Abstract
Somatically acquired genetic abnormalities lead to the salient features that define myelodysplastic syndromes (MDS): clonal hematopoiesis, aberrant differentiation, peripheral cytopenias, and risk of progression to acute myeloid leukemia. Although specific karyotypic abnormalities have been linked to MDS for decades, more recent findings have demonstrated the importance of mutations within individual genes, focal alterations that are not apparent by standard cytogenetics, and aberrant epigenetic regulation of gene expression. The spectrum of genetic abnormalities in MDS implicates a wide range of molecular mechanisms in the pathogenesis of these disorders, including activation of tyrosine kinase signaling, genomic instability, impaired differentiation, altered ribosome function, and changes in the bone marrow microenvironment. Specific alterations present in individual patients with MDS may explain much of the heterogeneity in clinical phenotype associated with this disease and can predict prognosis and response to therapy. Elucidation of the full complement of genetic causes of MDS promises profound insight into the biology of the disease, improved classification and prognostic scoring schemes, and the potential for novel targeted therapies with molecular predictors of response.
INTRODUCTION
Myelodysplastic syndromes (MDS) are clonal disorders of hematopoiesis characterized by inefficient hematopoiesis, peripheral blood cytopenias, and risk of progression to acute myeloid leukemia (AML). The clinical phenotype of patients with MDS are diverse with respect to the number and severity of cytopenias, cellularity and blast count in the bone marrow, rate of progression to AML, overall survival, and response to treatment. Much of this phenotypic heterogeneity is likely due to the variety of genetic lesions that contribute to disease pathogenesis. Unraveling the genetic complexity of MDS promises to elucidate the pathophysiology, refine the taxonomy and prognostic scoring systems, and provide novel therapeutic targets.
The known set of genetic lesions that cause MDS include copy number changes (genetic amplifications or deletions), mutations that alter the sequence or expression of individual genes, and epigenetic abnormalities. although balanced translocations are rare, chromosomal abnormalities evident by standard karyotypic analysis are present in approximately half of patients with MDS. The most common of these are loss of 5q (5q-), loss of 7 or 7q (-7/7q-), trisomy 8 (+8), loss of 20q (20q-), and loss of Y (-Y; Table 1).1 More sensitive technologies such as single nucleotide polymorphism microarrays can detect copy number changes or acquired uniparental disomy in as many as 75% of patients with MDS.2,3 Moreover, the majority of patients have mutations that alter the sequence and function of oncogenes or tumor suppressor genes (Fig 1).4 In addition, patients with MDS commonly have abnormal epigenetic profiles, resulting in aberrant gene expression.5
Table 1.
Genetic Abnormalities in MDS
| Genetic Abnormality | Frequency (%) | Pathogenic Mechanism | Clinical Consequence |
|---|---|---|---|
| Chromosomal abnormalities | |||
| 5q- | 15 | Haploinsufficiency for RPS14, miR-145/146a, CTNNA1, EGR1, APC, NPM | Better prognosis, high rate of response to lenalidomide |
| -7/7q- | 5-10 | Unknown | Poor prognosis, more common in therapy-related MDS |
| Trisomy 8 | 5-8 | Unknown | Can help predict response to immunosuppression, some evidence as a marker of progression to AML |
| 20q- | 2-5 | Unknown | Better prognosis |
| -Y | 2-4 | Age-related phenomenon that may not be disease related | May be useful as a marker of clonal hematopoiesis |
| Complex (three or more abnormalities) | 10-15 | Various; often abnormal chromosome 17 (TP53 locus) | Poor prognosis |
| -13/13q-, 11q-, 12p-, 9q-, idic(X)(q13), i(17q), t(11;16), t(3;21), t(1;3), t(2;11), inv(3), t(6;9) | Rare | Various, mostly unknown; chromosome 3q26 lesions alter expression of EVI1 | Presumptive evidence of MDS in patients with otherwise unexplained refractory cytopenia and no morphologic evidence of dysplasia |
| Gene mutations | |||
| TET2 | Approximately 20 | Unknown | Unknown |
| RUNX1 | 15-20 | Mutations typically alter DNA-binding domain or disrupt protein-binding domain | Increased risk of progression to AML; more common in therapy-related MDS |
| TP53 | 5-10 | Loss of function of p53 tumor suppressor activity; associated with chromosomal instability | Often complex cytogenetics, relative resistance to therapy, poor prognosis |
| ASXL1 | 10-15 | Unknown; most mutations are distal heterozygous frame shifts, suggesting a dominant negative function | Unknown |
| NRAS/KRAS | Approximately 10 | Loss of GTPase activity leads to constitutive activation of serine/threonine kinase | Increased risk of progression to AML |
| EZH2 | 6 | Loss of histone 3 lysine 27 methyltransferase activity | Poor prognosis |
| CBL/CBLB | Rare | Loss of ubiquitin ligase activity; mutants can inhibit wild-type enzymatic function | Unknown; increased risk of progression to leukemia in MPN and in CMML where this mutation is more common |
| JAK2 | 5% of RA, 50% of RARS-T | Constitutive activation of tyrosine kinase | Unknown; does not appear to alter prognosis |
| MPL | 5% of RARS-T | Constitutive activation of tyrosine kinase | Unknown |
| ATRX | Rare | Loss of function leads to decreased alpha-globin expression, likely through epigenetic dysregulation | Acquired alpha-thalassemia, often with very severe anemia |
| NPM1 | Rare | Terminal frame shift disrupts nucleolar localization signal leading to cytoplasmic redistribution of protein | Unknown; this mutation is very common in AML with normal cytogenetics |
| IDH1, IDH2 | Rare | Missense mutations alter catalytic function to convert α-ketoglutarate into 2-hydroxyglutarate while consuming NADPH | Associated with more advanced disease and progression to AML |
| CEBPA | Rare | Loss of function known to impair granulopoiesis | Unknown; germline mutations associated with risk of AML, not MDS |
| WT1 | Very rare | Impairment of transcription factor activity | Unknown; mutations are more common in AML and associated with poor outcomes |
| PTPN11 | Very rare | Alters function of gene product SHP2, an adaptor protein with tyrosine phosphatase activity | More common in JMML, rare in CMML |
| FLT3, CSF1R, CKIT | Very rare | Constitutive activation of tyrosine kinase | Associated with more advanced disease and progression to AML |
NOTE. Rare mutations are present in 2% to 5% of patients; very rare refers to individual reports or < 2% of patients.
Abbreviations: MDS, myelodyplastic syndromes; CTNNA1, alpha-catenin 1; EGR1, early growth response 1; APC, adenomatous polyposis coli; NPM1, nucleophosmin 1; AML, acute myeloid leukemia; GTPase, guanosine triphosphatase; MPN, myeloproliferative neoplasms; CMML, chronic myelomonocytic leukemia; NADPH, nicotinamide adenine dinucleotide phosphate; JMML, juvenile myelomonocytic leukemia.
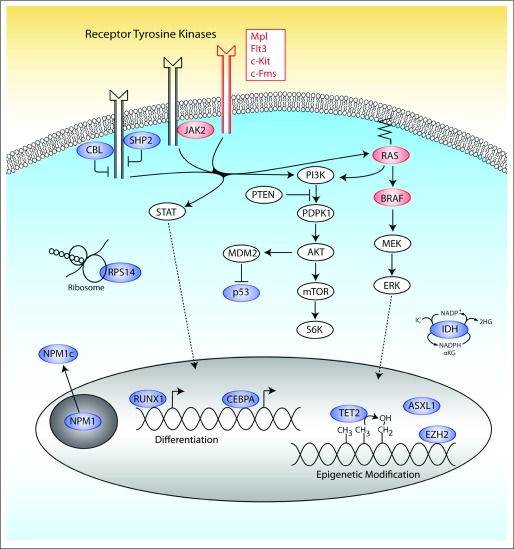
Fig 1.
Targets of point mutations and deletions in myelodysplastic syndromes. Mutations in multiple pathways have been implicated in the pathogenesis of myelodysplastic syndromes. Features shown in red are targets of activating mutations, whereas mutations or deletions of features in blue result in a loss of function. Mutations that impair function may also cause a gain of function as is believed to be the case for a subset of mutations in CBL, ASXL1, NPM1c, IDH1/ IDH2, TP53, and RUNX1.
Molecular lesions are already used to guide the diagnosis, prognosis, and treatment of MDS. The International Prognostic Scoring System (IPSS) incorporates the common karyotypic abnormalities.6 The use of lenalidomide is guided by the presence of a chromosome 5q deletion because this karyotypic abnormality increases the likelihood of cytogenetic and hematologic responses.7 However, genetic predictors of response to hypomethylating agents or bone marrow transplantation have not yet been identified and validated. Point mutations, epigenetic states, and gene expression profiles are not currently integrated into diagnostic classifications or prognostic scoring systems.
The catalog of genes that play a role in the pathophysiology of MDS is rapidly expanding, and studies are underway to investigate the biologic and clinical consequences of each genetic lesion. With the advent of whole-genome sequencing in human malignancies, the rate of discovery is likely to accelerate. A more complete genetic characterization of MDS has great potential to elucidate the molecular basis for the clinical heterogeneity of these disorders and identify disease subtypes with shared outcomes and responses to therapy.
MODEL OF PATHOGENESIS
To manifest clinically, an MDS clone must pass several biologic milestones.8 Individual mutations may contribute to more than one step in the transformation process, although most cases of cancer appear to require the acquisition of multiple genetic abnormalities. In addition, each step in the process can be achieved by alterations in one of several genes, providing a multitude of molecular routes to disease development.
