Abstract
Sodium caprylate was added to a pharmaceutical-grade human serum albumin (HSA) to stabilize the product. In this study we have aimed to establish how caprylate ligand protects HSA from thermal degradation. The fatty acid stabilizer was first removed from commercial HSA by charcoal treatment. Cleaned HSA was made to 10% w/v in pH 7.4 buffered solutions and doped with sodium caprylate in serial concentrations up to 0.16 mmol/g-protein. These solutions as well as a commercial HSA, human serum, and enriched-albumin fraction were subjected to differential scanning calorimetry (DSC) within the temperature range of 37–90°C at a 5.0°C/min scanning rate. The globular size of the cleaned HSA solutions was measured by dynamic light scattering. The denaturing temperatures for albumin with sodium caprylate and a commercial one were significantly higher than for albumin only. It was found that the protein globules of cleaned HSA were not as stable as that of the native one due to aggregation, and the caprylate ion may reduce the aggregation by enlarging the globules’ electrical double layer. A rational approximation of the Lumry-Eyring protein denaturation model was used to treat DSC denaturing endotherms. The system turned from irreversible dominant Scheme: 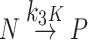 to reversible dominant Scheme:
to reversible dominant Scheme: 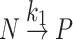 with the increase in caprylate concentration from null to ~0.08 mmol/g-protein. It was postulated that the caprylate ligand may decrease the rate of reversible unfolding as it binds to the IIIA domain which is prone to reversible unfolding/refolding and causes further difficulty for irreversible denaturation which, in turn, HSA can be stabilized.
with the increase in caprylate concentration from null to ~0.08 mmol/g-protein. It was postulated that the caprylate ligand may decrease the rate of reversible unfolding as it binds to the IIIA domain which is prone to reversible unfolding/refolding and causes further difficulty for irreversible denaturation which, in turn, HSA can be stabilized.
KEY WORDS: differential scanning calorimetry, human serum albumin, Lumry-Eyring model, protein denaturation, sodium caprylate
INTRODUCTION
Human serum albumin (HSA) is a monomeric multi-domain protein that possesses an extraordinary capacity for binding. It plays an important role in keeping and transporting endogenous substances, metabolites, and drugs throughout the human circulatory system. The protein can affect the pharmacokinetics of several drugs, has been shown to reduce the detrimental effects of various toxins, and can modify some metabolic processes. It also provides antioxidant activity and has some of the enzymatic activities of human plasma (1). Clinically, HSA is used to treat a variety of diseases such as hypovolemia, shock, burns, hemorrhage, and trauma in critically ill patients (2). The pharmaceutical-grade HSA is prepared by a cold alcohol fractionation method from pooled human serum, and thermally treated at 60°C for a suitably long period of time to terminate the activities of infectious viruses. However, during the heat sterilization process HSA may loose some of its activities. Fatty acids such as sodium caprylate have been added to stabilize the HSA against heat treatment (for example: 3). It has been reported that sodium caprylate plays a significant role in the resistance against heat while sodium acetyltryptophanate, a co-stabilizer, may protect the protein in HSA from oxidative stress (4). Arakawa and Kita (5) hypothesized that the caprylate ion may diminish the irreversible aggregation via protein binding but any definitive mechanism is still unclear. The aim of the study was to provide some insight on the role of sodium caprylate in stabilizing the HSA. By using and testing a simple model for protein denaturation under thermal stress, it may be possible to understand how the caprylate ion stabilizes HSA.
Lumry-Eyring Model for Serum Albumin Denaturation
In 1954, Lumry and Eyring modeled a two-step mechanism that explained the conformational changes that occurred in proteins during heat denaturation (6). The model is simple and straightforward and has been very popular for studies on the denaturation of several proteins and the analysis of the model has been extensively described (7). During a temperature increment, the denaturation of proteins may occur in two steps including: (a) a reversible unfolding/refolding of the native protein, and (b) an irreversible denaturation of the unfolded protein. This two-step model may be described by Scheme 1. Where N, U, and P are native, unfolding intermediate, and the final state of the protein respectively . It is assumed that all the sub-processes (arrows labeled by the rate parameters in Scheme 1) obey first-order kinetics having the temperature-dependent rate parameters k1 and k2, and k3 for reversible unfolding, refolding, and irreversible denaturation, respectively. Chemical equilibrium between N and U is always established and shows equilibrium parameters namely; K = k1/k2. The K value is temperature-dependent. Using these rate equations, Sanchez-Ruiz (7) derived and described the mole fractions of the native protein: xN, unfolded intermediate: xU, and final stage denatured protein: xP as being exponential functions of the absolute temperature, T:
 |
1 |
 |
2 |
 |
3 |
Scheme 1.

