Abstract
Although epigenetic regulation plays a critical role in embryonic development, few studies have examined the relationship of epigenome-wide methylation with fetal growth. Using the Infinium HumanMethylation450 BeadChip (Illumina, Inc., San Diego, California) in a substudy of 1,046 infants from the Norwegian Mother and Child Cohort Study (MoBa) enrolled between 1999 and 2008, we examined epigenome-wide cord blood DNA methylation in relation to birth weight. In multivariable-adjusted robust linear regression models, we identified differential methylation at 19 cytosine-guanine dinucleotides (CpGs) associated with either decreased (AT-rich interactive domain 5B (MRF1-like) (ARID5B), 2 CpGs) or increased (x-ray repair complementing defective repair in Chinese hamster cells 3 (XRCC3), 4 CpGs) birth weight. ARID5B knockout mice have less adipose tissue and significantly lower weight in the postnatal period. XRCC3 plays a key role in the maintenance of chromosome stability and the repair of DNA damage. Although there are fewer data on the other implicated genes, many of these genes have been shown to have roles in developmental processes. This constitutes the largest and most robust study of birth weight using an epigenome-wide methylation platform and offers potential insights into epigenetic mechanisms of fetal growth.
Keywords: birth weight, cord blood, epigenetics, methylation, MoBa, Norwegian Mother and Child Cohort Study
Epigenetic pathways regulate fetal development by controlling the expression of genes (1), facilitating both precisely timed and highly coordinated developmental processes (2). The most well-characterized of these epigenetic pathways is DNA methylation, the addition of a methyl group usually to cytosines in cytosine-guanine dinucleotide (CpG) sites (3). The relationship between CpG methylation and gene expression is complex and incompletely understood. Recent studies indicate that methylation at promoter and island regions tends to result in gene silencing; however, methylation in gene bodies tends to enhance gene expression (4–6). Loss of methylation at specific imprinted regions leads to serious growth-related congenital anomalies, such as Beckwith-Wiedemann and Silver-Russell syndromes (7, 8). However, there are limited data in humans on the role of more modest variability in DNA methylation status in the growth and development of the fetus.
Although some portion of epigenetic lability is under genetic control (9), epigenomic consequences of exposures experienced in utero (10–12) have been documented in humans. For example, maternal depression (13) and smoking during pregnancy (11), both of which are predictors of reduced birth weight, have been associated with altered methylation profiles in either gene-specific (13) or epigenome-scale (11) investigations. In particular, Joubert et al. (11) identified significant associations between maternal smoking in pregnancy and differential methylation in genes involved in fundamental developmental processes. Together, these results support the hypothesis that birth weight, and/or pathways leading to birth weight, may be affected by differences in methylation.
A few studies have begun to examine the associations of gene-specific methylation with birth weight. Targeted investigations have involved genes hypothesized to play key roles in growth (e.g., insulinlike growth factor 2), and/or that may be sensitive to famine exposure in pregnancy (GNAS antisense RNA 1 (GNASAS), INS-IGF2 readthrough (INS-IGF2), and leptin (LEP)). However, studies have thus far not provided consistent evidence of an association with birth weight in humans (14–16). Recently, 2 epigenome-scale investigations of methylation in relation to birth weight have been published, although both had relatively small study populations and lacked adjustment for potentially important confounders. None of the principal findings in these studies overlap (17, 18).
We undertook an investigation of the relationship between CpG-specific cord blood DNA methylation and birth weight using the Infinium HumanMethylation450 BeadChip (Illumina, Inc., San Diego, California) among 1,046 newborns from the Norwegian Mother and Child Cohort Study (MoBa).
METHODS
Study population
MoBa enrolled more than 100,000 women between 1999 and 2008. Study design and selection characteristics have been described in detail elsewhere (19, 20). Women were invited by mail to participate prior to their routine ultrasonography examinations at their local hospitals, usually scheduled at approximately 18 weeks' gestation. Participation rates varied by study year (20) but averaged 38.5%. Exposure-related information was collected by questionnaire at the first enrollment visit and then again at approximately 30 weeks' gestation. Information on dietary folate intake was collected using a semiquantitative food frequency questionnaire returned by the mothers at approximately 18–22 gestational weeks. The food frequency questionnaire consisted of 263 questions about 255 food items and was designed to capture dietary habits and intakes of dietary supplements during the first 4–5 months of pregnancy (21, 22). Methods regarding calculation of nutrient and energy intakes have been previously described (21, 22). Briefly, nutrient and energy intakes were calculated using FoodCalc (http://www.ibt.ku.dk/jesper/FoodCalc/Default.htm) and the Norwegian Food Composition Table (23). We adjusted for folate intake from foods. Measurement of plasma folate status was obtained from maternal blood samples collected at the enrollment visit (at approximately 18 weeks' gestation). Plasma folate was measured using a microbiological assay with a chloramphenicol-resistant strain of Lactobacillus casei (24). The assay determines biologically active folate species, predominantly 5-methyl-tetrahydrofolate, and has a coefficient of variation of 4% within day and 5% between days at population median concentration (24). The Medical Birth Registry of Norway receives mandatory information on all deliveries at hospitals using a standardized birth notification form (25). This form includes demographic information about the mother and father, information about the mother's health before and during pregnancy, including chronic diseases and pregnancy complications, and information on delivery characteristics.
