Abstract
Background. Cryptosporidium is emerging as 1 of the 4 leading diarrheal pathogens in children in developing countries. Its infections in patients with AIDS can be fatal, whereas fully effective treatments are unavailable. The major goal of this study is to explore parasite fatty acyl-coenzyme A synthetase (ACS) as a novel drug target.
Methods. A colorimetric assay was developed to evaluate biochemical features and inhibitory kinetics of Cryptosporidium parvum ACSs using recombinant proteins. Anticryptosporidial efficacies of the ACS inhibitor triacsin C were evaluated both in vitro and in vivo.
Results. Cryptosporidium ACSs displayed substrate preference toward long-chain fatty acids. The activity of parasite ACSs could be specifically inhibited by triacsin C with the inhibition constant Ki in the nanomolar range. Triacsin C was highly effective against C. parvum growth in vitro (median inhibitory concentration, 136 nmol/L). Most importantly, triacsin C effectively reduced parasite oocyst production up to 88.1% with no apparent toxicity when administered to Cryptosporidium-infected interleukin 12 knockout mice at 8–15 mg/kg/d for 1 week.
Conclusions. The findings of this study not only validated Cryptosporidium ACS (and related acyl-[acyl-carrier-protein]-ligases) as pharmacological targets but also indicate that triacsin C and analogues can be explored as potential new therapeutics against the virtually untreatable cryptosporidial infection in immunocompromised patients.
Keywords: Cryptosporidium parvum, cryptosporidiosis, long chain fatty acyl-CoA synthetase, triacsin C, anti-cryptosporidial efficacy
Cryptosporidium parvum is a unicellular zoonotic pathogen that can cause severe watery diarrhea in both humans and animals [1]. It is emerging as 1 of the 4 top diarrheal pathogens in children <5 years old in developing countries [2]. Cryptosporidial infections in immunocompromised individuals can be prolonged and life-threatening. In the United States however, Food and Drug Administration–approved treatments remain unavailable to treat this opportunistic infection in patients with AIDS, whereas only nitazoxanide is approved for use in immunocompetent individuals. Therefore, there is an urgent need for new drugs, particularly those that can be safely used in immunocompromised persons. The slow progress in developing anticryptosporidial drugs is largely related to the unique metabolic features in this parasite, which are represented by a highly streamlined metabolism and inability to synthesize nutrients de novo [3, 4]. This parasite has completely lost the plastid-derived apicoplast present in many other apicomplexans, and the remnant mitochondrion lacks the citrate cycle and cytochrome-based respiratory chain. Therefore, many classic drug targets are unavailable in Cryptosporidium, and novel targets need to be identified for drug development.
However, essential core metabolic pathways, including energy metabolism and lipid synthesis are present in this parasite. Many enzymes within these pathways may serve as new drug targets because they are either absent in, or highly divergent from humans and animals. Within lipid metabolism, adenosine monophosphate (AMP)-binding long-chain (LC) and very long-chain fatty acyl coenzyme A (CoA) synthetases (ACSs; also known as fatty acid-CoA ligases [ACLs]; EC 6.2.1.3) are a large family of enzymes that catalyze the thioesterification between free fatty acids and CoA to form fatty acyl-CoAs via a 2-step reaction (Figure 1A) [5]. ACS may also function as fatty acid transporters in some organisms [6, 7]. It is indispensible in all organisms, because fatty acids have to be activated by this family of enzymes to form acyl-CoA thioesters before they can enter various catabolic and anabolic pathways. Fatty acyl-CoA thioesters serve as key intermediates in the biosynthesis of cellular lipids and in energy metabolism via β-oxidation [8]. Additionally, acyl-CoAs are also involved in biological processes such as protein modification [9, 10], cell proliferation [11], intracellular protein transport [12], and cell signaling [13, 14].
Figure 1.
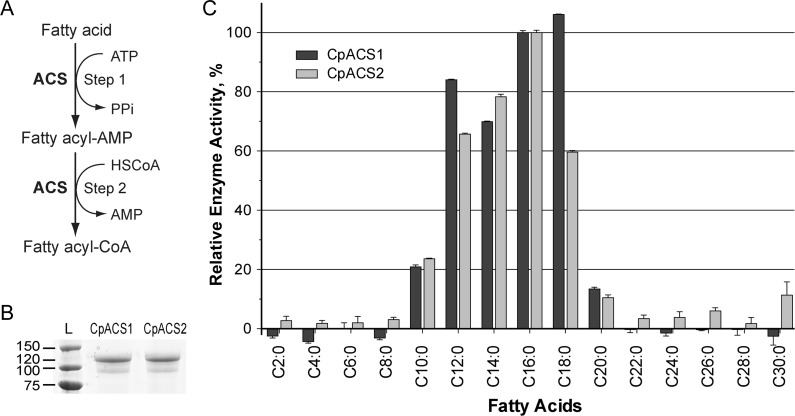
Activation of long chain fatty acids by Cryptosporidium parvum acyl-coenzyme A (CoA) synthetase (ACS). A, Illustration of ACS-catalyzed 2-step reaction to form fatty acyl CoA from free fatty acid, adenosine triphosphate (ATP) and CoA in reduced form (HSCoA). B, Purified recombinant CpACS1 and CpACS2 proteins fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. C, Substrate preferences of recombinant CpACS1 and CpACS2 proteins toward variable carbon chain–length saturated fatty acids as determined by 5,5′-dithio-bis-(2-nitrobenzoate) (DTNB) assay. Activities are expressed relative to C16 palmitic acid L, protein ladder (kDa).
The C. parvum genome encodes 3 LC-ACSs (CpACS1, CpACS2, and CpACS3), and differs from other apicomplexans, such as Plasmodium falciparum, which has at least 11 ACS genes [15]. In the current study, we successfully expressed recombinant CpACS1 and CpACS2 as enzymatically active, maltose-binding protein (MBP) fusions, and assayed their detailed enzyme kinetics. We also demonstrated that triacsin C, a potent ACS inhibitor, not only could inhibit CpACS enzyme activity but was also efficacious against C. parvum growth, both in cell culture and in mice.
