Abstract
AmpC β-lactamases are clinically important cephalosporinases encoded on the chromosomes of many Enterobacteriaceae and a few other organisms, where they mediate resistance to cephalothin, cefazolin, cefoxitin, most penicillins, and β-lactamase inhibitor/β-lactam combinations. The increase in antibiotic resistance among Gram-negative bacteria is a notable example of how bacteria can procure, maintain and express new genetic information that can confer resistance to one or several antibiotics. Detection of organisms producing these enzymes can be difficult, because their presence does not always produce a resistant phenotype on conventional disc diffusion or automated susceptibility testing methods. These enzymes are often associated with potentially fatal laboratory reports of false susceptibility to β-lactams phenotypically. With the world-wide increase in the occurrence, types and rate of dissemination of these enzymes, their early detection is critical. AmpC β-lactamases show tremendous variation in geographic distribution. Thus, their accurate detection and characterization are important from epidemiological, clinical, laboratory, and infection control point of view. This document describes the methods for detection for AmpC β-lactamases, which can be adopted by routine diagnostic laboratories.
Keywords: AmpC β-lactamases, disk approximation test, gram-negative bacteria, three-dimensional extract test
INTRODUCTION
Drug resistance poses a therapeutic problem not only in the hospital settings, but also in the community as most of the bacteria have acquired resistance to multiple antibiotics.[1,2] The various mechanisms of drug resistance in Gram-negative bacteria include extended spectrum beta-lactamases (ESBL) production, AmpC β-lactamase production, efflux mechanism and porin deficiency. In the clinical laboratory settings, the commonly detected enzymes causing resistance are AmpC β-lactamases and ESBLs. Clinical relevance of AmpC β-lactamases lies in the fact that they confer resistance to both narrow and broad spectrum cephalosporins, beta-lactam/beta-lactamase inhibitor combinations and aztreonam.[3]
AmpC β-lactamases can be chromosomally or plasmid mediated. The plasmid mediated AmpC β-lactamases hydrolyze all β-lactam antibiotics except cefepime and carbapenems. The plasmid-mediated AmpC genes are derived from inducible chromosomal genes that have been mobilized among various organisms. The commonly reported genotypes are ACC, FOX, MOX, DHA, CMY, CIT and EBC.[4,5,6] These mobilized plasmid mediated enzymes confer a resistance pattern similar to the overproduction of chromosomal AmpC β-lactamases, which also involve all β-lactam antibiotics except for carbapenems and cefepime.[7]
Detection of AmpC is important to improve the clinical management of patients suffering from infections and would also provide us with sound epidemiological data. However, there are no Clinical and Laboratory Standards Institute guidelines for detection of AmpC mediated resistance in Gram-negative clinical isolates and hence, it usually poses a problem due to misleading results, especially so in phenotypic tests.[8]
Personnel qualifications
The test performer should be having a diploma in laboratory technologies and preferably university graduate in biological sciences with sufficient experience.
Education and training
Personnel are required to be knowledgeable of the procedures in the microbiology laboratory. The laboratory staff should confirm that they can properly perform the procedures before commencing work. The details are given in Table 1.
Table 1.
Education and training must be given on the following topics

PROCEDURE
Techniques to identify AmpC β-lactamase-producing isolates are available but are still evolving and are not yet optimized for the clinical laboratory, which leads to the underestimation of these resistance mechanisms. Carbapenems can usually be used to treat infections due to AmpC-producing bacteria, but carbapenem resistance can arise in some organisms by mutations that reduce influx (outer membrane porin loss) or enhance efflux (efflux pump activation).
REQUIREMENTS
The list of all the requirements are given in Table 2.
Table 2.
List of all the requirements for performing the test
Isolates
Gram-negative bacterial isolates recovered from clinical samples.
Preparation of reagents and chemicals
-
300 μg phenylboronic acid
- Weigh 150 mg phenylboronic acid and dissolve in 10 ml sterile distilled water
- Use it as 15 μg/μl stock
- Store at 4°C
-
0.5 M ethylenediaminetetraacetic acid (EDTA) buffer
- Weigh 18.6 g EDTA and add distilled water to 90 ml
- Adjust pH to 8.0
- Add distilled water and make final volume 100 ml
- Store at 4°C
-
Tris-EDTA (TE) buffer
- 50X TE (stock solution)
- Weigh 121 g Tris base and dissolve in 400 ml distilled water
- Add 50 ml 0.5 M EDTA
- Add distilled water and make final volume 500 ml
- Store at 4°C
-
TE buffer (working)
- Dilute 50X master stock to 1X with distilled water.
