Abstract
Rafflesia is a genus of holoparasitic plants endemic to Southeast Asia that has lost the ability to undertake photosynthesis. With short-read sequencing technology, we assembled a draft sequence of the mitochondrial genome of Rafflesia lagascae Blanco, a species endemic to the Philippine island of Luzon, with ∼350× sequencing depth coverage. Using multiple approaches, however, we were only able to identify small fragments of plastid sequences at low coverage depth (<2×) and could not recover any substantial portion of a chloroplast genome. The gene fragments we identified included photosynthesis and energy production genes (atp, ndh, pet, psa, psb, rbcL), ribosomal RNA genes (rrn16, rrn23), ribosomal protein genes (rps7, rps11, rps16), transfer RNA genes, as well as matK, accD, ycf2, and multiple nongenic regions from the inverted repeats. None of the identified plastid gene sequences had intact reading frames. Phylogenetic analysis suggests that ∼33% of these remnant plastid genes may have been horizontally transferred from the host plant genus Tetrastigma with the rest having ambiguous phylogenetic positions (<50% bootstrap support), except for psaB that was strongly allied with the plastid homolog in Nicotiana. Our inability to identify substantial plastid genome sequences from R. lagascae using multiple approaches—despite success in identifying and developing a draft assembly of the much larger mitochondrial genome—suggests that the parasitic plant genus Rafflesia may be the first plant group for which there is no recognizable plastid genome, or if present is found in cryptic form at very low levels.
Keywords: holoparasite, Tetrastigma, plastid, gene loss, horizontal gene transfer, NUPTs
Introduction
The ability to conduct photosynthesis is one of the defining features of plants. They owe this capacity to endosymbiotic chloroplasts, once free-living cyanobacteria that were assimilated by an ancestral protist ∼1.5 billion years ago (Gould et al. 2008). Chloroplasts, one of several plastids that develop from meristematic proplastids, are the site of production and storage of key plant metabolites (Wise 2006). Plastid organelles—which include chromoplasts and amyloplasts—are also involved in fatty acid synthesis, production of tetrapyrroles and aromatic substances, and pigment and starch storage (Neuhaus and Emes 2000).
Chloroplasts possess circular DNA genomes, which range size from ∼107 to 217 kb (mean of ∼152 kb) in the 220 photosynthetic angiosperms that have been examined to date (http://www.ncbi.nlm.nih.gov/genome, last accessed February 8, 2014). Chloroplast genomes (or plastomes) typically encode ∼85 proteins and ∼45 transfer RNA (tRNA) and ribosomal RNA (rRNA). The genome is a relic of the endosymbiotic origin of this organelle and is reduced in size from its bacterial ancestor, having lost some genes as well as transferring others to the nucleus (Martin 2003). As a consequence of this evolutionary relocation of genes, most proteins required for chloroplast function (∼2,500–3,500) are encoded by nuclear loci (Blanchard and Schmidt 1995).
Chloroplast genome structure is highly conserved across flowering plants, with the interesting exception of parasitic plants. Plant parasitism is an interesting evolutionary adaptation, arising independently at least 12–13 times in flowering plants, with about 1% of all known angiosperm species being parasitic plants (Barkman et al. 2007; Westwood et al. 2010). Hemiparasites, which depend on their hosts only for water and inorganic nutrients, still retain much of their chloroplast genomes (Braukmann and Stefanovic 2012). Achlorophyllous holoparasites, however, have undergone evolutionary reduction in genome size associated with gene loss. Epifagus virginiana, for example, has a much smaller plastid genome (∼70 kb) and is about half of the expected plastome size in autotrophic land plants (Wolfe et al. 1992). This reduced chloroplast (or plastid) genome contains only 42 genes and has lost the loci for photosynthesis and chlororespiration (Wolfe et al. 1992). Despite these reductions in genome size, all plants examined to date, including the nonphotosynthetic parasitic plants, as well as apicomplexan parasites such as Plasmodium, continue to retain even a vestige of a plastid genome (McFadden et al. 1996; Maréchal and Cesbron-Delauw 2001; Krause 2008; Li et al. 2013).
The genus Rafflesia, which belongs to the family Rafflesiaceae (order Malpighiales), is one of the eight known genera of plant holoparasites. Rafflesia is unique to the tropics of Southeast Asia, with some species in the genus producing the largest single flowers in the world, growing up to a meter in diameter. It has no stems, roots, or leaves, with only its massive flower protruding from the roots or stems of its sole host plant, the tropical vine Tetrastigma (Vitaceae) (Nais 2001) (see fig. 1). Nearly one-third of the 30 known Rafflesia species are endemic to the Philippines (Nickrent et al. 1997; Barcelona et al. 2009). Other members of the Rafflesiaceae family include Sapria and Rhizanthes, which are also holoparasites of Tetrastigma (Nais 2001).
Fig. 1.
Open flower of R. lagascae Blanco. The flower is 15–20 cm in diameter.
Attempts to isolate highly conserved plastid genes from members of the holoparasite genus Rafflesia (Rafflesiaceae) (Nickrent et al. 1997; Davis et al. 2007) have failed, and another study has indicated the possibility of plastid genome loss in Rafflesia leonardi (Nickrent DL, Molina J, Geisler M, Bamber AR, Pelser PB, Barcelona JF, Inovejas SAB, Uy I, Purugganan MD, unpublished data). Here we provide a strong evidence that suggests that the chloroplast/plastid genome is entirely absent in R. lagascae Blanco (see fig. 1), a species found only on the Philippine island of Luzon but nevertheless the most widespread of all the Philippine Rafflesia species (Pelser et al. 2013). With data from Illumina next-generation sequencing, we employ multiple separate techniques for organellar genome assembly. We are able to assemble a draft of the R. lagascae mitochondrial genome at high coverage. We cannot, however, identify an intact plastid genome in R. lagascae, which indicates that members of the parasitic plant genus Rafflesia may be the first plant group shown to have lost its plastid genome.
Results
Draft Sequence Assembly of a Mitochondrial Genome in Rafflesia
A floral bud of R. lagascae was collected from Cagayan province in the Philippines. Attached only at its base to its Tetrastigma host, Rafflesia tissue was carefully dissected from the host plant, and genomic DNA was extracted from the disk distant from host tissue and enclosed in layers of bracts. Both 100-bp and 3-kb insert libraries were made from genomic DNA and sequenced using Illumina next-generation technology.
