB-MM (Fig. 3). Deletion of zupT had a modest, but clearly discernible, effect on S. Typhimurium growth in this medium, whereas the absence of znuABC resulted in a much more severe growth defect, as previously shown17,23. Remarkably, the znuABC zupT mutant was hardly able to grow in this medium. When VB-MM was supplemented with 3μM zinc, a metal concentration largely adequate to repress znuA expression,17, 11 the growth of all mutant strains was restored to levels comparable to those of the wild type strain (Fig. 3, panel B). Nonetheless, we observed slight but significant differences in the lag and exponential phases of growth among wild type and znuABC mutant strains. It is likely that such growth retardation of mutant strains is due to an alteration in the rate of zinc entry within cells due to the absence of zinc transporters. Manganese or iron were not able to rescue the growth of the znuABC zupT mutant strain in VB-MM (Fig. S3, ESI).
Fig. 3.
Growth curves of S. Typhimurium wild type (■), znuABC (▲), zupT (▼), znuABCzupT (◆). Strains were grown in VB-MM alone (panel A) and VB-MM supplemented with 3 μM zinc (panel B). OD595, optical density at 595 nm.
Lack of ZupT determines changes in intracellular zinc concentration
Previous studies have shown that zinT and znuABC are coregulated by Zur and that ZinT accumulation is strongly induced by EDTA and repressed by zinc.23 The addition of 0.5 μM ZnSO4 to the culture medium causes the complete abrogation of ZinT accumulation in a wild type Salmonella strain, but this expression profile is changed in a strain lacking the znuA gene due to a reduced intracellular zinc influx. In line with the finding that znuA and znuABC mutant strains have similar phenotypes,23 we demonstrated the same zinT expression pattern in strains lacking either znuA alone or the whole znuABC operon. In this work, we set out to compare ZinT accumulation in the znuA mutant and in the znuABC zupT mutant strains, as a means to indirectly infer whether the lack of zupT causes a decrease in the intracellular zinc content. Consistent with these early studies, ZinT accumulation was abolished when the znuA mutant was grown in a defined medium (VB-MM) containing approximately 40 μM of zinc sulfate (Fig. 4). Conversely, at this zinc concentration zinT was clearly expressed in the znuABC zupT strain and low protein levels were detected even in bacteria grown in presence of 60 μM zinc (Fig. 4). As zinT expression is under the direct control of Zur, the main transcriptional regulator activated in response to zinc deprivation, the observed differences in the ZinT accumulation pattern in the two mutant strains strongly support the hypothesis that the absence of zupT further reduces the ability of Salmonella enterica to import zinc from the environment.
Fig. 4.
ZinT::3xFLAG accumulation in the znuA deleted strain (PP137, upper panel) and in znuABC zupT deleted strain (MC113, intermediate panel) grown in VB-MM alone or supplemented with ZnSO4 as indicated. The lower panel shows the accumulation of CAT-3xFLAG in the control strain SA212
ZupT enhances S. Typhimurium resistance to H2O2
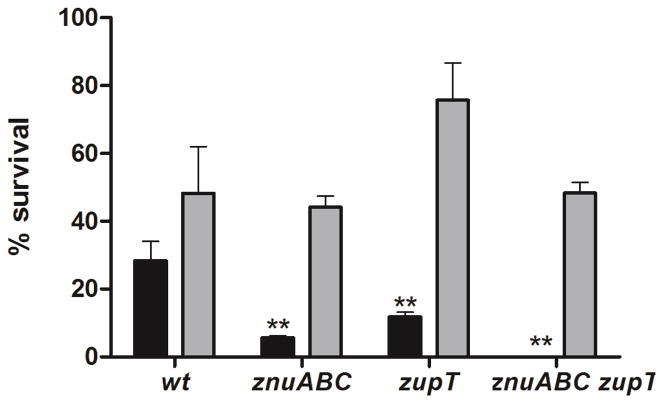
Previous studies carried out in uropathogenic E. coli19, Corynebacterium diphtheriae,39 Bacillus subtilis,40 and Lactococcus lactis 41 have suggested that zinc uptake mediates bacterial resistance to oxidative stress and that inactivation of zinc transporters enhances the susceptibility of these microorganisms to exogenous H2O2-mediated killing. These observations may be at least partially explained by the requirement of zinc as a cofactor in proteins involved in the response to reactive oxygen species, such as PerR42 and SodCI/SodCII43 and by the role of this metal in protecting protein thiols from oxidation.41,40 We thus set out to determine whether ZupT enhances S. Typhimurium resistance to H2O2-mediated killing. To this end, we monitored the survival of S. Typhimurium wild type or mutant strains grown overnight in M9-MM with or without zinc supplementation, after a 1 hour incubation with 0.5 mM H2O2 (Fig. 5). In comparison to S. Typhimurium wild type, all mutant strains showed increased susceptibility to H2O2, although the zupT mutant strain was less susceptible to H2O2 the znuABC mutant. Moreover, the absence of both znuABC and zupT rendered S. Typhimurium hypersensitive to hydrogen peroxide. The enhanced sensitivity of mutant strains to H2O2 is dependent on their reduced ability to import zinc, as the growth of the same strains in a defined medium supplemented with zinc re-established resistance to oxidative damage comparable to that of the wild type strain. These results indicate that ZupT also cooperates with ZnuABC in conferring resistance to oxidative stress.