Within MoBa, a nested case-cohort subset was established to examine prenatal risk factors for asthma at 3 years of age (26), which included 507 singleton asthma cases and a reference population of 1,455 singletons randomly selected from the MoBa population with 3-year follow-up data available. The current study is based on the subset of these singletons for whom cord blood and valid methylation data and data on covariates relevant to analysis of birth weight were available (n = 1,046) (11). This study was approved by the University of North Carolina Chapel Hill Office of Human Research Ethics (Chapel Hill, North Carolina).
DNA methylation
DNA methylation at 485,577 cytosine positions (CpG sites) was measured in cord blood from the MoBa samples using the Infinium HumanMethylation450 BeadChip (27, 28). Bisulfite conversion was performed using the EZ-96 DNA methylation kit (Zymo Research Corporation, Irvine, California) according to manufacturer instructions and was checked per methods previously described (27). For each sample, the methylation level at each CpG was calculated in Illumina's GenomeStudio methylation module as β = intensity of the methylated allele (M) / (intensity of the unmethylated allele (U) + intensity of the methylated allele (M) + 100) (27). The β values were logit transformed to obtain the log ratio [log ratio = log(β / 1 − β)], which may have better statistical properties than the β values (29). Bisulfite conversion and DNA methylation were performed at Illumina, Inc. (San Diego, California) within a single month according to methods previously described (27). Quality control has been previously described (11). Briefly, duplicate samples and control samples were included on each plate in a blinded fashion, and a series of methylation controls (10%, 35%, 60%, and 85% methylated) was included on the first and last plates. The distribution of birth weight by analytical plate was similar, with overlapping means and interquartile ranges (data not shown). After adjustment for the covariates' associations, birth weight was not found to be different across DNA plates (Kruskal-Wallis P = 0.30). Detection P values and multidimensional scaling were used to identify erroneous samples, and CpGs were excluded on the basis of missing data and genomic position. After quality control procedures, 1,068 subjects and 485,492 CpGs, including X and Y probes, were available for analysis.
Statistical analysis of birth weight
We excluded 14 in vitro fertilization subjects and 9 subjects with missing covariates, leaving 1,046 subjects for analysis. We conducted an epigenome-wide analysis in which we evaluated the association between birth weight and methylation in cord blood at each of the 485,492 CpG sites, 1 at a time. Specifically, using birth weight as the dependent variable and the log ratio of methylation as the predictor, we applied robust linear regression with sandwich-based estimators of the covariance to accommodate potential outliers and heteroscedasticity (30). We adjusted for child sex, maternal plasma cotinine (11), parity, maternal age, dietary folate not including supplements (24), asthma, gestational age at delivery, gestational age at delivery squared, preeclampsia, season of birth, and leukocyte cell–type proportions (31, 32) as potential confounders. Analyses using plasma instead of dietary folate yielded similar results. Given the number of statistical tests conducted, we report only CpGs that are significant after Bonferroni correction (485,492 statistical tests, P < 1.03 × 10−7).
RESULTS
The majority of the mothers were between the ages of 25 and 34 years. Approximately 40% were nulliparous. The distribution of maternal plasma cotinine levels suggested that approximately 13% were active smokers (plasma cotinine >56.8 nmol/L (33)) at the time of blood collection, which occurred at approximately 18 weeks’ gestation. The vast majority of births occurred at term and were of a healthy birth weight (≥2,500 g). In total, 54% of the infants were boys. As described above (26), this cohort was designed to overrepresent asthma at 3 years, which accounted for approximately 33% of this study population (Table 1).
Table 1.