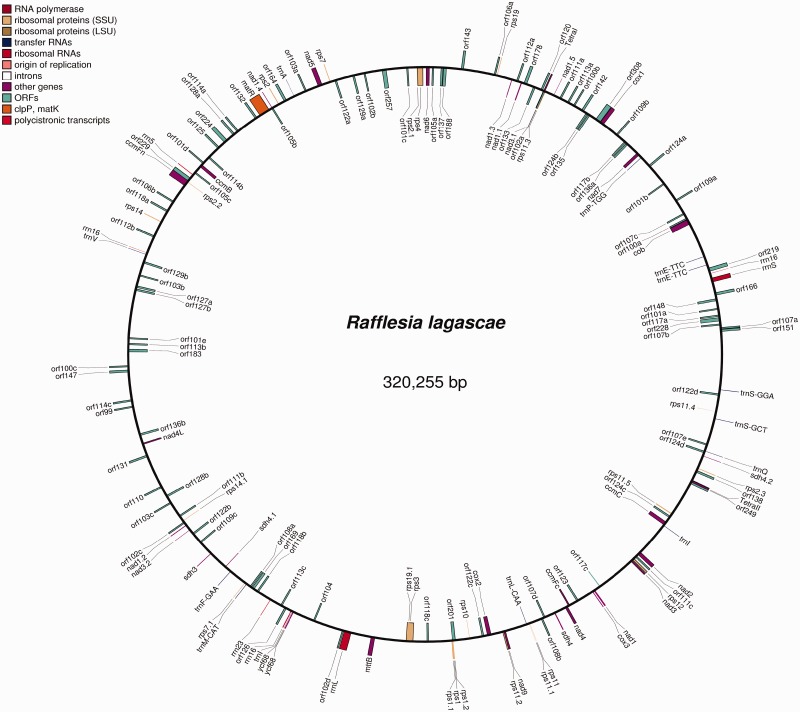
Of the approximately 440 million Illumina paired-end (PE) sequencing reads from R. lagascae from both insert libraries, we used two distinct methods to assemble a draft sequence of the mitochondrial genome. First, we used a bait mapping approach by employing previously published data for R. cantleyi (Xi et al. 2013) to assemble the mitochondrion from this species using SOAPdenovo (Luo et al. 2012), and used this assembled R. cantleyi sequence as bait to identify mitochondrial genome sequences from the R. lagascae Illumina reads. These identified reads were then assembled by SOAPdenovo to provide a draft genome assembly of the R. lagascae mitochondrial genome. We were able to identify and assemble ∼320.3 kb of the R. lagascae mitochondrial genome (N50 = 4.45 kb); this constitutes a draft assembly with 213 gaps (see fig. 2). The mean sequencing depth coverage across the reference is 349.7×, with a standard deviation of 173.02× (see supplementary fig. S1, Supplementary Material online, for sequence depth coverage across the draft-assembled genome).
Fig. 2.
Draft structure of R. lagascae mitochondrial genome. The gene positions indicated are based on the assumption of synteny with Ricinus (GenBank accession number HQ874649; Rivarola et al. 2011). The coordinates and encoded products of the specific genes are shown in supplementary table S1, Supplementary Material online. Although depicted as a complete circular genome, it should be stressed that this is a draft assembly with 213 gaps and that portions of the mitochondrial sequence remain unassembled.
Additionally, we used a de novo assembly approach (without any reference genome) to obtain ∼1,447,235 sequence contigs of the R. lagascae sequence data using CLC Genomics Workbench (CLC Bio, Aarhus, Denmark), with an N50 = 182 bp. The low N50 for the entire data set is due to either the large size of the R. lagascae nuclear genome or its high repeat content. Despite this, we were able to readily identify ∼4,000 sequence contigs containing mitochondrial genome sequence through Blast analysis of assembled sequence contigs against 15 plant mitochondrial genomes. The largest 49 sequence contigs were shown to have about 210× sequence coverage. These contigs ranged from 1.0 to 17.3 kb, with an average size of 6.3 ± 5.0 kb; this represents a total sequence length of 382 kb.
We identified 42 known plant protein-coding mitochondrial genes, as well as 60 unique open reading frames (ORFs) (see fig. 2 and supplementary table S1, Supplementary Material online). Phylogenetic analyses of the mitochondrial protein-coding genes show that 7 of 24 genes with clear phylogenetic placement (29%) have these Rafflesia loci allied to Vitis vinifera (Vitaceae) or other plant groups, instead of the more closely related Ricinus communis (Euphorbiaceae, Malpighiales) (results not shown). The other identified mitochondrial genes show equivocal placement in the phylogenies. These confirm previous findings of rampant horizontal gene transfer (HGT) in this genus (Xi et al. 2012, 2013; Nickrent DL, Molina J, Geisler M, Bamber AR, Pelser PB, Barcelona JF, Inovejas SAB, Uy I, Purugganan MD, unpublished data).
Searching for a Plastid Genome
We attempted to recover plastid sequences from R. lagascae using several approaches. First, we mapped the sequence contigs obtained from the CLC Genomics Workbench assembly method onto the chloroplast genomes of Ricinus and Vitis, using the program Geneious R6 (Biomatters, Auckland, New Zealand). This yielded 17 sequence contigs (G1–G17; table 1) that mapped to the plastid genomes of these two species, with a total length of ∼3.9 kb. We also conducted a BlastN search of all conserved plastid genes available from GenBank against the assembled CLC sequence contigs, which identified additional 26 contigs with significant (e-value < 1e−10) hits (table 1). In addition, we generated profile hidden Markov models (HMMs; Henderson et al. 1997) from alignments of conserved plastid genes. This identified three more contigs with significant hits (e-value < 1e−10) (table 1). Together, these approaches identified a total of 46 putative plastid sequence contigs with an average length of 242 bp and a total length of 11.5 kb.
Table 1.
Identified Plastid Sequences in Rafflesia lagascae Including Noncoding Sequences in Inverted Repeat (IR) Regions.