Fig. 5.
Zinc import and hydrogen peroxide resistance. S. Typhimurium wild-type, SA186 (znuABC), SA321 (zupT) and SA327 (znuABC zupT) were grown in M9-MM with (gray bars) or without (black bars) the addition of 0.01mM ZnSO4 and exposed to H2O2 for one hour. Samples were assayed in triplicate and the graph shows the average of survival for each strain (percent respect to H2O2 unexposed bacteria). P values: ** p < 0,01
ZupT contributes to Salmonella virulence in mice
To assess the contribution of the low affinity zinc transporter ZupT to Salmonella virulence, we carried out in vivo competition assays between S. Typhimurium wild type and mutant strains in different mouse models of infection. First, we tested whether ZupT contributes to S. Typhimurium systemic infection by using the typhoid mouse model. Intraperitoneal infections were performed both in Salmonella-resistant DBA-2 mice (Nramp1+/+) and in the highly susceptible BALB/c mice (Nramp1−/−). As recently observed,33 when infections were carried out in mice expressing a functional Nramp1 protein, the zupT mutant was significantly outcompeted by the wild type strain in the spleens of infected mice (Fig. 6A). Moreover, we found that a strain lacking both znuABC and zupT was significantly disadvantaged with respect to the single mutant znuABC.
Fig. 6.

Competition assay in intraperitoneally infected mice. Bacteria were recovered from the spleens of Nramp+/+ (DBA-2, panel A) and Nramp−/− (Balb/C, panelB) mice infected with mixed inocula (wild type vs zupT and znuABC vs. znuABCzupT, see legend). P values: * p < 0,05
In contrast, both wild type and the zupT mutant strains colonized the spleens of intraperitoneally-infected BALB/c mice to similar levels (Fig. 6B). Nonetheless, in mixed infections the znuABC strain outcompeted the znuABC zupT double mutant, revealing that ZupT contributes to metal uptake during S. Typhimurium infection also in Nramp1−/− mice.
To further evaluate the role of ZupT, competition assays were carried out also in the streptomycin-pretreated mouse colitis model of S. Typhimurium infection. C57BL/6 mice pretreated with streptomycin were orally infected with different mixtures of strains and the ability of these bacteria to colonize the cecum and the spleen was evaluated at 72 and 96 hours post infection. In line with previous studies,26 the S. Typhimurium mutant strains znuA and znuA zupT were poorly able to colonize the gut of infected mice. In contrast, the wild type and the zupT strains were equally able to colonize the cecum at 96 h, whereas the znuA zupT strain showed only a small reduction in the ability to colonize this intestinal tract with respect to the znuA mutant (Fig. 7A). Moreover, we found that inactivation of zupT did not alter the ability of S. Typhimurium to colonize the Peyer’s patches (data not shown), but was associated to a small but significant reduction in the number of bacteria recovered from the spleen and the mesenteric lymph nodes of orally infected mice (Fig. 7B). Taken together, these data further underline that ZnuABC plays a crucial role in the recruitment of Zn(II) in the infected host, but also suggest that ZupT contributes to zinc acquisition during Salmonella infection.