Primer reconstitution.
Primers are often shipped and received in a lyophilized state. First create a master 100X stock (for each primer and then dilute it to a 20X working stock).
Master stock, 100 μM
100 μM = X nmoles lyophilized primer + (X × 10 μl molecular grade H2O)
To determine the amount of H2O to add to the lyophilized primer simply multiply the number of nmol of primer in the tube by 10 and that will be the amount of H2O to add to make a 100 μM primer stock
Vortex tube and incubate at room temperature for 10 min.
Working stock, 20 μM
Dilute the primer master stock in a sterile microcentrifuge tube 1:5 with molecular grade H2O.
TEST PROCEDURE
What is unnecessary?
It is unnecessary to detect AmpC production in organisms that produce an inducible chromosomal AmpC β-lactamase because the organism identification is indicative of AmpC production; i.e. 100% isolates of Enterobacter cloacae, Enterobacter aerogenes, Citrobacter freundii, Serratia marcescens, Providencia sp., Morganella morganii, Hafnia alvei, Aeromonas sp., and P. aeruginosa can be assumed to be AmpC producers. Detection of an AmpC β-lactamase in Klebsiella sp., Citrobacter koseri or Proteus mirabilis is confirmatory for plasmid-mediated AmpC production because these organisms lack a chromosomal AmpC β-lactamases.[9,10]
Screening
-
Requirement
- Test organism
- Normal saline
- Blood agar plates
- Mueller-Hinton agar (MHA) plates
- 30-μg cefoxitin disk
-
Procedure
- Make 0.5 McFarland bacterial suspension in normal saline prepared from an overnight blood agar plate
- Inoculate surface of MHA plate with this suspension by swabbing
- Place 30-μg cefoxitin disk on inoculated MHA
- Invert the plate and incubate overnight at 35°C
-
Plate reading and interpretation
- After overnight incubation measure zone diameter around 30-μg cefoxitin disk
- Select the isolates with zone diameters less than 18 mm for confirmation of AmpC production.
Phenotypic confirmatory tests
Three-dimensional extract test
-
Requirement
- Test organism
- Escherichia coli ATCC 25922 or E. coli ATCC 11775
- Normal saline
- Blood agar plates
- MHA plates
- 12 ml brain heart infusion (BHI) broth
- 30-μg cefoxitin disk
- Sterile blade
-
Procedure
- Prepare 0.5 McFarland bacterial suspension from an overnight blood agar plate
- Inoculate 12 ml BHI broth with 50 μl of 0.5 McFarland bacterial suspension and incubate for 4 h at 37°C
- Concentrate cells by centrifugation and freeze-thaw 5 times to prepare crude enzyme
- Prepare 0.5 McFarland bacterial suspension using one of two E. coli ATCC 25922 or ATCC 11775 and inoculate surface of MHA plate by using this suspension
- Place 30-μg cefoxitin disk on the inoculated agar plate
- With a sterile scalpel blade, cut a slit beginning 5 mm from the edge of the disk in an outward radial direction
- By using a pipette, dispense 25-30 μl of enzyme preparation into the slit, beginning near the disk and moving outward, avoiding slit overfill
- Incubate inoculated media overnight at 37°C
-
Plate reading and interpretation
- After overnight incubation check the enhanced growth of the surface organism at the point where the slit intersected
- If there is a zone of inhibition of surface organism, the test is positive three-dimensional test [Figure 1].
Figure 1.