Characteristics of 1,046 Subjects in the Norwegian Mother and Child Cohort Study, 1999–2008
| Characteristic | No. | % |
|---|---|---|
| Maternal age, years | ||
| 15–24 | 130 | 12.4 |
| 25–29 | 406 | 38.8 |
| 30–34 | 384 | 36.7 |
| ≥35 | 126 | 12.0 |
| Parity | ||
| 0 | 425 | 40.6 |
| 1 | 432 | 41.3 |
| ≥2 | 189 | 18.1 |
| Prenatal plasma cotinine, nmol/L | ||
| Undetectable | 725 | 69.3 |
| >0–56.8 | 188 | 18.0 |
| 56.9–388 | 68 | 6.5 |
| ≥389 | 65 | 6.2 |
| Gestational age at delivery, weeks | ||
| <32 (<224 days) | 1 | 0.1 |
| 32–36 (224–258 days) | 36 | 3.4 |
| 37–41 (259–293 days) | 926 | 88.5 |
| ≥42 (≥294 days) | 83 | 7.9 |
| Infant birth weight, g | ||
| <2,500 | 20 | 1.9 |
| 2,500–3,999 | 770 | 73.6 |
| ≥4,000 | 256 | 24.5 |
| Infant sex | ||
| Male | 562 | 53.7 |
| Female | 484 | 46.3 |
| Asthma at age 3 years | 349 | 33.4 |
For the multivariable analysis of differential DNA methylation in relation to birth weight, we identified 19 CpGs that reached Bonferonni significance (P < 1.03 × 10−7) (Table 2, Web Figure 1 available at http://aje.oxfordjournals.org/). Of these, 13 were within genes and 6 were within intergenic regions. CpGs associated with birth weight were AT-rich interaction domain 5B (MRF1-like) (ARID5B) (2 CpGs); Kruppel-like factor 9 (KLF9); x-ray repair complementing defective repair in Chinese hamster cells 3 (XRCC3) (4 CpGs); phosphatidylethanolamine-binding protein 4 (PEBP4); sema domain, immunoglobulin domain, transmembrane domain, and short cytoplasmic domain, (semaphorin) 4C (SEMA4C); spondin 2, extracellular matrix protein (SPON2); ubiquitin-like with PHD and ring finger domains 1 (UHRF1); ankyrin repeat domain 11 (ANKRD11); and major facilitator superfamily domain containing 10 (MFSD10). The distributions of methylation values were transformed to approximate a normal distribution. Thus, the interpretation of the magnitude of the difference in birth weight in relation to methylation increases is as follows. For ARID5B, for each 1-logit increase in methylation of cg25953130 or cg02863179, birth weight was lower by 371 g or 341 g, respectively (Table 2, Figure 1). For XRCC3, for each 1-logit increase in methylation at cg02194129, birth weight was higher by approximately 727 g (Table 2, Figure 2). These models were adjusted for multiple confounders, including the estimated leukocyte cell–type proportions of each sample (31, 32). For the remaining probes found to be significant in Table 2, illustrations of the relationship between the untransformed methylation β value in relation to the birth weight residual after adjustment for important confounders can be found in Web Figure 2.
Table 2.
Associations of Selected CpGsa and Infant Birth Weight in the Norwegian Mother and Child Cohort Study, 1999–2008
| CpG by Birth Weight Difference | Gene | Adjusted Mean Difference in Birth Weight (SE)b | Bonferroni-Corrected P Valuec | Adjusted Mean Difference in Birth Weight (SE)d | Bonferroni-Corrected P Valuec |
|---|---|---|---|---|---|
| Decrease in birth weight | |||||
| cg25953130 | ARID5B | −371.26 (58.11) | 8.11 × 10−5 | −376.86 (53.93) | 1.35 × 10−6 |
| cg08005122 | −493.38 (83.70) | 1.82 × 10−3 | −345.57 (74.14) | 1.00 | |
| cg20076442 | −302.79 (51.62) | 2.17 × 10−3 | −304.93 (47.00) | 4.22 × 10−5 | |
| cg02863179 | ARID5B | −340.99 (59.30) | 4.33 × 10−3 | −340.46 (51.76) | 2.32 × 10−5 |
| cg25124943 | −480.24 (89.53) | 3.96 × 10−2 | −402.67 (77.99) | 0.12 | |
| cg00049440 | KLF9 | −314.29 (58.69) | 4.14 × 10−2 | −302.06 (61.59) | 0.45 |
| Increase in birth weight | |||||
| cg02194129 | XRCC3 | 726.57 (96.34) | 2.24 × 10−8 | 403.00 (81.84) | 0.41 |
| cg17836177 | PEBP4 | 558.39 (93.08) | 9.61 × 10−4 | 481.80 (78.73) | 4.56 × 10−4 |
| cg12798040 | XRCC3 | 326.29 (54.73) | 1.21 × 10−3 | 268.26 (53.75) | 0.29 |
| cg00605777 | SEMA4C | 728.27 (122.89) | 1.50 × 10−3 | 234.27 (83.10) | 1.00 |
| cg14172849 | XRCC3 | 660.38 (112.59) | 2.17 × 10−3 | 271.75 (76.66) | 1.00 |
| cg23127323 | SPON2 | 323.05 (56.72) | 5.97 × 10−3 | 257.23 (43.93) | 2.31 × 10−3 |
| cg25162533 | 382.67 (68.28) | 1.01 × 10−2 | 371.26 (65.87) | 8.40 × 10−3 | |
| cg23369670 | XRCC3 | 557.29 (99.73) | 1.12 × 10−2 | 144.55 (66.65) | 1.00 |
| cg17714703 | UHRF1 | 296.96 (54.11) | 1.97 × 10−2 | 322.68 (47.99) | 8.64 × 10−6 |
| cg08420923 | ANKRD11 | 493.22 (91.53) | 3.45 × 10−2 | 457.16 (75.05) | 5.44 × 10−4 |
| cg23237276 | 523.19 (97.67) | 4.12 × 10−2 | 382.82 (73.05) | 0.08 | |
| cg05993265 | MFSD10 | 442.00 (82.59) | 4.23 × 10−2 | 236.24 (67.26) | 1.00 |
| cg24693803 | 538.69 (101.12) | 4.84 × 10−2 | 457.97 (88.27) | 0.10 |
Abbreviations: ANKRD11, ankyrin repeat domain 11; ARID5B, AT-rich interaction domain 5B (MRF1-like); CpG, cytosine-guanine dinucleotide; KLF9, Kruppel-like factor 9; MFSD10, major facilitator superfamily domain containing 10; PEBP4, phosphatidylethanolamine-binding protein 4; SE, standard error; SEMA4C, sema domain, immunoglobulin domain, transmembrane domain, and short cytoplasmic domain, (semaphorin) 4C; SPON2, spondin 2, extracellular protein; UHRF1, ubiquitin-like with PHD and ring finger domains 1; XRCC3, x-ray repair complementing defective repair in Chinese hamster cells 3.