METHODS
Ethics Statement
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health under the Animal Welfare Assurance numbers A4168-01 (Atlanta VA Medical Center) and A3893-01 (Texas A&M University). Animal experiments were performed in accordance with procedures approved by the Institutional Animal Care and Use Committees of the Atlanta VA Medical Center (protocol V001-06) and Texas A&M University (protocol 2009-21).
Molecular Cloning and Heterologous Expression
The IOWA-1 strain of C. parvum maintained by infecting calves was purchased from Bunch Grass Farm, and fresh oocysts (≤3 months old since harvest) were used in this study. Oocysts were purified from calf feces by a sucrose-gradient centrifugation, followed by treatment with 10% Clorox (∼7.5% sodium hypochlorite) on ice for 7 minutes and repeated washes with pure water for 5–8 times [16, 17]. If minor debris was still present, oocysts were further purified by a Percoll gradient–based protocol and resuspended in phosphate-buffered saline (PBS) before use. Genomic DNA was isolated from oocysts or sporozoites with a DNeasy Blood & Tissue Kit (Qiagen).
The CpACS1, CpACS2, and CpACS3 genes have been annotated by the C. parvum genome-sequencing project (GenBank accession Nos. XM_626649, XM_626248. and XM_625917 for ACS1, ACS2, and ACS3, respectively). Their open reading frames were amplified from C. parvum DNA by polymerase chain reaction (PCR) using high-fidelity Pfu DNA polymerase (Stratagene) and cloned into a pCR2.1-TOPO vector (Invitrogen). Cloned genes were sequenced to confirm their identities and then subcloned into the pMAL-c2x expression vector (New England Biolabs) for expression as MBP-fused proteins. Primers used for cloning are listed in Table 1. Plasmids with correct sequences and orientation were transformed into a Rosetta 2 strain of Escherichia coli competent cells (Novagen). The expression of MBP-fused proteins and purification using amylose resin-based affinity chromatography followed standard procedures [18].
Table 1.
Primers Used in the Current Study
| Gene | Direction | Sequence of Primer pairs (5′–3′)a | Product Size, Base Pairs | Application |
|---|---|---|---|---|
| CpACS1 | Forward | gtatatggatccATGGAAGAGAAAATCGACA | 2058 | Expression |
| Reverse | ccacgatctagaTCAAATTTGTGATCTTAAAG | |||
| CpACS2 | Forward | gaattaggatccATGGGGAACATTACTTCA | 2052 | Expression |
| Reverse | gaacaatctagaTCATTCATTCTTAGGTTTTGAG | |||
| Cp18S | Forward | TTGTTCCTTACTCCTTCAGCAC | 175 | qRT-PCR |
| Reverse | TCCTTCCTATGTCTGGACCTG | |||
| Hs18S | Forward | GGC GCC CCC TCG ATG CTC TTA | 189 | qRT-PCR |
| Reverse | CCC CCG GCC GTC CCT CTT A |
Abbreviation: qRT-PCR, quantitative reverse-transcription polymerase chain reaction. aLowercase letters represent the linker regions of the sequence.
Biochemical Assays
All chemicals used in the current study were purchased from Sigma-Aldrich or as otherwise specified. Triacsin C was purchased from Fermentek. The activity of CpACS enzymes was determined by monitoring the reduction of CoA concentrations by a 5,5′-dithio-bis-(2-nitrobenzoate) (DTNB; Ellman's reagent) colorimetric assay. In this assay, free CoA in reduced form (CoA-SH) reacted with DTNB to form 5-thionitrobenzoic acid that could be measured at 412 nm [19, 20]. A typical assay was performed in 200 µL of reaction buffer (0.1 mol/L Tris-HCl; pH8.0) containing 10 mmol/L potassium chloride, 10 mmol/L magnesium chloride, 50 µmol/L CoA-SH, 200 µmol/L ATP and 100 µmol/L fatty acid. A panel of fatty acids with varied chain lengths (C2:0 to C30:0) was individually evaluated to determine the substrate preferences for CpACS enzymes. The reactions were started with the addition of 1 µg of freshly purified MBP-CpACS fusion proteins. After incubation for 10 minutes at 32°C, reactions were stopped by heating samples at 80°C for 5 minutes. After samples cooled down to room temperature, 4 µL of 5 mmol/L DTNB was added into each reaction, followed by 5 minutes of color development. The optical density at 412 nm was measured with a Multiskan Spectrum spectrophotometer (Thermo Scientific). The CoA-SH concentration was calculated according to the standard curve, using serially diluted concentrations of CoA-SH.
Enzyme kinetics were similarly assayed with varied substrate concentrations (ie, palmitic acid at 0–600 µmol/L and ATP at 0–3000 µmol/L). Guanosine triphosphate, cytidine triphosphate, and uridine triphosphate were used to replace ATP in the assay to test whether they could serve as alternative energy sources for CpACS enzymes. The inhibitory effects of triacsin C on CpACS enzymes were analyzed under similar reaction conditions, except that 1 to 32 µmol/L of triacsin C was added to test its effect on the reaction. In all experiments, the MBP-tag alone was used as control.
Drug Efficacy Against Parasite Growth In Vitro
A quantitative real-time reverse-transcription PCR (RT-PCR) assay was used to evaluate drug efficacy against C. parvum growth in vitro [17, 21]. Human HCT-8 cells (American Type Culture Collection CCL-225) were seeded into 24-well cell culture plates and allowed to grow overnight at 37°C under an atmosphere of 5% carbon dioxide in Roswell Park Memorial Institute 1640 medium (Sigma) containing 10% fetal bovine serum or until they reached approximately 80% confluence. For drug testing, the C. parvum oocysts were added into the cell culture at a parasite: host cell ratio of 1:2 (ie, 1 × 105 oocysts per well), and allowed to infect host cells for 3 hours. Parasites that failed to invade host cells were removed by a medium exchange and triacsin C was added in the medium (0.2% dimethyl sulfoxide). Parasite-infected cells were incubated at 37°C for another 41 hours. To test the effect of triacsin C on parasite invasion, excystated sporozoites were prepared by incubating oocysts in 0.25% trypsin and 0.5% taurodeoxycholic acid for 45–60 minutes at 37°C, followed by 5–8 washes with PBS. Sporozoites were incubated with triacsin C at specified concentrations for 30 minutes at 37°C, washed with PBS to remove drug, and allowed to invade HCT-8 cells for 3 hours. Before RNA isolation, monolayers were gently washed 3 times with nuclease-free PBS and lysed in 350 µL of lysis buffer.