| Gene Name | Size (bp) | Method of Recovery | Contig Numbera | Phylogenetic Allianceb |
|---|---|---|---|---|
| accD | 136 | BlastN | 763,474 | Vitis mt, cp, nucc |
| atpA | 158 | BlastN | 1,105,137 | — |
| atpB | 127 | BlastN | 568,017 | — |
| IR | 138 | BlastN | 690,602 | Hevea cp, Vitis cp |
| IR | 160 | BlastN | 552,527 | — |
| IR | 163 | Geneious | G14 | — |
| IR | 190 | Geneious | G11 | — |
| IR | 199 | Geneious | G3 | Vitis cp, nucc |
| IR | 207 | BlastN | 435,549 | Vitis nucc |
| IR | 219 | BlastN | 1,355,988 | — |
| IR | 227 | Geneious | G15 | Vitis nucc |
| IR | 274 | BlastN | 426,447 | Vitis cp, nucc |
| IR | 299 | Geneious | G6 | — |
| IR, trnA-UGC | 104 | Geneious | G8 | Vitis nuc |
| 113 | Geneious | G7 | Vitis nuc | |
| 168 | Geneious | G16 | Vitis nuc | |
| 330 | BlastN | 859,744 | Vitis nuc | |
| 484 | Geneious | G10 | Vitis nucc | |
| IR, trnI-GAU | 496 | BlastN | 12,131 | — |
| IR, trnV-GAC | 145 | Geneious | G13 | Vitis nuc |
| 178 | Geneious | G5 | Vitis nuc | |
| 178 | BlastN | 1,147,528 | Vitis nucc | |
| matK | 227 | BlastN | 710,409 | Vitis cp, mt, nucc |
| ndhB | 118 | Geneious | G4 | — |
| ndhJ | 247 | BlastN | 1,167,865 | Vitis cp, nucc |
| 247 | Geneious | G1 | Vitis cp, nuc | |
| petB | 140 | Geneious | G2 | — |
| petG | 116 | BlastN | 823,312 | — |
| psaB | 211 | BlastN | 76,772 | Nicotiana cp |
| 315 | BlastN | 662,630 | Nicotiana cp | |
| 324 | BlastN | 972,383 | Nicotiana cpc | |
| psbA | 161 | BlastN | 1,131,304 | Vitis mtc |
| psbD | 280 | BlastN | 39,796 | — |
| psbZ | 137 | BlastN | 1,038,399 | — |
| rbcL1 | 395 | BlastN | 127,053 | — |
| rbcL2 | 647 | BlastN | 170,975 | Vitis cp, mtc |
| rps11 | 113 | HMM | 322,006 | Vitis nucc |
| rps16 | 148 | BlastN | 641,235 | — |
| rps7 | 119 | HMM | 864,624 | — |
| rrn16 (IR) | 191 | Geneious | G12 | — |
| 203 | BlastN | 1,114,615 | — | |
| 317 | BlastN | 4,358 | — | |
| rrn23 (IR) | 139 | Geneious | G17 | Vitis cp |
| 766 | Geneious | G9 | Vitis cp | |
| 1026 | BlastN | 19,544 | Vitis cpc | |
| ycf2 (IR) | 203 | HMM | 113,164 | — |
Note.—Certain IR sequences may appear multiple times because they belong to the inverted repeats region of the plastid genome, whereas genes like ndhJ were identified by different methods.
aContig number is the specific contig out of the approximately 1.4 million contigs from CLC, except for those prefixed with “G,” which were derived from Geneious.
bTaxa and genome/s (mt, mitochondria; cp, chloroplast; nuc, nuclear) to which Rafflesia was shown strongly associated with (>50% BS, compare with fig. 4) in phylogenetic analyses; sequences marked with “—” had ambiguous phylogenetic positions. Multiple taxon/genome associations represent polytomous nodes >50% BS in which Rafflesia is embedded in.
cCorresponding phylogenies for these sequence contigs that show Rafflesia in unequivocal positions are provided as Supplementary Material (taxa in the rps11 phylogeny represent the only significant hits recovered).
To ensure that our failure to identify a plastid genome was not due to a problem of the genome assembly method, we identified 925 Illumina 100-bp PE reads (out of ∼214 million reads) that directly mapped to plastid sequences found in GenBank. These sequences were then de novo assembled using SOAPdenovo, another sequence assembly program for short-read sequencing data (Luo et al. 2012). Aside from those sequences already identified by the previous methods, we found additional five sequence fragments, but they appear embedded in the assembled mitochondrial genome of R. lagascae.
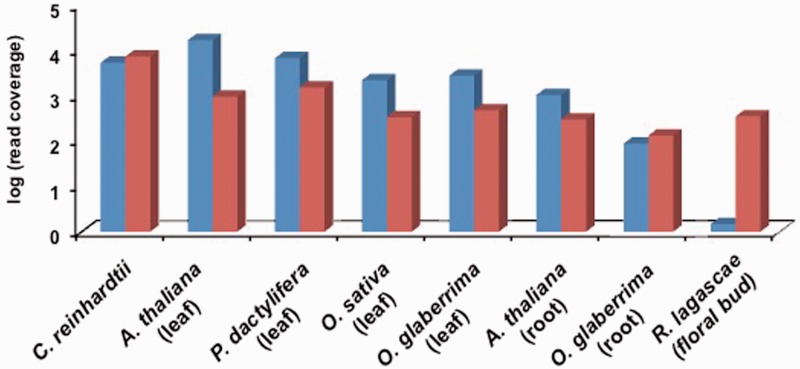
Unlike the high sequence coverage for the draft assembly of the mitochondrial genome (∼350×), the mean sequence coverage for the plastid sequence fragments we identified was substantially lower at 1.48× ± 1.26× reads. The anomalously low sequencing read depth coverage contrasts with the several hundred copies of plastid genomes that should exist within plant cells (Bock 2007). As a comparison with the normal expectation in different species, we used Illumina whole genome re-sequencing data to demonstrate that chloroplast/plastid genome sequencing depth coverage was nearly equal to or exceeded that of mitochondrial genome in leaf tissue in Oryza, Phoenix, and Arabidopsis, and light-grown single-cell culture in the algae Chlamydomonas (see fig. 3). The sequencing reads in these species covered >98% of the bases in these organellar genome sequences.
Fig. 3.
Whole-genome sequencing coverage for mitochondrial and chloroplast genomes in different species. Blue bars are for chloroplast genomes, whereas red bars are for mitochondrial genomes. The tissue source for the genomic DNA is indicated; for C. reinhardtii, it is single-cell culture under constant light.
Because the plastid copy number is reduced in nonphotosynthetic tissues like roots (Isono et al. 1997), it is possible that lower levels of plastid DNA in the nonphotosynthetic Rafflesia may be the source of our inability to identify a plastid genome in this species. As a further positive control, we therefore obtained ∼62 and ∼104 million 100-bp PE Illumina sequencing reads from root DNA in Oryza glaberrima and Arabidopsis thaliana, respectively, and examined relative levels of plastid and mitochondrial genome copies in these nonphotosynthetic tissues. Based on sequencing depth coverage from root genomic DNA (see fig. 3), the plastid genome in O. glaberrima is ∼66% that of mitochondrial genome levels, whereas in A. thaliana the plastid genome is found at ∼3.5-fold greater levels than mitochondrial genome (down from an ∼17-fold greater level in leaves). Our data demonstrate a reduction in plastid genome levels in the nonphotosynthetic root compared with leaf tissues but nevertheless show that plastid genome levels can be substantial in nonphotosynthetic tissues.
Fragments of the Plastid Genome
Although we could not identify an intact plastid genome, we did find small fragments of plastid genes that ranged in size from 104 to 1,026 bp. We recovered short segments of 17 protein-coding genes (including ribosomal proteins), two rRNA and three tRNA genes, as well as ten intergenic sequences that are found in inverted repeat regions of the chloroplast genome in other species (see table 1). No full gene sequences were identified. None of the plastid sequence contigs had intact reading frames when aligned with coding sequences of plastid genes from photosynthetic taxa (results available upon request).
To determine whether any of the recovered plastid sequences were expressed, we also mapped the sequencing reads from the R. cantleyi transcriptome library (accession SRA052224) to these R. lagascae sequence fragments (Xi et al. 2012). The greatest number of reads mapped were 23 singleton reads from the partial ribosomal rrn16 (∼150 bp) fragment. These results indicate that there is no significant expression for any of these putative plastid-derived sequences, at least in the actively developing floral bud tissue of this parasitic plant.
