Abstract
Gonadotropin-releasing hormone (GnRH) is a critical reproductive regulator in vertebrates. Homologous peptides are also found in invertebrates, with a variety of characterized functions. In the amphioxus, an invertebrate that provides the best model for the transition to vertebrates, four GnRH receptors (GnRHRs) were previously described, but their native ligands were not identified. Using a more sensitive search methodology with hidden Markov models, we identified the first GnRH-like peptide confirmed in the amphioxus Branchiostoma floridae. This peptide specifically activated one of the four GnRHRs. Although the primary structure of this peptide was divergent from any previously isolated GnRH peptide, the minimal conserved residues found in all other GnRH superfamily members were retained. The peptide was immunolocalized in proximity of the central canal of the anterior nerve cord, a region where other neuropeptides and receptors have been found. Additionally, the amphioxus GnRH-like gene was positioned in a locus surrounded by syntenic homologs of the human GnRH paralogon. The amphioxus GnRH-like peptide, with its distinct primary structure, activated a receptor with equal potency to multiple ligands that span the GnRH superfamily.
Keywords: hormone evolution, receptor evolution, GnRH superfamily, amphioxus genome, corazonin, adipokinetic hormone
Introduction
Vertebrates and amphioxi share a number of significant characteristics including a dorsal tubular central nervous system with an anterior brain and extended spinal cord (Wicht and Lacalli 2005). Although the other group of invertebrate chordates, tunicates, are now considered to be the closest living relatives to the vertebrates based on phylogenomic analyses (Bourlat et al. 2006; Delsuc et al. 2006), amphioxi form a sister group with the rest of the chordates and in many ways provide a better model for the comparison of invertebrate protochordates with vertebrates (Louis et al. 2012). Several features of amphioxus including the body plan, arrangement of hox genes, synteny of neighboring genes and embryonic development are more similar to the vertebrates than those same characteristics in tunicates.
Examination of the amphioxus (Branchiostoma floridae) genome (Putnam et al. 2008) indicated that the complement of hormones and receptors controlling reproduction shared a number of similarities with the vertebrate pattern (Holland et al. 2008). For neuroendocrine control of reproduction, kisspeptin receptor (GPR54) homologs (Holland et al. 2008) and kisspeptin-like sequences have been identified from the amphioxus genome, however, their function remains unknown (Mirabeau and Joly 2013). In vertebrates, the kisspeptin system is expressed in the brain and regulates the release of gonadotropin-releasing hormone (GnRH). More recently, proof that amphioxus also expresses GnRH-like receptors was provided by cloning and in vitro activation of receptors using vertebrate GnRH peptides, as the endogenous amphioxus GnRH ligand(s) remained elusive (Tello and Sherwood 2009).
In vertebrate reproduction, the two pituitary gonadotropin hormones, known as luteinizing hormone (LH) and follicle stimulating hormone (FSH), are stimulated by GnRH. Amphioxus and tunicates lack genes encoding any of the six common vertebrate pituitary hormones, including the gonadotropins or their receptors (Campbell et al. 2004; Holland et al. 2008). Both protochordate groups possess genes encoding the thyrostimulin subunits (Campbell et al. 2004; Holland et al. 2008; Tando and Kubokawa 2009b), which duplicated within the vertebrates to produce the glycoprotein hormone subunits of LH, FSH, and the related thyroid stimulating hormone (Sudo et al. 2005; Dos Santos et al. 2009). LH and FSH act on their receptors in the gonads to stimulate gametogenesis and sex steroid synthesis. The amphioxus genome encodes two nuclear receptors that are similar to the sex steroid receptors in vertebrates (Schubert et al. 2008) and a set of enzymes that act in the synthesis of estradiol, testosterone, and progesterone. Although these steroids have been identified in the amphioxus gonad (Callard et al. 1984; Mizuta and Kubokawa 2007), only estradiol has been shown to bind one of the steroidal receptors (Paris et al. 2008; Katsu et al. 2010). Nonetheless, amphioxus has considerable potential for reproductive cycle feedback at the level of the nervous system, gonads, and other tissues that may be targeted by the sex steroids.
The transition from invertebrate to vertebrate is hypothesized to have involved two rounds of whole-genome duplication in ancestral vertebrates before the radiation of lineages leading to the extant vertebrate classes (reviewed in Cañestro 2012). The resulting genomes contained up to four paralogous genes, although many losses occurred. The proposed four copies of the original vertebrate gnrh genes have been traced by identifying their gene neighbors in syntenic analyses. GnRH4 has not been identified in extant vertebrates and is thought to have been lost early in evolution of the vertebrates (Tostivint 2011; Decatur et al. 2013; Smith et al. 2013). Genomic analysis suggests GnRH3 was lost from the tetrapod lineage but is present throughout the teleost fish (Kuo et al. 2005) and may be homologous with lamprey GnRH-I and GnRH-III (Decatur et al. 2013; Smith et al. 2013). GnRH2 has been identified throughout the vertebrates including humans and has retained its conserved synteny (Gwee et al. 2009), although some mammals (e.g., mouse, rat, and chimp) have lost a functional GnRH2 gene (Stewart et al. 2009). GnRH1 is found from bony fish to humans; there are at least nine variant forms of GnRH1 that share structure and function (Roch et al. 2011). In the human genome, GnRH genes or their traces (ghosts) exist on four different chromosomes surrounded by genes with conserved synteny: GnRH1 is encoded on chromosome 8, GnRH2 on chr. 20, a lost GnRH3 (ghost) on chr. 10, and a GnRH4 ghost on chr. 5. These conserved GnRH paralogons belong to a shared chordate ancestral linkage group (ALG 7), which was defined by comparison with homologous genes found in the amphioxus B. floridae (Putnam et al. 2008).
The GnRH family of peptides and receptors has been proposed to be part of a larger superfamily including invertebrate-specific members (Roch et al. 2011). Initially, two invertebrate (Drosophila) receptors were shown to be structurally related to GnRH receptors (GnRHRs), but their ligands were identified as corazonin peptide and adipokinetic hormone (AKH) (Hauser et al. 1998; Cazzamali et al. 2002; Park et al. 2002; Staubli et al. 2002). Further analysis of the receptor families revealed that GnRH, corazonin, and AKH receptors were also closely related to the oxytocin, vasopressin, and crustacean cardioactive peptide (CCAP) receptors forming two large superfamilies with a shared ancestry that emerged before the origin of the bilaterians (Roch et al. 2011).
In this study, evidence is presented that an amphioxus GnRH-like peptide has been identified. The cloned amphioxus gnrh-like cDNA is characterized, and the putative peptide is aligned with vertebrate GnRHs and closely related invertebrate GnRH-like peptides, corazonins, and AKHs. The bioactivity of the new peptide is tested in vitro for activity with the four known amphioxus GnRHRs. Synteny analysis is used to determine the lineage of this ancestral-type peptide and its relationship to vertebrate GnRHs. Phylogenetic analysis of the amphioxus receptors demonstrates a long standing and complex relationship among the receptors of GnRH, corazonin, AKH, oxytocin/vasopressin, and neuropeptide S.
Results
Identification of a GnRH-Like Peptide in the Amphioxus B. Floridae
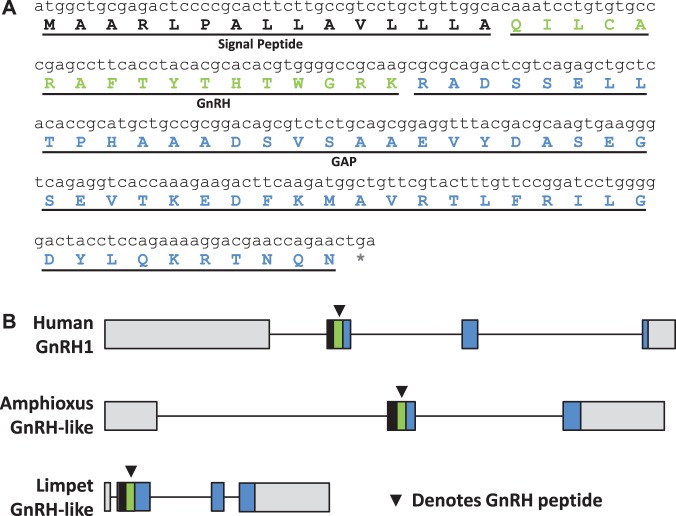
Two putative GnRH-like peptides from B. floridae sequence databases were uncovered using a hidden Markov Model (HMM) for a variety of vertebrate and invertebrate GnRH, AKH, and corazonin propeptides. Subsequently, only one peptide was revealed to be bioactive, and a cDNA clone representing the gene transcript was amplified and confirmed by sequencing (GenBank accession number KF601546). As seen in figure 1A, the amphioxus GnRH-like propeptide contains the basic elements found in GnRH family members: a signal peptide, mature peptide with an N-terminal glutamine (pyroglutamic acid in vivo) and C-terminal glycine (amidated in vivo) followed by a dibasic cleavage site, and a nonspecific gene-associated peptide (GAP) of 58 amino acids. The organization of the amphioxus gnrh gene as determined from the genome showed three exons with GnRH encoded on exon 2 and GAP on exons 2 and 3 (fig. 1B).
Fig. 1.