Total RNA was isolated from drug-treated and untreated cells infected with C. parvum by using an RNeasy Mini Kit (Qiagen) [17]. These RNA samples were also treated with RNase-free DNase (Qiagen) to remove DNA, according to the manufacturer's protocol. The quality and purity of RNA were determined using a NanoDrop ND-1000 spectrophotometer at 260/280 nm (Nano-Drop Technologies). A Qiagen 1-step RT-PCR QuantiTect SYBR Green RT-PCR kit was employed to evaluate parasite growth by detecting the relative levels of parasite 18S ribosomal RNA (rRNA) normalized using human 18S rRNA as a control, as described elsewhere [17, 21]. Primers used in the assay are listed in Table 1. Cytotoxic effects of triacsin C on host cells were evaluated by the relative levels of 18S rRNA in uninfected HCT-8 cells and by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay kit (Sigma), as described elsewhere [22].
Drug Efficacy In Vivo in a Mouse Model of Acute Cryptosporidiosis
The anticryptosporidial activity of triacsin C was assessed in the interleukin 12 (IL-12) knockout mouse model. Although this model, like other mouse models of cryptosporidiosis, does not demonstrate frank signs of diarrhea, it represents an acute model in that mice have rapidly intensifying parasite loads but then recover by day 14 [23]. C. parvum oocysts (IOWA bovine isolate) were collected, purified through discontinuous sucrose and cesium chloride gradients, and stored as described elsewhere [24]. Oocyst inocula were prepared by washing purified oocysts (stored <6 months) with PBS to remove potassium dichromate. Mice (6–8 weeks old) were inoculated with 1000 oocysts and treated by gavage with triacsin C (8 or 15 mg/kg/d), vehicle control (10% polyethylene glycol (PEG) or 1% dimethyl sulfoxide), or paromomycin (2 mg/kg/d; positive control). Treatment was given once daily for 7 days and mice were killed on day 8 (peak infection). At least 10 mice were used for each experimental condition.
Parasite burden was assessed by flow cytometry, as described elsewhere [25]. Briefly, fecal samples were collected from individual mice on postinfection days 5 and 7 and processed through microscale sucrose gradients in 2.0-mL microcentrifuge tubes. The partially purified stool concentrate containing oocysts was incubated for 30 minutes at 37°C with 5 µL of an oocyst-specific monoclonal antibody conjugated with fluorescein isothiocyanate (OW50-FITC) and then analyzed by means of flow cytometry. Absolute counts were calculated from the data files as oocysts per 100 µL of sample suspension [25].
Statistical Analyses
All biochemical and in vitro drug assays were performed at least in duplicate, and ≥2 independent assays were performed for each experiment. However, only data derived from single experiments are shown in the figures. In vivo drug testing was performed in 2 separate experiments. Data were typically analyzed using Microsoft Excel and GraphPad Prism software (version 5.0f; http://www.graphpad.com). Statistical significance was assessed with 2-tailed Student t tests, Mann-Whitney nonparametric tests (not assuming Gaussian distributions), and 2-way analysis of variance.
RESULTS
CpACS1- and CpACS2-Activated LC Fatty Acids
To test whether or not the CpACSs represent bona-fide fatty acid activating enzymes, we attempted to clone and express these proteins as MBP-fused proteins in bacteria and examine their activity against saturated fatty acids. All 3 CpACS genes could be amplified by PCR, cloned into the pCR2.1-TOPO cloning vector, and subcloned into the expression vector pMAL-c2x. However, only CpACS1 and CpACS2 have been successfully expressed as MBP-fused proteins (Figure 1B). Numerous attempts to express recombinant CpACS3 were unsuccessful, including the use of different expression conditions and the removal of the N-terminal signal peptide as described elsewhere for expressing P. falciparum ACS1 [26], suggesting potential cytotoxicity of this protein to E. coli. Therefore, biochemical analyses reported here were focused on the recombinant CpACS1 and CpACS2 enzymes.
Classic ACS assays use radioactive substrates coupled with isopropanol and/or heptane extractions [26, 27]. Here we adapted a DTNB colorimetric assay to assess enzyme kinetics and inhibitor effects by measuring the reduction of thiol groups of free CoA-SH in the reactions [28]. This method is more user friendly, safer, and suitable for adaptation to high-throughput screening. Both CpACS1 and CpACS2 displayed a clear preference toward 12- to 18-carbon fatty acids and lower activities toward C10:0 and C20:0 (Figure 1C). These 2 enzymes had no or little activity with saturated fatty acids <10 or >20 carbons in length, except for CpACS2, which displayed low activity toward some very long-chain fatty acids.