Phylogenetic Analysis of Remnant Rafflesia Plastid Gene Sequences
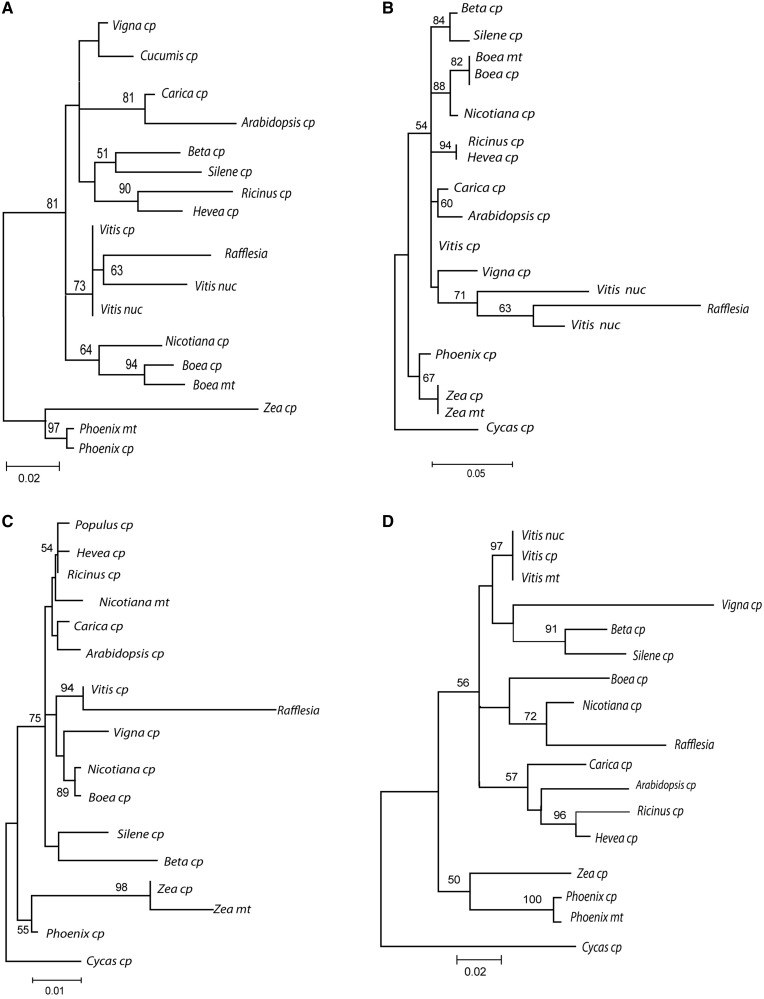
Phylogenetic analysis of the 46 plastid sequence fragments that we identified show that 15 of these Rafflesia sequence fragments are allied with Vitis with >50% BS (table 1, fig. 4 and supplementary data S2, Supplementary Material online), The rest of the plastid sequences show Rafflesia in equivocal positions (BS <50%).
Fig. 4.
Phylogenies of recovered plastid sequences from R. lagascae. Rafflesia sequences group with (A) both nuclear (nuc) and plastid (cp) sequences in Vitis; (B) only Vitis nuclear sequence; (C) only Vitis plastid sequences; (D) Nicotiana plastid sequence. Only bootstrap support >50% is indicated.
Four sequence fragments, including ndhJ (fig. 4A), depict Rafflesia plastid sequences associated with both Vitis’ nuclear and plastid sequences. Five other sequences such as a noncoding sequence from one of the inverted repeats were more similar to a Vitis’ nuclear sequence than they are to plastid sequences (fig. 4B; supplementary data S2, Supplementary Material online; table 1). Only Rafflesia’s rrn23 was solely with a Vitis chloroplast sequence (fig. 4C). A 136-bp fragment from the accD gene had Rafflesia grouping with nuclear, plastid and mitochondrial copies (76% BS; supplementary data S2, Supplementary Material online, and table 1).
No plastid sequence was found to be phylogenetically associated with Ricinus or Hevea, the closest relatives to Rafflesia with available organellar genome sequences. However, the Rafflesia psaB gene fragment grouped with the Nicotiana homolog with 80% BS (fig. 4D).
The anomalous phylogenetic placement of these remnant Rafflesia plastid genes may arise from contamination from the DNA of the host plant Tetrastigma. To test for contamination of Tetrastigma in the R. lagascae DNA extract, we used barcoding primers to amplify the rbcL gene (Kress et al. 2009) in these two species. We were unable to polymerase chain reaction (PCR)-amplify rbcL from R. lagascae, although this gene was easily amplified from the host Tetrastigma and from other evolutionary divergent photosynthetic taxa from the asterid and rosid families. Interestingly, we were able to recover two nonoverlapping segments of rbcL sequence (rbcL1, rbcL2) from the Illumina sequence contigs (∼1 kb in size) from R. lagascae (table 1), but these sequences are diverged in the barcoding primer sequence regions (Kress et al. 2009) that are normally conserved across multiple divergent autotrophic angiosperm taxa. Like the other recovered plastid sequences, Rafflesia’s rbcL contains premature stop codons.
Discussion
Possible Loss of the Plastid Genome in a Parasitic Plant
The parasitic plant lifestyle affords an intimate connection between a parasite and its host plant. As in many parasites, this can lead to subsequent relaxation of selection pressure to maintain key genes in the parasite as they become dependent on host plants for crucial functions (Bromham et al. 2013; Wicke et al. 2013). Moreover, the close connection to the host can also lead to genetic transfer of information to the parasite (Davis and Wurdack 2004; Xi et al. 2012, 2013).
The evolution of the chloroplast genome in parasitic plants, particularly nonphotosynthetic holoparasites, can lead to significantly reconfigured plastomes (Wicke et al. 2013). In these plants many photosystem and energy production genes are lost from the plastome (Krause 2008; Li et al. 2013; Wicke et al. 2013). The 45.6-kb plastome of Conopholis americana (Orobanchaceae) is the smallest published plastid genome to date (Colwell 1994; Wicke et al. 2013). Though devoid of genes expected to be present in autotrophic plants, it still maintains genes that are conserved in all previously sequenced plastomes, like genes for rRNA, some genes for ribosomal proteins, tRNA (e.g., trnE and trnfM) and the essential genes clpP and ycf2 (Wicke et al. 2013). Other parasitic plants and even the protist Plasmodium, descendant of the same photosynthetic protist as plants, still show remnant plastid genomes (McFadden et al. 1996; Maréchal and Cesbron-Delauw 2001; Krause 2008; Li et al. 2013).