Fig. 7.
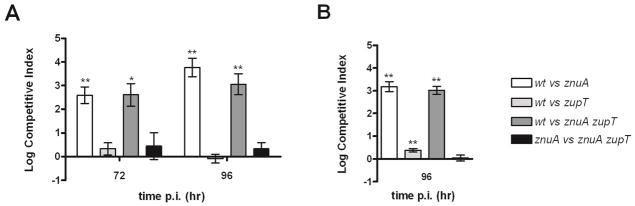
Competition assay in orally infected mice. Competitive index in cecal contents (panel A) and in the spleens (panel B) of C57BL6 mice infected with mixed inocula (wild type vs. znuA; wild type vs. zupT; wild type vs. znuA zupT; znuA vs. znuA zupT; see legend). P values: * p < 0,05; ** p < 0,01
Discussion
Bacterial pathogens have evolved several mechanisms to acquire essential metals even when they are limited by the host or in the environment. In this work, we investigated the contribution of the permease ZupT to zinc uptake in S. Typhimurium, showing that this transporter contributes to metal import both in vitro and during infection of mice. We found that strains lacking the zupT gene have a growth defect in a defined minimal medium poor of zinc (Fig. 3). Even though the observed growth disadvantage with respect to the wild type strains is rather small, it was consistently observed in many different experimental settings and was always reverted by the addition of zinc to the culture medium. These observations suggest that, under conditions of severe zinc limitation, the reduced zinc acquisition deriving by the absence of ZupT can not be fully compensated by the strong induction of znuABC.17 The requirement for two distinct zinc importers to ensure maximal efficiency in zinc recruitment is further underlined by the comparison of the growth rate of the znuABC and the znuABC zupT mutant strains in VB-MM or in agarose-LB plates. In these conditions, the growth of a strain lacking both ZupT and ZnuABC is barely detectable, unless zinc is added to the culture medium. Moreover, addition of other metals like manganese (Fig. 2 and Fig. S3 ESI) or iron (Fig. 2), which are known to be substrate of ZupT, do not rescue the growth defect of this mutant. Therefore, although bacteria possess several low affinity zinc importers,28 the severe growth defect of the znuABC zupT mutant strain in the absence of abundant environmental zinc suggests that Salmonella does not encode for other efficient zinc importers besides ZnuABC and ZupT.
The growth defect of a mutant strain lacking both ZnuABC and ZupT was particularly evident on agarose-LB plates. The ability of polysaccharides (i.e. pectin and other oligosaccharides) to chelate transition metals from aqueous solutions has been known for a long time.44,45 For example, it is well known that algae produce cell wall exopolysaccharides that protects them from heavy metal toxicity.46 As a matter of fact, polysaccharides are investigated as an inexpensive and effective tool for the removal of toxic metals from contaminated materials.47 Despite the chemical bases of zinc sequestration by agarose require further investigations, we propose that the use of agarose-LB plates represents a potential alternative to chelating agents for the analysis of the mechanisms of response to zinc limitation also in other bacterial strains.
Two other observations highlight the relevance of ZupT in zinc homeostasis. Unlike the high affinity zinc uptake transporter ZnuABC, which is produced exclusively under conditions of severe zinc limitation,17 ZupT is only moderately induced by metal shortage and is constitutively expressed in media containing abundant levels of metals. However, using ZinT as a reporter of available zinc inside the cell, we have also shown that in the absence of ZupT, the Zur-mediated response to zinc shortage is repressed at higher concentrations of environmental zinc. Taken together, the observations reported in this study indicate that ZupT has a central role in zinc homeostasis in Salmonella, either by ensuring zinc uptake under conditions of moderate zinc requirements or by participating in metal transport upon severe zinc starvation, a condition characterized by the induction of the Zur-regulated operon.