Steps associated with the pathogenesis of MDS include (1) enhanced self-renewal of a hematopoietic stem cell or acquisition of self-renewal in a progenitor cell, (2) increased proliferative capacity in the disease-sustaining clone and/or in its more differentiated progeny, (3) impaired or blocked differentiation, (4) genetic and epigenetic instability, (5) antiapoptotic mechanisms in the disease-sustaining cell, (6) evasion of the immune system, and (7) suppression of normal hematopoiesis.
The capacity for self-renewal must be present in the MDS disease-initiating cell.9 This cell may arise from a self-renewing hematopoietic stem cell or it may come from a more differentiated myeloid progenitor that acquires the ability to self-renew. Further clonal expansion may occur through increased proliferation or resistance to apoptosis, and an abnormal bone marrow microenvironment could favor the development of a neoplastic clone. MDS occur when at least one of the molecular lesions present in the dominant clone or in its microenvironment also causes dysplastic differentiation of one or more myeloid lineages giving rise to ineffective hematopoiesis (Fig 2). The degree to which each step is affected can determine how the disease manifests clinically, including the types and degree of cytopenias present and whether the disease is indolent or rapidly progressive.
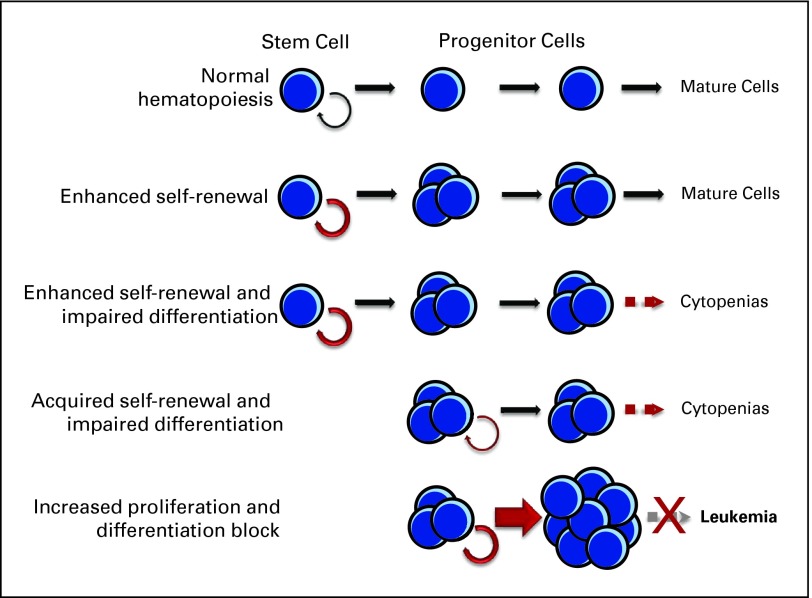
Fig 2.
Various pathways to transformation. The defining feature of myelodysplastic syndromes is clonal and inefficient hematopoiesis that causes peripheral cytopenias. A heterogeneous set of genetic and epigenetic abnormalities drive the cellular events that give rise to clinical phenotypes. Individual lesions may be responsible for one transformative step alone (eg, enhanced self-renewal or altered apoptosis) and therefore be clinically silent in isolation, or they could result in several abnormalities (eg, acquired self-renewal and impaired differentiation). Cooperation between two or more lesions is likely needed for the full expression of disease.
CHROMOSOMAL ABNORMALITIES
Chromosome 5q Deletions
With an incidence of roughly 15%, deletions of chromosome 5q (5q-) are the most common cytogenetic abnormality in patients with MDS, and isolated 5q- is associated with a relatively favorable prognosis. Deletions of 5q are universally heterozygous. Despite extensive sequencing, no mutations have been indentified in genes on the remaining intact allele, and uniparental disomy is not seen on 5q in patients with MDS.10–12 These findings indicate that instead of biallelic inactivation of a tumor suppressor gene, haploinsufficiency for one or more 5q genes is likely pathogenic. Careful analysis of the 5q- breakpoints in multiple patients with MDS localized two distinct commonly deleted regions (CDRs).13–15 The more distal CDR in 5q33.1 is associated with a clinical phenotype termed the 5q- syndrome, characterized by severe macrocytic anemia, relative thrombocythemia, female predominance, and a lower risk of progression to AML.16 The proximal CDR at 5q31 is associated with therapy-related MDS and a more aggressive MDS and AML phenotype.13–15
Multiple genes on 5q have been implicated in the pathogenesis of MDS. RPS14 was identified as a critical gene for the erythroid phenotype of the 5q- syndrome in a systematic functional screen of the 5q33 CDR.17 Congenital mutations or deletions causing haploinsufficiency for other ribosomal proteins cause Diamond-Blackfan anemia, a disease also characterized by a severe macrocytic anemia.18 Since patients with Diamond-Blackfan anemia do not have elevated platelet counts, deletion of RPS14 alone is unlikely to be responsible for the entire 5q- syndrome phenotype. Recent studies have demonstrated that haploinsufficiency for two microRNAs located on chromosome 5q33, miR-145 and miR-146, can cause elevated platelet counts and may provide a selective advantage to the 5q- clone.19,20
The proximal CDR at 5q31 also contains several candidate MDS genes. The early growth response gene (EGR1) increases stem-cell self-renewal when one copy is deleted.21 Alpha catenin, CTNNA1, is underexpressed in patients with 5q- syndrome, and hypermethylation of the remaining allele is associated with transformation to AML.14,22 Deletions of 5q are not limited to the CDRs and often encompass both of these regions and beyond. Other candidate MDS genes that lie outside the CDRs but are often lost with deletions of 5q include APC and NPM1.23–25
Deletion of some 5q genes may not be involved in the development of MDS but could sensitize cells to therapeutic agents. This may explain the robust response that patients with 5q- MDS have to lenalidomide, an analog of thalidomide that is approved by the US Food and Drug Administration for the treatment of 5q- MDS.26 Although nearly half of patients with low-risk MDS reduce their need for transfusions after lenalidomide therapy, those with 5q- MDS often have complete cytogenetic remissions, demonstrating the unique sensitivity this genetic lesion imparts to these cells. Two cell cycle–regulating phosphatases encoded by the genes on 5q, CDC25C and PP2A, have been implicated in the favorable response to lenalidomide.27
Chromosome 7 and 7q Deletions
Unlike 5q deletions, monosomy 7 or interstitial loss of 7q is associated with a relatively poor prognosis. Approximately 10% of patients with MDS carry an abnormality of chromosome 7, either in isolation or as part of a complex karyotype.1 This frequency approaches 50% in patients who have therapy-related MDS from a prior history of treatment with alkylating agents.28 At least three common deleted regions on 7q have been identified, but the underlying molecular lesions driving the development of MDS in these cases are not well characterized.29–31 The recent discovery of mutations in EZH2, a gene located at 7q36, may partially explain why this chromosomal region is often deleted. Conditional heterozygous deletion of a 2.5-Mb region syntenic to human 7q22 in murine hematopoietic stem cells had no phenotype indicating that this locus may not harbor a critical suppressor.32 In therapy-related MDS, chromosome 7 abnormalities co-occur with 5q- or mutations in RUNX1 more often than would be expected if these events were distributed randomly among all patients.33 This suggests that -7/7q- lesions drive distinct steps in MDS development from 5q deletions or RUNX1 mutations.
Trisomy 8 and Response to Immunosuppressive Therapy
Trisomy 8 is the only recurrent chromosomal amplification observed in MDS. It is present as sole abnormality in roughly 8% of patients and is considered to be an intermediate-risk cytogenetic abnormality. MDS patients with +8 have less than half the median expected survival of patients with a normal karyotype (22.0 v 53.4 months).1 However, a subset of patients with MDS and a sole +8 abnormality respond quite well to immunosuppressive therapy. These patients are more likely to be young, have refractory anemia of short duration, and carry HLA DR15.34 Patients with +8 MDS have been shown to have expansion of Vβ-restricted CD8+ T cells. After immunosuppressant therapy, Vβ representation in CD8+ T cells often returns to a more normal polyclonal distribution. Despite improvement in blood counts, examination of the bone marrow in responders demonstrates the persistence or even the expansion of the +8 clone. This suggests that the immune system targets diseased cells and, as a consequence, impairs normal hematopoiesis. Cells with trisomy 8 have been shown to express higher levels of antiapoptotic genes and are resistant to irradiation compared with normal cells. In the setting of autoimmunity, the +8 clone is likely to have a selective advantage over normal hematopoietic precursors. This mechanism of immune-mediated growth advantage and impairment of normal hematopoiesis is not restricted to +8 MDS. Patients without this abnormality, particularly those with low-risk disease, can also benefit from immunosuppression, albeit with a lower frequency of response.35
Chromosome Y and 20q Deletions
MDS patients with isolated 20q- or -Y abnormalities are considered to be in the same favorable cytogenetic risk group as patients with a normal karyotype. The loss of chromosome Y appears to be unrelated to disease pathogenesis.36 Isolated -Y is found in men without evident hematologic diseases, increasing in incidence with age.37 Interstitial deletion of 20q, on the other hand, does appear to be pathologic but is not restricted to cases of MDS. It is a common cytogenetic abnormality in myeloproliferative disorders and AML.38 The CDR of 20q deduced from patients with MDS and AML includes 19 genes, none of which have been conclusively linked to the pathogenesis of myeloid disorders.39,40 In patients with MDS, the favorable cytogenetic risk associated with 20q- suggests that it does little to modify the course of disease compared with patients with a normal karyotype, although late acquisition of 20q- implies clonal evolution and can precede progression of disease.41 As with other chromosomal abnormalities, both -Y and 20q- can be useful for confirming the presence of clonal hematopoiesis and for monitoring the response of patients to treatment. However, neither lesion is sufficient to make the diagnosis of MDS.