It is clearly challenging to prove the complete absence of a plastid genome (Keeling 2010). Nevertheless, our inability to identify substantial plastid genome sequences from R. lagascae using multiple approaches, despite success in identifying and developing a draft assembly of the much larger mitochondrial genome, strongly suggests that there may be no recognizable plastid genome in the parasitic plant R. lagascae, or if present in cryptic form, is at very low levels. Moreover, a similar result has also been observed for R. leonardi (Nickrent DL, Molina J, Geisler M, Bamber AR, Pelser PB, Barcelona JF, Inovejas SAB, Uy I, Purugganan MD, unpublished data), which our study now reinforces with multiple lines of evidence. These results together suggest that plastid genome loss may be shared across multiple Rafflesia species, and this putative loss is likely an evolutionary consequence of the very ancient onset of parasitism in the family (Xi et al. 2013), perhaps dating back to mid-Cretaceous (Molina J, unpublished data).
It is possible for organelles to lose their genomes. This has already been documented in some anaerobic ciliates, trichomonads, and fungi, which possess hydrogenosomes, genomeless organelles that produce molecular hydrogen and are derived from mitochondria (Van der Giezen et al. 2005). However, the loss of the plastome has not yet been demonstrated in plants but deemed possible (Palmer 1997). Plastids have been secondarily lost outside the flowering plant lineage, including the protozoan trypanosomes (Martin and Borst 2003). However, in a recent study by Braukmann et al. (2013) on some species of the parasitic flowering plant Cuscuta (Convolvulaceae), difficulty has been reported in detecting plastid rRNA (rrn) genes that are highly conserved elements in plant plastomes even in heterotrophic species (Krause 2011; Wicke et al. 2011; Li et al. 2013). Accordingly, Braukmann et al’s (2013, p. 9) observations led them to conclude that some holoparasitic Cuscuta may have “reached the same or similar evolutionary endpoint, where the very presence of a plastid genome is questionable.”
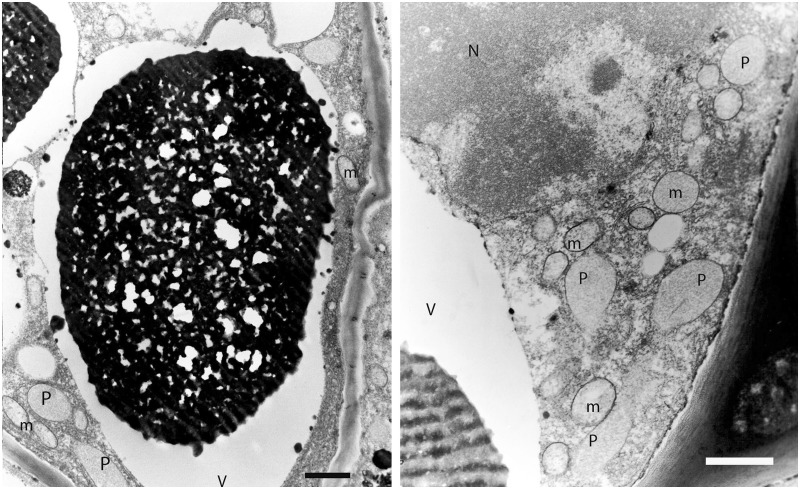
Despite the possible loss of the chloroplast and its genome, there still remain plastid-like structures in Rafflesia. Images from ultrastructural analysis from the congeneric R. philippensis using transmission electron microscopy (TEM) demonstrate that Rafflesia (Nickrent DL, Molina J, Geisler M, Bamber AR, Pelser PB, Barcelona JF, Inovejas SAB, Uy I, Purugganan MD, unpublished data; for additional images, see fig. 5) does contain plastid-like compartments with homogeneous stroma (Renzaglia K, personal communication). None of these structures have the distinctive lamellar/endomembrane system found in all types of plastids. These suggest that Rafflesia retains plastid compartments for certain metabolic functions, even in the apparent absence of a plastid genome.
Fig. 5.
Electron micrographs of cells from R. philippensis ramenta indicating plastid-like (P) structures (N, nucleus; m, mitochondria; V, vacuole). Scale bars = 1 µm.
There are two possible scenarios for the evolution of these genome-less plastids. One is that any relevant genes still necessary for metabolic function have relocated to the nucleus and/or mitochondria. Another possibility is that these Rafflesia plastids were originally obtained from the host, with subsequent translocation of host-encoded plastid genes to the nucleus and eventually degenerating as pseudogenes. The latter possibility may explain the large proportion of remnant nonfunctional plastid gene sequences in R. lagascae that are phylogenetically allied with Vitis nuclear sequences (table 1; supplementary data S2, Supplementary Material online). Such host-derived plastids have been observed in certain parasitic red algae (Goff and Coleman 1995) as well as in natural grafts of sexually incompatible species of Nicotiana (Stegemann et al. 2012). These two possibilities are not mutually exclusive, and the precise genetic basis for the maintenance of these Rafflesia plastid-like structures must await more detailed analysis of fully assembled nuclear genomes.
Interestingly, osmiophilic plastoglobules or carotenoid bodies, which are typical inclusions of chromoplasts in colored flowers (Camara et al. 1995), were also not seen in the Raffesia plastid-like structures, and thus, may not be the primary source of the bright red-orange coloration characteristic of Rafflesia species. Instead, there were very large vacuoles observable in Rafflesia ramenta filled with osmiophilic material, which may be phenolic compounds or terpenoids that also appear as electron-dense material in other TEM studies of trichomes (Sacchetti et al. 1999; Wen-Zhe et al. 2002). These plant terpenoids can serve as precursors for a diversity of plant metabolites such as carotenoid and anthocyanin pigments and volatiles responsible for flower color and odor (Tanaka et al. 2008).
Several hypotheses have been proposed with respect to what biochemical constraints on plastid genome size or its complete loss might exist owing to the need to retain particular genes required for metabolism (Bungard 2004; Barbrook et al. 2006). The essential tRNA hypothesis states that plastid-encoded trnE is considered essential for heme biosynthesis (a component of the mitochondrial P450 cytochromes) and could not be easily replaced by a cytosolic tRNA. It may be that Rafflesia continues to retain plastids for various metabolic functions, but the genes that encode for these are found in the nucleus or in the mitochondria as in the case of a trnE sequence recovered in R. leonardi (Nickrent DL, Molina J, Geisler M, Bamber AR, Pelser PB, Barcelona JF, Inovejas SAB, Uy I, Purugganan MD, unpublished data). The fates of other essential plastid genes, such as the clpP and ycf2 loci (Wicke et al. 2013), are unknown, and must await detailed genomic and biochemical studies on these parasitic plant species.
Vestiges of the Missing Plastid Genome
The small regions we could successfully identify in R. lagascae as putative plastid sequences represent less than 10% of the chloroplast genome of photosynthetic plants, and do not have intact reading frames and so are likely nonfunctional. These results would make Rafflesia arguably the plant with the smallest amount of plastid “genome” sequence that has been observed thus far (Krause 2008; Li et al. 2013). Given the low sequencing read coverage for these gene fragments (<2× coverage), it is likely that these remnant plastid sequences are located in the nuclear genome, similar to what has been observed for nuclear-integrated chloroplast genes and/or nuclear plastid DNAs (NUPTs) observed in other plant species (Blanchard and Schmidt 1995; Kleine et al. 2009).