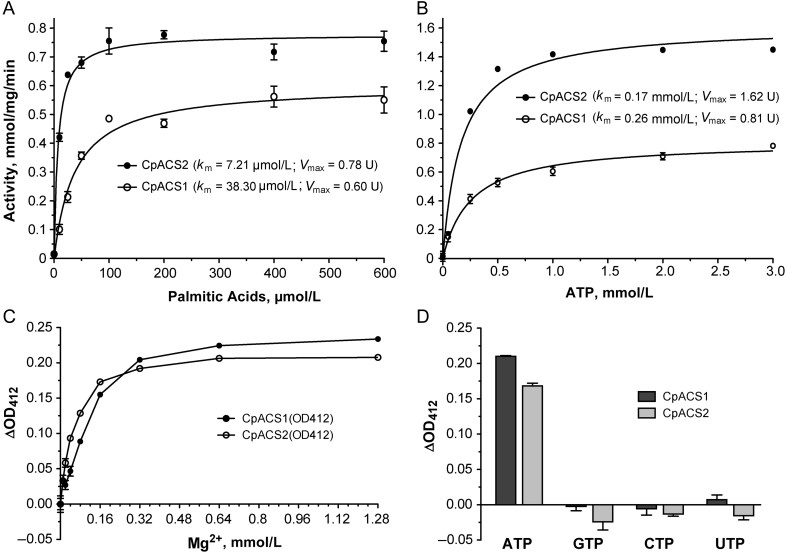
CpACS1 and CpACS2 followed Michaelis-Menten kinetics toward C16:0 (palmitic acid) and ATP with Michaelis constant (Km) values in the lower micromolar and nanomolar levels, respectively (Table 2 and Figure 2A and 2B). The 2 enzymes were Mg2+ dependent, and their activities peaked at an Mg2+ concentration of approximately 1 mmol/L (Figure 2C). Both enzymes could use only ATP as their energy source, because no activity was observed when guanosine triphosphate, cytidine triphosphate, and uridine triphosphate were used in the reactions (Figure 2D), similar to findings in the P. falciparum ACS1 and the ACL domain in the C. parvum polyketide synthase (CpPKS1) [26, 27].
Table 2.
CpACS1 and CpACS2 Enzyme Kinetics Toward Substrates and Inhibitor
| Substrate or Inhibitor and Parameter | CpACS1 | CpACS2 |
|---|---|---|
| Palmitic acid | ||
| Km, µmol/L | 38.30 | 7.21 |
| Vmax, mmol/mg/min | 0.60 | 0.78 |
| ATP | ||
| Km, µmol/L | 0.26 | 0.17 |
| Vmax, mmol/mg/min | 0.81 | 1.62 |
| Triacsin C | ||
| IC50, µmol/L | 3.70 | 2.32 |
| Ki, µmol/L | 0.60 | 0.11 |
Abbreviations: ATP, adenosine triphosphate; IC50, median inhibitory concentration; Ki, inhibition constant; Km, Michaelis constant; Vmax, maximum velocity.
Figure 2.
Biochemical features of CpACS1 and CpACS2, as determined using recombinant proteins. A, Enzyme kinetics using palmitic acid. Vmax, enzyme maximum velocity. B, Enzyme kinetics using adenosine triphosphate (ATP). C, Dose-dependent activity using Mg2+. ΔOD412, change in optical density at 412 nm. D, Enzyme activities using different nucleoside triphosphates (NTPs). Abbreviations: CTP, cytidine triphosphate; GTP, guanosine triphosphate; UTP, uridine triphosphate.
Triacsin C Inhibition CpACS Enzyme Activities and Effectiveness Against C. parvum Growth Both In Vitro and In Vivo
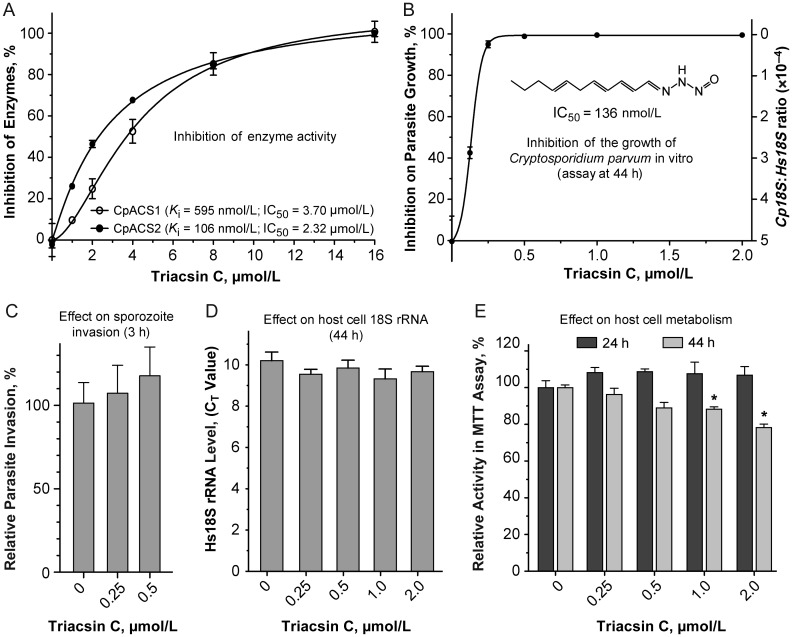
To assess whether or not CpACS enzymes are amenable to inhibition by small molecules, we examined enzyme activities in the presence of triacsin C (CAS 76896-80-5; synonym: 2,4,7-undecatrienal nitrosohydrazone), a fungal metabolite originally isolated from Streptomyces aureofaciens and identified as a vasodilator [29, 30]. It resembles a polyunsaturated fatty acid (see Figure 3B inset) and can differentially inhibit various LC-ACSs [31–36]. For example, in humans, it can inhibit LC-ACSs, but has little effect on short-chain (SC)-ACSs or mitochondrial MC-ACSs [34]. In rats, this compound was effective on ACSL1, ACSL3, and ACSL4 at low micromolar levels but was not effective against ACSL5 and ACSL6 [35, 36]. In the current study, we observed that triacsin C could inhibit CpACS1 and CpACS2 enzyme activities, with median inhibitory concentration (IC50) values at 3.70 and 2.32 µmol/L (Table 2 and Figure 3A), corresponding to inhibition constant (Ki) values at 595 nmol/L and 106 nmol/L, respectively, as calculated using a competitive inhibition model [37]. Triacsin C also displayed strong efficacy against the growth of C. parvum cultured with HCT-8 cells at nanomolar levels (ie, IC50, 136 nmol/L) (Figure 3B), although a 30-minute treatment on sporozoites did not affect parasite invasion (Figure 3C). Triacsin C had no significant effect on the HCT-8 host cells, based on the Hs18S rRNA levels (Figure 3D), and an MTT assay suggested a moderate decrease in host cell metabolism when this inhibitor was used at ≥1 µmol/L for 2 days (Figure 3E).