Abnormalities Involving Chromosome 3q26
Recurrent translocations and inversions involving 3q26 have been identified in AML and rare cases of MDS in which they are associated with a poorer prognosis.1,42 Breakpoints typically include the MDS1-EVI1 gene locus (MECOM).43 Alternative splicing can produce the full-length MDS1-EVI1 transcript or just the more distal EVI1 transcript. Chromosomal abnormalities at this locus preferentially activate expression of EVI1 alone.44 The products of these transcripts have different and potentially antagonistic functions. EVI1 can impair the activity of several transcription factors including MDS1-EVI1, GATA1, PU.1, and RUNX1 leading to impaired hematopoiesis.44–47 In mouse models, forced EVI1 expression results in an MDS-like phenotype and increased self-renewal of hematopoietic progenitors.48,49 The Mecom locus is a frequent transforming retroviral insertion site in mice, and retroviral EVI1 activation has been implicated as a cause of MDS in participants in a hematopoietic gene therapy trial.50–52 Even in patients without 3q26 abnormalities, EVI1 overexpression is associated with a poor outcome.53,54
Other Cytogenic Abnormalities
Several rare, but recurrent cytogenetic abnormalities have been associated with MDS. In the absence of an alternative diagnosis, these chromosomal lesions can be used as presumptive evidence of MDS in persistently cytopenic patients, even if they have little evidence of dysplasia.55 These include various abnormalities of chromosome 17 (presumably disrupting TP53), isodicentric chromosome Xq13 (often associated with the presence of ring sideroblasts), and the t(6;9)(p23;q34) translocation that generates the DEK-NUP214 fusion gene (also referred to as DEK-CAN). The prognostic implications of these rare karyotypes are not considered independently in common clinical prognostic scoring systems such as the IPSS. These lesions are lumped into the intermediate-risk group when they occur in isolation or with another single abnormality besides -7/7q-. Newer studies that associate outcome with cytogenetic abnormalities are more inclusive of these rarer lesions and may better predict the prognosis of patients with MDS.42
GENE MUTATIONS
TET2 Mutations
TET2 is the most frequently mutated gene identified in MDS to date. Mutations of TET2 are present in nearly 20% of patients with MDS and are also seen in myeloproliferative neoplasms (MPN) (10%), chronic myelomonocytic leukemia (CMML; 30% to 50%), and secondary AML (25%).56–59 Its homolog, the ten-eleven translocation gene (TET1), was named for its role as a fusion partner with the mixed-lineage leukemia (MLL) gene in rare cases of AML.60 TET1 encodes an alpha ketoglutarate (αKG) –dependent dioxygenase that converts 5-methylcystosine into 5-hydroxymethylcytosine, thereby altering the epigenetic mark created by DNA methyltransferases (DNMTs).61 TET2 shares a high degree of homology with TET1, including its catalytic domain, and has been shown to have the same enzymatic capability.61a
TET2 mutations are not tightly associated with other recurrent mutations or cytogenetic abnormalities. The presence of TET2 mutations in MPN indicates that they do not cause dysplasia since these disorders retain normal differentiation. Nor are TET2 mutations likely to cause the highly proliferative phenotype of AML and MPN, since they are frequently present in low-risk cases of MDS. Instead, TET2 mutations may drive a pathogenic step common to all of the myeloid neoplasms in which they are found, such as the establishment or enhancement of clonal dominance in the disease cell of origin.
In some patients with TET2 and JAK2 V617F mutant MPN, a clonal population of cells with mutant TET2 and unmutated JAK2 can be found, indicating that the JAK2 mutation occurred in an already expanded TET2 mutant clone. In other patients, TET2 mutations were clearly acquired after JAK2 V617F and were associated with progression of disease.62 In MDS, the impact of TET2 remains unclear. Early studies suggested that TET2 mutations were associated with improved prognosis independent of the IPSS or were more likely to occur in low-risk patients, but more recent studies suggest no impact on overall survival.58,63,63a In contrast, TET2 mutations in AML and CMML are associated with a relatively poor prognosis.57,64
ASXL1 Mutations
Mutations in the additional sex-comb like-1′ (ASXL1) gene have been described in roughly 10% of MDS and MPN, 17% of AML, and > 40% of patients with CMML.65–67 ASXL1 encodes a member of the polycomb family of chromatin-binding proteins and is involved in the epigenetic regulation of gene expression.68 It contains the plant homeo domain finger and nuclear receptor box domains and functions as a ligand-dependent coactivator of the retinoic acid receptor. It is has been shown to mediate its effects through a direct interaction with the histone acyltransferase encoded by NCOA1 or the histone demethylase encoded by LSD1.69,70 Unlike TET2, mutations in ASXL1 appear to be mutually exclusive of some of the other recurrent genetic abnormalities. For example, ASXL1 mutations were described in patients with essential thrombocythemia and primary myelofibrosis but only in those with wild-type JAK2.67 In a small study of patients with AML,71 mutations of ASXL1 were almost entirely exclusive of the common mutations in the terminal exon of NPM1. In contrast, ASXL1 mutations can co-occur with mutations of RUNX1 and TET2.65
Targeted disruption of Asxl1 in the germline of mice resulted in embryonic/perinatal lethality in a subset of animals. Mice deficient for Asxl1 who survived to birth had only mild hematopoietic defects with reduced numbers of lymphocytes and modest splenomegaly. However, Asxl1-deficient mice had no evidence of myeloid dysplasia, had no detectable progenitor or stem-cell abnormalities, and did not go on to develop leukemia or lymphoma.72 Unlike in these mice in which Asxl1 was deleted, most of the ASXL1 mutations seen in patients occur in the C-terminal portion of the protein. The result is deletion of the plant homeo domain protein interaction domain while sparing its N-terminal motifs, potentially indicating that ASXL1 mutations generate a dominant-negative protein that could inhibit its wild-type counterpart as well as other members of the polycomb multiprotein complex. The prognostic significance of ASXL1 mutations in MDS has yet to be determined.
RUNX1 Mutations
RUNX1 is a member of the transcriptional core-binding factor gene family (also known as CBFA2 or AML1) and is the second most commonly mutated gene in MDS. It was identified as the translocation partner to ETO (now called RUNX1T1) in cases of t(8;21) core-binding factor AML. As with other translocations, those involving RUNX1 are more common in or exclusive to AML compared with MDS. RUNX1 point mutations, however, are found in MDS, AML, CMML, and more rarely, in MPN. In MDS, they are present in 7% to 15% of de novo patients and at a higher frequency in therapy-related disease.28 In both MDS and AML, RUNX1 mutations are markers of poor prognosis.73–75
The RUNX1 protein contains a proximal Runt homology domain, important for DNA binding, and a distal transactivation domain responsible for protein-protein interactions and recruitment of cofactors. Missense mutations of RUNX1 are clustered in the Runt domain, whereas stop codon mutations and frame shifts are found throughout the length of the gene and almost always disrupt the transactivation domain.75,76 This distinction may be physiologically relevant since Runt domain mutants with impaired DNA binding can function as dominant negatives. These mutants would be capable of reducing Runx1 activity much more than simple loss-of-function mutations in one allele.77,78
Germline mutations of RUNX1 cause a rare human disease called familial platelet disorder with propensity to leukemia.79 Patients with this partially penetrant, genetically dominant disease have an MDS-like phenotype with thrombocytopenia and/or dysfunctional platelets.80 Roughly a third of affected patients with familial platelet disorder will develop AML, often after acquiring a second RUNX1 mutation.81 The long latency to leukemia in these individuals (median age of onset is 33 years) and in mice carrying mutant RUNX1 genes indicates that secondary mutations are required for progression to AML. In patients with MDS, RUNX1 mutations are often accompanied by activation of the Ras pathway or mutations in these genes.82,83
The genetic manipulation of Runx1 in mouse models has helped explain how acquired RUNX1 mutations might promote the development of MDS. Runx1 knockout mice are not viable and die as embryos with no evidence of definitive hematopoiesis.84–86 If Runx1 is excised in the adult hematopoietic compartment, mice develop extramedullary hematopoiesis, lymphoid defects, expansion of the myeloid progenitor pool, and inefficient platelet production, but do not progress to AML.87 In mice transplanted with bone marrow cells that overexpress a Runt domain mutant form of RUNX1, the phenotype is different. The mice have increased numbers of bone marrow blasts and splenomegaly, and they often succumb to an AML-like disease. If instead, RUNX1 with a more distal frame shift mutation sparing the Runt domain is used, the phenotype more closely resembles MDS with marked erythroid dysplasia, pancytopenia, and fewer cases of AML.88 These findings support a gain of function for RUNX1 mutations that affect the Runt domain.
IDH1 and IDH2 Mutations
Two isocitrate dehydrogenase genes (IDH1 and IDH2) were identified as mutated oncogenes in a high percentage of glioma and secondary glioblastoma samples.89 Analysis of nearly 500 solid tumor samples from tissues outside the CNS revealed no mutations in IDH1 and IDH2, suggesting that these were tissue-specific oncogenes. Therefore, it was unexpected when whole-genome sequencing of an AML sample led to the discovery of recurring mutations of IDH1 in 16 of 188 patients with AML.90 Mutations of IDH1 and IDH2 have been confirmed in AML, MPN at the time of leukemic transformation, and in rare cases of MDS.91–95
IDH1 and IDH2 are homodimeric, nicotinamide adenine dinucleotide phosphate (NADP+) dependent enzymes that convert isocitrate to αKG and reside in the cytoplasm and mitochondria, respectively. The mutations identified in these genes are specific missense mutations of conserved codons. No frame shift or early termination mutations are seen, and all mutations appear to be heterozygous to an intact wild-type allele. This suggests that IDH mutations cause either a dominant negative phenotype, in which the mutant protein inhibits the wild-type enzyme, or a gain of function, in which the mutant protein acts in a way that the wild-type form does not. Recent evidence suggests the latter. Metabolic profiling of cell lines expressing mutant IDH1 were found to contain high levels of 2-hydroxyglutarate (2HG).96 This abnormal metabolite can also be detected in primary leukemic cells harboring IDH1 or IDH2 mutations.97 It appears that mutant IDH enzymes have altered substrate specificity. Instead of generating a NADPH molecule by catalyzing the conversion of isocitrate to αKG, mutant IDH consumes NADPH while converting αKG into 2HG. Whether 2HG is directly oncogenic or whether NADPH and αKG depletion are drivers of leukemogenesis is not yet known.