Phylogenetic analyses of the remnant plastid gene fragments in Rafflesia show that many of these genes have anomalous but strongly supported (>50% BS support) phylogenetic placements that suggest they are the products of HGT. Most of these genes grouped phylogenetically with homologs in Vitis (which belongs to the same family as the host species Tetrastigma), rather than either Ricinus or Hevea, which together are more closely related to Rafflesia. There is a possibility that these may be due to contamination of the Rafflesia DNA, but control experiments using universal rbcL barcoding primers suggest that this is unlikely. The anomalous phylogenetic placement of remnant Rafflesia plastid sequences may also arise from a complex history of gene duplication and extinction, although we think that HGT is a more parsimonious explanation.
Although these other possibilities cannot be completely ruled out, we feel that our results suggest that ∼33% of the plastid loci we have identified in R. lagascae may have been horizontally acquired, most likely from its host as a result of parasitism. It has already been demonstrated in other parasitic plants that the plasmodesmatal continuity between host and parasite allows for molecular movement, including that of genetic material (Birschwilks et al. 2006; Roney et al. 2007; Talianova and Janousek 2011). Rampant HGT between Rafflesia and Tetrastigma involving several nuclear and mitochondrial genes has also been repeatedly shown (Xi et al. 2012, 2013). Individual phylogenies of chloroplast sequences recovered from the confamilial Sapria have also exhibited a closer relationship to Vitis than to Ricinus (Xi et al. 2013). A complete genome sequence of the Rafflesia nuclear genome will be necessary in order to conclusively determine the fate of the genes that would normally reside in the plastid as well as examine the other molecular evolutionary consequences of the obligate parasitic lifestyle of this enigmatic yet fascinating plant genus.
Materials and Methods
Illumina Whole-Genome Sequencing
A flower bud of R. lagascae was collected from Sitio Kanapawan, Barangay Bolos Point, Gattaran Municipality, Cagayan Province, Philippines in 2010 (collection number Nickrent 5791). All necessary collecting (Philippine Department of Environment and Natural Resources Gratuitous Permit 193, and subsequent renewals GP 202 and 217), transport, and export permits were obtained.
A sizeable Rafflesia bud, attached only at its base to its host, Tetrastigma, was carefully dissected from the host plant. Genomic DNA was extracted from a portion of the disk, which is sufficiently distant from host tissue and enclosed in layers of bracts. DNA extraction was performed following Nickrent et al. (2004). Contamination of R. lagascae DNA by Tetrastigma was tested using standard PCR amplification with degenerate rbcL barcoding primers (Kress et al. 2009). PCR amplification was positive in Tetrastigma but negative in R. lagascae (as well as in R. leonardi). The extracted R. lagascae DNA was submitted to Ambry Genetics (Aliso Viejo, CA) for Illumina next-generation sequencing, using both a 100-bp and a 3-kb insert size library on Illumina HiSeq 2000. Sequencing reads are deposited with the NCBI Sequence Read Archive (SRX434531).
Mitochondrial Genome Assembly and Analysis
Illumina sequencing reads from R. lagascae were assembled using two approaches. In one approach, we used a combination of SOAPdenovo (Luo et al. 2012) and reference-assisted mapping to assemble R. lagascae’s organellar genomes. We first assembled the mitochondrial gene sequence of the closely related species, R. cantleyi (Xi et al. 2013), whose Illumina reads were previously published and available in NCBI (accession SRR629613) and used this assembly of its mitochondrial genome as reference bait sequence. We then collected all the R. lagascae 100-bp PE reads that mapped to the R. cantleyi mitochondrial genome and used these to de novo assemble R. lagascae’s mitochondrial genome in SOAPdenovo. To annotate its mitochondrial genome, we used Mitofy (Alverson et al. 2010). Annotated ORFs with their accompanying numbers are based on the NCBI GenBank database. HGT of the mitochondrial genes was detected using phylogenetic analyses in MEGA5 (Tamura et al. 2011). In another approach, we did a de novo assembly of the R. lagascae sequence reads using CLC Genomics Workbench (ver. 5.5.1) (CLC Bio, Aarhus, Denmark).
Plastid Sequence Identification and Analysis
To identify sequence contigs comprising the plastid genome, we employed multiple approaches. First, sequence contigs from R. lagascae identified by the CLC Genomics Workbench were mapped using Geneious R6 (Biomatters Ltd., Auckland, New Zealand). These contigs were first mapped to the full mitochondrial genomes of V. vinifera (GenBank accession number FM179380) and Ri. communis (HQ874649) to eliminate mitochondrial sequences, and all unmapped reads were then collected and mapped to the chloroplast genomes of Vitis (DQ424856.1) and Ricinus (JF937588.1). Ricinus communis, like R. lagascae, is in the Malpighiales, and is the closest species to Rafflesia for which mitochondrial genome sequences are available. Vitis vinifera is the closest species to Tetrastigma for which both mitochondrial and chloroplast genome sequences are also available. Second, we conducted a BlastN search (e-value < e−10) of all the conserved plastid genes found in angiosperms available from GenBank against the CLC-assembled sequence contigs. We also generated profile HMM (Henderson et al. 1997) from alignments of conserved plastid genes using HMMER (http://hmmer.janelia.org/, last accessed February 8, 2014), which develops probabilistic models (profile HMMs) and can detect more remote similarities in sequence searches. We then mapped all these plastid sequence contigs identified (Geneious, BlastN, HMM) to the chloroplast genome of another species in the Malpighiales, Hevea brasiliensis (GenBank accession number NC_015308), using Geneious R6 (Biomatters, Ltd.). In a third approach, we mapped the 100-bp PE reads using BWA (Burrows–Wheeler alignment) (Li and Durbin 2010) and SAMtools (Sequence Alignment/Map) (Li et al. 2009) to angiosperm plastid sequences from GenBank, and then the resulting mapped reads were de novo assembled using SOAPdenovo (Luo et al. 2012).
Putative plastid sequences identified earlier were then aligned with homologous regions from other taxa whose chloroplast genomes are available in GenBank using default parameters in the Multiple Alignment using Fast Fourier Transform (MAFFT) program (Katoh and Toh 2008) and visually checked (supplementary data S1, Supplementary Material online). The alignments were analyzed phylogenetically in MEGA5 (Tamura et al. 2011) using maximum likelihood with 100 bootstrap replicates and applying the best substitution model with the lowest Bayesian information criterion scores (supplementary data S2, Supplementary Material online).
To determine whether any of these putative plastid sequences are expressed, we also mapped the reads from the R. cantleyi transcriptome library (accession SRA052224) (Xi et al. 2012) using Bowtie 1.0.0 (Langmead et al. 2009), TopHat2 (Kim et al. 2013), and Cufflinks (Trapnell et al. 2010) to the resulting contigs.