ᅟ
The exponent of each equation was the integration of a temperature-dependent parameter from a low temperature T0 where xP was negligible to a temperature T, and v was the rate of temperature increment.
By applying the Lumry-Eyring model to thermal denaturation of native HSA, Pico (8) found that the irreversible alteration was so slow that it was considered to be a rate limiting step, i.e., k3 < < k2. Also K was far less than 1 and this showed that the protein states N and P were significantly populated. Thus, Scheme 1 can be modified as Scheme 2.
Scheme 2.

ᅟ
Furthermore, notice that there was an additional case where k3 >> k2 while the assumption of K << 1 still held (7). In this situation, the rate determining step was no longer the irreversible denaturation but a reversible unfolding, i.e., step (a) of Scheme 1. Most of the protein molecules were converted to the final state and the amount of U was very low. This overall transition can therefore be described as Scheme 3.
Scheme 3.

ᅟ
The function describing the mole fraction of HSA, e.g., Eq. 1 can be modified as:
 |
4 |
where, kapp was an apparent first order rate parameter, i.e., kapp = Kk3 or kapp = k1 for Scheme 2, or Scheme 3, respectively.
According to the Arrhenius law, kapp can be described as an exponential function:
 |
5 |
where, Eapp is the apparent activation energy, Tk is the temperature at which k is equal to 1 min−1 corresponding to the Arrhenius constant (7). Introducing Eq. 5 to 4 yields:
 |
6 |
The integration of the exponent of Eq. 6 does not have an analytical solution and faces the difficulty in fitting the experimental data to obtain Eapp. Fortunately, it can be simplified by imposing a rational approximation. For T < 370 K and Eapp > 40 kcal/mol, as Khrapunov and Brenowitz (9) had suggested a simplified approximated exponential integral that had reasonably low errors;
 |
7 |
which, in turn, converts Eq. 6 to:
 |
8 |
Differential scanning calorimetry (DSC) has been a technique to gain information on the thermal energetics associated with an event that occurs in a system that was stimulated by a temperature change and the DSC directly measured the heat-flow through the system with a programmable temperature variation. Although DSC measures the irreversible transition, it is possible to extend its applicability to the Lumry-Eyring model where the equilibrium between native and unfolded forms of a protein is established. The reasonable assumption is that the step responsible for irreversible denaturation has far lower enthalpy than the cooperative reversible unfolding (7). And, it is because all protein molecules will eventually be in the final state upon temperature increments, the enthalpy of the final state will be equal to the calorimetric enthalpy of the DSC transition. Taking N as the reference state, the enthalpy of the transition at a temperature T; 〈ΔH〉T is directly proportional to the mole fractions of U and P.
 |
9 |
where, ΔH is the total enthalpy of the denaturing transition that is directly determined from a DSC endotherm. And, xN + xU + xP = 1. Therefore it can be deduced that:
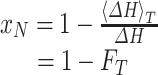 |
10 |
where, FT is the fraction of the endotherm at temperature T, i.e., the ratio between the amount of heat at T and the total heat of transition of the entire DSC endotherm. Equations 8 and 10 allow one to estimate the Eapp from the DSC data. As the Eapp could characterize the denaturing scheme of the HSA, e.g., by either Scheme 2 or 3, through tracking the changes of Eapp due to ligand interactions that may lead to a better understanding on how the caprylate ion stabilizes the protein.