Peptide, cDNA, and gene organization for GnRH-like hormone in amphioxus. (A) The cloned cDNA sequence from the beginning of the signal peptide to the stop codon is shown in black lowercase letters. The deduced amino acids are shown for the signal peptide in black capital letters; the GnRH sequence with a terminal amidation site (G) and dibasic cut site (RK) in green; and the GAP sequence in blue. The asterix (*) indicates the stop codon. (B) The organization of the amphioxus gnrh-like gene is compared with the human GNRH1 and limpet gnrh-like genes. The exon–intron data were obtained from the respective genome databases for amphioxus and human and from (Tsai and Zhang 2008) for limpet. The distances are drawn to scale according to the number of nucleotides. The signal peptides are shown by a black box, the GnRH region by green, GAP by blue, and the 5′- and 3′-untranslated regions in gray. The arrowhead (▾) denotes the GnRH peptide coding region.
The deduced mature sequence of amphioxus GnRH-like peptide was aligned with vertebrate GnRH2 along with a variety of invertebrate GnRH superfamily members. As seen in figure 2, even with a significant insertion of five amino acids near the N-terminal region of amphioxus GnRH-like peptide, key residues are conserved among all superfamily members (highlighted in blue). These include the N-terminal pyroglutamate, a dibasic aromatic-serine/threonine motif (F/W/Y-S/T), and a C-terminal aromatic residue, typically tryptophan (W). Five out of 10 residues in the pan-vertebrate GnRH2 peptide are similar in the amphioxus peptide. The second putative GnRH-like peptide, which was nonfunctional in B. floridae GnRHR assays, is also shown at the bottom of figure 2 for comparison.
Fig. 2.
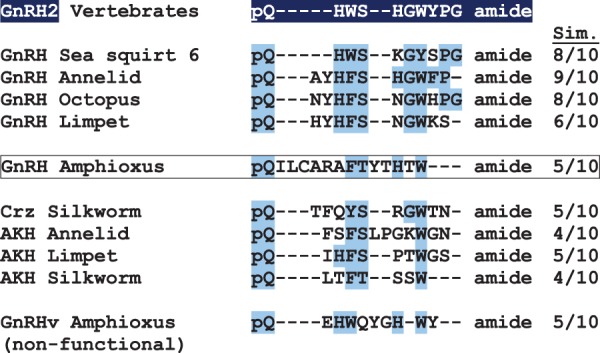
Alignment of GnRH superfamily peptide homologs from a variety of bilaterians. Putative full-length peptide sequences from representative species were manually aligned for GnRH, corazonin (Crz), and adipokinetic hormone (AKH), which are members of the GnRH superfamily of peptides. The vertebrate GnRH2, which is present throughout the vertebrates, is used as the standard for comparison of amino acid similarity among the peptides. The number of similar amino acids is shown on the right under the column marked Sim. The novel amphioxus GnRH is shown with its amino acid similarity to GnRH2 (five amino acids are similar to the GnRH2 standard, which has 10 amino acids). A second GnRH peptide (GnRHv, variant) that was identified in the amphioxus genome is shown below (this synthesized peptide did not activate any of the four known amphioxus GnRHRs).
In Vitro Functional Assays with Amphioxus GnRHRs
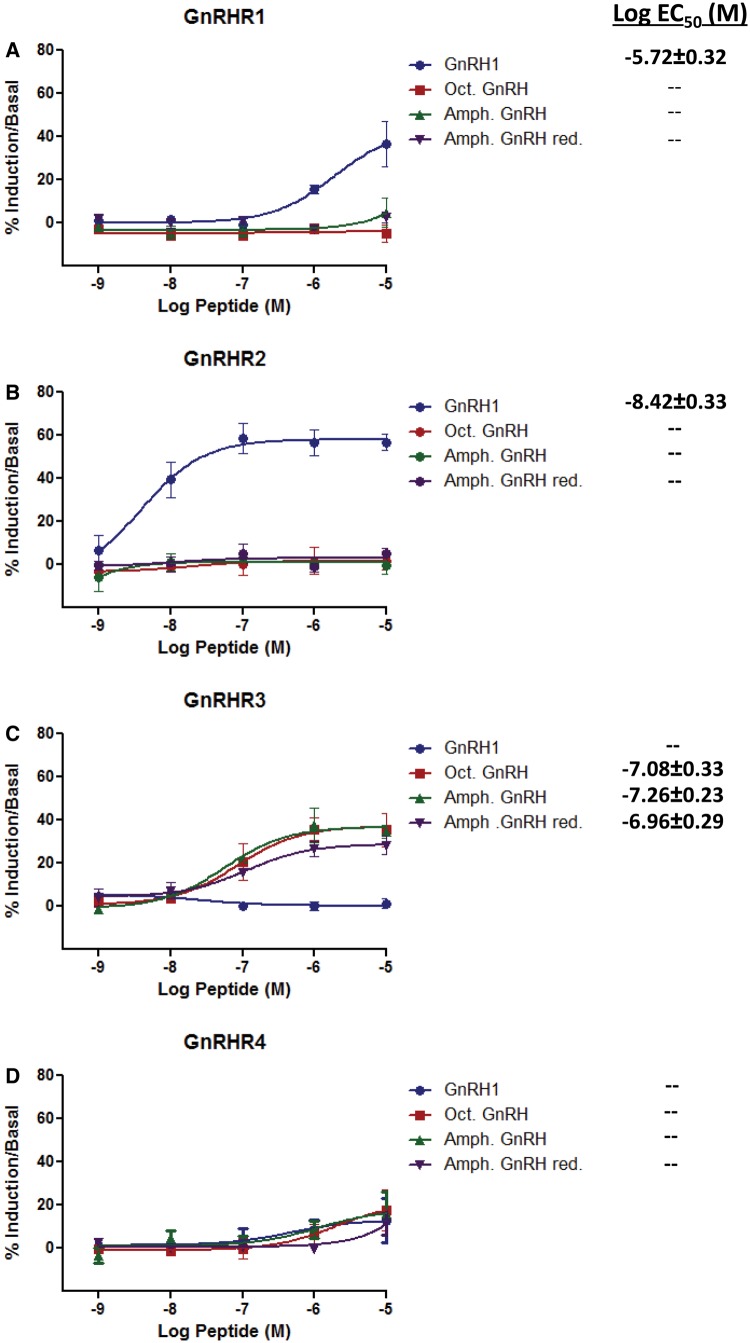
The putative amphioxus GnRH-like peptides were tested for bioactivity in vitro with the previously identified B. floridae GnRHRs (Tello and Sherwood 2009). Because of the cysteine residue in the amphioxus GnRH-like peptide (see in fig. 1), reducing conditions were tested alongside the standard peptide. For comparison, the octopus GnRH-like peptide and vertebrate GnRH1 were used. Receptors were tested for activation by both the Gq/11 pathway (inositol phosphate 1 or IP1 accumulation) and the Gs/Gi pathway (cyclic adenosine monophosphate or cAMP stimulation/inhibition). Only Gq/11 activation was detected for any of the receptors. As seen in figure 3, only vertebrate GnRH1 activated amphioxus GnRHRs 1 and 2. The amphioxus GnRH-like peptide only activated GnRHR3 to a significant degree (log EC50 of −7.26 ± 0.23 M). Both the reduced amphioxus GnRH-like peptide and the octopus peptide also activated GnRHR3 to a similar degree (log EC50 of −6.96 ± 0.29 M and −7.08 ± 0.33 M, respectively). The resulting log EC50 values were not statistically different (P > 0.05) by one-way analysis of variance (ANOVA). None of the peptides activated GnRHR4 within the limits tested. The second putative amphioxus GnRH peptide did not activate any of the receptors (not shown).
Fig. 3.
Response of four amphioxus GnRHRs to human GnRH1, octopus, and amphioxus GnRH-like peptide, both oxidized and reduced. The percent induction of inositol phosphate one (IP1) accumulation compared with basal levels in COS-7 cells expressing amphioxus GnRHRs (R) is shown after induction with graded concentrations of peptide for 1 h. Cells were transfected with a vector containing (A) GnRHR1, (B) GnRHR2, (C) GnRHR3, or (D) GnRHR4 followed 48 h later by peptide addition. Each dose-response curve was generated by three independent experiments; error bars signify the mean ± standard error of means (SEM) of replicates. The concentration of peptide that stimulated the log EC50 IP1 response is shown on the right. A dash (––) indicates that a response was not detected.