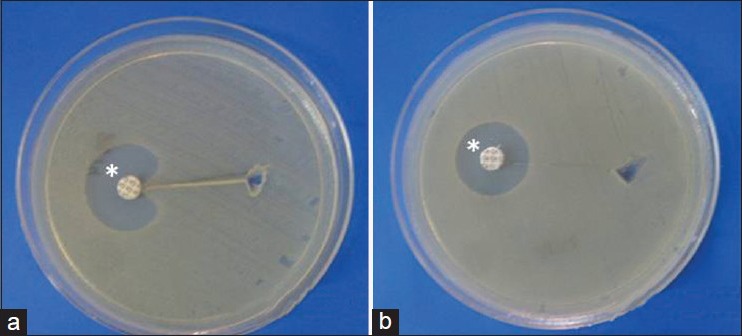
Representation of three dimensional extract test. (a) Zone of inhibition showing positive test, (b) no zone of inhibition showing negative test. *30 μg cefoxitin disk
AmpC disk test
-
Requirements
- AmpC disks (filter paper disks containing TE)
- 30 μg cefoxitin disk
- Blood agar plates
- MHA plates
- Test organisms
- E. coli ATCC 25922
- Normal saline
-
Procedure
- Prepare 0.5 McFarland bacterial suspension of E. coli ATCC 25922
- Inoculate surface of MHA plate using this suspension as per standard disk diffusion method
- Immediately prior to use, rehydrate AmpC disk with 20 μl of saline and several colonies of each test organism apply to a disk
- Place a 30 μg cefoxitin disk on the inoculated surface of the MHA
- Place inoculated AmpC disk almost touching the antibiotic disk with the inoculated disk face in contact with the agar surface
- Invert the plate and incubate overnight at 35°C in ambient air
-
Plate reading and interpretation
- After overnight incubation, examine the plate for either an indentation or a flattening of the zone of inhibition
- If there is any zone of inhibition, it indicates enzymatic inactivation of cefoxitin (positive result)
- If no zone inhibition, indicates no significant inactivation of cefoxitin (negative result) [Figure 2].
Figure 2.
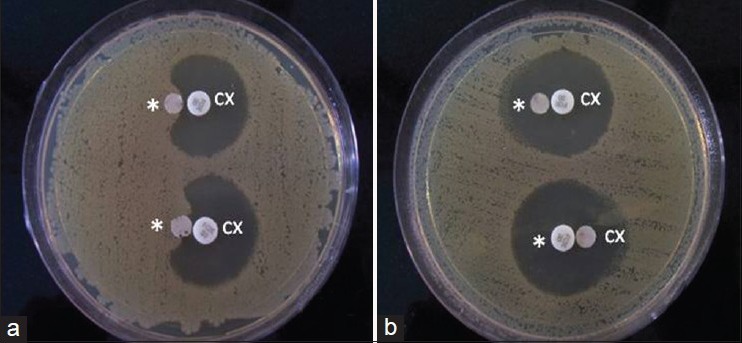
Representation of AmpC disk test. (a) Zone of inhibition showing positive test results, (b) no zone of inhibition showing negative test results, *AmpC disks (filter paper containing tris-EDTA), CX: 30μg cefoxifin disks
Boronic acid disk test method
-
Requirements
- Test organism
- 30 μg cefoxitin disk
- Phenylboronic acid
- MHA plates.
-
Procedure
- Prepare 0.5 McFarland bacterial suspension from an overnight blood agar plate
- Inoculate surface of MHA plate using this suspension as per standard disk diffusion method
- Place a 30 μg cefoxitin disk on the inoculated surface of the MHA
- Using sterile tips, dispense 20 μl of 15 μg/ml phenylboronic acid onto the disk
- Let the disk absorb it
- Invert the plate and incubate overnight at 35°C.
-
Plate reading and interpretation
- After overnight incubation, compare the zone diameter around the antibiotic disk with added boronic acid and the antibiotic-containing disk alone
Figure 3.