Our results also indicate that ZupT plays a role in promoting S. Typhimurim colonization of host tissues. In line with a recent study,33 we have found that a zupT mutant strain of S. Typhimurim is attenuated in Nramp1+/+, but not in Nramp1−/− mice. However, it is difficult to directly correlate these observations to the ability of Salmonella to recruit zinc in vivo. Nramp1 is a macrophage metal transporter likely showing significant higher affinity for iron and manganese than for zinc.48 As ZupT may also transport iron and manganese in addition to zinc, it is possible that the reduced ability of the zupT mutant to colonize the spleen of Nramp1+/+ mice is related to its reduced ability to acquire these metals in the Salmonella-containing vacuole. Nonetheless, our findings that the znuABC zupT double mutant is less able to colonize the spleen of intraperitonelly infected mice than the znuABC mutant strain suggest that ZupT is active during infections and likely contributes to zinc uptake. The involvement of ZupT in the ability of Salmonella to colonize the host was further confirmed by oral infections in streptomycin pre-treated mice. In fact, although ZnuABC is much more important than ZupT for gut colonization (Fig. 7A), we observed that disruption of zupT significantly affects the ability of Salmonella to infect the spleen (Fig. 7B) or the mesenteric lymph nodes (data not shown), thus suggesting that the ability to successfully colonize these organs requires redundant mechanisms to resist to metal starvation. In addition, it is worth noting that mutant strains lacking zupT show reduced resistance to oxidative stress and that a znuABC zupT is hypersensitive to hydrogen peroxide (Fig. 5). It is likely that this feature contributes to the in vivo attenuation of Salmonella mutant strains defective in zinc homeostasis.
Most of the studies on the importance of zinc in host-pathogen interaction have been focused on the ZnuABC transporter. Undoubtedly the ZnuABC transporter plays a key role in the ability of bacteria to rapidly adapt to conditions of severe zinc paucity, such as those encountered in the infected host. Inactivation of ZnuABC determines phenotypic effects which are much more marked than those observed in zupT mutant strains and this likely explain why relatively few studies have focused on the role of ZupT in metal homeostasis. However, this study suggests that the contribution of the ZupT permease to the acquisition of zinc is much more significant than previously thought. In fact, the results presented here show that ZupT remarkably contributes to zinc acquisition under condition of severe zinc shortage either in vitro or in vivo and suggest the induction of ZnuABC is not sufficient to completely compensate for the reduction in metal uptake that is consequent to zupT inactivation. These observations demonstrate that ZupT is critical in the process of acquisition of zinc in Salmonella (and, most likely, in the majority of Gram-negative bacteria), where it likely represents a central route of zinc entry, whereas ZnuABC is the necessary complement to the basal activity of ZupT to adapt bacteria to conditions of zinc limitation.
Supplementary Material
Acknowledgments
This work was supported by Fondazione Roma (A.B.), Istituto Superiore di Sanita Intramural Research Project 11 US 24 (P.P. and A.B.) and the European Union Eranet-EMIDA project T99 (P.P. and A.B.). Work in M.R. lab is supported by Public Health Service Grant AI083663 and by the Pacific Southwest Regional Center of Excellence for Biodefense and Infectious Disease Research funds (Supported by award number U54 AI065359 from the National Institute of Allergy and Infectious Diseases). J. Z. L. was supported by the National Institute of Health Immunology Research Training Grant T32 AI60573 and by a predoctoral fellowship from the American Heart Association.