Localization of the GnRH-Like Peptide within the Amphioxus Nervous System
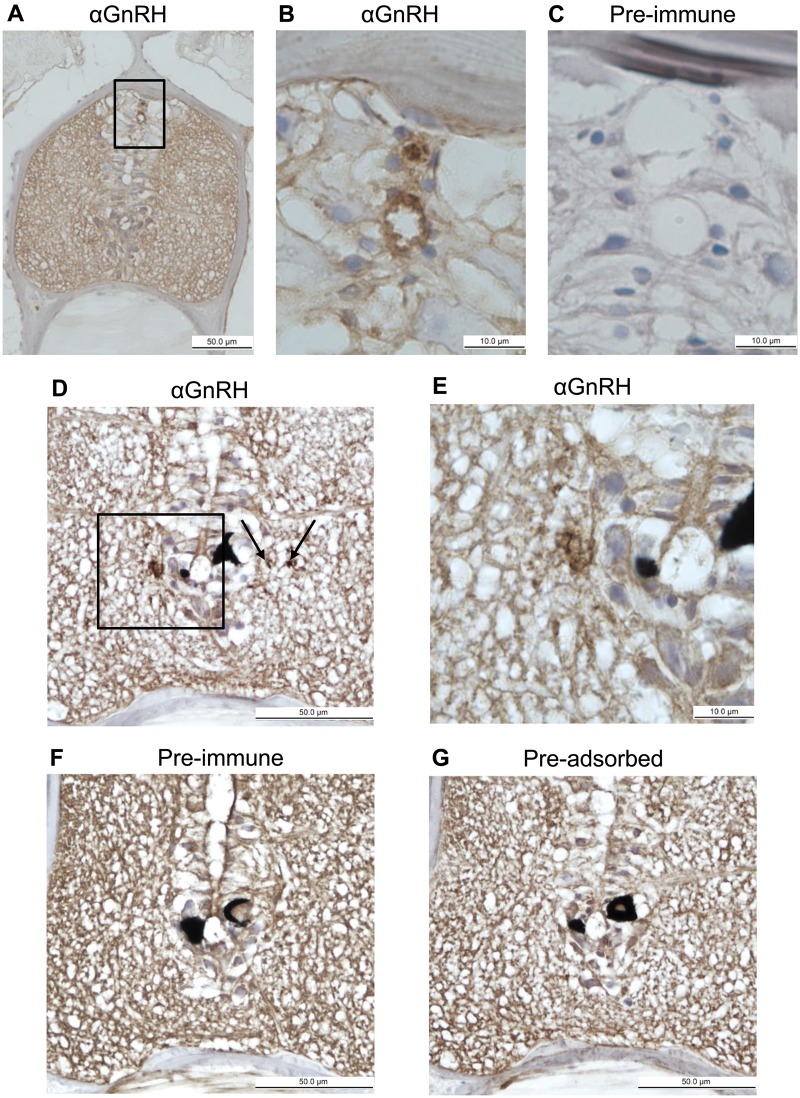
A sexually mature male B. floridae specimen was sectioned transversally in 4.0 µm increments from the anterior end of the animal throughout the brain-like region (Wicht and Lacalli 2005). Specific labeling for amphioxus GnRH-like peptide was observed in proximity of the central canal of the amphioxus anterior nerve cord (brain-like region), as shown in figure 4. Labeling was observed in both the dorsal central canal (fig. 4A and B) and near the ventral central canal (fig. 4D and E). GnRH labeling appeared to be associated with cell bodies clustered around this ventricle, as hematoxylin-stained nuclei can be seen surrounding the space. The location of the displayed sections coincides with the amphioxus-specific organ known as Hatschek’s pit, which does not make contact with the nerve cord. No labeling was observed in the control sections, which were treated exactly the same as sections incubated with amphioxus GnRH-like antibody. Controls included preimmune serum taken from the same rabbit before it was inoculated with amphioxus GnRH-like peptide (fig. 4C and F) and primary antibody preadsorbed with amphioxus GnRH-like peptide (fig. 4G).
Fig. 4.
Immunolabeling of GnRH in the central nervous system (CNS) by an amphioxus-specific GnRH antiserum. (A) The transverse section through the head region of amphioxus Branchiostoma floridae shows the location of the brain-like anterior nerve cord (brown). A boxed area shows the site of GnRH labeling in the dorsal central canal. (B) Magnification of the boxed area shows GnRH label surrounding the open central canal (ventricle) or filling the canal (above) in the CNS. The cells surrounding the outside of the canal are identified by their nuclei, which are stained blue by hematoxylin. (C) A transverse section 0.2 mm posterior to (B), incubated with preimmune serum and showing the same region. (D) Cross section of amphioxus CNS approximately 0.7 mm posterior to that in (A). In the box and next to the arrowheads, labeling is observed near the ventral central canal. (E) An enlargement of the CNS from (D). A pair of black pigment cells is shown. (F) A transverse section adjacent to that of (D), labeled with preimmune serum instead of primary antibody, without specific labeling. (G) A transverse section adjacent to (F), labeled with preadsorbed primary antibody, without specific labeling. Typically, many cells (each identified by a blue nucleus) are clustered near the central canal as the CNS is not vascularized.
Syntenic Relationship among Amphioxus, Limpet, and Human GnRH Paralogons
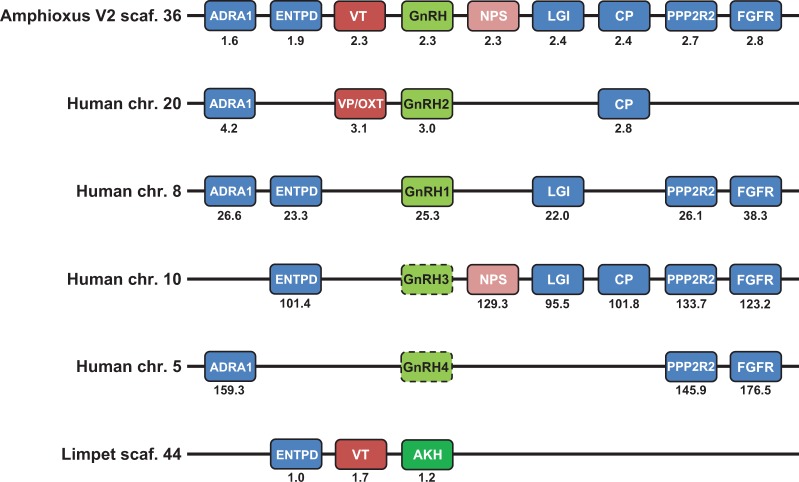
The syntenic relationship of neighboring genes surrounding the B. floridae GnRH-like gene model is compared with both human GNRH genes and limpet akh gene in figure 5. Also listed are the anticipated locations of “ghost” GNRH3 and 4 on human chromosomes 10 and 5, respectively, which were lost at different periods of vertebrate evolution (Kim et al. 2011; Tostivint 2011). The amphioxus gnrh-like gene was located on scaffold 36 in the second version of the genome assembly and was flanked by gene models for a vasotocin homolog and a neuropeptide S homolog. Vasotocin family (VT: vasopressin/oxytocin/mesotocin/isotocin in vertebrates) genes are often found immediately flanking GnRH genes in vertebrates, as is the case for both vasopressin and oxytocin adjacent to GnRH2 on chromosome 20 in humans (fig. 5). Homologous genes on amphioxus scaffold 36 that are found on human chromosomes corresponding to the GnRH paralogon (regions of chromosomes 5, 8, 10, and 20) are also shown. This chordate GnRH paralogon also shares more limited synteny with the protostome limpet scaffold that contains the AKH and VT gene models, although this is the only example of association between a GnRH superfamily member and VT found in protostomes thus far.
Fig. 5.
Synteny of amphioxus GnRH with human GnRH paralogons and limpet AKH. The amphioxus genome, version 2 (V2), scaffold (scaf.) 36 contains the GnRH gene (green) with its nearest neighbors of vasotocin (VT: red) and neuropeptide S (NPS: pink) on either side. Further along the scaffold are a number of selected genes (blue) that also show synteny with the human GnRH paralogon. Four human chromosomes (chr.) have GnRH genes (GnRH1 and GnRH2) that are functional or genes (“ghost” GnRH3 and GnRH4) that were lost (dotted lines around gene) during the evolution of the vertebrates, but the retained neighboring genes mark the location of the lost genes. An important comparison is between the conserved neighboring genes of GnRH and VT: amphioxus GnRH-VT, human GnRH2-VP/OXT (vasopressin/oxytocin), and the limpet AKH-VT. The association of amphioxus GnRH and NPS and the same in human GnRH3 and NPS is also shown. The neighboring genes are aligned for ease of comparison, but distances (shown below) and order may vary.