Figure 3.
Inhibition of enzyme activity and parasite growth in vitro. A, Inhibition of CpACS1 and CpACS2 activities by triacsin C. IC50, median inhibitory concentration; Ki, inhibition constant.B, Efficacy of triacsin C in inhibiting the growth of Cryptosporidium parvum cultured in vitro with HCT-8 cells, determined using a quantitative reverse-transcription polymerase chain reaction assay at 44 hours after infection. C, Relative level of invasion by sporozoites receiving 30-minute treatment of triacsin C, determined at 3 hours after infection. D, Effect of triacsin C on 18S ribosomal RNA (rRNA) levels in uninfected HCT-8 cells (at 44 hours after infection). CT, cycle threshold. E, Effect of triacsin C on uninfected HCT-8 cell metabolism as determined by MTT (3-(4,5-dimethylthia-zol-2-yl)-2,5-diphenyltetrazolium bromide) assay 24 and 44 hours after infection. *P < .05 (comparison with untreated controls).
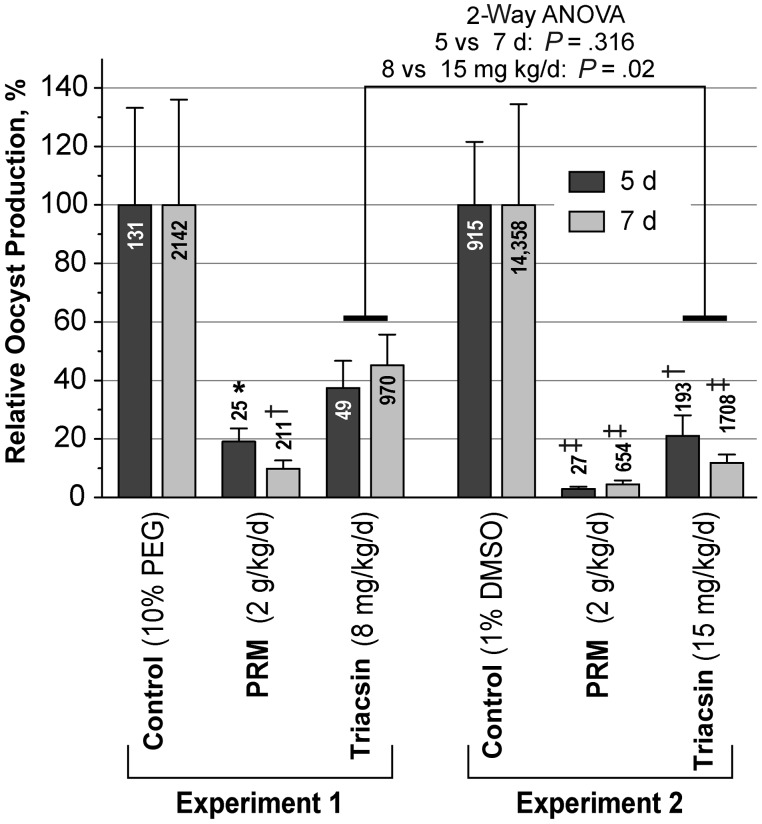
The promising results of triacsin C against both CpACS enzyme activity and parasite growth in vitro motivated us to examine how triacsin C would affect the pathogenesis of disease in a mouse model of acute cryptosporidiosis. In vivo drug testing using an IL-12 knockout mouse model of acute cryptosporidial infection revealed that triacsin C could effectively inhibit parasite growth at suitable therapeutic doses. At 8 mg/kg/d, triacsin C reduced C. parvum oocyst production in feces by 62.50% and 54.73% on postinfection days 5 and 7, respectively, compared with 78.91% and 88.11%, respectively, for 15 mg/kg/d (Figure 4). Both Student's t and nonparametric tests indicated that the reductions were significant in all treated groups, except for the group receiving 8 mg/kg/d. Two-way analysis of variance also showed significant differences in the percentage of inhibition between the 2 doses of triacsin C treatments (P = .02) but not between postinfection days 5 and 7 (P = .32) (Figure 4). In addition, gross histology of IL-12 knockout mice given triacsin C at 8 or 15 mg/kg/d revealed no observable pathological changes in the major organs (data not shown). Moreover, triacsin C treatments at the 2 doses resulted in a general improvement in weight gain in mice infected with C. parvum, although the improvement was not statistically significant by Student's t or Mann-Whitney tests (data not shown).
Figure 4.
Efficacy of triacsin C or paromomycin (PRM) on cryptosporidial infection in interleukin 12 knockout mice, as measured by the relative production of Cryptosporidium parvum oocysts on postinfection days 5 and 7. Numbers in or above bars indicate average oocyst counts (per 100 µL) in each experimental group. ANOVA, analysis of variance; PEG, polyethylene glycol. *P < .05; †P < .01; ‡P < .001 (significant differences by nonparametric Mann-Whitney test for comparison with controls; Student t test produced the same result, albeit with smaller P values [all <.05]).
The observed effective dose of triacsin C (<0.008 to 0.015 g/kg/d) against cryptosporidial infection in mice was >10 times lower than nitazoxanide (the only Food and Drug Administration–approved treatment for human cryptosporidiosis) and paromomycin (a “gold standard” anticryptosporidial control drug), because these 2 drugs could effectively inhibit cryptosporidial infections in various animal models only at doses between 0.2 and 2 g/kg/d [38] (see also Figure 4). Nitazoxanide and paromomycin are known for their low toxicity in humans and animals (eg, oral median lethal dose values for acute toxicity in mice at 1.35 and 23.5 g/kg/d, based on the Registry of Toxic Effects of Chemical Substances database) (http://ccinfoweb.ccohs.ca/rtecs/search.html). Although toxicity of triacsin C in mice remains to be determined, this compound was not toxic to mice at the low dosage levels used here and in other studies (eg, [39, 40]).