MATERIALS AND METHODS
Albumin (Human) USP was obtained as a 20% solution form (Albutein®, Grifols International, Barcelona, Spain). Its content was confirmed to meet USP specification by an in-house reversed phase HPLC method (data not showed). Pooled human serum in the frozen state was a gift from the research project entitled: “Detection of in vivo quantitation of circulating tumor cells in patients with breast and ovarian cancers by molecular cytogenetics”. The project had been approved by the Ethics Committee of the Faculty of Medicine, Prince of Songkla University. It was obtained from a group of healthy volunteers enrolled as the control subjects in the above mentioned project. Sodium caprylate (≥99%: capillary GC; analytical standard) was from Sigma-Aldrich, Saint Louis, MO. Other chemicals were AR grade or equivalent. The substances were used without further purification.
The Preparation of Albumin-Enriched Fraction
The albumin-enriched fraction of human serum was prepared from pooled human serum using the method modified from Gundry et al. (10). Briefly, after thawing, human serum was mixed with 42% ethanol/100 mM sodium chloride in equal volume and incubated at 4°C for 1 h. The mixture was then centrifuged at 16,000×g for 45 min. The clear supernatant which was the albumin-enriched fraction was collected. This fraction was stored at 4°C overnight prior to the DSC experiment.
Removal of the Stabilizers from Commercial HSA by Charcoal Treatment
The method for removal of the stabilizers from commercial HSA was taken from Chen (11). Briefly, 10 mL of human albumin USP 20% was mixed with 10 mL of distilled water. One gram of activated charcoal was added to the solution and it was acidified to pH 3.0 using 0.2 N HCl. The mixture was then cooled to ~2–4°C and allowed to mix thoroughly for 1 h. Activated charcoal was removed by centrifugation at 20,200×g for 20 min under the same cool conditions. The supernatant was brought to pH 7.0 by the addition of 0.2 N NaOH. The clear solution of the cleaned HSA was lyophilized and yielded a solid powder and was subjected to further studies. In addition, the residual sodium caprylate was analyzed by an HPLC method modified from (12) to make sure that it was completely removed. And the caprylate ion residue in the charcoal-treated HSA could not be quantified by the method used.
The human serum, albumin-enriched fraction, and commercial HSA were diluted twice with a pH 7.4 phosphate-buffered solution prior to the DSC experiment. The charcoal-treated HSA in the lyophilized powder form was accurately weighed and dissolved in the pH 7.4 phosphate-buffered solution to yield a 10% w/v HSA solution. The charcoal-treated HSA solutions were doped with sodium caprylate in serial concentrations of 0, 0.02, 0.04, 0.06, 0.08, and 0.16 mmol/g-HSA before being subjected to the DSC experiment.
The DSC Temperature Scanning Experiment
The Perkin-Elmer differential scanning calorimeter (DSC 8000, Perkin-Elmer Crop., Norwalk, CT, USA) equipped with a cooling accessory (Intercooler II) as well as being purged by nitrogen gas was employed. Prior to operation, the machine was calibrated with standard zinc and indium to ensure the accuracy and precision of the obtained heat of transitions as well as for the corresponding temperatures. Approximately 15 μL of liquid sample was accurately weighed and placed in a tightly sealed aluminum pan. The individual sample pan was subjected to run against an empty pan as a reference. With a loading temperature of 25°C, the temperature scanning program comprised (1) pre-heating from 25°C to 37°C, (2) equilibrating at 37°C for 1 min, and (3) heating from 37°C to 90°C at a rate of 5.0°C/min. Three replicates were prepared for each of the samples. All the DSC thermograms were analyzed using Pyris® software (version 11.0.0.0449, Perkin-Elmer Perkin-Elmer Crop., Norwalk, CT, USA).
Dynamic Light Scattering Measurement
Although it was confirmed (2,11) that the amount and the binding capability of HSA after charcoal treatment were fully recovered, its globular structure after reconstitution might not be as stable as that of the original. To examine the globular aggregation, the hydrodynamic diameter of a sample solution was determined using dynamic light scattering (DLS) measurements (Zetasizer® Nano ZS ZEN3600, Malvern Instruments Ltd., Worcestershire, UK). This machine measures the time-dependent intensity of the fluctuations of the photons scattered by the protein globules in a sample solution at the scattering angle of 173°C. The intensity of the fluctuation data were inputted to the instrument’s software (Malvern Zetasizer version 6.34, Malvern Instruments Ltd., Worcestershire, UK) to estimate the hydrodynamic diameters. Six replications were prepared for each of the sample solutions to meet the criteria of the measurement. At least three replications that satisfied the criteria were taken to calculate the reported mean as well as its standard deviation. In addition, the zeta potential (ζ) for each of the samples was also measured.