Tyrosine Kinases, RAS Genes, and CBL Mutations
Members of the tyrosine kinase (TK) signaling pathways are mutated commonly in myeloid malignancies but infrequently in MDS. For example, mutations of FLT3 and KIT occur frequently in AML, and MPL mutations and platelet-derived growth factor receptor (PDGFR) rearrangements are found in various MPN. In MDS, rare mutations of CSF1R have been described in addition to rare mutations in the TKs more commonly mutated in other myeloid neoplasms. Patients with MDS are more commonly characterized by activating mutations of the downstream RAS genes. NRAS mutations are present in 10% to 15% of patients with another 1% to 2% having KRAS mutations.98,99 Considered in isolation, these mutations are associated with a poor prognosis and progression of MDS to AML.100,101 Other members of this signaling pathway that are mutated in rare cases of MDS include BRAF, PTPN11, and CBL.99,102–104
The CBL gene product is a TK-associated ubiquitin ligase that negatively regulates signaling through these receptors by targeting them for degradation. It is mutated in roughly 15% of CMML and in patients with juvenile myelomonocytic leukemia but is found in fewer than 5% of MDS. CBL mutations result in increased receptor TK levels and phosphorylation of STAT5, which are believed to mediate the hypersensitivity of these mutant cells to a wide variety of growth factors and cytokines.104–106
Mutations of CBL are often biallelic, suggesting that CBL is a tumor suppressor gene and that loss of wild-type function is advantageous. However, frame shift and early truncation mutants are uncommon. Instead, CBL mutations cluster in exons 8 and 9 and usually leave the rest of the gene intact, suggesting a gain-of-function effect of these mutations. The common CBL mutations have been shown to encode a dominant negative protein that inhibits the ubiquitin ligase activity of the wild-type gene product as well as the product of its homolog, CBLB.104 Mouse cells deficient for Cbl show modest cytokine hypersensitivity.107 This is greatly enhanced by the introduction of a mutated CBL gene into these cells, perhaps through the inhibition of partially compensatory CBLB activity.
Across myeloid disorders, CBL mutations appear to be mutually exclusive of several other commonly mutated genes including FLT3, KIT, NPM1, CEBPA, PTPN11, and NRAS.108,109 In AML, they are more frequently associated with the t(8;21) abnormality, suggesting a cooperative advantage to this pairing.110 If CBL mutations are equivalent to RAS mutations in MDS, then they will likely be associated with a negative prognosis, although this has yet to be described.
Activating mutations of JAK2 form part of the diagnostic criteria for polycythemia vera and are found in roughly half of patients with essential thrombocythemia and primary myelofibrosis. Only the most common mutation, V617F, has been described in MDS. It is present in just 5% of unselected patients and does not appear to have prognostic significance.111 The MDS subtype defined by refractory anemia with ring sideroblasts and thrombocythemia (RARS-T) is the exception. JAK2 V617F mutations are present in 50% of these patients, as are rare mutations of MPL, another MPN-associated gene.112,113 This prompts the possibility that RARS-T is actually a frustrated form of MPN and not a subtype of MDS.111,114 However, cases of JAK2 wild-type RARS have been described in which thrombocythemia occurs only at the time that the V617F mutation is acquired.115 Given the high rate of JAK2 mutation in RARS-T, there would appear to be a cooperative advantage between those abnormalities responsible for the ring sideroblast phenotype and the constitutive activation of JAK2. The nature of this interaction is not yet understood.
TP53 Activation and Mutation
The TP53 gene, located on chromosome 17p, is a prototypical tumor suppressor gene. Its gene product, p53, is activated by a variety of cellular stressors and can cause cell cycle arrest, induce the DNA repair response, and drive cells into apoptosis. In cases of 5q- MDS, activation of p53 appears to be essential for the erythropoietic defect associated with haploinsufficiency for RPS14.116
TP53 mutations have been found in nearly every tumor type and are often associated with genomic instability. In MDS, TP53 is mutated in 5% to 15% of de novo patients and more frequently in patients who have had prior exposure to alkylating agents or radiation.33,117 Loss of wild-type p53, either through point mutations or larger abnormalities of 17p, is associated with advanced disease, complex karyotype, and resistance to treatment.118,119 Mutations of TP53 are among the few single-gene mutations to have been associated with a poor prognosis in MDS after adjusting IPSS risk group.120,121 The detection of TP53 mutations in small subclones of MDS may prove to be important, because this treatment-resistant population could drive progression or relapse despite the presence of otherwise favorable prognostic markers.122
EPIGENETICS IN THE PATHOGENESIS AND TREATMENT OF MDS
The term “epigenetic” refers to the heritable component of cellular phenotypes that are not mediated by changes to the genomic DNA sequence. Since most cells in an organism share an identical genetic code, it is the epigenetic state of each cell and its interaction with the environment that are the greatest determinants its behavior. The most relevant molecular mediators of the epigenetic state in MDS are gene-expression patterns maintained by methylation of cytosine residues in DNA and covalent modification of histones.
DNA methyl transferases convert cytosine bases into 5-methylcytosines, particularly when they form the first base in a CpG dinucleuotide. These CpG islands are commonly found clustered in and around gene promoters, consistent with their role in the regulation of gene expression. Methylation of CpG islands can alter their interaction with DNA-binding proteins, such as transcription factors and histone-modifying enzymes. Typically, methylation of CpG islands in promoters leads to silencing of neighboring genes and represents a mechanism for loss of tumor suppressor gene expression. In MDS and AML, several genes have been described as targets of DNA hypermethylation. These include the cell cycle regulators CDKN2A (p14 and p16) and CDKN2B (p15), CTNNA1, E-cadherin (CDH1), and many others. Genome-wide increases in promoter hypermethylation predict survival, even after taking into consideration age, sex, and IPSS risk group, and are seen during progression to AML.123,124
These observations provide a rationale for the use of demethylating agents in MDS. The nucleoside analog azacitidine and its 2′-deoxy counterpart decitabine are inhibitors of DNMTs and have been approved for the treatment of MDS in the United States. Both medications have good response rates in MDS with 30% to 73% of patients experiencing a 50% or better decrease in transfusion dependence. Low-risk patients have response rates comparable to those of higher-risk patients, and no cytogenetic or molecular marker has been validated as a predictor of response in MDS. Surprisingly, the degree of promoter hypermethylation does not predict response to DNMT inhibitors. In practice, decitabine or azacitidine are offered mainly to higher-risk patients, particularly since azacitidine has been shown to prolong survival in this group.124a
Inhibition of histone-modifying enzymes represents another potential epigenetic target for MDS therapy. Histones are protein multimers associated with DNA that help form chromatin structure. They can be covalently modified in several ways that alter how they interact with DNA and with other chromatin-binding proteins. Taken together, these modifications represent the histone code, which influences the level of transcription of nearby genes. The acetylation of lysine residues on histones is associated with an open chromatin conformation and increased transcription. Histone deacetylases (HDACs) can remove these acetyl groups and silence nearby genes. This is a critical mechanism for the regulation of gene expression, but when gone awry, can lead to changes that promote tumor development. As with DNMT inhibitors, drugs that inhibit HDACs theoretically work by allowing for the re-expression of inappropriately silenced tumor suppressor genes. This explanation is certainly too simplistic since these drugs and their targets have multiple effects, and dramatic changes in gene expression are not observed after treatment with HDAC inhibitors. Given the success of DNMT-targeted therapy and the interaction between DNA methylation and histone modification, HDAC inhibitors have been considered promising for the treatment of myeloid malignancies.
ROLE OF THE BONE MARROW MICROENVIRONMENT
Abnormalities of the bone marrow microenvironment are well documented in MDS and other myeloid malignancies. Levels of vascular endothelial growth factor and several inflammatory cytokines are elevated in the bone marrow of patients with MDS.125,126 These changes are thought to be the result of the complex interplay between the abnormal hematopoietic cells and the adaptive immune response they instigate: activation of the innate immune system and autocrine or cell contact–mediated interactions with the stroma. Such alterations to the microenvironment can negatively impact normal hematopoiesis, providing a potential explanation for why cytopenias can occur even when MDS cells occupy only a fraction of the bone marrow.
Stromal changes need not always be a consequence of abnormal hematopoiesis, however. A recent study has demonstrated how a primary stromal cell abnormality can create dysplasia in normal hematoipoietic cells and even drive the clonal evolution of these cells into a malignant state.127 Selective deletion of the microRNA processing gene, dicer, in osteoprogenitor cells of mice causes a phenotype that closely resembles MDS. These mice have cytopenias, multilineage dysplasia, bone marrow hypercellularity, and increased apoptosis. After a long latency, three mice developed an AML-like disease characterized by clonal chromosomal abnormalities. Additional support for primary stromal changes in MDS comes from studies that have identified acquired chromosomal abnormalities in nonhematopoietic bone marrow cells from patients with MDS.128 These lesions were distinct from those seen in the MDS clones. Whether genetic abnormalities in mesenchymal cells provide a clonal advantage that allows them to outgrow normal stroma is not known.