DISCUSSION
We identified 3 LC-ACS genes in the C. parvum genome. Their protein sequences were 98% (ACS3) to 100% (ACS1 and ACS2) identical to the orthologs in Cryptosporidium hominis, or 73%–76% identical to those from Cryptosporidium muris. CpACS1 and CpACS2 were successfully expressed as MBP-fused proteins. Although different strategies are still being explored to express CpACS3, our biochemical analysis of CpACS1 and CpACS2 provides sufficient insights on the biochemical features of this family of enzymes in C. parvum. The DTNB-based colorimetric assay not only permits the measurements of enzyme activity and the effect of inhibitors, but it can also be easily adapted for high-throughput screening of drugs against ACS.
Although LC-ACS enzymes are essential in fatty acid metabolism and present in multiple forms in various apicomplexans, little is known about their biochemical features. In fact, only 1 earlier study assayed the ACS activity in a crude extract of Plasmodium-infected simian erythrocytes [41], and a relatively simple biochemical analysis was performed specifically on a single P. falciparum ACS (ie, PfACS1) protein [26]. In C. parvum, 1 of the 3 ACS genes (corresponding to CpACS3) was previously identified by screening an expression library with a monoclonal antibody [42], but no biochemical analysis was performed. The lack of functional studies on apicomplexan ACSs might be due in part to the technical difficulties in expressing and/or obtaining enzymatically active recombinant proteins, as noted in expressing CpACS3 (this study) and PfACS1 [26]. Therefore, our observation that recombinant CpACS1 and CpACS2 proteins retain activity for only a very short time and need to be assayed immediately after purification is critical and should be kept in mind when studying other recombinant apicomplexan ACS enzymes.
The most significant observation is the excellent efficacy of the ACS inhibitor triacsin C against cryptosporidial infection in mice. Because of the lack of completely effective drugs for cryptosporidiosis in humans and animals, particularly in patients with AIDS, great efforts have been devoted to the discovery of drugs against this parasite. Drug screenings have, in fact, identified many compounds highly effective against the growth of C. parvum in vitro, but most (if not all), including nitazoxanide and paromomycin, failed to achieve comparable efficacies when tested in animals [38, 43]. The current study shows that triacsin C effectively inhibited the specific drug targets CpACS1 and CpACS2, displayed excellent anticryptosporidial efficacy in vitro (IC50, 136 nmol/L), and could reduce C. parvum oocyst production in mice by approximately 50%–88% at pharmaceutical levels (ie, 8–15 mg/kg/d).
It is known that triacsin C may also inhibit LC-ACS enzymes in humans and animals, thus affecting certain aspects of lipid metabolism. For example, treatment of human hepatocytes with triacsin C reduced the formation of lipid droplets [44], whereas in human fibroblast cells, it could block de novo synthesis of glycerolipids and cholesterol esters [45]. However, triacsin C did not affect the recycling of fatty acids into phospholipid, which not only suggests the presence of functionally separate pools of acyl-CoA, but also explains why no apparent toxic effects occurred in the animals used here and in previous studies [45].
Although triacsin C has not been tested or therapeutically used in humans, several studies in animals have shown the benefits of triacsin C treatment. For example, in vitro and in vivo experiments in chickens have shown that triacsin C could rescue palmitic acid–induced cytotoxicity of follicle granulosa cells in fuel-overloaded broiler hens [46]. In mice, triacsin C displayed activities to control cancer growth and enhance the efficacy of etoposide at a nontoxic dose (4 mg/kg/d) [39] and antiatherosclerotic activity at a dose of 10 mg/kg/d [40]. These observations suggest that, although pharmacokinetics and toxicity tests are needed to further evaluate safety, triacsin C is generally safe in animals when administrated at low dosages and that its potential as a new anticryptosporidial drug should be explored. In fact, triacsin C is also an effective inhibitor against the acyl (acyl-carrier protein)-ligase domains in the unique, multifunctional type I fatty acid synthase (FAS1) and polyketide synthase (PKS1) in C. parvum [27]. Therefore, triacsin C has the potential to hit multiple targets and to block both the fatty acid activation by discrete ACSs and the elongation of fatty acids by FAS1 and PKS1.
Finally, whether triacsin C could be truly developed into a new anticryptosporidial drug may require further pharmacokinetic, preclinical, and clinical tests, but our current findings strongly support the notion that the discrete ACSs (and ACL domains in FAS1 and PKS1) can serve as novel drug targets. New triacsin C analogues can be synthesized, and new classes of anti-ACS/ACL compounds may be identified by drug screening for the development of more-selective and safer anticryptosporidial drugs.
Notes
Author contributions. F. G., H. Z., J. R. M., and G. Z. designed research; F. G., H. Z., J. M. F., S. D. R., L. X., N. N. M., and G. Z. performed research; F. G., H. Z., J. R. M., and G. Z. analyzed the data; F. G., H. Z., J. M. F., S. D. R., J. R. M., and G. Z. wrote the manuscript.
Financial support. The work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant R01 AI44594 to G. Z.) and by the Atlanta Research and Education Foundation and Atlanta VA Medical Center (J. R. M.).