a From Infinium HumanMethylation450 BeadChip (Illumina, Inc., San Diego, California).
b Per logit increase in methylation fraction, including covariates for child sex, maternal plasma cotinine, parity, maternal age, dietary folate not including supplements, asthma, gestational age at delivery, gestational age at delivery squared, preeclampsia, season of birth, and leukocyte cell–type proportions.
c Unadjusted P values multiplied by 485,492 (number of tests). The threshold for significance remains P < 0.05 (5 × 10−2).
d Per logit increase in methylation fraction, including covariates for factors listed above with leukocyte cell–type proportion removed.
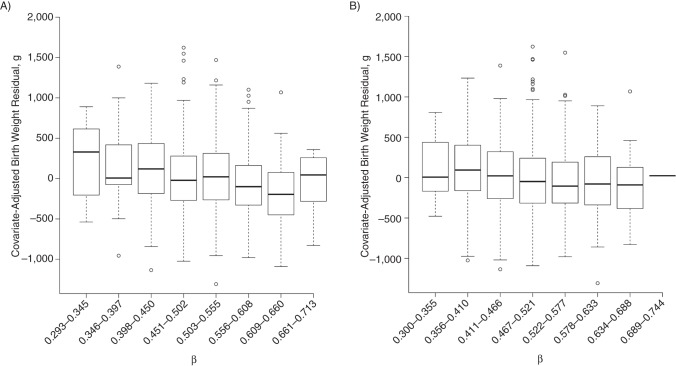
Figure 1.
Cytosine-guanine dinucleotide methylation β values for significant AT-rich interactive domain 5B (MRF1-like) (ARID5B) probes in relation to birth weight residual in the Norwegian Mother and Child Cohort Study, 1999–2008. Untransformed methylation β values for ARID5B A) cg25953130 (P = 8.11 × 10−5) and B) cg02863179 (P = 4.33 × 10−3) are plotted against the birth weight residual after adjustment for child sex, maternal plasma cotinine, parity, maternal age, dietary folate not including supplements, asthma, gestational age at delivery, gestational age at delivery squared, preeclampsia, season of birth, and leukocyte cell–type proportions. Methylation β values were divided into 8 equally spaced intervals. Both show a roughly linearly declining trend in birth weight with increasing methylation fraction.
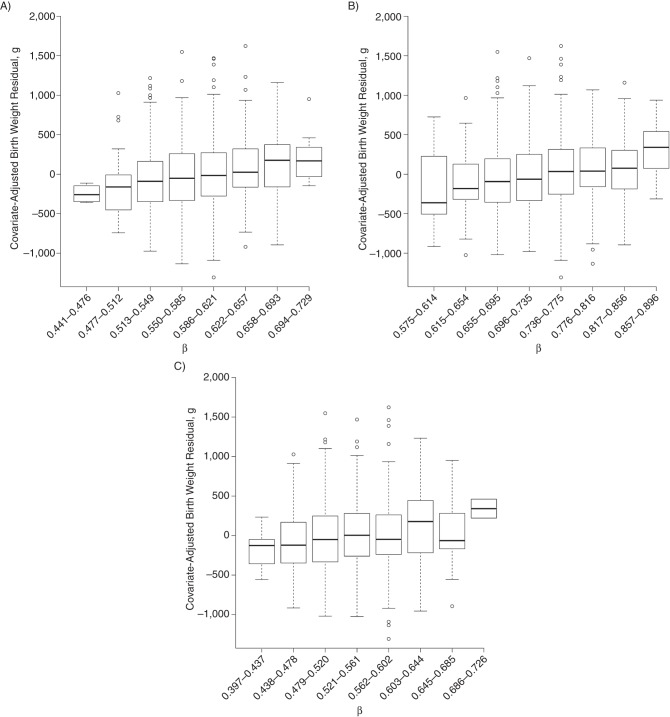
Figure 2.
Cytosine-guanine dinucleotide methylation β values for significant x-ray repair complementing defective repair in Chinese hamster cells 3 (XRCC3) probes in relation to birth weight residual. Untransformed methylation β values of XRCC3 A) cg02194129 (P = 2.24 × 10−8), B) cg12798040 (P = 1.21 × 10−3), and C) cg14172849 (P = 2.17 × 10−3) are plotted against the birth weight residual after adjustment for child sex, maternal plasma cotinine, parity, maternal age, dietary folate not including supplements, asthma, gestational age at delivery, gestational age at delivery squared, preeclampsia, season of birth, and leukocyte cell–type proportions. Methylation β values were divided into 8 equally spaced intervals. All show a roughly linearly increasing trend in birth weight with increasing methylation fraction.