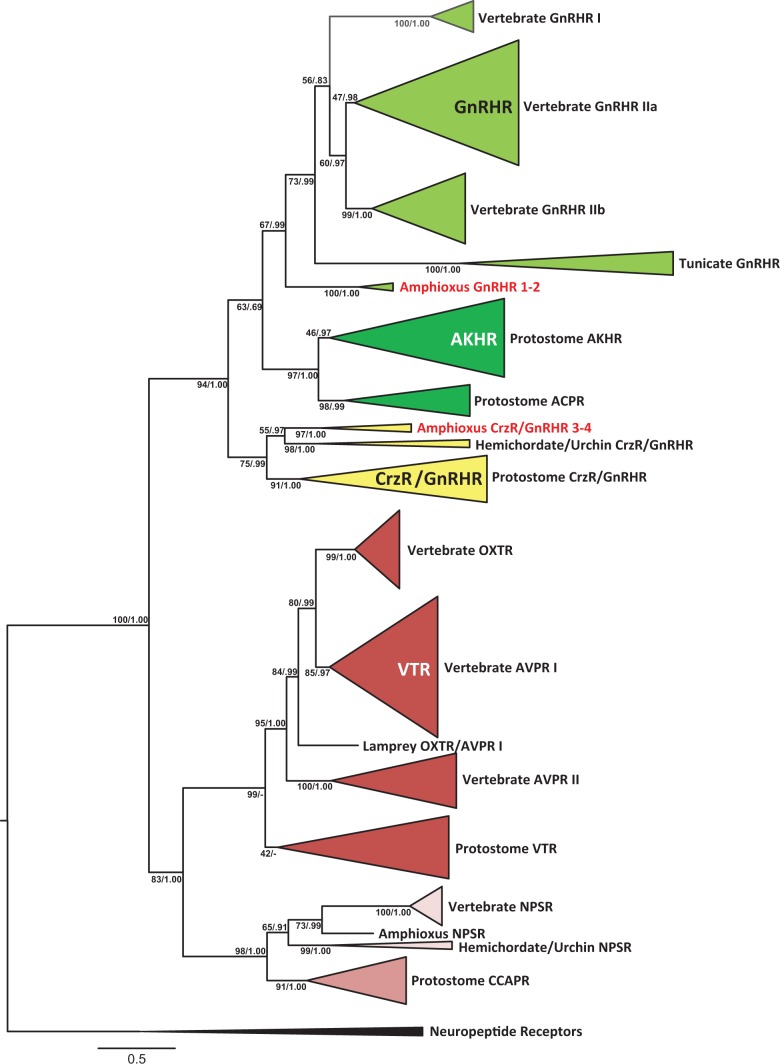
Phylogenetic Relationship of GnRHRs with Related Bilaterian GPCRs
Phylogenetic analysis of the GnRH superfamily receptors (fig. 6 and supplementary fig. S2, Supplementary Material online) places amphioxus GnRHRs 1 and 2 basal to the vertebrate/tunicate GnRHR group, whereas amphioxus GnRHRs 3 and 4 segregate with the invertebrate corazonin receptor (CrzR)/GnRHR clade. The relationship of the GnRHR “superfamily” is defined, with the chordate GnRHRs grouped together with the protostome AKHR/ACPRs. The CrzR/GnRHRs form a basal sister group with this clade.
Fig. 6.
Phylogenetic tree of GnRH and VP receptor superfamily from vertebrates and invertebrates. Homologs of receptors used to generate a maximum likelihood tree are gonadotropin-releasing hormone (GnRHR: light green), adipokinetic hormone (AKHR: dark green), AKHR/Corazonin-related peptides (ACPR: dark green), corazonin (Crz: yellow), vasopressin family receptors (red) including oxytocin (OXTR), arginine vasopressin (AVPR), and vasotocin (VT), along with neuropeptides S (NPSR: pink), and crustacean cardioactive peptide (CCAPR: dark pink). The outgroup included a variety of neuropeptide receptors including those for tachykinin (TACR), somatostatin (SSTR), and galanin (GALR). The receptors are listed in supplementary table S1, Supplementary Material online. Nodes were compressed to represent animal lineages. The tree was prepared from a degapped alignment of receptors, using the conserved region of the seven-transmembrane domain including the intracellular and extracellular loops. RaxML 7.4.2 was used to generate the maximum likelihood tree topology, using the combined tree-search/fast bootstrap method under the PROTGAMMALG model (four discrete rate categories) with 1,000 fast bootstraps. Posterior probabilities are also listed at each node after the bootstrap value, from two converging chains generated in PhyloBayes MPI 1.4f under the same model constraints as the maximum likelihood tree. The two chains were sampled every 10 trees for 20,000 cycles (2,000 trees per chain) after an initial burn-in of 5,000 cycles was discarded.
The relationship of vasotocin receptors (VTRs) is also shown in figure 6, with vertebrate oxytocin receptor (OXTR) grouping with vasopressin receptor I (AVPR I) and vertebrate AVPR II a sister group to both. The protostome VTR group forms a sister group with the entire vertebrate VTR family, with weak support. All the bilaterian VTRs form a sister group with the deuterostome neuropeptide S (NPS) receptors and protostome crustacean cardioactive peptide receptors (CCAPRs), which appear to share a more recent common ancestor. Although we have previously noted the relationship between the vasopressin and GnRH superfamilies (Roch et al. 2011), neuropeptide S receptors (NPSRs) are now included based on their structural homology (Pitti and Manoj 2012) and on the syntenic relationship of the NPS peptide with GnRHs and VTs.
Discussion
Evolution of a Structurally Novel GnRH-Like Peptide in the Amphioxus
We have documented the first confirmed native peptide from the amphioxus lineage with GnRH-like features. The propeptide shares the basic overall structure (signal peptide, mature peptide, and GAP peptide) with other GnRH-like peptides (fig. 1A). The predicted gene structure is also conserved with both the human GNRH1 and limpet gnrh-like genes, as seen in figure 1B, although the amphioxus gnrh-like gene has three exons instead of the canonical four, missing the third exon that typically encodes a region of the GAP peptide. Regardless, the overall pattern of the coding and noncoding elements, especially the signal peptide, mature peptide, and start of the GAP peptide on exon 2, is conserved in the amphioxus.
The primary structure of the mature peptide, as seen in figure 2, illustrates both the unique features of the amphioxus GnRH-like ligand and conserved amino acids found throughout the superfamily members in very distantly related bilaterians. The amphioxus GnRH-like peptide has a novel insertion of five amino acids near the N-terminus, including a cysteine residue at position four, which makes it unlike any superfamily peptide characterized to date except for a GnRH identified in the tunicate Chelyosoma productum; the latter was shown by mass spectrometry to form a disulfide-linked homodimer (Powell et al. 1996). However, amphioxus GnRH-like peptide is unlikely to form a dimer as receptor activation was not changed in the reduced state (fig. 3), and the synthetic peptide had a mass consistent with the monomer as observed by mass spectrometry (not shown). Along with an additional two-amino acid insertion close to the C-terminal, amphioxus GnRH-like peptide is the longest superfamily member characterized to date, at 14 amino acids, compared with the 10 amino acid peptides found in vertebrates and 8–12 amino acid peptides found in other invertebrates. It lacks several residues conserved in the vertebrate peptides, including the histidine at position two, the glycine at position six, the proline at position nine, and the glycine at the C-terminus. These residues have been inferred as critical components for the vertebrate peptides’ three-dimensional structure and receptor-binding capability in a variety of studies (reviewed in Millar et al. 2004). In the case of amphioxus GnRH-like peptide and several other members of the superfamily found in invertebrates, however, a significantly relaxed pattern of conservation is observed (fig. 2).
It appears that the minimal conservation of structure necessary to activate receptors within the GnRH peptide superfamily is the N-terminal pyroglutamate, an aromatic and threonine/serine dipeptide motif (W/F/Y-S/T), an additional aromatic residue, typically tryptophan, found close to the C-terminus, and an amidated C-terminus. The only exception to this pattern of conservation observed in all peptides characterized to this date is a replacement of the tryptophan residue close to the C-terminus with a leucine (or V/M) in vertebrate GnRH1 and in four tunicate GnRHs (Roch et al. 2011). Every individual receptor has additional constraints depending on other amino acids in its cognate peptide and has been detailed for vertebrate peptides and receptors (reviewed in Millar et al. 2004). Confirmation of this minimal pattern of necessary conservation was provided by the second putative GnRH-like peptide from amphioxus (GnRHv, fig. 2) that we tested and found nonfunctional. In that case, the GnRHv peptide had a W-Q dipeptide motif instead of W-S/T, while retaining the other three conserved elements. Earlier we tested six potential amphioxus GnRH decapeptides that proved to be nonfunctional; five peptides lacked W-S/T and the sixth lacked an aromatic W/Y/F near the C-terminus (unpublished). A comparison of functional GnRH-like peptides found across the bilaterians, including the novel peptide found in amphioxus, redefines the minimal primary structure necessary for this superfamily of peptides.
An Unexpected Functional Profile of Receptor Activation
The initial characterization of amphioxus GnRHR-like receptors was carried out using heterologous ligands from vertebrates, octopus and silkworm, as the native peptide was unknown (Tello and Sherwood 2009). That study, which was partially repeated here, found that amphioxus receptors were activated in a pattern consistent with their phylogenetic position. Receptors 1 and 2, which group with the vertebrate and tunicate GnRHRs (fig. 6), were activated by mammalian GnRH1 and vertebrate GnRH2 exclusively. In the case of amphioxus GnRHR2, mammalian GnRH1 was able to activate the receptor within a vertebrate physiological range (log EC50 −8.42 ± 0.33 M in this study). Another group claimed to have isolated a peptide identical to mammalian GnRH1 from B. lanceolatum, an amphioxus closely related to B. floridae (Chambery et al. 2009). This peptide has not been confirmed in the genome by us or others. Considering the GnRH1 standard they used on their column may have contaminated it, the legitimacy of this peptide is questionable. The amphioxus GnRH-like peptide we isolated and characterized, however, does not appear to be the native peptide for amphioxus GnRHR1 or 2, as neither were stimulated in receptor activation assays for either Gq/11 or Gs/Gi activity (fig. 3). This leads us to hypothesize that there is still another, more vertebrate-like peptide to be found in amphioxus that is missing from the current nucleotide and sequence databases, as amphioxus expresses these two functional vertebrate-like receptors.