Comparative Levels of Mitochondrial and Plastid Genomes in Algal and Flowering Plant Species
To compare our results in Rafflesia with the relative levels of mitochondrial and chloroplast genome DNA in various species, we used whole genome resequencing data from three monocot species (Phoenix dactylifera, O. sativa, and O. glaberrima), one eudicot species (A. thaliana), and one photosynthetic algae (Chlamydomonas reinhardtii). DNA from these species was isolated from single-cell cultures (for C. reinhardtii), leaf or shoot tissue (P. dactylifera, O. sativa, O. glaberrima, and A. thaliana), or nonphotosynthetic root tissue (O. glaberrima and A. thaliana). Libraries were constructed with 100-bp insert sizes using Illumina Standard DNA Library or Nextera kits (Illumina, San Diego, CA) and were sequenced as either 50-bp or 100-bp PE reads using Illumina Hiseq 2000 at NYU Center for Genomics and Systems Biology in New York or Abu Dhabi to obtain between 50 and 250 million reads per sample. For all these sequences, we mapped the reads using BWA (Li and Durbin 2010) and SAMtools (Sequence Alignment/Map) (Li et al. 2009), with the species genome sequence available in GenBank as a reference sequence.
Supplementary Material
Supplementary data S1 and S2, figure S1, and table S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The sequencing data have been deposited in the NCBI Sequence Read Archive (accession number SRX434531). The authors are grateful to Mary Ann Cajano, Salud Pangan, Filipinas Natividad, Karen Renzaglia, Steve Rounsley, Joseph Morin, Sandra Yap, Neda Barghi, Steven Sullivan, Jane Carlton, Arturo Lluisma, and Cynthia Saloma for their help and support in various aspects of the project. The authors are also grateful for the help provided by the Department of Environment and Natural Resources-Protected Areas and Wildlife Bureau (DENR-PAWB) of the Philippines, and thank Director Theresa Mundita S. Lim, Josefina de Leon, and Cecile Garcia for facilitating issuance of collecting permits. For assistance in collecting R. lagascae, they thank Jovencio Payba, Provincial Environment and Natural Resources Office (PENRO)/DENR-Region 2, Apolinario U. Unarce, Sumper Aresta, Tabuc, Rolando Echanique and Alfredo Gabriel and The Cagayan Valley Partners in People Development (CAVAPPED) through its President/CEO Perla A. Visorro. For assistance in collecting R. philippensis (Mt. Banahaw), the authors thank Jerry R. Mendua, Ananias M. Cahilo Sr., Romeo R. Diala, Angeles Coronado, Salud Pangan, and MaryAnn O. Cajano. This work was supported by a start-up grant from LIU and a Visiting Professorship grant from the University of the Philippines to J.M., and grants from the US National Science Foundation Plant Genome Research Program (IOS 1126971) and the NYU Abu Dhabi Institute to M.D.P. This article is dedicated to the memory of Leonard Co, a leading Philippine botanist and Rafflesia investigator, who passed away while conducting fieldwork.
References
- Alverson AJ, Wei X, Rice DW, Stern DB, Barry K, Palmer JD. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae) Mol Biol Evol. 2010;27:1436–1448. doi: 10.1093/molbev/msq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbrook AC, Howe CJ, Purton S. Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci. 2006;11:101–108. doi: 10.1016/j.tplants.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Barcelona JF, Pelser PB, Balete DS, Co LL. Taxonomy, ecology, and conservation status of Philippine Rafflesia (Rafflesiaceae) Blumea. 2009;54:77–93. [Google Scholar]
- Barkman TJ, Mcneal JR, Lim SH, Coat G, Croom HB, Young ND, dePamphilis CW. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evol Biol. 2007;7:248. doi: 10.1186/1471-2148-7-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birschwilks M, Haupt S, Hofius D, Neumann S. Transfer of phloem-mobile substances from the host plants to the holoparasite Cuscuta sp. J Exp Bot. 2006;57:911–921. doi: 10.1093/jxb/erj076. [DOI] [PubMed] [Google Scholar]
- Blanchard JL, Schmidt GW. Pervasive migration of organellar DNA to the nucleus in plants. J Mol Evol. 1995;41:397–406. doi: 10.1007/BF00160310. [DOI] [PubMed] [Google Scholar]
- Bock R. Structure, function, and inheritance of plastid genomes. In: Bock R, editor. Cell and molecular biology of plastids: topics in current genetics. Berlin: Springer; 2007. pp. 29–63. [Google Scholar]
- Braukmann T, Kuzmina M, Stefanovic S. Plastid genome evolution across the genus Cuscuta (Convolvulaceae): two clades within subgenus Grammica exhibit extensive gene loss. J Exp Bot. 2013;64:977–989. doi: 10.1093/jxb/ers391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braukmann T, Stefanovic S. Plastid genome evolution in mycoheterotrophic Ericaceae. Plant Mol Biol. 2012;79:5–20. doi: 10.1007/s11103-012-9884-3. [DOI] [PubMed] [Google Scholar]
- Bromham L, Cowman PF, Lanfear R. Parasitic plants have increased rates of molecular evolution across all three genomes. BMC Evol Biol. 2013;13:126. doi: 10.1186/1471-2148-13-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bungard RA. Photosynthetic evolution in parasitic plants: insight from the chloroplast genome. BioEssays. 2004;26:235–247. doi: 10.1002/bies.10405. [DOI] [PubMed] [Google Scholar]
- Camara B, Hugueney P, Bouvier F, Kuntz M, Moneger R. Biochemistry and molecular biology of chromoplast development. Int Rev Cytol. 1995;163:175–247. doi: 10.1016/s0074-7696(08)62211-1. [DOI] [PubMed] [Google Scholar]
- Colwell AE. [St Louis (WA)]: Washington University; 1994. Genome evolution in a nonphotosynthetic plant, Conopholis americana [PhD dissertation] [Google Scholar]
- Davis CC, Latvis M, Nickrent DL, Wurdack KJ, Baum DA. Floral gigantism in Rafflesiaceae. Science. 2007;315:1812. doi: 10.1126/science.1135260. [DOI] [PubMed] [Google Scholar]
- Davis C, Wurdack K. Host to parasite transfer in flowering plants: phylogenetic evidence from Malpighiales. Science. 2004;305:676–678. doi: 10.1126/science.1100671. [DOI] [PubMed] [Google Scholar]
- Goff LJ, Coleman AW. Fate of parasite and host organelle DNA during cellular transformation of red algae by their parasites. Plant Cell. 1995;7:1899–1911. doi: 10.1105/tpc.7.11.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SB, Waller RF, McFadden GI. Plastid evolution. Annu Rev Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- Henderson J, Salzberg S, Fasman KH. Finding genes in DNA with a hidden Markov model. J Comput Biol. 1997;4:127–141. doi: 10.1089/cmb.1997.4.127. [DOI] [PubMed] [Google Scholar]