We conducted a sensitivity analysis in which we adjusted for caffeinated beverage intake in early pregnancy, maternal prepregnancy body mass index (weight (kg)/height (m)2), and maternal weight gain up until her completion of the third-trimester questionnaire. Inclusion of these covariates resulted in the loss of 128 participants. Adjustment for these additional covariates had little impact on the overall results when compared with a model of the same sample size with those covariates excluded (Web Figure 3A), although the following probes were no longer significant with this reduced sample size, which was likely primarily due to a loss of power: cg20076442, cg25124943, cg00605777, cg23127323, cg25162533, cg23369670, cg23237276, cg05993265, and cg24693803. Of the probes that remained significant, the magnitude of the regression coefficient in all cases preserved direction (increase or decrease in birth weight), with only small changes in magnitude (mean change = 6.5%). In addition, when we adjusted for measured plasma folate instead of dietary folate, there was no substantial change in the estimated P values (Web Figure 3B).
It is not yet clear how appropriate the available methods for correcting for leukocyte cell–type proportions (31) are in the setting of cord blood methylation, given that reference data sets are available for only adult peripheral blood (32). Therefore, we conducted analyses with and without this adjustment factor (Table 2). In these models, 22 CpG probes were significant at a Bonferroni-corrected threshold (Web Table 1). However, the CpGs did not overlap perfectly with those that were significant after adjustment for cell type. Among the 19 probes that were significant in models adjusted for cell-type proportions, 8 CpGs remained significant without adjustment for cell type (Table 2). Interestingly, associations with all of the XRCC3 CpGs were significant only in the models with cell-type adjustment.
DISCUSSION
In this analysis of a homogeneous Norwegian population, we report significant associations between CpGs in cord blood DNA and birth weight. We identified associations in 9 genes, 2 of which (ARID5B (2 CpGs) and XRCC3 (4 CpGs)) had multiple significant CpGs. This is one of the first studies to examine DNA methylation across the genome in relation to birth weight and the first to have used the 450K platform. Additionally, our use of a large, well-defined birth cohort provides essential covariate data to adjust for confounders of the methylation–birth weight relationships.
We found an inverse association between ARID5B methylation and birth weight (higher methylation fraction associated with lower birth weight) (Figure 1). ARID5B (also known as MRF2/MRF-2), is a transcriptional coactivator that plays a key role in adipogenesis. ARID5B knockout mice have a high rate of neonatal death and reduced lipid accumulation, which manifests in significantly lower weight from postnatal day 5 onward (34). Both full and partial ARID5B knockouts are resistant to weight gain and obesity, even after high-fat dietary challenges (34). One possible mechanism explaining these findings is the modulation of leptin levels, given that down-regulation of ARID5B results in increased expression of leptin, an important regulator of energy balance, insulin resistance, and metabolism (35).
The ARID5B CpGs associated with birth weight in our study are located in the gene body, which typically results in enhanced expression (4–6). However, using gene expression and methylation data from 41 normal breast tissue samples in The Cancer Genome Atlas (http://cancergenome.nih.gov), we observed an inverse relationship between methylation at the 2 significant CpGs in the ARID5B gene and ARID5B expression. Although the effect size was large, the correlation was not statistically significant (P = 0.15). Thus, further systematic functional studies are required to describe the relationship between methylation at this site and ARID5B gene expression in cord blood samples. Of potential interest, polymorphisms in the ARID5B gene have been associated with increased risk for acute lymphoblastic leukemia (36, 37), coronary artery disease (38), and type 2 diabetes (39).
XRCC3 is a critical DNA repair gene, whose function during development is ensuring the accurate duplication of cells through homologous recombination (40, 41). In XRCC3-deficient hamster cell lines, repair of DNA double-strand breaks is decreased 25-fold (41). Infante-Rivard et al. (42) examined a coding polymorphism (Thr241Met) in XRCC3 in relation to small for gestational age and found no overall increased risk of small for gestational age related to genotype, although women who smoked in the third trimester and carried the variant that encodes threonine were at somewhat increased risk of having offspring that were small for gestational age. However, there are no other studies that have reported an association of XRCC3 with any fetal growth parameter. In our study, 4 XRCC3 CpGs were associated with increased birth weight, suggesting an inverse correlation between XRCC3 expression (via increased methylation) and birth weight. However, these associations were significant only in models adjusted for peripheral blood leukocyte cell–type proportion and, thus, require more research to fully understand.
Other genes highlighted in our study have been shown to be important in developmental processes, although their relation to fetal growth in particular has not been addressed. UHRF1 has multiple domains that bind chromatin and has been shown to be important in the maintenance of DNA methylation (43). In mice, UHRF1 is highly expressed in pluripotent stem cells, as opposed to differentiated tissue (44), and genetic ablation of UHRF1 has been shown to result in genomic hypomethylation (45, 46). Yippee-like 3 (Drosophila) (YPEL3) is regulated by p53 and induces cellular senescence. It has growth-inhibitory effects in both normal and tumor cell lines (47). In our data, increased methylation (presumably negatively regulating gene expression) was associated with increased birth weight, which would represent a logically consistent direction of association. ANKRD11 enhances the transcriptional activity of p53 (48) and is of increasing interest in relation to autism (49). Mutations in ANKRD11 are associated with KBG syndrome, a rare genetic disease characterized by short stature, typical facial dysmorphism, dental issues, and developmental delay (50). In KBG syndrome, birth weight is typically normal but birth length is often below the third percentile.