RESULTS AND DISCUSSION
DSC Thermograms for HSA Denaturation
Figure 1 compares the DSC thermograms among human serum proteins: enriched albumin fractionated from serum, commercial HSA, and HSA with its stabilizers removed by charcoal treatment. It was reported that the thermal unfolding/refolding of the HSA would be reversible up to ~55–60°C but beyond this temperature the denaturation was irreversible (13). As the temperature scanning underwent a further increase past 60°C, all the DSC thermograms in the study passed the steps of reversible and irreversible denaturation that fulfilled Scheme 1. As seen in Fig. 1, a multi-modal endotherm was observed during the denaturation of the human serum protein (Endotherm (a)). This is because the endotherm was made up of the thermal denaturing traces of the many different proteins present in serum, for examples, albumin, IgG, and haptoglobin. Albumin is the most plentiful protein and exhibits the highest denaturing peak among others (14). Table I summarizes the denaturing temperatures of albumins from the DSC endothermic traces obtained from Fig. 1. After alcohol/NaCl fractionation of human serum, it becomes enriched with albumin (showed as Endotherm (b)). As a result, Endotherm (b) exhibited a less irregular shoulder whereas the peak was invariant (67.23°C cf. 66.24°C for albumin in the serum proteins; statistically, a non-significant two-sided tested at a p value of 0.05). In contrast, the endotherm of HSA with stabilizers removed (Endotherm (c)) shifted downward to the temperature where albumin was practically denatured it was deduced that the HSA became reversibly destabilized after the stabilizer was removal. However, as seen in Table I, the denaturing temperature of HSA with sodium caprylate (at 0.08 mmol/g-protein level) increased from 67.23°C to 72.85°C, i.e., it was again stabilized by the addition of sodium caprylate. The denaturing temperature of the commercial HSA (Endotherm (d)) was substantially higher compared to the HSA after the stabilizers were removed (denaturing temperatures of 77.16°C vs. 67.23°C for the commercial HSA vs. that of the enriched-albumin fraction: Table I, and Fig. 1) as it was stabilized by sodium caprylate and the findings are consistent with previous studies (4,5,15). The Endotherm (d) also exhibited a shoulder after the curve reached a maximum. Generally, the phenomenon could be interpreted as a serial unfolding of different domains within a macromolecule (16). However, this might not be the case because Endotherm (c) did not show any shoulder even through the material sources and the DSC scanning conditions were identical. Shrake and Ross (15) suggested that this type of transition would arise from a significant stability of the remaining HSA during denaturation due to the stabilizers’ redistribution rather than to a sequential unfolding of the subdomains of the protein.
Fig. 1.

DSC thermograms of the denaturing transitions for serum proteins (a), enriched-albumin serum fraction (b), charcoal-treated commercial HSA (c), and commercial HSA (d)
Table I.