Potential conflict of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Chen XM, Keithly JS, Paya CV, LaRusso NF. Cryptosporidiosis. N Engl J Med. 2002;346:1723–31. doi: 10.1056/NEJMra013170. [DOI] [PubMed] [Google Scholar]
- 2.Kotloff KL, Nataro JP, Blackwelder WC, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 3.Abrahamsen MS, Templeton TJ, Enomoto S, et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–5. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- 4.Rider SD, Jr, Zhu G. Cryptosporidium: genomic and biochemical features. Exp Parasitol. 2010;124:2–9. doi: 10.1016/j.exppara.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersson CS, Lundgren CA, Magnusdottir A, et al. The Mycobacterium tuberculosis very-long-chain fatty acyl-CoA synthetase: structural basis for housing lipid substrates longer than the enzyme. Structure. 2012;20:1062–70. doi: 10.1016/j.str.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Weimar JD, DiRusso CC, Delio R, Black PN. Functional role of fatty acyl-coenzyme A synthetase in the transmembrane movement and activation of exogenous long-chain fatty acids: amino acid residues within the ATP/AMP signature motif of Escherichia coli FadD are required for enzyme activity and fatty acid transport. J Biol Chem. 2002;277:29369–76. doi: 10.1074/jbc.M107022200. [DOI] [PubMed] [Google Scholar]
- 7.Zou Z, Tong F, Faergeman NJ, Borsting C, Black PN, DiRusso CC. Vectorial acylation in Saccharomyces cerevisiae. Fat1p and fatty acyl-CoA synthetase are interacting components of a fatty acid import complex. J Biol Chem. 2003;278:16414–22. doi: 10.1074/jbc.M210557200. [DOI] [PubMed] [Google Scholar]
- 8.Fujino T, Kang MJ, Suzuki H, Iijima H, Yamamoto T. Molecular characterization and expression of rat acyl-CoA synthetase 3. J Biol Chem. 1996;271:16748–52. doi: 10.1074/jbc.271.28.16748. [DOI] [PubMed] [Google Scholar]
- 9.Berthiaume L, Deichaite I, Peseckis S, Resh MD. Regulation of enzymatic activity by active site fatty acylation: a new role for long chain fatty acid acylation of proteins. J Biol Chem. 1994;269:6498–505. [PubMed] [Google Scholar]
- 10.Marin EP, Derakhshan B, Lam TT, Davalos A, Sessa WC. Endothelial cell palmitoylproteomic identifies novel lipid-modified targets and potential substrates for protein acyl transferases. Circ Res. 2012;110:1336–44. doi: 10.1161/CIRCRESAHA.112.269514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomoda H, Igarashi K, Cyong JC, Omura S. Evidence for an essential role of long chain acyl-CoA synthetase in animal cell proliferation: inhibition of long chain acyl-CoA synthetase by triacsins caused inhibition of Raji cell proliferation. J Biol Chem. 1991;266:4214–9. [PubMed] [Google Scholar]
- 12.Glick BS, Rothman JE. Possible role for fatty acyl-coenzyme-A in intracellular protein-transport. Nature. 1987;326:309–312. doi: 10.1038/326309a0. [DOI] [PubMed] [Google Scholar]
- 13.Korchak HM, Kane LH, Rossi MW, Corkey BE. Long chain acyl coenzyme A and signaling in neutrophils: an inhibitor of acyl coenzyme A synthetase, triacsin C, inhibits superoxide anion generation and degranulation by human neutrophils. J Biol Chem. 1994;269:30281–7. [PubMed] [Google Scholar]
- 14.Faergeman NJ, Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem J. 1997;323(Pt 1):1–12. doi: 10.1042/bj3230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matesanz F, Tellez MM, Alcina A. The Plasmodium falciparum fatty acyl-CoA synthetase family (PfACS) and differential stage-specific expression in infected erythrocytes. Mol Biochem Parasitol. 2003;126:109–12. doi: 10.1016/s0166-6851(02)00242-6. [DOI] [PubMed] [Google Scholar]
- 16.Arrowood MJ, Sterling CR. Isolation of Cryptosporidium oocysts and sporozoites using discontinuous sucrose and isopycnic percoll gradients. J Parasitol. 1987;73:314–319. [PubMed] [Google Scholar]
- 17.Zhang H, Guo F, Zhu G. Involvement of host cell integrin alpha2 in Cryptosporidium parvum infection. Infect Immun. 2012;80:1753–8. doi: 10.1128/IAI.05862-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo F, Zhu G. Presence and removal of a contaminating NADH oxidation activity in recombinant maltose-binding protein fusion proteins expressed in Escherichia coli. BioTechniques. 2012;52:247–53. doi: 10.2144/0000113822. [DOI] [PubMed] [Google Scholar]
- 19.Bernson VSM. Acetyl-CoA hydrolase: activity, regulation and physiological significance of enzyme in brown adipose-tissue from hamster. Eur J Biochem. 1976;67:403–410. doi: 10.1111/j.1432-1033.1976.tb10705.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhuravleva E, Gut H, Hynx D, et al. Acyl coenzyme A thioesterase Them5/Acot15 is involved in cardiolipin remodeling and fatty liver development. Mol Cell Biol. 2012;32:2685–97. doi: 10.1128/MCB.00312-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai X, Woods KM, Upton SJ, Zhu G. Application of quantitative real-time reverse transcription-PCR in assessing drug efficacy against the intracellular pathogen Cryptosporidium parvum in vitro. Antimicrob Agents Chemother. 2005;49:4437–42. doi: 10.1128/AAC.49.11.4437-4442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fritzler JM, Zhu G. Novel anti-Cryptosporidium activity of known drugs identified by high-throughput screening against parasite fatty acyl-CoA binding protein (ACBP) J Antimicrob Chemother. 2012;67:609–17. doi: 10.1093/jac/dkr516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell LD, Stewart JN, Mead JR. Susceptibility to Cryptosporidium parvum infections in cytokine- and chemokine-receptor knockout mice. J Parasitol. 2002;88:1014–6. doi: 10.1645/0022-3395(2002)088[1014:STCPII]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 24.Arrowood MJ, Hurd MR, Mead JR. A new method for evaluating experimental cryptosporidial parasite loads using immunofluorescent flow cytometry. J Parasitol. 1995;81:404–9. [PubMed] [Google Scholar]