References
- 1.Waldron KJ, Robinson NJ. Nat Rev Microbiol. 2009;7:25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- 2.Andreini C, Banci L, Bertini I, Rosato A. J Proteome Res. 2006;5:196–201. doi: 10.1021/pr050361j. [DOI] [PubMed] [Google Scholar]
- 3.Andreini C, Banci L, Bertini I, Rosato A. J Proteome Res. 2008;7:209–216. doi: 10.1021/pr070480u. [DOI] [PubMed] [Google Scholar]
- 4.Colvin RA, Holmes WR, Fontaine CP, Maret W. Metallomics. 2010;2:306–317. doi: 10.1039/b926662c. [DOI] [PubMed] [Google Scholar]
- 5.Patzer SI, Hantke K. Mol Microbiol. 1998;28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 6.Outten CE, Tobin DA, Penner-Hahn JE, O’Halloran TV. Biochemistry. 2001;40:10417–10423. doi: 10.1021/bi0155448. [DOI] [PubMed] [Google Scholar]
- 7.Panina EM, Mironov AA, Gelfand MS. Proc Natl Acad Sci U S A. 2003;100:9912–9917. doi: 10.1073/pnas.1733691100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham AI, Sanguinetti G, Bramall N, McLeod CW, Poole RK. Microbiology. 2012;158:284–292. doi: 10.1099/mic.0.053843-0. [DOI] [PubMed] [Google Scholar]
- 9.Lim CK, Hassan KA, Penesyan A, Loper JE, Paulsen IT. Environ Microbiol. 2013;15:702–715. doi: 10.1111/j.1462-2920.2012.02849.x. [DOI] [PubMed] [Google Scholar]
- 10.Pawlik MC, Hubert K, Joseph B, Claus H, Schoen C, Vogel U. J Bacteriol. 2012;194:6594–6603. doi: 10.1128/JB.01091-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciavardelli D, Ammendola S, Ronci M, Consalvo A, Marzano V, Lipoma M, Sacchetta P, Federici G, Di Ilio C, Battistoni A, Urbani A. Mol Biosyst. 2011;7:608–619. doi: 10.1039/c0mb00140f. [DOI] [PubMed] [Google Scholar]
- 12.Lewis DA, Klesney-Tait J, Lumbley SR, Ward CK, Latimer JL, Ison CA, Hansen EJ. Infect Immun. 1999;67:5060–5068. doi: 10.1128/iai.67.10.5060-5068.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campoy S, Jara M, Busquets N, Perez De Rozas AM, Badiola I, Barbe J. Infect Immun. 2002;70:4721–4725. doi: 10.1128/IAI.70.8.4721-4725.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrido ME, Bosch M, Medina R, Llagostera M, Pérez de Rozas AM, Badiola I, Barbé J. FEMS Microbiol Lett. 2003;221:31–37. doi: 10.1016/S0378-1097(03)00131-9. [DOI] [PubMed] [Google Scholar]
- 15.Kim S, Watanabe K, Shirahata T, Watarai M. J Vet Med Sci. 2004;66:1059–1063. doi: 10.1292/jvms.66.1059. [DOI] [PubMed] [Google Scholar]
- 16.Yang X, Becker T, Walters N, Pascual DW. Infect Immun. 2006;74:3874–3879. doi: 10.1128/IAI.01957-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ammendola S, Pasquali P, Pistoia C, Petrucci P, Petrarca P, Rotilio G, Battistoni A. Infect Immun. 2007;75:5867–5876. doi: 10.1128/IAI.00559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis LM, Kakuda T, DiRita VJ. J Bacteriol. 2009;191:1631–1640. doi: 10.1128/JB.01394-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabri M, Houle S, Dozois CM. Infect Immun. 2009;77:1155–1164. doi: 10.1128/IAI.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gabbianelli R, Scotti R, Ammendola S, Petrarca P, Nicolini L, Battistoni A. BMC Microbiol. 2011;11:36. doi: 10.1186/1471-2180-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corbett D, Wang J, Schuler S, Lopez-Castejon G, Glenn S, Brough D, Andrew PW, Cavet JS, Roberts IS. Infect Immun. 2012;80:14–21. doi: 10.1128/IAI.05904-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hood MI, Mortensen BL, Moore JL, Zhang Y, Kehl-Fie TE, Sugitani N, Chazin WJ, Caprioli RM, Skaar EP. PLoS Pathog. 2012;8:e1003068. doi: 10.1371/journal.ppat.1003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrarca P, Ammendola S, Pasquali P, Battistoni A. J Bacteriol. 2010;192:1553–1564. doi: 10.1128/JB.01310-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pesciaroli M, Aloisio F, Ammendola S, Pistoia C, Petrucci P, Tarantino M, Francia M, Battistoni A, Pasquali P. Vaccine. 2011;29:1783–1790. doi: 10.1016/j.vaccine.2010.12.111. [DOI] [PubMed] [Google Scholar]