Amphioxus GnRHR3 was the only receptor that was activated by the native GnRH-like peptide. This receptor has an interesting functional and phylogenetic profile. Both amphioxus GnRHR3 and 4 phylogenetically group with members of the protostome CrzR/GnRHR receptor family, as seen in figure 6 and supplementary figure S2, Supplementary Material online. The corazonins, unlike the GnRHs, are found in insects and crustaceans and appear to be associated with a variety of nonreproductive functions including cardiostimulatory effects (Veenstra 1989) and the modulation of stress (Veenstra 2009; Boerjan et al. 2010). The GnRH-like peptides characterized in mollusks, which bind a CrzR/GnRHR, have demonstrated divergent effects. Both octopus and Aplysia (sea hare) GnRH-like peptide stimulated nonreproductive responses related to motor control (Kanda et al. 2006; Tsai et al. 2010), but the octopus peptide additionally exhibited reproductive functions (Kanda et al. 2006). As we did not assess any physiological impacts of amphioxus GnRH in vivo, we can only speculate that reproductive or nonreproductive functions are possible in this basal chordate.
The amphioxus GnRH-like peptide activated GnRHR3 specifically for the Gq/11 pathway and not the Gs/Gi pathway, as seen in figure 3. The predominate pathway for the GnRH superfamily receptors is Gq/11. The notable exception to this functional profile was demonstrated when the tunicate GnRHRs were characterized and found almost entirely to activate the Gs pathway (Tello et al. 2005). Amphioxus GnRHR4, which was not activated by any of the tested peptides, may be nonfunctional or, as is the case for tunicate GnRHR4, may not bind GnRH but form heterodimers with the other receptors to modulate their actions (Sakai et al. 2010).
A striking feature of the activation of GnRHR3 by amphioxus GnRH-like peptide was that the dose responses were similar between the native and reduced peptide and also that of octopus GnRH (fig. 3). The EC50 values in each case were not statistically different. Indeed, the initial receptor GnRHR3 characterization, which also tested silkworm AKH, found EC50 values that were statistically the same with the octopus peptide (Tello and Sherwood 2009). As well, vertebrate GnRH2 activated amphioxus GnRHR3 within the vertebrate physiological range. Given the significant differences found in the primary structures of amphioxus GnRH-like peptide, octopus GnRH, silkworm AKH, and vertebrate GnRH2 (fig. 2), it seems that the binding and activation specificity of amphioxus GnRHR3 must be less selective than other CrzR/GnRHR family members characterized to date (Cazzamali et al. 2002; Park et al. 2002; Belmont et al. 2006; Kanda et al. 2006). This may help to explain why amphioxus GnRH-like peptide, with such a divergent primary structure, can still activate this receptor at vertebrate physiological concentrations.
GnRH-Like Peptide Localization along the Nerve Cord Central Canal
The localization of amphioxus GnRH-like peptide in limited areas around the central canal of the brain-like region of the anterior nerve cord provides further evidence of its conserved role as a neuropeptide hormone. As seen in figure 4, immunolabeling with an antibody raised against amphioxus GnRH-like peptide revealed localization near or within the central canal. This ventricle runs the length of the nerve cord, to the posterior end of the amphioxus notochord, and is surrounded by the majority of neuronal cell bodies in the nervous system (Wicht and Lacalli 2005). The central canal provides a potential distribution system for secreted neuropeptides. Expression of other neuropeptide transcripts have been identified proximal to the central canal by in situ hybridization, including the thyrostimulin subunits gpa2 (Tando and Kubokawa 2009a) and gpb5 (Tando and Kubokawa 2009b), as well as the transcript for vasotocin (Kubokawa et al. 2010). Additionally, the expression of amphioxus gnrhr1 and gnrhr2 was detected near the anterior central canal, although localization of gnrhr3 was not demonstrated (Tando 2009). A previous immunohistochemical profile of the amphioxus nerve cord using a heterologous polyclonal antibody raised against lamprey GnRHI labeled Hesse photoreceptor cells in the amphioxus nerve cord, located ventral to the central canal (Castro et al. 2006). The significance of this finding cannot be confirmed, however, as our antibody is designed against the amphioxus GnRH that activates receptor GnRHR3. Both previous and present antibody or in situ labeling were not associated with the organ known as Hatschek’s pit, which has been suggested as an amphioxus homolog of the vertebrate pituitary (Nozaki and Gorbman 1992; Candiani and Pestarino 1998; Gorbman et al. 1999; Candiani et al. 2008).
Synteny Reveals a Deep Genomic Relationship Conserved between GnRH, VT, and NPS
The conserved synteny of the amphioxus gnrh locus, found on scaffold 36 in version 2 of the genome, is detailed in figure 5. As displayed, the gene encoding amphioxus GnRH is flanked by the gene encoding vasotocin (VT), which is a genomic feature conserved throughout the vertebrates with gnrh2 and oxt/avp (oxytocin/vasopressin) in close proximity (Gwee et al. 2009; Kim et al. 2011; Tostivint 2011). Additionally, a novel gene encoding a neuropeptide S-like peptide, as well as a neurophysin-like peptide, was recently described by Mirabeau and Joly (2013) and is found immediately downstream of the amphioxus gnrh-like gene. Although neuropeptide S is not found in proximity to human GNRH1 or GNRH2, it is located within the “ghost” GNRH3 paralogon on chromosome 10, suggesting the genomic organization of all three peptides has been conserved throughout chordate evolution. The gene for GnRH-like peptide in amphioxus was not predicted and therefore omitted from two previous studies that investigated this locus (Gwee et al. 2009; Mirabeau and Joly 2013). Although the functional significance of the conserved syntenic relationship between vt and gnrh is unknown, it has been suggested that clusters of genes maintained in syntenic relationships share regulatory elements driving coexpression and other selective constraints (reviewed in Michalak 2008).
The shared synteny of genes encoding GnRH-like peptides and vasotocins appears to have been preserved throughout the vertebrates, amphioxus, and limpet, supported by the close proximity of akh and vt in the limpet genome (fig. 5). Many other genes in this syntenic locus are conserved on amphioxus scaffold 36 and multiple GNRH-bearing chromosomes of the human genome, including homologs of protein phosphatase 2 (PPP2R2), which has been previously identified as a conserved syntenic element in the gnrh paralogon of vertebrates (Kim et al. 2011; Tostivint 2011). Our analysis covered a wider region of the chordate ancestral linkage group 7 that includes the GNRH paralogon in the human genome, and so we have identified genes encoding other orthologs conserved in humans and amphioxus including adrenoreceptor alpha 1, leucine-rich glioma inactivated, carboxypeptidase, and fibroblast growth factor receptor (fig. 5 and supplementary table S1, Supplementary Material online). The gene encoding ectonucleoside triphosphate diphosphohydrolase is not only conserved in the vertebrate gnrh paralogon (Tostivint 2011) and amphioxus but also on the limpet scaffold containing akh/vt. Taken together, it is clear that limited microsynteny has been conserved on the paralogon throughout bilaterian evolution, with a close relationship between genes encoding GnRH-like peptides and vasotocins.
GnRH-Like Peptide and Its Receptor in Amphioxus—An Evolutionary Novelty?
The discovery of an amphioxus GnRH-like peptide and characterization of its functional profile raises questions about its place within the larger GnRH superfamily. As previously discussed, the peptide specifically binds GnRHR3 in amphioxus, which bears a closer structural homology to the invertebrate CrzR/GnRHR family than to the vertebrate GnRHR family (fig. 6). To date, we have not been able to identify a peptide that clearly resembles corazonin in the amphioxus, hemichordate, or sea urchin sequence databases. Synteny analysis, however, demonstrates that the amphioxus gnrh-like gene locus is directly related to both the vertebrate gnrh and the limpet akh gene loci. Because of the primary structure of amphioxus GnRH-like peptide, its relative homology to the different members of the superfamily is ambiguous (fig. 2), and without any other native peptides to compare it to, we are left to hypothesize about its origin. A graphical overview of GnRHR superfamily members identified and characterized in various animal lineages is presented in figure 7. Chordates and possibly deuterostomes that have been characterized to date have a receptor that binds vertebrate GnRHs, whereas protostomes have receptors that bind AKHs specifically. The invertebrate CrzR/GnRHR family appears to have arisen at the origin of the bilaterians and has been maintained in most lineages; however, it was lost independently by nematodes and by the ancestor of tunicates and vertebrates. Homologous receptors can still be found in the other invertebrate deuterostomes (amphioxi, hemichordates, and echinoderms), but their native ligands have not been characterized, and so it is unknown whether unique structural characteristics are shared with the amphioxus GnRH-like peptide.
Fig. 7.