Denaturing temperatures, for HSA samples, with apparent activation energies for some selected endotherms, among albumins obtained from different DSC thermograms. For each of the samples, three replications were conducted and a mean value with standard deviations (S.D.) are presented
| Source | Peak temperature °C (Mean, S.D.) | E app a kcal/mol (mean ± SE, r 2) |
|---|---|---|
| Albumin in serumb | 66.24, 0.38 | – |
| Enriched-albumin fractionc | 67.23, 0.56 | – |
| Stabilizer-removed HSAd | 65.01*, 0.13 | 151.3 ± 4.5, 0.9900 |
| Commercial HSA | 77.16*, 0.83 | 69.0 ± 2.7, 0.9949 |
| Stabilizer-removed HSA with caprylate ione | 72.85*, 0.31 | 76.6 ± 7.9, 0.9567 |
*p value of 0.025; statistically significantly different from the denaturing temperature of albumin in serum
aApparent activation energy (E app) of particular DSC endotherms obtained from data fitting according to Eqs. 8 and 10
bDefined as the highest peak among DSC endothermic tracings (14)
cObtained from hydro-alcoholic fractionation from serum proteins
dCommercial HSA after treatment by charcoal to remove the stabilizers
eSodium caprylate was added to charcoal-treated commercial HSA at a concentration of 0.08 mmol/g-HSA to match that of commercial HSA
Effect of Sodium Caprylate on the Hydrodynamic Characteristics of the HSA Globule in a pH 7.4 Solution
Figure 2 illustrates the effect of sodium caprylate on the hydrodynamic characteristics of HSA in a pH 7.4 solution. The charcoal-treated HSA forms nano-size aggregates that yielded more than one scattering intensity peak in the DLS measurement, or in other words, the co-existence of aggregated and dispersed HSA was observed. It is deduced that HSA may undergo conformational alteration, e.g. unfolding/denaturing, to some degrees after charcoal treatment. These unfolded/denatured molecules might not be able to refold into their native structure but form aggregation after reconstitution. The average aggregated diameter as well as the dispersed one was plotted against the sodium caprylate concentration level (Fig. 2a). At the temperature of the measurement (25°C), the hydrodynamic diameter of the dispersed HSA had a size of ~10 nm that was consistent with the globular tertiary structure of the native HSA reported by Mitra and coworkers (17). In addition during charcoal treatment, HSA solution became cloudy at ~pH 5 and disappeared where the solution passed to pH 7. Jachimska et al. (18) found the isoelectric point of HSA to be at pH 5.1. The protein precipitated due to the lack of electrostatic repulsion at pH approaching isoelectric point and reversibly dissociated when they regained the electrostatic charges at higher pH. As observed by DLS experiment, the dissociated particles may be both aggregated and dispersed HSA. Furthermore, the aggregation might exhibit the effect on subsequent DSC measurement as the denaturing temperature of the charcoal-treated HSA was lower than that of HSA in serum by ~1–2°C (Table I and Fig. 1). And, addition of caprylate ligand to charcoal-treated HSA can only increase the denaturing temperature to 72.85°C whereas the commercial HSA denatured at 77.16°C (Table I). The discrepancy may be due to the presence of the irreversible aggregates yielding some HSA molecules not available to be stabilized by caprylate ions. As a result, the stability of the protein’s globular structure was not fully recovered.
Fig. 2.

Plots of the hydrodynamic size a and zeta potentials, ζ b of HSA globules after reconstitution with a pH 7.4-buffered solution against the concentration level in millimoles per grams protein of sodium caprylate
A slight increase in diameter (from 8.2 to 13.8 nm) with the increasing caprylate ion concentration was observed (Fig. 2a). Several lines of evidence have indicated that the caprylate ion strongly binds to human albumin (19,20). With a high binding affinity, e.g., the associate constant (Ka) was as high as 3 × 105 M−1 (21), it was found that the caprylate ion binds to Sudlow’s site II on the subdomain IIIA which is predominantly a hydrophobic cleft (18). After the binding site has been occupied, the ligand ion may invite more of the surrounding water molecules to make the subdomain less hydrophobic, i.e., it becomes more hydrated. As a result the protein may swell to a higher hydrodynamic size. It was further observed that the average aggregated size reduced from ~300 to ~50 nm with the increased caprylate ion concentration up to 0.08 mmol/g-HSA whereas the ζ value decreased from −7.8 to −11.1 mV (Fig. 2b). Thus, we suggest that the caprylate ligand ion may enlarge the electrical double layer (with the evidence of “a more negative” ζ value) of the protein globules and, in turn, repulse each other so as to form smaller aggregates.
The Effect of the Sodium Caprylate on the Apparent Energetic Information During HSA Denaturation
The protection by the physical barrier, i.e., an increased electrical double layer that surrounded the protein globules described in the previous section seems not to be sufficient for addressing the protein stabilization by sodium caprylate. However, it could not explain how the HSA complex was more resistant to thermal stress but partially protected the protein globules from aggregation. The analysis of the energetic information during HSA denaturation may be useful to provide insight into this mechanism: According to the simplified approximated protein denaturation model, Eqs. 8 and 10 allowed for a non-linear fitting between xN and 1/T by the in-house software. As the treatment of the DSC thermograms, e.g. Eqs. 9–10, was based on the assumption that the enthalpy of reversible unfolding was much greater than that of the irreversible transition, it was assumed further that the enthalpy of the irreversible step was negligible (7). Thus, the influence of the protein aggregation on DSC data treatment may be minimal. It was found that the regression of the data from the DSC endotherms of the system of the HSA in pH 7.4-buffered solutions with or without the caprylate ion was successful with an r2 of between 0.9844 and 0.9949. An example of the agreement between the model fitting and the actual data is illustrated in Fig. 3. The fitted Eapp values of the commercial and stabilizers-removed HSA with and without sodium caprylate at 0.08 mmol/g-protein are also tabulated in Table I.