Genes are turned on and off dynamically during development (51); thus, methylation, as a means of controlling gene expression and the activation of biological cascades, is important for fetal development and parturition. However, in the current and previous studies (17, 18, 52, 53), methylation status is measured only once (at delivery), and therefore may not always generalize to antecedent periods or to births that occur remote from term. Indeed, all studies of cord blood methylation and birth outcomes are, by their nature, cross-sectional (i.e., exposure (methylation) and outcome (birth weight) are measured simultaneously). The methylation status of cord blood CpGs may be the consequence of exposures experienced during pregnancy that result in changes in the timing of delivery or growth of the fetus, or alternatively, it may reflect the developmental process ongoing at the time of delivery. Because of the temporal uncertainty and influence of other factors on cord blood methylation patterns, we have presented our results as associations without making strong causal arguments.
The strengths of our study are many. First, we used the Illumina 450K technology, which provides state-of-the-art epigenome-wide coverage. Second, our study was nested within MoBa, a population-based cohort of Norwegian women recruited early in pregnancy. The MoBa cohort is racially homogeneous, which is beneficial in the setting of birth weight analyses given the strong and consistent racial disparities in mean birth weight across the spectrum of gestational age (47). Norwegians also have nationalized health care, ensuring the adequacy of free antenatal care across the socioeconomic spectrum. These beneficial characteristics of MoBa would tend to reduce the likelihood of residual confounding by any unmeasured factors associated with race and access to care.
A significant strength of our study is that we were able to adjust for strong known correlates of birth weight (such as prenatal smoking, as measured by plasma cotinine levels in the second trimester), parity, maternal age, and other factors associated with the probability of selection into this nested study. Although the original study was sampled to overrepresent asthma cases at 3 years, our results adjusted for this selection factor and were found to be robust on the basis of analyses of only the random subcohort (data not shown). Moreover, ours is the largest study to date to examine cord blood methylation in relation to birth weight. We observed somewhat different associations when we adjusted for estimated leukocyte cell–type proportions, which have been shown to exhibit important variation in methylation profiles (31, 32). However, it is not yet clear how appropriate these methods are in the setting of cord blood. For example, cord blood contains more nucleated red blood cells than does adult blood, and these might vary by gestational age, although the percentage of nucleated red blood cells of all nucleated cells remains low in cord blood (3.2%) (54). Further research examining variability in cord blood cell population methylation profiles is required.
In conclusion, using cord blood DNA, we identified a small number of CpGs in 9 genes, including multiple significant CpGs in ARID5B and XRCC3, which are associated with birth weight in a large population-based cohort. Some biological plausibility for a role for ARID5B in particular in fetal growth is provided by associations with postnatal growth and lipid accumulation in mice. The other genes have been less well studied, but many are involved in developmental processes. Although the underlying mechanism and associated causal processes are unclear, these findings may provide novel insights into the role of epigenetic modifications, including methylation, in the regulation of fetal growth and parturition.
Supplementary Material
ACKNOWLEDGMENTS
Author affiliations: Department of Epidemiology, Gillings School of Global Public Health, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina (Stephanie M. Engel, Andrew F. Olshan); Epidemiology Branch, Division of Intramural Research, National Institute of Environmental Health Sciences, Research Triangle Park, North Carolina (Bonnie R. Joubert, Stephanie J. London); Department of Biostatistics, Gillings School of Global Public Health, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina (Michael C. Wu); Institute Management and Staff, Norwegian Institute of Public Health, Oslo, Norway (Siri E. Håberg); Department of Clinical Science, University of Bergen, Bergen, Norway (Per Magne Ueland); Laboratory of Clinical Biochemistry, Haukeland University Hospital, Bergen, Norway (Per Magne Ueland); Norwegian Institute of Public Health, Division of Epidemiology, Oslo, Norway (Wenche Nystad, Stein Emil Vollset); Department of Global Public Health and Primary Care, University of Bergen, Bergen, Norway (Roy M. Nilsen, Stein Emil Vollset); and Biostatistics Branch, Division of Intramural Research, National Institute of Environmental Health Sciences, Research Triangle Park, North Carolina (Shyamal D. Peddada)
This research was supported in part by the intramural research program of the National Institute of Environmental Health Sciences (grant Z01-ES-49019) and the National Institute of Environmental Health Sciences (grant P30ES010126). The Norwegian Mother and Child Cohort Study is supported by the Norwegian Ministry of Health and the Ministry of Education and Research and the National Institute of Environmental Health Sciences (contract NO-ES-75558), the National Institute of Neurological Disorders and Stroke (grant 1 UO1 NS 047537-01), and the Norwegian Research Council/Functional Genomics (grant 151918/S10). Support for Drs. Engel and Wu was provided by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (grant R01HD058008).
S.M.E., B.R.J., and M.C.W. contributed equally to the paper and should each be considered first author.