- 25.Arrowood MJ, Donaldson K. Improved purification methods for calf-derived Cryptosporidium parvum oocysts using discontinuous sucrose and cesium chloride gradients. J Eukaryot Microbiol. 1996;43:89S. doi: 10.1111/j.1550-7408.1996.tb05015.x. [DOI] [PubMed] [Google Scholar]
- 26.Matesanz F, Duran-Chica I, Alcina A. The cloning and expression of Pfacs1, a Plasmodium falciparum fatty acyl coenzyme A synthetase-1 targeted to the host erythrocyte cytoplasm. J Mol Biol. 1999;291:59–70. doi: 10.1006/jmbi.1999.2964. [DOI] [PubMed] [Google Scholar]
- 27.Fritzler JM, Zhu G. Functional characterization of the acyl-[acyl carrier protein] ligase in the Cryptosporidium parvum giant polyketide synthase. Int J Parasitol. 2007;37:307–16. doi: 10.1016/j.ijpara.2006.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ichihara K, Shibasaki Y. An enzyme-coupled assay for acyl-CoA synthetase. J Lipid Res. 1991;32:1709–12. [PubMed] [Google Scholar]
- 29.Yoshida K, Okamoto M, Umehara K, et al. Studies on new vasodilators, WS-1228 A and B. I. Discovery, taxonomy, isolation and characterization. J Antibiot (Tokyo) 1982;35:151–6. doi: 10.7164/antibiotics.35.151. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka H, Yoshida K, Itoh Y, Imanaka H. Studies on new vasodilators, WS-1228 A and B. II. Structure and synthesis. J Antibiot (Tokyo) 1982;35:157–63. doi: 10.7164/antibiotics.35.157. [DOI] [PubMed] [Google Scholar]
- 31.Tomoda H, Igarashi K, Omura S. Inhibition of acyl-CoA synthetase by triacsins. Biochim Biophys Acta. 1987;921:595–8. [PubMed] [Google Scholar]
- 32.Omura S, Tomoda H, Xu QM, Takahashi Y, Iwai Y. Triacsins, new inhibitors of acyl-CoA synthetase produced by Streptomyces sp. J Antibiot (Tokyo) 1986;39:1211–8. doi: 10.7164/antibiotics.39.1211. [DOI] [PubMed] [Google Scholar]
- 33.Hartman EJ, Omura S, Laposata M. Triacsin C: a differential inhibitor of arachidonoyl-CoA synthetase and nonspecific long chain acyl-CoA synthetase. Prostaglandins. 1989;37:655–71. doi: 10.1016/0090-6980(89)90103-2. [DOI] [PubMed] [Google Scholar]
- 34.Vessey DA, Kelley M, Warren RS. Characterization of triacsin C inhibition of short-, medium-, and long-chain fatty acid: CoA ligases of human liver. J Biochem Mol Toxicol. 2004;18:100–6. doi: 10.1002/jbt.20009. [DOI] [PubMed] [Google Scholar]
- 35.Kim JH, Lewin TM, Coleman RA. Expression and characterization of recombinant rat acyl-CoA synthetases 1, 4, and 5. Selective inhibition by triacsin C and thiazolidinediones. J Biol Chem. 2001;276:24667–73. doi: 10.1074/jbc.M010793200. [DOI] [PubMed] [Google Scholar]
- 36.Van Horn CG, Caviglia JM, Li LO, Wang S, Granger DA, Coleman RA. Characterization of recombinant long-chain rat acyl-CoA synthetase isoforms 3 and 6: identification of a novel variant of isoform 6. Biochemistry. 2005;44:1635–42. doi: 10.1021/bi047721l. [DOI] [PubMed] [Google Scholar]
- 37.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 38.Stockdale HD, Spencer JA, Blagburn BL. In: Prophylaxis and chemotherapy. 2nd ed. Fayer R, Xiao L, editors. Boca Raton, FL: CRC Press; 2007. pp. 255–88. Cryptosporidium and cryptosporidiosis. [Google Scholar]
- 39.Mashima T, Sato S, Okabe S, et al. Acyl-CoA synthetase as a cancer survival factor: its inhibition enhances the efficacy of etoposide. Cancer Sci. 2009;100:1556–62. doi: 10.1111/j.1349-7006.2009.01203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuda D, Namatame I, Ohshiro T, Ishibashi S, Omura S, Tomoda H. Anti-atherosclerotic activity of triacsin C, an acyl-CoA synthetase inhibitor. J Antibiot (Tokyo) 2008;61:318–21. doi: 10.1038/ja.2008.45. [DOI] [PubMed] [Google Scholar]
- 41.Beaumelle BD, Vial HJ. Acyl-CoA synthetase activity in Plasmodium knowlesi-infected erythrocytes displays peculiar substrate specificities. Biochim Biophys Acta. 1988;958:1–9. doi: 10.1016/0005-2760(88)90239-1. [DOI] [PubMed] [Google Scholar]
- 42.Camero L, Shulaw WP, Xiao L. Characterization of a Cryptosporidium parvum gene encoding a protein with homology to long chain fatty acid synthetase. J Eukaryot Microbiol. 2003;50(Suppl):534–8. doi: 10.1111/j.1550-7408.2003.tb00621.x. [DOI] [PubMed] [Google Scholar]
- 43.Theodos CM, Griffiths JK, D'Onfro J, Fairfield A, Tzipori S. Efficacy of nitazoxanide against Cryptosporidium parvum in cell culture and in animal models. Antimicrob Agents Chemother. 1998;42:1959–65. doi: 10.1128/aac.42.8.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujimoto Y, Onoduka J, Homma KJ, et al. Long-chain fatty acids induce lipid droplet formation in a cultured human hepatocyte in a manner dependent of acyl-CoA synthetase. Biol Pharm Bull. 2006;29:2174–80. doi: 10.1248/bpb.29.2174. [DOI] [PubMed] [Google Scholar]
- 45.Igal RA, Wang P, Coleman RA. Triacsin C blocks de novo synthesis of glycerolipids and cholesterol esters but not recycling of fatty acid into phospholipid: evidence for functionally separate pools of acyl-CoA. Biochem J. 1997;324(Pt 2):529–34. doi: 10.1042/bj3240529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie YL, Pan YE, Chang CJ, et al. Palmitic acid in chicken granulosa cell death-lipotoxic mechanisms mediate reproductive inefficacy of broiler breeder hens. Theriogenology. 2012;78:1917–28. doi: 10.1016/j.theriogenology.2012.07.004. [DOI] [PubMed] [Google Scholar]