Fig. 3.

The plot of x N as a functison of 1/T for the DSC endotherm of charcoal-treated HSA. x N was obtained from the fraction of the endotherm at temperature T as shown by Eq. 10. The broken line is the result from a non-linear fitting according to Eq. 8 (9)
Eapp is the energetic information obtained for each of the DSC endotherms obtained from the rational approximation of the Lumry-Eyring model in which Scheme 1 is estimated to Scheme 2 or 3. Figure 4 illustrates the plot of Eapp as well as the denaturing temperature from the DSC endotherms of HSA against the caprylate ion. As seen in Fig. 4, Eapp decreases with an increase of the caprylate ion concentration and flattens off when the ligand approaches ~0.08 mmol/g-HSA whereas the denaturing temperature increases from 65.01°C and reaches a plateau (72.85°C) at about identical ligand concentrations.
Fig. 4.

Plots of the apparent activation energy (E app: circle legends) and denaturing temperature (midpoint of denaturing endotherm: diamond legends) for the charcoal-treated HSA against caprylate ions. E app is the energetic information with regard to the protein denaturation obtained from the fitting in accord with Eq. 8. As a guide for the eyes, the trend lines were drawn by empirical best fit having an r 2 of more than 0.81
The Mechanism of HSA Stabilization According to Lumry-Eyring Model
The ligand binding on HSA may affect the protein denaturation that might yield a good approximation of the process to favor either Scheme 2 or 3 depending on the rate kinetics of the steps in the Lumry-Eyring model (Scheme 1). It has been previously reported for HSA that the kinetic state of Scheme 2 was successful in describing the denaturation of the native HSA at a temperature below 74°C (8). Thus the kinetics of the HSA thermal denaturation without caprylate ion in the current study may be controlled by an irreversible step that can be approximated as Scheme 2. In this case with a non-linear fitting revealing an r2 value as high as 0.9900, Eapp is estimated to be 151 kcal/mol (Table I). Based on this derivation, Eapp was attributed to the product between K and k3. It was therefore contributed to two energy states including reversible folding/unfolding and the irreversible denaturation that corresponded to the equilibrium parameter, K and the kinetic parameter, k3, respectively;
 |
11 |
where ΔHU and Ea,3 are the enthalpy of equilibrium folding/unfolding, and the activation energy of the irreversible denaturing step. By investigating the thermal effect on the fluorescence of native HSA, Pico (8) estimated the ΔHU at pH 7.4 to be 88.9 kcal/mol. In addition the DSC derived data showed a thermal unfolding enthalpy of the protein at pH 7.2 of 115 kcal/mol (22). Thus, from Eq. 11, Ea,3 for the native HSA denaturation at neutral pH may be roughly 36–62 kcal/mol. If the caprylate ion could further inhibit the irreversible step in the denaturation model as approximated in Scheme 2, the energy curve showed in Fig. 4 would have undergone continuously a monotonic change. But this was not the case as the curve exhibited a continuous stepwise alteration (Fig. 4). In addition, it has been observed that when it comes to hydration beyond 55°C the HSA suffers irreversible denaturation with a rapid loss of its α-helicity of domain IIA which is not the targeted binding site for the caprylate ligand (17). Thus, the caprylate ligand is not likely to be involved with the irreversible step as suggested by Arakawa and Kita (5). As observed in Fig. 4, the curve reveals two different energy levels between two extremes that corresponded to the energy ranges of 140–160 kcal/mol and 60–80 kcal/mol, respectively. It may be anticipated that the former level is designated as the energy state of Scheme 2 whereas the latter one is that of the other scheme.