Conflict of interest: none declared.
REFERENCES
- 1.Wolffe AP, Matzke MA. Epigenetics: regulation through repression. Science. 1999;286(5439):481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- 2.Bernal AJ, Jirtle RL. Epigenomic disruption: the effects of early developmental exposures. Birth Defects Res A Clin Mol Teratol. 2010;88(10):938–944. doi: 10.1002/bdra.20685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heijmans BT, Tobi EW, Lumey LH, et al. The epigenome: archive of the prenatal environment. Epigenetics. 2009;4(8):526–531. doi: 10.4161/epi.4.8.10265. [DOI] [PubMed] [Google Scholar]
- 4.Ball MP, Li JB, Gao Y, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27(4):361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 6.Maunakea AK, Nagarajan RP, Bilenky M, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466(7303):253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eggermann T. Silver-Russell and Beckwith-Wiedemann syndromes: opposite (epi)mutations in 11p15 result in opposite clinical pictures. Horm Res. 2009;71(suppl 2):30–35. doi: 10.1159/000192433. [DOI] [PubMed] [Google Scholar]
- 8.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152(6):1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 9.Bjornsson HT, Sigurdsson MI, Fallin MD, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299(24):2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoyo C, Murtha AP, Schildkraut JM, et al. Methylation variation at IGF2 differentially methylated regions and maternal folic acid use before and during pregnancy. Epigenetics. 2011;6(7):928–936. doi: 10.4161/epi.6.7.16263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joubert BR, Håberg SE, Nilsen RM, et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect. 2012;120(10):1425–1431. doi: 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy SK, Adigun A, Huang Z, et al. Gender-specific methylation differences in relation to prenatal exposure to cigarette smoke. Gene. 2012;494(1):36–43. doi: 10.1016/j.gene.2011.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Murphy SK, Murtha AP, et al. Depression in pregnancy, infant birth weight and DNA methylation of imprint regulatory elements. Epigenetics. 2012;7(7):735–746. doi: 10.4161/epi.20734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoyo C, Fortner K, Murtha AP, et al. Association of cord blood methylation fractions at imprinted insulin-like growth factor 2 (IGF2), plasma IGF2, and birth weight. Cancer Causes Control. 2012;23(4):635–645. doi: 10.1007/s10552-012-9932-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.St-Pierre J, Hivert MF, Perron P, et al. IGF2 DNA methylation is a modulator of newborn's fetal growth and development. Epigenetics. 2012;7(10):1125–1132. doi: 10.4161/epi.21855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tobi EW, Heijmans BT, Kremer D, et al. DNA methylation of IGF2, GNASAS, INSIGF and LEP and being born small for gestational age. Epigenetics. 2011;6(2):171–176. doi: 10.4161/epi.6.2.13516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon L, Joo JE, Powell JE, et al. Neonatal DNA methylation profile in human twins is specified by a complex interplay between intrauterine environmental and genetic factors, subject to tissue-specific influence. Genome Res. 2012;22(8):1395–1406. doi: 10.1101/gr.136598.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turan N, Ghalwash MF, Katari S, et al. DNA methylation differences at growth related genes correlate with birth weight: A molecular signature linked to developmental origins of adult disease? BMC Med Genomics. 2012;5:10. doi: 10.1186/1755-8794-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magnus P, Irgens LM, Haug K, et al. Cohort profile: the Norwegian Mother and Child Cohort Study (MoBa) Int J Epidemiol. 2006;35(5):1146–1150. doi: 10.1093/ije/dyl170. [DOI] [PubMed] [Google Scholar]
- 20.Nilsen RM, Vollset SE, Gjessing HK, et al. Self-selection and bias in a large prospective pregnancy cohort in Norway. Paediatr Perinat Epidemiol. 2009;23(6):597–608. doi: 10.1111/j.1365-3016.2009.01062.x. [DOI] [PubMed] [Google Scholar]
- 21.Brantsaeter AL, Haugen M, Alexander J, et al. Validity of a new food frequency questionnaire for pregnant women in the Norwegian Mother and Child Cohort Study (MoBa) Matern Child Nutr. 2008;4(1):28–43. doi: 10.1111/j.1740-8709.2007.00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haugen M, Brantsaeter AL, Alexander J, et al. Dietary supplements contribute substantially to the total nutrient intake in pregnant Norwegian women. Ann Nutr Metab. 2008;52(4):272–280. doi: 10.1159/000146274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norwegian Food Safety Authority. Norwegian Food Composition Database 2013. Oslo, Norway: Norwegian Directorate of Health and University of Oslo; 2013. [Google Scholar]
- 24.Nilsen RM, Vollset SE, Monsen AL, et al. Infant birth size is not associated with maternal intake and status of folate during the second trimester in Norwegian pregnant women. J Nutr. 2010;140(3):572–579. doi: 10.3945/jn.109.118158. [DOI] [PubMed] [Google Scholar]
- 25.Irgens LM. The Medical Birth Registry of Norway. Epidemiological research and surveillance throughout 30 years. Acta Obstet Gynecol Scand. 2000;79(6):435–439. [PubMed] [Google Scholar]