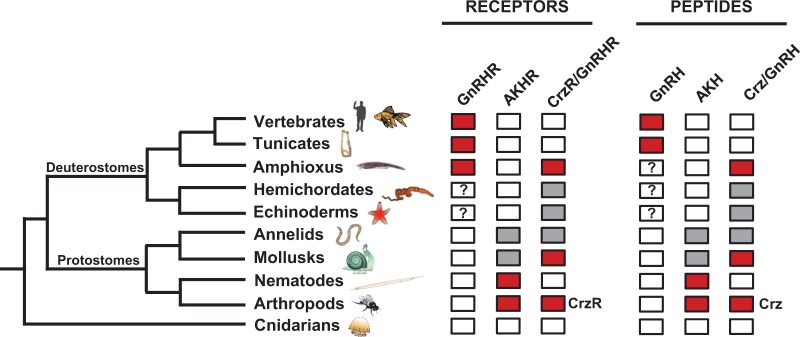
Diagram showing the identification of GnRHR superfamily members and ligands. The presence of three families of receptors is shown in colored boxes for invertebrates and vertebrates from cnidarians to humans. The receptors (R) include chordate-type gonadotropin-releasing hormone (GnRHR), adipokinetic hormone (AKHR), and corazonin/invertebrate GnRH (CrzR/GnRHR). Also included are the corresponding peptides for the receptors. Red boxes indicate that receptors and peptides from respective groups have been isolated and are functional. Gray boxes indicate that gene predictions and/or transcripts have been identified for receptors and peptides, but they have not been characterized. The question marks in the hemichordate and echinoderm GnRHR and peptide boxes indicate that a group of GnRHR-like receptor and peptide sequences have been identified in Saccoglossus kowalevskii and Strongylocentrotus purpuratus (sea urchin), but their phylogenetic position is ambiguous. The question mark in the amphioxus GnRH box indicates that the peptide(s) activating GnRHR1 and 2 has been predicted but not identified.
Given that amphioxus expresses receptors that group with the vertebrate GnRHRs and are activated by vertebrate peptides, it is likely that at least one other amphioxus GnRH-like peptide is expressed, with an unknown genomic location. For an obscure reason, selection was likely relaxed on the peptide characterized in this study, allowing for the divergent primary structure. This pattern of selection is often seen in gene duplications and can result in the neofunctionalization of one copy for a new role (Force et al. 1999; Lynch and Conery 2000; Pegueroles et al. 2013). Selection on the binding constraints of GnRHR3 was also likely relaxed, supported by its multiple ligand activation by other GnRH superfamily peptides that share limited structural homology (fig. 3).
Conclusion
Amphioxus is often regarded as the best prevertebrate model before the transition to the vertebrates as it has a well-conserved complement of the genes lacking the two rounds of whole-genome duplication present in the vertebrate ancestor (Putnam et al. 2008; Louis et al. 2012). For the GnRH superfamily, this would appear to be the case as B. floridae has both vertebrate-type GnRHRs and invertebrate-type corazonin/GnRHRs, supported by phylogenetics and nonnative peptide activation. The native GnRH-like peptide we have isolated in this study has a unique structure and is one of several peptides tested that activates the amphioxus invertebrate-type GnRHR3. Synteny analysis of the region surrounding the amphioxus gnrh-like gene indicated for the first time a conserved gene neighborhood between the vertebrate GnRH paralogon and an invertebrate. The tightly conserved genomic relationship of genes expressing GnRH and vasotocin was also reinforced and a new relationship with neuropeptide S was found.
Materials and Methods
Animals
Adult (sexually mature) and juvenile B. floridae specimens were collected in the Gulf of Mexico (Gulf Specimen Marine Lab, Panacea, FL) and shipped to Victoria, BC. Upon arrival, animals were euthanized and either frozen in liquid nitrogen followed by storage at −80 °C for RNA extraction or immersed in 4% paraformaldehyde in phosphate buffered saline (PBS), followed by storage in 100% methanol at −20 °C, for immunohistochemistry. RNA isolation and cDNA synthesis were carried out as described previously (Tello and Sherwood 2009).
Data Mining for GnRH Homologs and Sequence Confirmation
Initial searches for GnRH homologs in B. floridae protein and nucleotide databases (http://genome.jgi-psf.org/Brafl1/Brafl1.home.html, last accessed January 2, 2014) using Blast (Altschul et al. 1997) failed to uncover any bonafide GnRH-like candidates. An alternative strategy employing HMMs with HMMER v3.0 (http://hmmer.janelia.org/, last accessed January 2, 2014) was employed with a custom HMM model for propeptide GnRH/AKH/corazonin peptides against B. floridae protein and translated nucleotide databases, revealing two potential GnRH-like sequences. Only one sequence was shown to be functional in receptor activation assays. To confirm the expression and coding sequence of this GnRH-like transcript, polymerase chain reaction (PCR) on adult and juvenile B. floridae tissues was conducted with the following primers (IDT DNA, Coralville, IA): fwd. 5′-CGAGTAGAACTCATCTGAGCACCA-3′, rev. 5′-GTTGTAGCTATCCTTTCAATACGCA-3′. PCR was carried out with 1U of Platinum Taq DNA Polymerase (Life Technologies, Burlington, ON), 1.5 mM MgCl2, and 0.4 µM of each primer according to the following program: 94 °C for 2 min, followed by 35 cycles of 94 °C for 15 s, 55 °C for 15 s, and 68 °C for 30 s, with a final extension at 68 °C for 5 min. Amplicons were separated by electrophoresis on a 1.2% agarose gel, visualized with ethidium bromide, and representative bands were excised and gel extracted using a QIAquick gel extraction kit (Qiagen, Mississauga, ON), according to the manufacturer’s protocol. Purified amplicons were ligated into the pGEM-T cloning vector (Promega, Madison, WI), electroporated into competent Escherichia coli (Lucigen, Middleton WI) and plated. Colonies were screened for positive clones, grown up in lysogeny broth, miniprepped with an EZ-10 Spin Column Plasmid DNA Kit (Bio Basic, Markham, ON) and sequenced on ABI 3720xl DNA sequencers (Eurofins MWG Operon, Huntsville, AL).
Receptor Assays
GnRHRs from B. floridae previously isolated by Tello and Sherwood (2009) were transfected into early passage COS-7 cells (ATCC, Manassas, VA) grown in DMEM medium with 10% FBS. Briefly, cells were grown to confluence and passaged onto 12-well plates at a density of 1 × 105 cells/well. The following day, at 90–95% confluence, cells were transfected with 0.8 µg purified B. floridae GnRHR expression plasmids and 2.5 µl of Lipofectamine 2000 (Life Technologies) according to the manufacturer’s protocol. Two days posttransfection, purified B. floridae GnRH-like peptides (Genscript, Piscataway, NJ), human GnRH1 peptide, and octopus GnRH-like peptide were added at various concentrations to the transfected wells in a stimulation assay for either Gq/11 activity (IP-One assay, Cisbio, Bedford, MA) or Gs/Gi activation (cAMP Paramater assay, R&D Systems, Minneapolis, MN) according to the manufacturer’s protocols. Additionally, the amphioxus GnRH-like peptide was tested in reducing conditions by diluting the peptide in 10 mM dithiothreitol before stimulation. All assays were performed in duplicate or triplicate with three separate experiments for each treatment. Absorbance values were read on a Synergy HT plate reader (BioTek, Winooski, VT), with curve fitting and statistical analysis (one-way ANOVA followed by a Tukey’s posttest) performed in Prism 5 (GraphPad, La Jolla, CA). P < 0.05 was considered statistically significant.
Immunohistochemistry
A primary polyclonal rabbit antiserum was raised against B. floridae GnRH-like peptide by inoculating two rabbits with peptide and adjuvant over six injection cycles, testing the titer after each cycle (Genscript). After the titer for one of the antisera against the peptide reached a specific threshold (enzyme-linked immunosorbent assay [ELISA] detection at 1:250,000), the antibody was purified on an affinity column and resuspended at 1 mg/ml. An adult B. floridae specimen was embedded in methyl methacrylate/butyl methacrylate (MBM) and sectioned at 4.0 µm using the same methodology as in von Schalburg et al. (2013). MBM was removed from sections with acetone immersion, followed by distilled water rinses, and antigen retrieval in boiling 0.01 M citrate buffer, pH 6.0 for 10 min. The rest of the procedure was conducted at room temperature, except where noted. Endogenous peroxidases were blocked with 0.3% hydrogen peroxide in methanol for 30 min, and then primary antibody blocking was performed in phosphate buffered saline with 1% ovalbumin for 30 min. Primary antibody was incubated in blocking solution at a dilution of 1:100 for 60 min and then overnight at 4 °C, followed by PBS washes and the addition of biotinylated goat anti-rabbit secondary antibody (Sigma-Aldrich, Oakville, ON) in blocking solution for 60 min. Following further PBS washes, streptavidin-conjugated peroxidase (Sigma) in blocking buffer was incubated at a dilution of 1:100 for 30 min, washed again, and sections were immersed in the chromogen 3,3′-diaminobenzidine tetrahydrochloride until color developed. Sections were counterstained with Harris hematoxylin. Negative controls were also performed on sections adjacent to those incubated with primary antibody, using antibody that was preadsorbed with amphioxus GnRH-like peptide in a 10:1 peptide:antibody molar ratio overnight at 4 °C. Preimmune serum, extracted before the first inoculation with amphioxus GnRH-like peptide, was used as an additional control. Negative controls were diluted at the same ratio as the primary antibody (1:100) and followed the same immunolabeling procedure. Sectioning and immunohistochemistry were performed at the Electron Microscopy Lab, Department of Biology, University of Victoria (Victoria, BC).