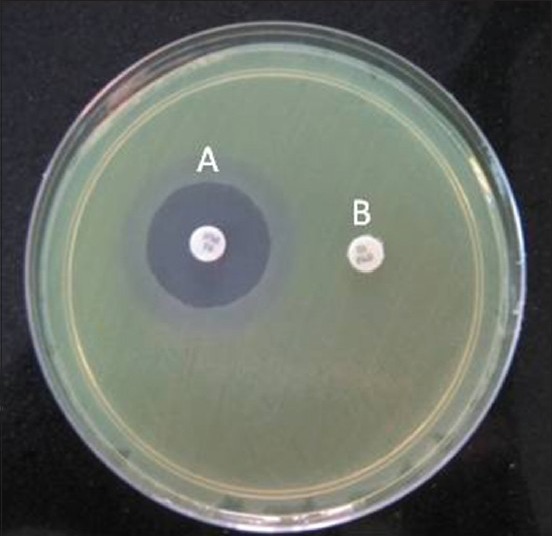
Representation of boronic acid disk test. A: 30 μg cefoxitin disk supplemented with 300μg of phenyl boronic acid. B: 30 μg cofoxitin disk alone
Disk approximation test
- Requirements
- Test organism
- Normal saline
- 10 μg imipenem disk
- 30 μg cefoxitin disk
- 20/10 μg amoxicillin-clavulanate disk
- 30 μg ceftazidime disk
- MHA Plates
-
Procedure
- Prepare 0.5 McFarland bacterial suspension from an overnight blood agar plate
- Inoculate surface of MHA plate using this suspension as per standard disk
- diffusion method
- Place a 30 μg ceftazidime disk at the center on the plate
- Place 10 μg imipenem, 30 μg cefoxitin, and 20/10 μg amoxicillin-clavulanate disks at a distance of 20 mm from ceftazidime disk
- Invert the plate and incubate overnight at 35°C
-
Plate reading and interpretation
- After overnight incubation, examine the plate for any obvious blunting or flattening of the zone of inhibition between the ceftazidime disk and the inducing substrates (imipenem, cefoxitin and amoxicillin-clavulanate disk)
- If there is any blunting or flattening of the zone, consider as a positive result for AmpC production [Figure 4].
Figure 4.
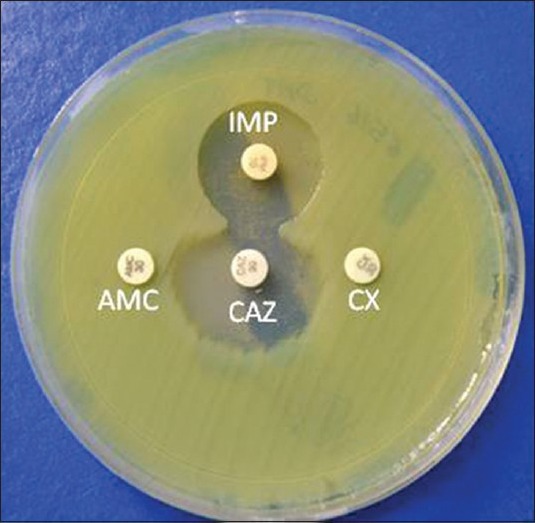
Representation of disk approximation test. Flattening of zone of ceftazidime toward imipenem disk (inducing substrate) showing positive result. IMP: Imipenem (10 μg), CAZ: Ceftazidime (10 μg), AMC: Amoxillin-clavulanate (20/10 μg)
Multiplex polymerase chain reaction for plasmid mediated AmpC genes
Phenotypic tests do not differentiate between chromosomal and plasmid mediated AmpC β-lactamases. Plasmid-mediated AmpC β-lactamases are most accurately detected with the multiplex AmpC PCR test.
Preparation of template deoxyribonucleic acid (DNA)
- Requirements
- Test organism
- Molecular biology grade water
- Microcentrifuge tubes
- Absolute ethanol
- DNA extraction kit
-
Procedure
- Inoculate a single colony of each organism into 5 ml of Luria-Bertani broth and incubate for 20 h at 37°C with shaking
- Harvest cells from 1.5 ml of the overnight culture by centrifugation at 10,000 × g for 5 min
- Discard supernatant, re-suspend the pellet in 500 μl of distilled water
- Extract total DNA by using DNA extraction kit according to manufacturer's instructions
- Quantify total DNA prior to the multiplex PCR using spectrophotometer
Multiplex PCR
- Requirements
- 0.5-ml thin-walled PCR tubes
- Molecular biology grade water
- Taq DNA polymerase (5U/μl)
- 10X Taq buffer with KCL
- Tris-HCl (pH 8.4)
- 25 mM MgCl2
- DNTPS 10Mm
-
Procedure
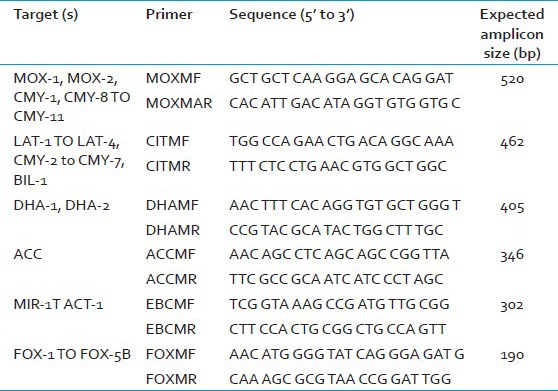
- Make a master mix containing 20 mM Tris-HCl (pH 8.4); 50 mM KCl; 0.2 mM each dNTPs; 1.5 mM MgCl2; 0.6 μM primers MOXMF, MOXMR, CITMF, CITMR, DHAMF, and DHAMR; 0.5 μM primers ACCMF, ACCMR, EBCMF, and EBCMR; 0.4 μM primers FOXMF and FOXMR; and 1.25 U of Taq DNA polymerase. Add 2 µl DNA template. The list of all the primers are given in Table 3.