- Isono K, Niwa Y, Satoh K, Kobayashi H. Evidence for transcriptional regulation of plastid photosynthesis genes in Arabidopsis thaliana roots. Plant Physiol. 1997;114:623–630. doi: 10.1104/pp.114.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Keeling PJ. The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci. 2010;365:729–748. doi: 10.1098/rstb.2009.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Maier UG, Leister D. DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu Rev Plant Biol. 2009;60:115–138. doi: 10.1146/annurev.arplant.043008.092119. [DOI] [PubMed] [Google Scholar]
- Krause K. From chloroplasts to “cryptic” plastids: evolution of plastid genomes in parasitic plants. Curr Genet. 2008;54:111–121. doi: 10.1007/s00294-008-0208-8. [DOI] [PubMed] [Google Scholar]
- Krause K. Piecing together the puzzle of parasitic plant plastome evolution. Planta. 2011;234:647–656. doi: 10.1007/s00425-011-1494-9. [DOI] [PubMed] [Google Scholar]
- Kress WJ, Erickson DL, Jones AF, Swenson NG, Perez R, Sanjur O, Bermingham E. Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proc Natl Acad Sci U S A. 2009;106:18621–18626. doi: 10.1073/pnas.0909820106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang TC, Qiao Q, Ren Z, Zhao J, Yonezawa T, Hasegawa M, Crabbe MJ, Li J, Zhong Y. Complete chloroplast genome sequence of holoparasite Cistanche deserticola (Orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (Chenopodiaceae) PLoS One. 2013;8:e58747. doi: 10.1371/journal.pone.0058747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal E, Cesbron-Delauw MF. The apicoplast: a new member of the plastid family. Trends Plant Sci. 2001;6:200–205. doi: 10.1016/s1360-1385(01)01921-5. [DOI] [PubMed] [Google Scholar]
- Martin W. Gene transfer from organelles to the nucleus: frequent and in big chunks. Proc Natl Acad Sci U S A. 2003;100:8612–8614. doi: 10.1073/pnas.1633606100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Borst P. Secondary loss of chloroplast in trypanosomes. Proc Natl Acad Sci U S A. 2003;100:765–767. doi: 10.1073/pnas.0437776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden GI, Reith ME, Munholland J, Lang-Unnasch N. Plastid in human parasites. Nature. 1996;381:482. doi: 10.1038/381482a0. [DOI] [PubMed] [Google Scholar]
- Nais J. Borneo (Kota Kinabalu): Natural History Publications; 2001. Rafflesia of the world. [Google Scholar]
- Neuhaus HE, Emes MJ. Nonphotosynthetic metabolism in plastids. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:111–140. doi: 10.1146/annurev.arplant.51.1.111. [DOI] [PubMed] [Google Scholar]
- Nickrent DL, Blarer A, Qiu Y-L, Vidal-Russell R, Anderson FE. Phylogenetic inference in Rafflesiales: the influence of rate heterogeneity and horizontal gene transfer. BMC Evol Biol. 2004;4:e40. doi: 10.1186/1471-2148-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickrent DL, Duff J, Ouyang Y, dePamphilis CW. Do holoparasitic Santalales possess a plastid genome? Plant Mol Biol. 1997;34:717–729. doi: 10.1023/a:1005860632601. [DOI] [PubMed] [Google Scholar]
- Palmer JD. The mitochondrion that time forgot. Nature. 1997;387:454–455. doi: 10.1038/387454a0. [DOI] [PubMed] [Google Scholar]
- Pelser PB, Nickrent DL, Callado JRC, Barcelona JF. Mt. Banahaw reveals: the resurrection and neotypification of the name R. lagascae (Rafflesiaceae) and clues to the dispersal of Rafflesia seeds. Phytotaxa. 2013;131:35–40. [Google Scholar]
- Rivarola M, Foster JT, Chan AP, Williams AL, Rice DW, Liu X, Melake-Berhan A, Creasy HH, Puiu D, Rosovitz MJ, et al. Castor bean organelle genome sequencing and worldwide genetic diversity analysis. PLoS ONE. 2011;6:e21743. doi: 10.1371/journal.pone.0021743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roney J, Khatibi P, Westwood J. Cross-species translocation of mRNA from host plants into the parasitic plant dodder. Plant Physiol. 2007;143:1037–1043. doi: 10.1104/pp.106.088369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchetti G, Romagnoli C, Nicoletti M, Di Fabio A, Bruni A, Poli F. Glandular trichomes of Calceolaria adscendens Lidl. (Scrophulariaceae): histochemistry, development and ultrastructure. Ann Bot. 1999;83:87–92. [Google Scholar]
- Stegemann S, Keuthe M, Greiner S, Bock R. Horizontal transfer of chloroplast genomes between plant species. Proc Natl Acad Sci U S A. 2012;109:2434–2438. doi: 10.1073/pnas.1114076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talianova M, Janousek B. What can we learn from tobacco and other Solanaceae about horizontal DNA transfer? Am J Bot. 2011;98:1231–1242. doi: 10.3732/ajb.1000370. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Sasaki N, Ohmiya A. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 2008;54:733–749. doi: 10.1111/j.1365-313X.2008.03447.x. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Giezen M, Tovar J, Clark CG. Mitochondrion-derived organelles in protists and fungi. Int Rev Cytol. 2005;244:175–225. doi: 10.1016/S0074-7696(05)44005-X. [DOI] [PubMed] [Google Scholar]
- Wen-Zhe L, Hong-Fei LB, Zheng-Hai H. Ultrastructure of the multicellular nodules in Hypericum perforatum leaves. Acta Bot Sin. 2002;44:649–656. [Google Scholar]
- Westwood JH, Yoder JI, Timko MP, dePamphilis CW. The evolution of parasitism in plants. Trends Plant Sci. 2010;15:227–235. doi: 10.1016/j.tplants.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, Müller KF, dePamphilis CW, Quandt D, Wickett NJ, Zhang Y, Renner SS, Schneeweiss GM. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell. 2013;25:3711–3725. doi: 10.1105/tpc.113.113373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RR. The diversity of plastid form and function. In: Wise RR, Hoober JK, editors. The structure and function of plastids. Vol. 23. Dordrecht (The Netherlands): Springer; 2006. pp. 3–26. [Google Scholar]
- Wolfe KH, Morden CW, Palmer JD. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci U S A. 1992;89:10648–10652. doi: 10.1073/pnas.89.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Bradley RK, Wurdack KJ, Wong K, Sugumaran M, Bomblies K, Davis CC. Horizontal transfer of expressed genes in a parasitic flowering plant. BMC Genomics. 2012;13:227. doi: 10.1186/1471-2164-13-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Wang Y, Bradley RK, Sugumaran M, Marx CJ, Rest JS, Davis CC. Massive mitochondrial gene transfer in a parasitic flowering plant clade. PLoS Genet. 2013;9:e1003265. doi: 10.1371/journal.pgen.1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.