It has been reported that subdomain IIIA is about to be reversibly unfolded/refolded due to relatively fewer interactions with other parts of the albumin molecule (23). The caprylate-protein binding interaction then may be involved with the subdomain IIIA conformation that is prone to be part of the reversible step, rather than to the irreversible step. It may cause the rate of the reversible unfolding to become less and less. As a result, the rate of protein unfolding in the equilibrium during the transition is relatively slow and becomes a rate determining step driving Scheme 2 kinetically to evolve to Scheme 3. Thus, it is postulated that the caprylate ion may govern the kinetics of the HSA denaturing transition by delaying the native protein unfolding within the reversible step to cause a difficulty in the irreversible transition to a subsequent step. As a result, the denaturing temperature of albumin was markedly increased (e.g., from 65.01°C to 72.85°C: Table I). By this means HSA could be stabilized. This supports the suggestion of Cordes et al. (24) who said that caprylate binding may provide the best conformation that stabilizes the HSA. It appears that Scheme 3 kinetics fully predominates when the added ion reached 0.08 mmol/g-HSA at which the energy curve leveled off (Fig. 4). At this point, Eapp may be interpreted as the activation energy for the kinetic parameter k1, i.e., Ea,1 that was estimated to be ~76 kcal/mol (fitted r2 of 0.9567: Table I). It was of interest that the concentration level of the added caprylate ion (0.08 mmol/g-HSA) was identical to that present in the commercial HSA product. This concentration level is said to be optimal for stabilization of HSA.
CONCLUSION
Although there is evidence that the caprylate ligand binding may protect HSA from aggregation by increasing the electrical double layer that surrounds the protein globule, it cannot address the problem of how the protein solution resists thermal agitation. The DSC experiment in the current study demonstrated that the thermal kinetic driven process according to the Lumry-Eyring model may explain the phenomenon: by binding to the protein’s domain that are prone to be the reversible folding/unfolding sites, the caprylate ligand may decrease the rate of the reversible unfolding rather than by directly inhibiting the irreversible transition. As a consequence, a higher temperature may be required to produce irreversible protein denaturation. The mechanism of albumin stabilization by the caprylate ligand ion may be summarized as Scheme 4.
Scheme 4.

ᅟ
ACKNOWLEDGMENTS
The authors are grateful to the Royal Golden Jubilee Ph. D. program for partially financial support and Prince of Songkla University (PSU) for the research grant. Special thanks go to PSU Scientific Equipment Center, and Drug Delivery System Excellent Center, Faculty of Pharmaceutical Sciences for Lab facilities. And, the authors would like to thank Dr. Brian Hodgson for English language revision.
REFERENCES
- 1.Fanali G, di Masi A, Trezza V, Marino M, Fasano M, Ascenzi P. Human serum albumin: from bench to bedside. Mol Aspects Med. 2012;33:209–90. doi: 10.1016/j.mam.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Olsen H, Andersen A, Kongsgaard UE, Bormer OP. Pharmaceutical-grade albumin: impaired drug-binding capacity in vitro. BMC Clin Pharmacol. 2004 doi: 10.1186/1472-6904-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albumin (Human) USP 20% solution: Albutein® prescribing information, Grifols Barcelona, Spain, 2008.