- 25.Pasquali P, Ammendola S, Pistoia C, Petrucci P, Tarantino M, Valente C, Marenzoni ML, Rotilio G, Battistoni A. Vaccine. 2008;26:3421–3426. doi: 10.1016/j.vaccine.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 26.Liu JZ, Jellbauer S, Poe AJ, Ton V, Pesciaroli M, Kehl-Fie TE, Restrepo NA, Hosking MP, Edwards RA, Battistoni A, Pasquali P, Lane TE, Chazin WJ, Vogl T, Roth J, Skaar EP, Raffatellu M. Cell Host Microbe. 2012;11:227–239. doi: 10.1016/j.chom.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ammendola S, Pasquali P, Pacello F, Rotilio G, Castor M, Libby SJ, Figueroa-Bossi N, Bossi L, Fang FC, Battistoni A. J Biol Chem. 2008;283:13688–13699. doi: 10.1074/jbc.M710499200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hantke K. Curr Opin Microbiol. 2005;8:196–202. doi: 10.1016/j.mib.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Gaither LA, Eide DJ. Biometals. 2001;14:251–270. doi: 10.1023/a:1012988914300. [DOI] [PubMed] [Google Scholar]
- 30.Grass G, Wong MD, Rosen BP, Smith RL, Rensing C. J Bacteriol. 2002;184:864–866. doi: 10.1128/JB.184.3.864-866.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grass G, Franke S, Taudte N, Nies DH, Kucharski LM, Maguire ME, Rensing C. J Bacteriol. 2005;187:1604–1611. doi: 10.1128/JB.187.5.1604-1611.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taudte N, Grass G. Biometals. 2010;23:643–656. doi: 10.1007/s10534-010-9319-z. [DOI] [PubMed] [Google Scholar]
- 33.Karlinsey JE, Maguire ME, Becker LA, Crouch ML, Fang FC. Mol Microbiol. 2010;78:669–685. doi: 10.1111/j.1365-2958.2010.07357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Datsenko KA, Wanner BL. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maloy Stanley SVJR, Taylor Ronald K. Genetic Analysis of Pathogenic Bacteria: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1996. [Google Scholar]
- 36.Casadaban MJ, Chou J, Cohen SN. J Bacteriol. 1980;143:971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: 1972. [Google Scholar]
- 38.Connolly EL, Fett JP, Guerinot ML. Plant Cell. 2002;14:1347–1357. doi: 10.1105/tpc.001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith KF, Bibb LA, Schmitt MP, Oram DM. J Bacteriol. 2009;191:1595–1603. doi: 10.1128/JB.01392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaballa A, Helmann JD. Mol Microbiol. 2002;45:997–1005. doi: 10.1046/j.1365-2958.2002.03068.x. [DOI] [PubMed] [Google Scholar]
- 41.Scott C, Rawsthorne H, Upadhyay M, Shearman CA, Gasson MJ, Guest JR, Green J. FEMS Microbiol Lett. 2000;192:85–89. doi: 10.1111/j.1574-6968.2000.tb09363.x. [DOI] [PubMed] [Google Scholar]
- 42.Lee JW, Helmann JD. J Biol Chem. 2006;281:23567–23578. doi: 10.1074/jbc.M603968200. [DOI] [PubMed] [Google Scholar]
- 43.Pacello F, Ceci P, Ammendola S, Pasquali P, Chiancone E, Battistoni A. Biochim Biophys Acta. 2008;1780:226–232. doi: 10.1016/j.bbagen.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 44.Kaplan D, Christiaen D, Arad SM. Appl Environ Microbiol. 1987;53:2953–2956. doi: 10.1128/aem.53.12.2953-2956.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cataldo SG, Antonio, Pettignano Alberto, Piazzese Daniela, Sammartano Silvio. International Journal of Electrochemical Science. 2012;7:6722–6737. [Google Scholar]
- 46.Andrade LR, Leal RN, Noseda M, Duarte MER, Pereira MS, Mourão PAS, Farina M, Amado Filho GM. Marine Pollution Bulletin. 2010;60:1482–1488. doi: 10.1016/j.marpolbul.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 47.Davis TA, Volesky B, Mucci A. Water Research. 2003;37:4311–4330. doi: 10.1016/S0043-1354(03)00293-8. [DOI] [PubMed] [Google Scholar]
- 48.Forbes JR, Gros P. Blood. 2003;102:1884–1892. doi: 10.1182/blood-2003-02-0425. [DOI] [PubMed] [Google Scholar]
- 49.Stojiljkovic I, Baumler AJ, Heffron F. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.