THE CLINICAL ROLE OF GENETIC TESTING IN MDS
Metaphase karyotyping is the sole genetic analysis currently performed in routine clinical practice for the diagnosis of MDS. Fluorescent in situ hybridization with markers that detect common cytogenetic abnormalities need only be performed if a bone marrow aspirate cannot be obtained or if metaphase karyotyping is unsuccessful. MDS-specific fluorescent in situ hybridization panels are available commercially and are often performed in addition to metaphase karyotyping. In practice, however, these tests rarely add clinically relevant information beyond that obtained with a standard karyotype.129 More sensitive techniques are available in the research setting, including spectral karyotyping and single nucleotide polymorphism array genotyping. However, the clinical significance of small amplifications, deletions, or loss of heterozygosity that are missed by standard karyotyping has not been determined.
Assays for single-gene mutations are becoming increasingly available but are not yet clinically used in MDS. However, experience with these types of tests in other myeloid neoplasms suggests that they could become routine. In AML, for example, assays for FLT3 and NPM1 mutations have prognostic relevance and can inform the decision to proceed to transplant in first complete remission.130 In chronic myeloid leukemia and acute promyelocytic leukemia, quantitative detection of the translocation gene product is used to measure response to treatment and to monitor for early recurrence, and JAK2/MPL mutations are an important component of the diagnostic evaluation of MPN. In the future, the evaluation of individual genes for mutations will likely be useful for diagnosis, prediction of prognosis, and selection of therapy in patients with MDS.
DISCUSSION
The clinical heterogeneity of MDS is a reflection of the various pathogenic mechanisms responsible for their development. Single genetic lesions are unlikely to be the sole disease-causing abnormalities in these disorders. Instead, a combination of two or more mutations may be needed in cooperation with more global changes in epigenetic states and cellular environment. In a reductionist model of myeloid leukemogenesis, cooperation has been postulated between type I mutations that drive proliferation and survival and type II mutations that impair differentiation.131 This model provides a framework in which one can interpret the observation that in AML, type I and type II mutations are often mutually exclusive of other mutations within their own type. Some genes, however, have more complex effects on self-renewal, clonal expansion, and differentiation that may be difficult to assign to this binary classification. Given the emerging complexity of genetic abnormalities in MDS, it may be clearer to consider how each mutation contributes to the pathogenesis of these disorders and how they cooperate with other mutations and microenvironmental changes. Mutations of TP53, for example, are difficult to categorize as type I or type II abnormalities since they do not appear to be mutually exclusive with either type in AML and often occur in isolation in MDS.33 Instead, we can describe TP53 mutations as contributing to genomic instability and antiapoptotic features of advanced MDS. Similarly, TET2 mutations have not been found to be mutually exclusive of any other abnormality and thus are difficult to categorize. They do not severely alter differentiation in MPN and do not appear to generate a proliferative phenotype in MDS. This suggests that TET2 mutations drive other pathogenic mechanisms such as epigenetic dysregulation or a clonal advantage for the disease-initiating cell.
Detailed analyses of a large number of MDS patient samples are needed to define the range of potential mutations and the frequency with which they co-occur. The identification of mutually exclusive lesions suggests either that these are alternative means of activating a common pathogenic mechanism or that the combination of these mutations leads to a more advanced, non-MDS phenotype such as AML. Finding frequently co-occurring mutated genes or pathways implies the activation of synergistic pathologic mechanisms, each driving different steps in the transformation of a normal hematopoietic cell into a diseased clone.
In conclusion, a complete definition of the genetic abnormalities associated with MDS will do more than provide us with a molecular taxonomy of these disorders. Characteristic mutations will help diagnose MDS in patients with refractory cytopenias and ambiguous morphologic features. The ability to detect these mutations reliably in peripheral blood will be useful when clinicians have a patient they are reluctant to biopsy or as a tool to monitor response to therapy or disease progression. Associations with clinical features will identify mutations that are prognostic and potentially predictive of therapeutic responses. The application of unbiased whole-genome assays may uncover additional mutations that point to novel therapeutic targets, increasing the range of treatment options for patients with MDS. With a more complete catalog of pathogenic mutations and their clinical consequences, genetic tests may play a growing role in the diagnosis, prediction of prognosis, and selection of therapies for patients with MDS.
Note Added in Proof
EZH2 Mutations.
The recent discovery of mutations in the enhancer of zeste homolog 2 (EZH2) gene expands the list of MDS genes that are involved in the regulation of epigenetic state. EZH2 resides on the distal portion of chromosome 7q and encodes a histone methyltransferase that serves as the catalytic subunit of the polycomb repressive complex 2 (PRC2). Trimethylation of lysine 27 on histone 3 by PRC2 is an epigenetic mark that is associated with gene silencing. In solid tumors, increased EZH2 expression has been linked to more aggressive disease and worse outcomes. However, in hematologic malignancies, EZH2 is often mutated, which leads to a loss of catalytic activity.132–135 Loss of PRC2 function has been shown to increase hematopoietic stem cell activity and expansion, which may explain how EZH2 mutations promote the development of myeloid neoplasms.136 EZH2 mutations have been reported in 6% of MDS cases and over 10% of MDS/MPN overlap disorders. These abnormalities seem to confer a poor prognosis. Despite its chromosomal location, loss of EZH2 does not seem to be the sole driver for the deletion of chromosome 7q. Several patients with UPD at 7q did not have an apparent EZH2 mutation, which suggests that duplication of a distinct lesion in that region has yet to be found.132,133
Footnotes
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Rafael Bejar, Foundation Medicine (C) Stock Ownership: None Honoraria: None Research Funding: Benjamin L. Ebert, GlaxoSmithKline Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Manuscript writing: Rafael Bejar, Ross Levine, Benjamin L. Ebert
Final approval of manuscript: Rafael Bejar, Ross Levine,Benjamin L. Ebert
REFERENCES
- 1.Haase D, Germing U, Schanz J, et al. New insights into the prognostic impact of the karyotype in MDS and correlation with subtypes: Evidence from a core dataset of 2124 patients. Blood. 2007;110:4385–4395. doi: 10.1182/blood-2007-03-082404. [DOI] [PubMed] [Google Scholar]
- 2.Look AT: Molecular pathogenesis of MDS. Hematology Am Soc Hematol Educ Program. 2005:156–160. doi: 10.1182/asheducation-2005.1.156. [DOI] [PubMed] [Google Scholar]
- 3.Gondek LP, Haddad AS, O'Keefe CL, et al. Detection of cryptic chromosomal lesions including acquired segmental uniparental disomy in advanced and low-risk myelodysplastic syndromes. Exp Hematol. 2007;35:1728–1738. doi: 10.1016/j.exphem.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Bejar R, Ebert BL. The genetic basis of myelodysplastic syndromes. Hematol Oncol Clin North Am. 2010;24:295–315. doi: 10.1016/j.hoc.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Issa JP. Epigenetic changes in the myelodysplastic syndrome. Hematol Oncol Clin North Am. 2010;24:317–330. doi: 10.1016/j.hoc.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenberg P, Cox C, LeBeau MM, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89:2079–2088. [PubMed] [Google Scholar]
- 7.List A, Dewald G, Bennett J, et al. Lenalidomide in the myelodysplastic syndrome with chromosome 5q deletion. N Engl J Med. 2006;355:1456–1465. doi: 10.1056/NEJMoa061292. [DOI] [PubMed] [Google Scholar]
- 8.Chao MP, Seita J, Weissman IL. Establishment of a normal hematopoietic and leukemia stem cell hierarchy. Cold Spring Harb Symp Quant Biol. 2008;73:439–449. doi: 10.1101/sqb.2008.73.031. [DOI] [PubMed] [Google Scholar]
- 9.Nimer SD. MDS: A stem cell disorder—But what exactly is wrong with the primitive hematopoietic cells in this disease? Hematology Am Soc Hematol Educ Program. 2008:43–51. doi: 10.1182/asheducation-2008.1.43. [DOI] [PubMed] [Google Scholar]