Synteny Analysis
Synteny analysis of the B. floridae GnRH-like peptide was performed against both the human genome (http://www.ncbi.nlm.nih.gov/genome/guide/human/, last accessed January 2, 2014) and the limpet (Lottia gigantea) genome (http://genome.jgi-psf.org/Lotgi1/Lotgi1.home.html, last accessed January 2, 2014). All peptide models from the B. floridae v2.0 assembly scaffold on which the GnRH-like peptide was found were retrieved and BlastP was used to identify homologs for both humans and limpets. A relaxed substitution matrix (BLOSUM45) and short word size (2) were employed to identify divergent homologs, which were considered if they had an E value ≤ 1e-20 and an identity/similarity of 30%/50%, respectively, along with greater than 50% coverage. Branchiostoma floridae protein models were considered homologous if they met these criteria, and there were no more than four closely related human paralogs, with the majority belonging to the same ancestral chordate linkage group or ALG (Putnam et al. 2008). Amphioxus homologs found in the human genome that had multiple copies in the GnRH paralogon, previously identified as ALG 7, were included. Homologs identified on L. gigantea scaffold 44 that had been previously confirmed in both amphioxus and human were also included. A supplementary table S1, Supplementary Material online, listing the amphioxus gene models on scaffold 36 that had human and limpet homologs is provided.
Phylogenetic Analysis
Phylogenetic analysis of GnRH superfamily receptors was conducted on receptor protein sequences from a variety of bilaterian species. Receptor sequences were first trimmed to their seven-transmembrane region using the “‘–trim” command in the HMMER program hmmalign and a HMM model for the transmembrane region found in rhodopsin-family G protein-coupled receptors (GPCRs) (http://pfam.sanger.ac.uk/family/PF00001/hmm, last accessed January 2, 2014). Sequences were then aligned using MSAProbs (Liu et al. 2010) with default values and degapped using BMGE (Criscuolo and Gribaldo 2010) with a BLOSUM45 matrix, entropy score cut off of 1 (“-h 1”), and block size of 1 (“-b 1”). The alignment is provided in supplementary figure S1, Supplementary Material online. ProtTest 3 (Abascal et al. 2005) was used to determine the best substitution model for further analysis using the AICc criteria, determined to be LG+G (estimated gamma value, no invariant sites, and model-based equilibrium frequencies). Maximum likelihood and Bayesian analyses were conducted on the Western Canadian Research Grid (http://www.westgrid.ca/, last accessed January 2, 2014). Maximum likelihood analysis employed RaxML 7.4.2 (Stamatakis 2006) with the combined tree-search/fast bootstrap method (“-f a”) under the PROTGAMMALG model (four discrete rate categories) with 1,000 fast bootstraps. The resulting tree topology was compressed and branches reordered in FigTree 1.4 (http://tree.bio.ed.ac.uk/software/figtree/, last accessed January 2, 2014). The original, noncompressed tree is also provided in supplementary figure S2A, Supplementary Material online, and the sequences used to construct the tree are listed in supplementary table S2, Supplementary Material online. Bayesian posterior probabilities are also included, from a consensus tree generated in PhyloBayes MPI 1.4f (Lartillot et al. 2013). The Bayesian tree was constructed using the same constraints as the maximum likelihood tree (LG substitution model, four estimated gamma categories), with two chains that had the first 5,000 trees discarded as burn-in, and the next 20,000 trees sampled at every 10th tree (2,000 total trees per chain). The original Bayesian consensus tree is presented in supplementary figure S2B, Supplementary Material online.
Supplementary Material
Supplementary figures S1 and S2 and tables S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors acknowledge the assistance of Brent Gowen for performing sectioning and immunohistochemical labeling. This work was supported by a grant from the Canadian Natural Sciences and Engineering Council (NSERC).
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont M, Cazzamali G, Williamson M, Hauser F, Grimmelikhuijzen CJP. Identification of four evolutionarily related G protein-coupled receptors from the malaria mosquito Anopheles gambiae. Biochem Biophys Res Commun. 2006;344:160–165. doi: 10.1016/j.bbrc.2006.03.117. [DOI] [PubMed] [Google Scholar]
- Boerjan B, Verleyen P, Huybrechts J, Schoofs L, De Loof A. In search for a common denominator for the diverse functions of arthropod corazonin: a role in the physiology of stress? Gen Comp Endocrinol. 2010;166:222–233. doi: 10.1016/j.ygcen.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Bourlat SJ, Juliusdottir T, Lowe CJ, Freeman R, Aronowicz J, Kirschner M, Lander ES, Thorndyke M, Nakano H, Kohn AB, et al. Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature. 2006;444:85–88. doi: 10.1038/nature05241. [DOI] [PubMed] [Google Scholar]
- Callard GV, Pudney JA, Kendall SL, Reinboth R. In vitro conversion of androgen to estrogen in amphioxus gonadal tissues. Gen Comp Endocrinol. 1984;56:53–58. doi: 10.1016/0016-6480(84)90060-1. [DOI] [PubMed] [Google Scholar]
- Campbell RK, Satoh N, Degnan BM. Piecing together evolution of the vertebrate endocrine system. Trends Genet. 2004;20:359–366. doi: 10.1016/j.tig.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Candiani S, Holland ND, Oliveri D, Parodi M, Pestarino M. Expression of the amphioxus Pit-1 gene (AmphiPOU1F1/Pit-1) exclusively in the developing preoral organ, a putative homolog of the vertebrate adenohypophysis. Brain Res Bull. 2008;75:324–330. doi: 10.1016/j.brainresbull.2007.10.023. [DOI] [PubMed] [Google Scholar]
- Candiani S, Pestarino M. Expression of the tissue-specific transcription factor Pit-1 in the lancelet, Branchiostoma lanceolatum. J Comp Neurol. 1998;392:343–351. [PubMed] [Google Scholar]
- Cañestro C. Two rounds of whole-genome duplication: evidence and impact on the evolution of vertebrate innovations. In: Soltis PS, Soltis DE, editors. Polyploidy and genome evolution. Berlin (Germany): Springer; 2012. pp. 309–339. [Google Scholar]
- Castro A, Becerra M, Manso MJ, Sherwood NM, Anadon R. Anatomy of the Hesse photoreceptor cell axonal system in the central nervous system of amphioxus. J Comp Neurol. 2006;494:54–62. doi: 10.1002/cne.20783. [DOI] [PubMed] [Google Scholar]
- Cazzamali G, Saxild N, Grimmelikhuijzen C. Molecular cloning and functional expression of a Drosophila corazonin receptor. Biochem Biophys Res Commun. 2002;298:31–36. doi: 10.1016/s0006-291x(02)02398-7. [DOI] [PubMed] [Google Scholar]
- Chambery A, Parente A, Topo E, Garcia-Fernandez J, D'Aniello S. Characterization and putative role of a type I gonadotropin-releasing hormone in the cephalochordate amphioxus. Endocrinology. 2009;150:812–820. doi: 10.1210/en.2008-1066. [DOI] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S. BMGE (block mapping and gathering with entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur WA, Hall JA, Smith JJ, Li W, Sower SA. Insight from the lamprey genome: glimpsing early vertebrate development via neuroendocrine-associated genes and shared synteny of gonadotropin-releasing hormone (GnRH) Gen Comp Endocrinol. 2013;192:237–245. doi: 10.1016/j.ygcen.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Dos Santos S, Bardet C, Bertrand S, Escriva H, Habert D, Querat B. Distinct expression patterns of glycoprotein hormone-alpha2 and -beta5 in a basal chordate suggest independent developmental functions. Endocrinology. 2009;150:3815–3822. doi: 10.1210/en.2008-1743. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbman A, Nozaki M, Kubokawa K. A brain-Hatschek's pit connection in amphioxus. Gen Comp Endocrinol. 1999;113:251–254. doi: 10.1006/gcen.1998.7193. [DOI] [PubMed] [Google Scholar]
- Gwee PC, Tay BH, Brenner S, Venkatesh B. Characterization of the neurohypophysial hormone gene loci in elephant shark and the Japanese lamprey: origin of the vertebrate neurohypophysial hormone genes. BMC Evol Biol. 2009;9:47. doi: 10.1186/1471-2148-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser F, Sondergaard L, Grimmelikhuijzen CJ. Molecular cloning, genomic organization and developmental regulation of a novel receptor from Drosophila melanogaster structurally related to gonadotropin-releasing hormone receptors for vertebrates. Biochem Biophys Res Commun. 1998;249:822–828. doi: 10.1006/bbrc.1998.9230. [DOI] [PubMed] [Google Scholar]