- 4.Anraku M, Tsurusaki Y, Watanabe H, Maruyama T, Kragh-Hansen U, Otagiri M. Stabilizing mechanisms in commercial albumin preparations: octanoate and N-acetyl-l-tryptophanate protect human serum albumin against heat and oxidative stress. Biochim Biophys Acta. 2004;1702:9–17. doi: 10.1016/j.bbapap.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Arakawa T, Kita Y. Stabilizing effects of caprylate and acetyltryptophanate on heat-induced aggregation of bovine serum albumin. Biochim Biophys Acta. 2000;1479:32–6. doi: 10.1016/S0167-4838(00)00061-3. [DOI] [PubMed] [Google Scholar]
- 6.Lumry R, Eyring H. Conformation changes of proteins. J Phys Chem. 1954;58:110–20. doi: 10.1021/j150512a005. [DOI] [Google Scholar]
- 7.Sanchez-Ruiz JM. Theoretical analysis of Lumry-Eyring models in differential scanning calorimetry. Biophys J. 1992;61:921–35. doi: 10.1016/S0006-3495(92)81899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pico GA. Thermodynamic feature of the thermal unfolding of human serum albumin. Int J Biol Macromol. 1997;20:63–73. doi: 10.1016/S0141-8130(96)01153-1. [DOI] [PubMed] [Google Scholar]
- 9.Khrapunov S, Brenowitz M. Stability, denaturation and refolding of Mycobacterium tuberculosis MfpA, a DNA mimicking protein that confers antibody resistant. Biophys Chem. 2011;159:33–40. doi: 10.1016/j.bpc.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gundry RL, Fu Q, Jelinek CA, Van Eyk JE, Cotter RJ. Investigation of an albumin-enriched fraction of human serum and its albuminome. Proteomics Clin Appl. 2007;1:73–88. doi: 10.1002/prca.200600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen RF. Removal of fatty acid from serum albumin by charcoal treatment. J Biol Chem. 1967;242:173–81. [PubMed] [Google Scholar]
- 12.Dengler T, Kellner S, Furst G. Quantitative determination of sodium-octanoate in human serum albumin preparations. Infusionstherapie. 1988;15:273–4. doi: 10.1159/000222305. [DOI] [PubMed] [Google Scholar]
- 13.Flora K, Brennan JD, Baker GA, Doody MA, Bright FV. Unfolding of acrylodan-labeled human serum albumin probed by steady- state and time-resolved fluorescence methods. Biophys J. 1998;75:1084–96. doi: 10.1016/S0006-3495(98)77598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garbett NC, Mekmaysy CS, Helm CW, Jenson AB, Chaires JB. Differential scanning calorimetry of blood plasma for clinical diagnosis and monitoring. Exp Mol Pathol. 2009;86:186–91. doi: 10.1016/j.yexmp.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Shrake A, Ross PD. Biphasic denaturation of human albumin due to ligand redistribution during unfolding. J Biol Chem. 1988;263:15392–9. [PubMed] [Google Scholar]
- 16.Privalov PL. Stability of proteins. Proteins which do not present a single cooperative system. Adv Protein Chem. 1982;35:1–104. doi: 10.1016/S0065-3233(08)60468-4. [DOI] [PubMed] [Google Scholar]
- 17.Mitra RK, Sinha SS, Pal SK. Hydration in protein folding: thermal unfolding/refolding of human serum albumin. Langmuir. 2007;23:10224–9. doi: 10.1021/la7014447. [DOI] [PubMed] [Google Scholar]
- 18.Jachimska B, Wasilewska M, Adamczyk Z. Characterization of globular protein solutions by dynamic light scattering, electrophoretic mobility, and viscosity measurements. Langmuir. 2008;24:6866–72. doi: 10.1021/la800548p. [DOI] [PubMed] [Google Scholar]
- 19.Kragh-Hansen U. Octanoate binding to the indole- and benzodiazepine-binding region of human seruim albumin. Biochem J. 1991;273:641–4. doi: 10.1042/bj2730641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varshney A, Sen P, Ahmad E, Rehan M, Subbarao N, Khan RH. Ligand binding strategies of human serum albumin: how can the cargo be utilized? Chirality. 2010;22:77–87. doi: 10.1002/chir.20709. [DOI] [PubMed] [Google Scholar]
- 21.Lee IY, McMenamy RH. Location of the medium chain fatty acid site on human serum albumin. Residues involved and relationship to the indole site. J Biol Chem. 1980;255:6121–7. [PubMed] [Google Scholar]
- 22.Celej MS, Dassie SA, Gonzalez M, Bianconi ML, Fidelio GD. Differential scanning calorimetry as a tool to estimate binding parameters in multiligand binding proteins. Anal Biochem. 2006;350:277–84. doi: 10.1016/j.ab.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 23.Sugio S, Kashima A, Mochizuki S, Noda M, Kobayashi K. Crystal structure of human serum albumin at 2.5 Å resolution. Protein Eng. 1999;12:439–46. doi: 10.1093/protein/12.6.439. [DOI] [PubMed] [Google Scholar]
- 24.Cordes AA, Platt CW, Carpenter JF, Randolph TW. Selective domain stabilization as a strategy to reduce fusion protein aggregation. J Pharm Sci. 2012;101:1400–9. doi: 10.1002/jps.23049. [DOI] [PubMed] [Google Scholar]