- 26.Håberg SE, London SJ, Nafstad P, et al. Maternal folate levels in pregnancy and asthma in children at age 3 years. J Allergy Clin Immunol. 2011;127(1):262–264. doi: 10.1016/j.jaci.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bibikova M, Barnes B, Tsan C, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98(4):288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Sandoval J, Heyn H, Moran S, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6(6):692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 29.Du P, Zhang XA, Huang CC, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huber PJ. Robust regression: asymptotics, conjectures and Monte Carlo. Ann Stat. 1973;1(5):799–821. [Google Scholar]
- 31.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reinius LE, Acevedo N, Joerink M, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One. 2012;7(7):e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw GM, Carmichael SL, Vollset SE, et al. Mid-pregnancy cotinine and risks of orofacial clefts and neural tube defects. J Pediatr. 2009;154(1):17–19. doi: 10.1016/j.jpeds.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Whitson RH, Tsark W, Huang TH, et al. Neonatal mortality and leanness in mice lacking the ARID transcription factor Mrf-2. Biochem Biophys Res Commun. 2003;312(4):997–1004. doi: 10.1016/j.bbrc.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 35.Dong J, Ishimori N, Paigen B, et al. Role of modulator recognition factor 2 in adipogenesis and leptin expression in 3T3-L1 cells. Biochem Biophys Res Commun. 2008;366(2):551–555. doi: 10.1016/j.bbrc.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 36.Healy J, Richer C, Bourgey M, et al. Replication analysis confirms the association of ARID5B with childhood B-cell acute lymphoblastic leukemia. Haematologica. 2010;95(9):1608–1611. doi: 10.3324/haematol.2010.022459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Treviño LR, Yang W, French D, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet. 2009;41(9):1001–1005. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang G, Watanabe M, Imai Y, et al. Genetic variations of Mrf-2/ARID5B confer risk of coronary atherosclerosis in the Japanese population. Int Heart J. 2008;49(3):313–327. doi: 10.1536/ihj.49.313. [DOI] [PubMed] [Google Scholar]
- 39.Wang G, Watanabe M, Imai Y, et al. Associations of variations in the MRF2/ARID5B gene with susceptibility to type 2 diabetes in the Japanese population. J Hum Genet. 2012;57(11):727–733. doi: 10.1038/jhg.2012.101. [DOI] [PubMed] [Google Scholar]
- 40.Griffin CS, Simpson PJ, Wilson CR, et al. Mammalian recombination-repair genes XRCC2 and XRCC3 promote correct chromosome segregation. Nat Cell Biol. 2000;2(10):757–761. doi: 10.1038/35036399. [DOI] [PubMed] [Google Scholar]
- 41.Pierce AJ, Johnson RD, Thompson LH, et al. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13(20):2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Infante-Rivard C, Weinberg CR, Guiguet M. Xenobiotic-metabolizing genes and small-for-gestational-age births: interaction with maternal smoking. Epidemiology. 2006;17(1):38–46. doi: 10.1097/01.ede.0000187669.34003.b1. [DOI] [PubMed] [Google Scholar]
- 43.Rothbart SB, Krajewski K, Nady N, et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol. 2012;19(11):1155–1160. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pichler G, Wolf P, Schmidt CS, et al. Cooperative DNA and histone binding by Uhrf2 links the two major repressive epigenetic pathways. J Cell Biochem. 2011;112(9):2585–2593. doi: 10.1002/jcb.23185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bostick M, Kim JK, Esteve PO, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317(5845):1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 46.Sharif J, Muto M, Takebayashi S, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450(7171):908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 47.Hauck FR, Tanabe KO, Moon RY. Racial and ethnic disparities in infant mortality. Semin Perinatol. 2011;35(4):209–220. doi: 10.1053/j.semperi.2011.02.018. [DOI] [PubMed] [Google Scholar]
- 48.Neilsen PM, Cheney KM, Li CW, et al. Identification of ANKRD11 as a p53 coactivator. J Cell Sci. 2008;121(Pt 21):3541–3552. doi: 10.1242/jcs.026351. [DOI] [PubMed] [Google Scholar]
- 49.Marshall CR, Noor A, Vincent JB, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82(2):477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sirmaci A, Spiliopoulos M, Brancati F, et al. Mutations in ANKRD11 cause KBG syndrome, characterized by intellectual disability, skeletal malformations, and macrodontia. Am J Hum Genet. 2011;89(2):289–294. doi: 10.1016/j.ajhg.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447(7143):425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 52.Lee H, Jaffe AE, Feinberg JI, et al. DNA methylation shows genome-wide association of NFIX, RAPGEF2 and MSRB3 with gestational age at birth. Int J Epidemiol. 2012;41(1):188–199. doi: 10.1093/ije/dyr237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schroeder JW, Conneely KN, Cubells JC, et al. Neonatal DNA methylation patterns associate with gestational age. Epigenetics. 2011;6(12):1498–1504. doi: 10.4161/epi.6.12.18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Proytcheva MA. Issues in neonatal cellular analysis. Am J Clin Pathol. 2009;131(4):560–573. doi: 10.1309/AJCPTHBJ4I4YGZQC. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.