- Holland LZ, Albalat R, Azumi K, Benito-Gutiérrez E, Blow MJ, Bronner-Fraser M, Brunet F, Butts T, Candiani S, Dishaw LJ, et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 2008;18:1100–1111. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda A, Takahashi T, Satake H, Minakata H. Molecular and functional characterization of a novel gonadotropin-releasing-hormone receptor isolated from the common octopus (Octopus vulgaris) Biochem J. 2006;395:125–135. doi: 10.1042/BJ20051615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsu Y, Kubokawa K, Urushitani H, Iguchi T. Estrogen-dependent transactivation of amphioxus steroid hormone receptor via both estrogen and androgen response elements. Endocrinology. 2010;151:639–648. doi: 10.1210/en.2009-0766. [DOI] [PubMed] [Google Scholar]
- Kim DK, Cho EB, Moon MJ, Park S, Hwang JI, Kah O, Sower SA, Vaudry H, Seong JY. Revisiting the evolution of gonadotropin-releasing hormones and their receptors in vertebrates: secrets hidden in genomes. Gen Comp Endocrinol. 2011;170:68–78. doi: 10.1016/j.ygcen.2010.10.018. [DOI] [PubMed] [Google Scholar]
- Kubokawa K, Tando Y, Roy S. Evolution of the reproductive endocrine system in chordates. Integr Comp Biol. 2010;50:53–62. doi: 10.1093/icb/icq047. [DOI] [PubMed] [Google Scholar]
- Kuo MW, Lou SW, Postlethwait J, Chung BC. Chromosomal organization, evolutionary relationship, and expression of zebrafish GnRH family members. J Biomed Sci. 2005;12:629–639. doi: 10.1007/s11373-005-7457-z. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Rodrigue N, Stubbs D, Richer J. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst Biol. 2013;62:611–615. doi: 10.1093/sysbio/syt022. [DOI] [PubMed] [Google Scholar]
- Liu Y, Schmidt B, Maskell DL. MSAProbs: multiple sequence alignment based on pair hidden Markov models and partition function posterior probabilities. Bioinformatics. 2010;26:1958–1964. doi: 10.1093/bioinformatics/btq338. [DOI] [PubMed] [Google Scholar]
- Louis A, Roest Crollius H, Robinson-Rechavi M. How much does the amphioxus genome represent the ancestor of chordates? Brief Funct Genomics. 2012;11:89–95. doi: 10.1093/bfgp/els003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Michalak P. Coexpression, coregulation, and cofunctionality of neighboring genes in eukaryotic genomes. Genomics. 2008;91:243–248. doi: 10.1016/j.ygeno.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Millar RP, Lu ZL, Pawson AJ, Flanagan CA, Morgan K, Maudsley SR. Gonadotropin-releasing hormone receptors. Endocr Rev. 2004;25:235–275. doi: 10.1210/er.2003-0002. [DOI] [PubMed] [Google Scholar]
- Mirabeau O, Joly JS. Molecular evolution of peptidergic signaling systems in bilaterians. Proc Natl Acad Sci U S A. 2013;110:E2028–E2037. doi: 10.1073/pnas.1219956110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta T, Kubokawa K. Presence of sex steroids and cytochrome P450 genes in amphioxus. Endocrinology. 2007;148:3554–3565. doi: 10.1210/en.2007-0109. [DOI] [PubMed] [Google Scholar]
- Nozaki M, Gorbman A. The question of functional homology of Hatschek's pit of amphioxus (Branchiostoma belcheri) and the vertebrate adenohypophysis. Zool Sci. 1992;9:387–395. [Google Scholar]
- Paris M, Pettersson K, Schubert M, Bertrand S, Pongratz I, Escriva H, Laudet V. An amphioxus orthologue of the estrogen receptor that does not bind estradiol: insights into estrogen receptor evolution. BMC Evol Biol. 2008;8:219. doi: 10.1186/1471-2148-8-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Kim YJ, Adams ME. Identification of G protein-coupled receptors for Drosophila PRXamide peptides, CCAP, corazonin, and AKH supports a theory of ligand-receptor coevolution. Proc Natl Acad Sci U S A. 2002;99:11423–11428. doi: 10.1073/pnas.162276199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegueroles C, Laurie S, Alba MM. Accelerated evolution after gene duplication: a time-dependent process affecting just one copy. Mol Biol Evol. 2013;30:1830–1842. doi: 10.1093/molbev/mst083. [DOI] [PubMed] [Google Scholar]
- Pitti T, Manoj N. Molecular evolution of the neuropeptide S receptor. PLoS One. 2012;7:e34046. doi: 10.1371/journal.pone.0034046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JF, Reska-Skinner SM, Prakash MO, Fischer WH, Park M, Rivier JE, Craig AG, Mackie GO, Sherwood NM. Two new forms of gonadotropin-releasing hormone in a protochordate and the evolutionary implications. Proc Natl Acad Sci U S A. 1996;93:10461–10464. doi: 10.1073/pnas.93.19.10461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Roch GJ, Busby ER, Sherwood NM. Evolution of GnRH: diving deeper. Gen Comp Endocrinol. 2011;171:1–16. doi: 10.1016/j.ygcen.2010.12.014. [DOI] [PubMed] [Google Scholar]
- Sakai T, Aoyama M, Kusakabe T, Tsuda M, Satake H. Functional diversity of signaling pathways through G protein-coupled receptor heterodimerization with a species-specific orphan receptor subtype. Mol Biol Evol. 2010;27:1097–1106. doi: 10.1093/molbev/msp319. [DOI] [PubMed] [Google Scholar]
- Schubert M, Brunet F, Paris M, Bertrand S, Benoit G, Laudet V. Nuclear hormone receptor signaling in amphioxus. Dev Genes Evol. 2008;218:651–665. doi: 10.1007/s00427-008-0251-y. [DOI] [PubMed] [Google Scholar]
- Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, Campbell MS, Yandell MD, Manousaki T, Meyer A, Bloom OE, et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. 2013;45:415–421. doi: 10.1038/ng.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Staubli F, Jorgensen TJ, Cazzamali G, Williamson M, Lenz C, Sondergaard L, Roepstorff P, Grimmelikhuijzen CJ. Molecular identification of the insect adipokinetic hormone receptors. Proc Natl Acad Sci U S A. 2002;99:3446–3451. doi: 10.1073/pnas.052556499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart AJ, Katz AA, Millar RP, Morgan K. Retention and silencing of prepro-GnRH-II and type II GnRH receptor genes in mammals. Neuroendocrinology. 2009;90:416–432. doi: 10.1159/000233303. [DOI] [PubMed] [Google Scholar]
- Sudo S, Kuwabara Y, Park JI, Hsu SY, Hsueh AJ. Heterodimeric fly glycoprotein hormone-alpha2 (GPA2) and glycoprotein hormone-beta5 (GPB5) activate fly leucine-rich repeat-containing G protein-coupled receptor-1 (DLGR1) and stimulation of human thyrotropin receptors by chimeric fly GPA2 and human GPB5. Endocrinology. 2005;146:3596–3604. doi: 10.1210/en.2005-0317. [DOI] [PubMed] [Google Scholar]
- Tando Y. Studies on the origin of the hypothalamus-pituitary endocrine axis and the evolution of glycoprotein hormones in amphioxus [thesis] [Tokyo (Japan)]: The University of Tokyo; 2009. p. 155. [Google Scholar]
- Tando Y, Kubokawa K. A homolog of the vertebrate thyrostimulin glycoprotein hormone alpha subunit (GPA2) is expressed in amphioxus neurons. Zool Sci. 2009a;26:409–414. doi: 10.2108/zsj.26.409. [DOI] [PubMed] [Google Scholar]
- Tando Y, Kubokawa K. Expression of the gene for ancestral glycoprotein hormone beta subunit in the nerve cord of amphioxus. Gen Comp Endocrinol. 2009b;162:329–339. doi: 10.1016/j.ygcen.2009.04.015. [DOI] [PubMed] [Google Scholar]
- Tello JA, Rivier JE, Sherwood NM. Tunicate gonadotropin-releasing hormone (GnRH) peptides selectively activate Ciona intestinalis GnRH receptors and the green monkey type II GnRH receptor. Endocrinology. 2005;146:4061–4073. doi: 10.1210/en.2004-1558. [DOI] [PubMed] [Google Scholar]
- Tello JA, Sherwood NM. Amphioxus: beginning of vertebrate and end of invertebrate type GnRH receptor lineage. Endocrinology. 2009;150:2847–2856. doi: 10.1210/en.2009-0028. [DOI] [PubMed] [Google Scholar]
- Tostivint H. Evolution of the gonadotropin-releasing hormone (GnRH) gene family in relation to vertebrate tetraploidizations. Gen Comp Endocrinol. 2011;170:575–581. doi: 10.1016/j.ygcen.2010.11.017. [DOI] [PubMed] [Google Scholar]
- Tsai PS, Sun B, Rochester JR, Wayne NL. Gonadotropin-releasing hormone-like molecule is not an acute reproductive activator in the gastropod, Aplysia californica. Gen Comp Endocrinol. 2010;166:280–288. doi: 10.1016/j.ygcen.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Tsai PS, Zhang LH. The emergence and loss of gonadotropin-releasing hormone in protostomes: orthology, phylogeny, structure, and function. Biol Reprod. 2008;79:798–805. doi: 10.1095/biolreprod.108.070185. [DOI] [PubMed] [Google Scholar]
- Veenstra JA. Isolation and structure of corazonin, a cardioactive peptide from the American cockroach. FEBS Lett. 1989;250:231–234. doi: 10.1016/0014-5793(89)80727-6. [DOI] [PubMed] [Google Scholar]
- Veenstra JA. Does corazonin signal nutritional stress in insects? Insect Biochem Mol Biol. 2009;39:755–762. doi: 10.1016/j.ibmb.2009.09.008. [DOI] [PubMed] [Google Scholar]
- von Schalburg KR, Gowen BE, Rondeau EB, Johnson NW, Minkley DR, Leong JS, Davidson WS, Koop BF. Sex-specific expression, synthesis and localization of aromatase regulators in one-year-old Atlantic salmon ovaries and testes. Comp Biochem Physiol B Biochem Mol Biol. 2013;164:236–246. doi: 10.1016/j.cbpb.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Wicht H, Lacalli TC. The nervous system of amphioxus: structure, development, and evolutionary significance. Can J Zool. 2005;83:122–150. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.