- 10.Graubert TA, Payton MA, Shao J, et al. Integrated genomic analysis implicates haploinsufficiency of multiple chromosome 5q31.2 genes in de novo myelodysplastic syndromes pathogenesis. PLoS One. 2009;4:e4583. doi: 10.1371/journal.pone.0004583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gondek LP, Tiu R, O'Keefe CL, et al. Chromosomal lesions and uniparental disomy detected by SNP arrays in MDS, MDS/MPD, and MDS-derived AML. Blood. 2008;111:1534–1542. doi: 10.1182/blood-2007-05-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heinrichs S, Kulkarni RV, Bueso-Ramos CE, et al. Accurate detection of uniparental disomy and microdeletions by SNP array analysis in myelodysplastic syndromes with normal cytogenetics. Leukemia. 2009;23:1605–1613. doi: 10.1038/leu.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horrigan SK, Arbieva ZH, Xie HY, et al. Delineation of a minimal interval and identification of 9 candidates for a tumor suppressor gene in malignant myeloid disorders on 5q31. Blood. 2000;95:2372–2377. [PubMed] [Google Scholar]
- 14.Liu TX, Becker MW, Jelinek J, et al. Chromosome 5q deletion and epigenetic suppression of the gene encoding alpha-catenin (CTNNA1) in myeloid cell transformation. Nat Med. 2007;13:78–83. doi: 10.1038/nm1512. [DOI] [PubMed] [Google Scholar]
- 15.Boultwood J, Fidler C, Strickson AJ, et al. Narrowing and genomic annotation of the commonly deleted region of the 5q- syndrome. Blood. 2002;99:4638–4641. doi: 10.1182/blood.v99.12.4638. [DOI] [PubMed] [Google Scholar]
- 16.Van den Berghe H, Cassiman JJ, David G, et al. Distinct haematological disorder with deletion of long arm of no. 5 chromosome. Nature. 1974;251:437–438. doi: 10.1038/251437a0. [DOI] [PubMed] [Google Scholar]
- 17.Ebert BL, Pretz J, Bosco J, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narla A, Ebert BL. Ribosomopathies: Human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Starczynowski DT, Kuchenbauer F, Argiropoulos B, et al. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med. 2010;16:49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 20.Kumar M, Narla A, Nonami A, et al. Coordinate loss of a microRNA Mir 145 and a protein-coding gene RPS14 cooperate in the pathogenesis of 5q- syndrome. 51st American Society of Hematology Annual Meeting and Exposition; December 5-8, 2009; New Orleans, LA. abstr 947. [Google Scholar]
- 21.Joslin JM, Fernald AA, Tennant TR, et al. Haploinsufficiency of EGR1, a candidate gene in the del(5q), leads to the development of myeloid disorders. Blood. 2007;110:719–726. doi: 10.1182/blood-2007-01-068809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye Y, McDevitt MA, Guo M, et al. Progressive chromatin repression and promoter methylation of CTNNA1 associated with advanced myeloid malignancies. Cancer Res. 2009;69:8482–8490. doi: 10.1158/0008-5472.CAN-09-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lane SW, Sykes SM, Al-Shahrour F, et al. The Apc(min) mouse has altered hematopoietic stem cell function and provides a model for MPD/MDS. Blood. 2010;115:3489–3497. doi: 10.1182/blood-2009-11-251728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Fernald AA, Anastasi J, et al. Haploinsufficiency of Apc leads to ineffective hematopoiesis. Blood. 2010;115:3481–3488. doi: 10.1182/blood-2009-11-251835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grisendi S, Bernardi R, Rossi M, et al. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437:147–153. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- 26.Komrokji RS, List AF. Lenalidomide for treatment of myelodysplastic syndromes: Current status and future directions. Hematol Oncol Clin North Am. 2010;24:377–388. doi: 10.1016/j.hoc.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 27.Wei S, Chen X, Rocha K, et al. A critical role for phosphatase haplodeficiency in the selective suppression of deletion 5q MDS by lenalidomide. Proc Natl Acad Sci U S A. 2009;106:12974–12979. doi: 10.1073/pnas.0811267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christiansen DH, Andersen MK, Pedersen-Bjergaard J. Mutations of AML1 are common in therapy-related myelodysplasia following therapy with alkylating agents and are significantly associated with deletion or loss of chromosome arm 7q and with subsequent leukemic transformation. Blood. 2004;104:1474–1481. doi: 10.1182/blood-2004-02-0754. [DOI] [PubMed] [Google Scholar]
- 29.Le Beau MM, Espinosa R, 3rd, Davis EM, et al. Cytogenetic and molecular delineation of a region of chromosome 7 commonly deleted in malignant myeloid diseases. Blood. 1996;88:1930–1935. [PubMed] [Google Scholar]
- 30.Döhner K, Brown J, Hehmann U, et al. Molecular cytogenetic characterization of a critical region in bands 7q35-q36 commonly deleted in malignant myeloid disorders. Blood. 1998;92:4031–4035. [PubMed] [Google Scholar]
- 31.Asou H, Matsui H, Ozaki Y, et al. Identification of a common microdeletion cluster in 7q21.3 subband among patients with myeloid leukemia and myelodysplastic syndrome. Biochem Biophys Res Commun. 2009;383:245–251. doi: 10.1016/j.bbrc.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 32.Wong JC, Zhang Y, Lieuw KH, et al. Use of chromosome engineering to model a segmental deletion of chromosome band 7q22 found in myeloid malignancies. Blood. 2010;115:4524–4532. doi: 10.1182/blood-2009-07-232504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pedersen-Bjergaard J, Christiansen DH, Desta F, et al. Alternative genetic pathways and cooperating genetic abnormalities in the pathogenesis of therapy-related myelodysplasia and acute myeloid leukemia. Leukemia. 2006;20:1943–1949. doi: 10.1038/sj.leu.2404381. [DOI] [PubMed] [Google Scholar]
- 34.Sloand EM, Wu CO, Greenberg P, et al. Factors affecting response and survival in patients with myelodysplasia treated with immunosuppressive therapy. J Clin Oncol. 2008;26:2505–2511. doi: 10.1200/JCO.2007.11.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim ZY, Killick S, Germing U, et al. Low IPSS score and bone marrow hypocellularity in MDS patients predict hematological responses to antithymocyte globulin. Leukemia. 2007;21:1436–1441. doi: 10.1038/sj.leu.2404747. [DOI] [PubMed] [Google Scholar]
- 36.Wiktor A, Rybicki BA, Piao ZS, et al. Clinical significance of Y chromosome loss in hematologic disease. Genes Chromosomes Cancer. 2000;27:11–16. [PubMed] [Google Scholar]
- 37.Wong AK, Fang B, Zhang L, et al. Loss of the Y chromosome: An age-related or clonal phenomenon in acute myelogenous leukemia/myelodysplastic syndrome? Arch Pathol Lab Med. 2008;132:1329–1332. doi: 10.5858/2008-132-1329-LOTYCA. [DOI] [PubMed] [Google Scholar]
- 38.Skoda R. The genetic basis of myeloproliferative disorders. Hematology Am Soc Hematol Educ Program 1-10. 2007 doi: 10.1182/asheducation-2007.1.1. [DOI] [PubMed] [Google Scholar]
- 39.Bench AJ, Nacheva EP, Hood TL, et al. Chromosome 20 deletions in myeloid malignancies: Reduction of the common deleted region, generation of a PAC/BAC contig and identification of candidate genes—UK Cancer Cytogenetics Group (UKCCG) Oncogene. 2000;19:3902–3913. doi: 10.1038/sj.onc.1203728. [DOI] [PubMed] [Google Scholar]
- 40.Wang PW, Eisenbart JD, Espinosa R, 3rd, et al. Refinement of the smallest commonly deleted segment of chromosome 20 in malignant myeloid diseases and development of a PAC-based physical and transcription map. Genomics. 2000;67:28–39. doi: 10.1006/geno.2000.6215. [DOI] [PubMed] [Google Scholar]
- 41.Liu YC, Ito Y, Hsiao HH, et al. Risk factor analysis in myelodysplastic syndrome patients with del(20q): Prognosis revisited. Cancer Genet Cytogenet. 2006;171:9–16. doi: 10.1016/j.cancergencyto.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 42.Schanz J, Tuechler H, Sole F, et al. Cytogenetic risk features in MDS-update and present state. 51st American Society of Hematology Annual Meeting and Exposition; December 5-8, 2009; New Orleans, LA. abstr 2772. [Google Scholar]
- 43.Poppe B, Dastugue N, Vandesompele J, et al. EVI1 is consistently expressed as principal transcript in common and rare recurrent 3q26 rearrangements. Genes Chromosomes Cancer. 2006;45:349–356. doi: 10.1002/gcc.20295. [DOI] [PubMed] [Google Scholar]
- 44.Soderholm J, Kobayashi H, Mathieu C, et al. The leukemia-associated gene MDS1/EVI1 is a new type of GATA-binding transactivator. Leukemia. 1997;11:352–358. doi: 10.1038/sj.leu.2400584. [DOI] [PubMed] [Google Scholar]
- 45.Laricchia-Robbio L, Fazzina R, Li D, et al. Point mutations in two EVI1 Zn fingers abolish EVI1-GATA1 interaction and allow erythroid differentiation of murine bone marrow cells. Mol Cell Biol. 2006;26:7658–7666. doi: 10.1128/MCB.00363-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laricchia-Robbio L, Premanand K, Rinaldi CR, et al. EVI1 impairs myelopoiesis by deregulation of PU.1 function. Cancer Res. 2009;69:1633–1642. doi: 10.1158/0008-5472.CAN-08-2562. [DOI] [PubMed] [Google Scholar]
- 47.Senyuk V, Sinha KK, Li D, et al. Repression of RUNX1 activity by EVI1: A new role of EVI1 in leukemogenesis. Cancer Res. 2007;67:5658–5666. doi: 10.1158/0008-5472.CAN-06-3962. [DOI] [PubMed] [Google Scholar]
- 48.Buonamici S, Li D, Chi Y, et al. EVI1 induces myelodysplastic syndrome in mice. J Clin Invest. 2004;114:713–719. doi: 10.1172/JCI21716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laricchia-Robbio L, Nucifora G. Significant increase of self-renewal in hematopoietic cells after forced expression of EVI1. Blood Cells Mol Dis. 2008;40:141–147. doi: 10.1016/j.bcmd.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morishita K, Parker DS, Mucenski ML, et al. Retroviral activation of a novel gene encoding a zinc finger protein in IL-3-dependent myeloid leukemia cell lines. Cell. 1988;54:831–840. doi: 10.1016/s0092-8674(88)91175-0. [DOI] [PubMed] [Google Scholar]