- Set the PCR program on an initial denaturation step at 94°C for 3 min, followed by 25 cycles of DNA denaturation at 94°C for 30s, primer annealing at 64°C for 30s, and primer extension at 72°C for 1 min. After the last cycle, a final extension step at 72°C for 7 min
- Set the tube in the PCR machine and run the program.
Table 3.
Primers for amplification of AmpC genes
Electrophoresis
- Requirements
- Agarose
- Ethidium bromide
- Loading die
- 100-bp DNA ladder
-
Procedure
- Prepare 2% agarose gel in 1X TE buffer
- Analyze 5 μl PCR product mixed with 1 μl 6X loading die
- Use 100-bp DNA ladder as a marker
- Stain gel with ethidium bromide (10 μg/ml) and analyze the presence of bands in ultraviolet transilluminator
- Use the PCR mixtures with the addition of water in place of template DNA as negative control.
Plate disposal
Keep all culture plates sealed inside blue plastic bags and seal in an autoclave bag
Autoclave at 121°C for 30 min
Discard the sealed sterilized bags in the site designed for this purpose.
ACKNOWLEDGMENT
We acknowledge the financial support of ICMR for the performance of this study.
Footnotes
Source of Support: We acknowledge the financial support of ICMR for the performance of this study
Conflict of Interest: None declared.
REFERENCES
- 1.Kaurthe J. Increasing antimicrobial resistance and narrowing therapeutics in typhoidal salmonellae. J Clin Diagn Res. 2013;7:576–9. doi: 10.7860/JCDR/2013/4765.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laxminarayan R, Klugman KP. Communicating trends in resistance using a drug resistance index. BMJ Open. 2011;1:e000135. doi: 10.1136/bmjopen-2011-000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drawz SM, Bonomo RA. Three decades of beta-lactamase inhibitors. Clin Microbiol Rev. 2010;23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanson ND. AmpC beta-lactamases: What do we need to know for the future? J Antimicrob Chemother. 2003;52:2–4. doi: 10.1093/jac/dkg284. [DOI] [PubMed] [Google Scholar]
- 5.Pérez-Pérez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40:2153–62. doi: 10.1128/JCM.40.6.2153-2162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papanicolaou GA, Medeiros AA, Jacoby GA. Novel plasmid-mediated beta-lactamase (MIR-1) conferring resistance to oxyimino-and alpha-methoxy beta-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1990;34:2200–9. doi: 10.1128/aac.34.11.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mirelis B, Rivera A, Miró E, Mesa RJ, Navarro F, Coll P. A simple phenotypic method for differentiation between acquired and chromosomal AmpC beta-lactamases in Escherichia coli. Enferm Infecc Microbiol Clin. 2006;24:370–2. doi: 10.1157/13089690. [DOI] [PubMed] [Google Scholar]
- 8.Handa D, Pandey A, Asthana AK, Rawat A, Handa S, Thakuria B. Evaluation of phenotypic tests for the detection of AmpC beta-lactamase in clinical isolates of Escherichia coli. Indian J Pathol Microbiol. 2013;56:135–8. doi: 10.4103/0377-4929.118686. [DOI] [PubMed] [Google Scholar]
- 9.Wayne, PA: Clinical and Laboratory Standards Institute; 2010. Performance Standards for Antimicrobial Susceptibility Testing, 20th Informational Supplement. M100-S 21. [Google Scholar]
- 10.Thomson KS. Extended-spectrum-beta-lactamase, AmpC, and Carbapenemase issues. J Clin Microbiol. 2010;48:1019–25. doi: 10.1128/JCM.00219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]