- 51.Du Y, Jenkins NA, Copeland NG. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005;106:3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stein S, Ott MG, Schultze-Strasser S, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat Med. 2010;16:198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- 53.Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, van Putten WL, et al. High EVI1 expression predicts poor survival in acute myeloid leukemia: A study of 319 de novo AML patients. Blood. 2003;101:837–845. doi: 10.1182/blood-2002-05-1459. [DOI] [PubMed] [Google Scholar]
- 54.Gröschel S, Lugthart S, Schlenk RF, et al. High EVI1 expression predicts outcome in younger adult patients with acute myeloid leukemia and is associated with distinct cytogenetic abnormalities. J Clin Oncol. 2010;28:2101–2107. doi: 10.1200/JCO.2009.26.0646. [DOI] [PubMed] [Google Scholar]
- 55.Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: Rationale and important changes. Blood. 2009;114:937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 56.Delhommeau F, Dupont S, Della Valle V, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 57.Kosmider O, Gelsi-Boyer V, Ciudad M, et al. TET2 gene mutation is a frequent and adverse event in chronic myelomonocytic leukemia. Haematologica. 2009;94:1676–1681. doi: 10.3324/haematol.2009.011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langemeijer SM, Kuiper RP, Berends M, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 59.Mullighan CG. TET2 mutations in myelodysplasia and myeloid malignancies. Nat Genet. 2009;41:766–767. doi: 10.1038/ng0709-766. [DOI] [PubMed] [Google Scholar]
- 60.Lorsbach RB, Moore J, Mathew S, et al. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia. 2003;17:637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 61.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61a.Ko M, Huang Y, Jankowska AM, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. doi: 10.1038/nature09586. epub ahead of print on November 7, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schaub FX, Looser R, Li S, et al. Clonal analysis of TET2 and JAK2 mutations suggests that TET2 can be a late event in the progression of myeloproliferative neoplasms. Blood. 2010;115:2003–2007. doi: 10.1182/blood-2009-09-245381. [DOI] [PubMed] [Google Scholar]
- 63.Kosmider O, Gelsi-Boyer V, Cheok M, et al. TET2 mutation is an independent favorable prognostic factor in myelodysplastic syndromes (MDSs) Blood. 2009;114:3285–3291. doi: 10.1182/blood-2009-04-215814. [DOI] [PubMed] [Google Scholar]
- 63a.Smith AE, Mohamedali AM, Kulasekararaj A, et al. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood. 2010;116:3923–3932. doi: 10.1182/blood-2010-03-274704. [DOI] [PubMed] [Google Scholar]
- 64.Abdel-Wahab O, Mullally A, Hedvat C, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114:144–147. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gelsi-Boyer V, Trouplin V, Adélaïde J, et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol. 2009;145:788–800. doi: 10.1111/j.1365-2141.2009.07697.x. [DOI] [PubMed] [Google Scholar]
- 66.Boultwood J, Perry J, Pellagatti A, et al. Frequent mutation of the polycomb-associated gene ASXL1 in the myelodysplastic syndromes and in acute myeloid leukemia. Leukemia. 2010;24:1062–1065. doi: 10.1038/leu.2010.20. [DOI] [PubMed] [Google Scholar]
- 67.Carbuccia N, Murati A, Trouplin V, et al. Mutations of ASXL1 gene in myeloproliferative neoplasms. Leukemia. 2009;23:2183–2186. doi: 10.1038/leu.2009.141. [DOI] [PubMed] [Google Scholar]
- 68.Fisher CL, Randazzo F, Humphries RK, et al. Characterization of Asxl1, a murine homolog of additional sex combs, and analysis of the Asx-like gene family. Gene. 2006;369:109–118. doi: 10.1016/j.gene.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 69.Cho YS, Kim EJ, Park UH, et al. Additional sex comb-like 1 (ASXL1), in cooperation with SRC-1, acts as a ligand-dependent coactivator for retinoic acid receptor. J Biol Chem. 2006;281:17588–17598. doi: 10.1074/jbc.M512616200. [DOI] [PubMed] [Google Scholar]
- 70.Lee SW, Cho YS, Na JM, et al. ASXL1 represses retinoic acid receptor-mediated transcription through associating with HP1 and LSD1. J Biol Chem. 2009;285:18–29. doi: 10.1074/jbc.M109.065862. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Carbuccia N, Trouplin V, Gelsi-Boyer V, et al. Mutual exclusion of ASXL1 and NPM1 mutations in a series of acute myeloid leukemias. Leukemia. 2010;24:469–473. doi: 10.1038/leu.2009.218. [DOI] [PubMed] [Google Scholar]
- 72.Fisher CL, Pineault N, Brookes C, et al. Loss-of-function additional sex combs like 1 mutations disrupt hematopoiesis but do not cause severe myelodysplasia or leukemia. Blood. 2010;115:38–46. doi: 10.1182/blood-2009-07-230698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen CY, Lin LI, Tang JL, et al. RUNX1 gene mutation in primary myelodysplastic syndrome: The mutation can be detected early at diagnosis or acquired during disease progression and is associated with poor outcome. Br J Haematol. 2007;139:405–414. doi: 10.1111/j.1365-2141.2007.06811.x. [DOI] [PubMed] [Google Scholar]
- 74.Steensma DP, Gibbons RJ, Mesa RA, et al. Somatic point mutations in RUNX1/CBFA2/AML1 are common in high-risk myelodysplastic syndrome, but not in myelofibrosis with myeloid metaplasia. Eur J Haematol. 2005;74:47–53. doi: 10.1111/j.1600-0609.2004.00363.x. [DOI] [PubMed] [Google Scholar]
- 75.Tang JL, Hou HA, Chen CY, et al. AML1/RUNX1 mutations in 470 adult patients with de novo acute myeloid leukemia: Prognostic implication and interaction with other gene alterations. Blood. 2009;114:5352–5361. doi: 10.1182/blood-2009-05-223784. [DOI] [PubMed] [Google Scholar]
- 76.Harada H, Harada Y, Niimi H, et al. High incidence of somatic mutations in the AML1/RUNX1 gene in myelodysplastic syndrome and low blast percentage myeloid leukemia with myelodysplasia. Blood. 2004;103:2316–2324. doi: 10.1182/blood-2003-09-3074. [DOI] [PubMed] [Google Scholar]
- 77.Matheny CJ, Speck ME, Cushing PR, et al. Disease mutations in RUNX1 and RUNX2 create nonfunctional, dominant-negative, or hypomorphic alleles. EMBO J. 2007;26:1163–1175. doi: 10.1038/sj.emboj.7601568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Harada Y, Harada H. Molecular pathways mediating MDS/AML with focus on AML1/RUNX1 point mutations. J Cell Physiol. 2009;220:16–20. doi: 10.1002/jcp.21769. [DOI] [PubMed] [Google Scholar]
- 79.Ho CY, Otterud B, Legare RD, et al. Linkage of a familial platelet disorder with a propensity to develop myeloid malignancies to human chromosome 21q22.1-22.2. Blood. 1996;87:5218–5224. [PubMed] [Google Scholar]
- 80.Owen C. Insights into familial platelet disorder with propensity to myeloid malignancy (FPD/AML) Leuk Res. 2010;34:141–142. doi: 10.1016/j.leukres.2009.07.037. [DOI] [PubMed] [Google Scholar]
- 81.Preudhomme C, Renneville A, Bourdon V, et al. High frequency of RUNX1 biallelic alteration in acute myeloid leukemia secondary to familial platelet disorder. Blood. 2009;113:5583–5587. doi: 10.1182/blood-2008-07-168260. [DOI] [PubMed] [Google Scholar]
- 82.Pedersen-Bjergaard J, Andersen MK, Andersen MT, et al. Genetics of therapy-related myelodysplasia and acute myeloid leukemia. Leukemia. 2008;22:240–248. doi: 10.1038/sj.leu.2405078. [DOI] [PubMed] [Google Scholar]
- 83.Niimi H, Harada H, Harada Y, et al. Hyperactivation of the RAS signaling pathway in myelodysplastic syndrome with AML1/RUNX1 point mutations. Leukemia. 2006;20:635–644. doi: 10.1038/sj.leu.2404136. [DOI] [PubMed] [Google Scholar]
- 84.Okuda T, van Deursen J, Hiebert SW, et al. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 85.Wang Q, Stacy T, Binder M, et al. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci U S A. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dowdy CR, Xie R, Frederick D, et al. Definitive hematopoiesis requires Runx1 C-terminal-mediated subnuclear targeting and transactivation. Hum Mol Genet. 2010;19:1048–1057. doi: 10.1093/hmg/ddp568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Growney JD, Shigematsu H, Li Z, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Watanabe-Okochi N, Kitaura J, Ono R, et al. AML1 mutations induced MDS and MDS/AML in a mouse BMT model. Blood. 2008;111:4297–4308. doi: 10.1182/blood-2007-01-068346. [DOI] [PubMed] [Google Scholar]
- 89.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Abdel-Wahab O, Manshouri T, Patel J, et al. Genetic analysis of transforming events that convert chronic myeloproliferative neoplasms to leukemias. Cancer Res. 2010;70:447–452. doi: 10.1158/0008-5472.CAN-09-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Paschka P, Schlenk RF, Gaidzik VI, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 93.Tefferi A, Lasho TL, Abdel-Wahab O, et al. IDH1 and IDH2 mutation studies in 1473 patients with chronic-, fibrotic- or blast-phase essential thrombocythemia, polycythemia vera or myelofibrosis. Leukemia. 2010;24:1302–1309. doi: 10.1038/leu.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thol F, Weissinger EM, Krauter J, et al. IDH1 mutations in patients with myelodysplastic syndromes are associated with an unfavorable prognosis. Haematologica. 2010;95:1668–1674. doi: 10.3324/haematol.2010.025494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kosmider O, Gelsi-Boyer V, Slama L, et al. Mutations of IDH1 and IDH2 genes in early and accelerated phases of myelodysplastic syndromes and MDS/myeloproliferative neoplasms. Leukemia. 2010;24:1094–1096. doi: 10.1038/leu.2010.52. [DOI] [PubMed] [Google Scholar]
- 96.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bacher U, Haferlach T, Kern W, et al. A comparative study of molecular mutations in 381 patients with myelodysplastic syndrome and in 4130 patients with acute myeloid leukemia. Haematologica. 2007;92:744–752. doi: 10.3324/haematol.10869. [DOI] [PubMed] [Google Scholar]
- 99.Christiansen DH, Andersen MK, Desta F, et al. Mutations of genes in the receptor tyrosine kinase (RTK)/RAS-BRAF signal transduction pathway in therapy-related myelodysplasia and acute myeloid leukemia. Leukemia. 2005;19:2232–2240. doi: 10.1038/sj.leu.2404009. [DOI] [PubMed] [Google Scholar]
- 100.Constantinidou M, Chalevelakis G, Economopoulos T, et al. Codon 12 ras mutations in patients with myelodysplastic syndrome: Incidence and prognostic value. Ann Hematol. 1997;74:11–14. doi: 10.1007/s002770050248. [DOI] [PubMed] [Google Scholar]
- 101.Paquette RL, Landaw EM, Pierre RV, et al. N-ras mutations are associated with poor prognosis and increased risk of leukemia in myelodysplastic syndrome. Blood. 1993;82:590–599. [PubMed] [Google Scholar]
- 102.Loh ML, Martinelli S, Cordeddu V, et al. Acquired PTPN11 mutations occur rarely in adult patients with myelodysplastic syndromes and chronic myelomonocytic leukemia. Leuk Res. 2005;29:459–462. doi: 10.1016/j.leukres.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 103.Dunbar AJ, Gondek LP, O'Keefe CL, et al. 250K single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutations, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res. 2008;68:10349–10357. doi: 10.1158/0008-5472.CAN-08-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sanada M, Suzuki T, Shih LY, et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature. 2009;460:904–908. doi: 10.1038/nature08240. [DOI] [PubMed] [Google Scholar]
- 105.Sargin B, Choudhary C, Crosetto N, et al. Flt3-dependent transformation by inactivating c-Cbl mutations in AML. Blood. 2007;110:1004–1012. doi: 10.1182/blood-2007-01-066076. [DOI] [PubMed] [Google Scholar]
- 106.Saur SJ, Sangkhae V, Geddis AE, et al. Ubiquitination and degradation of the thrombopoietin receptor c-Mpl. Blood. 2010;115:1254–1263. doi: 10.1182/blood-2009-06-227033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rathinam C, Thien CB, Langdon WY, et al. The E3 ubiquitin ligase c-Cbl restricts development and functions of hematopoietic stem cells. Genes Dev. 2008;22:992–997. doi: 10.1101/gad.1651408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Loh ML, Sakai DS, Flotho C, et al. Mutations in CBL occur frequently in juvenile myelomonocytic leukemia. Blood. 2009;114:1859–1863. doi: 10.1182/blood-2009-01-198416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Reindl C, Quentmeier H, Petropoulos K, et al. CBL exon 8/9 mutants activate the FLT3 pathway and cluster in core binding factor/11q deletion acute myeloid leukemia/myelodysplastic syndrome subtypes. Clin Cancer Res. 2009;15:2238–2247. doi: 10.1158/1078-0432.CCR-08-1325. [DOI] [PubMed] [Google Scholar]
- 110.Abbas S, Rotmans G, Löwenberg B, et al. Exon 8 splice site mutations in the gene encoding the E3-ligase CBL are associated with core binding factor acute myeloid leukemias. Haematologica. 2008;93:1595–1597. doi: 10.3324/haematol.13187. [DOI] [PubMed] [Google Scholar]
- 111.Steensma DP, Tefferi A. JAK2 V617F and ringed sideroblasts: Not necessarily RARS-T. Blood. 2008;111:1748. doi: 10.1182/blood-2007-11-121608. [DOI] [PubMed] [Google Scholar]
- 112.Boissinot M, Garand R, Hamidou M, et al. The JAK2-V617F mutation and essential thrombocythemia features in a subset of patients with refractory anemia with ring sideroblasts (RARS) Blood. 2006;108:1781–1782. doi: 10.1182/blood-2006-03-008227. [DOI] [PubMed] [Google Scholar]
- 113.Szpurka H, Tiu R, Murugesan G, et al. Refractory anemia with ringed sideroblasts associated with marked thrombocytosis (RARS-T), another myeloproliferative condition characterized by JAK2 V617F mutation. Blood. 2006;108:2173–2181. doi: 10.1182/blood-2006-02-005751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wardrop D, Steensma DP. Is refractory anaemia with ring sideroblasts and thrombocytosis (RARS-T) a necessary or useful diagnostic category? Br J Haematol. 2009;144:809–817. doi: 10.1111/j.1365-2141.2008.07526.x. [DOI] [PubMed] [Google Scholar]
- 115.Malcovati L, Della Porta MG, Pietra D, et al. Molecular and clinical features of refractory anemia with ringed sideroblasts associated with marked thrombocytosis. Blood. 2009;114:3538–3545. doi: 10.1182/blood-2009-05-222331. [DOI] [PubMed] [Google Scholar]
- 116.Barlow JL, Drynan LF, Hewett DR, et al. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q- syndrome. Nat Med. 2010;16:59–66. doi: 10.1038/nm.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Christiansen DH, Andersen MK, Pedersen-Bjergaard J. Mutations with loss of heterozygosity of p53 are common in therapy-related myelodysplasia and acute myeloid leukemia after exposure to alkylating agents and significantly associated with deletion or loss of 5q, a complex karyotype, and a poor prognosis. J Clin Oncol. 2001;19:1405–1413. doi: 10.1200/JCO.2001.19.5.1405. [DOI] [PubMed] [Google Scholar]
- 118.Kaneko H, Misawa S, Horiike S, et al. TP53 mutations emerge at early phase of myelodysplastic syndrome and are associated with complex chromosomal abnormalities. Blood. 1995;85:2189–2193. [PubMed] [Google Scholar]
- 119.Wattel E, Preudhomme C, Hecquet B, et al. p53 mutations are associated with resistance to chemotherapy and short survival in hematologic malignancies. Blood. 1994;84:3148–3157. [PubMed] [Google Scholar]
- 120.Horiike S, Kita-Sasai Y, Nakao M, et al. Configuration of the TP53 gene as an independent prognostic parameter of myelodysplastic syndrome. Leuk Lymphoma. 2003;44:915–922. doi: 10.1080/1042819031000067620. [DOI] [PubMed] [Google Scholar]
- 121.Kita-Sasai Y, Horiike S, Misawa S, et al. International prognostic scoring system and TP53 mutations are independent prognostic indicators for patients with myelodysplastic syndrome. Br J Haematol. 2001;115:309–312. doi: 10.1046/j.1365-2141.2001.03073.x. [DOI] [PubMed] [Google Scholar]
- 122.Jädersten M, Saft L, Pellagatti A, et al. Clonal heterogeneity in the 5q- syndrome: p53 expressing progenitors prevail during lenalidomide treatment and expand at disease progression. Haematologica. 2009;94:1762–1766. doi: 10.3324/haematol.2009.011528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shen L, Kantarjian H, Guo Y, et al. DNA methylation predicts survival and response to therapy in patients with myelodysplastic syndromes. J Clin Oncol. 2010;28:605–613. doi: 10.1200/JCO.2009.23.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jiang Y, Dunbar A, Gondek LP, et al. Aberrant DNA methylation is a dominant mechanism in MDS progression to AML. Blood. 2009;113:1315–1325. doi: 10.1182/blood-2008-06-163246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124a.Fenaux P, Mufti GJ, Hellstrom-Lindberg E, et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: A randomised, open-label, phase III study. Lancet Oncol. 2009;10:223–232. doi: 10.1016/S1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bellamy WT, Richter L, Sirjani D, et al. Vascular endothelial cell growth factor is an autocrine promoter of abnormal localized immature myeloid precursors and leukemia progenitor formation in myelodysplastic syndromes. Blood. 2001;97:1427–1434. doi: 10.1182/blood.v97.5.1427. [DOI] [PubMed] [Google Scholar]
- 126.Tsimberidou AM, Estey E, Wen S, et al. The prognostic significance of cytokine levels in newly diagnosed acute myeloid leukemia and high-risk myelodysplastic syndromes. Cancer. 2008;113:1605–1613. doi: 10.1002/cncr.23785. [DOI] [PubMed] [Google Scholar]
- 127.Raaijmakers MH, Mukherjee S, Guo S, et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464:852–857. doi: 10.1038/nature08851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lopez-Villar O, Garcia JL, Sanchez-Guijo FM, et al. Both expanded and uncultured mesenchymal stem cells from MDS patients are genomically abnormal, showing a specific genetic profile for the 5q- syndrome. Leukemia. 2009;23:664–672. doi: 10.1038/leu.2008.361. [DOI] [PubMed] [Google Scholar]
- 129.Pitchford CW, Hettinga AC, Reichard KK. Fluorescence in situ hybridization testing for -5/5q, -7/7q, +8, and del(20q) in primary myelodysplastic syndrome correlates with conventional cytogenetics in the setting of an adequate study. Am J Clin Pathol. 2010;133:260–264. doi: 10.1309/AJCPZ4JL5ZMRPFTD. [DOI] [PubMed] [Google Scholar]
- 130.Schlenk RF, Döhner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 131.Kelly LM, Gilliland DG. Genetics of myeloid leukemias. Annu Rev Genomics Hum Genet. 2002;3:179–198. doi: 10.1146/annurev.genom.3.032802.115046. [DOI] [PubMed] [Google Scholar]
- 132.Ernst T, Chase AJ, Score J, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 133.Makishima H, Jankowska AM, Tiu RV, et al. Novel homo- and hemizygous mutations in EZH2 in myeloid malignancies. Leukemia. 2010;24:1799–1804. doi: 10.1038/leu.2010.167. [DOI] [PubMed] [Google Scholar]
- 134.Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nikoloski G, Langemeijer SM, Kuiper RP, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 136.Majewski IJ, Ritchie ME, Phipson B, et al. Opposing roles of polycomb repressive complexes in hematopoietic stem and progenitor cells. Blood. 2010;116:731–739. doi: 10.1182/blood-2009-12-260760. [DOI] [PubMed] [Google Scholar]