Abstract
Background:
In osteosarcoma tissue, both MMP-2 and MMP-9 are over expressed compared to their expression in non-affected stromal tissue. Hence, gelatinases are attractive targets for anti-osteosarcoma drugs.
Objective:
To study the inhibitory activity of compounds isolated from Ageratum houstonianum against MMP-2 and MMP-9 by in-silico approach.
Material and Methods:
We performed docking study using ‘Autodock 4.2’ between 1,2-benzenedicarboxylic acid-bis (2-ethylhexyl) ester; squalene; 3,5-bis (1,1-dimethylethyl) phenol; pentamethyl tetrahydro-5H-chromene; (1, 4-cyclohexylphenyl) ethanone and 6-vinyl-7-methoxy-2,2-dimethylchromene with MMP-2 and MMP-9.
Results:
Among all six compounds isolated from Ageratum houstonianum, (1, 4-cyclohexylphenyl) ethanone showed the maximum potential as a putative inhibitor of both MMP-2 and MMP-9 enzymes with reference to ΔG (−7.95 and −8.2 kcal/mol, respectively) and Ki (1.48 and 0.98 μM, respectively) values. Total intermolecular energy of docking for (1, 4-cyclohexylphenyl) ethanone-MMP catalytic domain-interaction was found to be −8.55 kcal/mol for MMP-2 and −9.21 kcal/mol for MMP-9.
Conclusion:
This study explores molecular interactions between human MMPs (MMP-2 and MMP-9) and six natural compounds. This study predicts that (1,4-cyclohexylphenyl) ethanone is a more efficient inhibitor of human MMP-2 and MMP-9 enzymes compared to the other natural compounds used in this study with reference to Ki and ΔG values.
Keywords: Ageratum houstonianum; docking; matrix metalloproteinases; matrix metalloproteinase 2; matrix metalloproteinase 9; oncoinformatics; (1, 4-cyclohexylphenyl) ethanone
INTRODUCTION
Matrix metalloproteinases (MMPs) are a family of 26 calcium dependent zinc-containing human endopeptidases that are responsible for the tissue remodeling and degradation of the extracellular matrix such as gelatin, elastins, collagens, matrix glycoproteins and proteoglycan. These metalloproteinases are secreted by a number of connective tissues and pro-inflammatory cells in “Zymogen” form. These zymogens get converted into their active forms by other proteolytic enzymes (e.g., serine proteases) via removal of pro-domain/pro-peptide and signal peptide. Most MMPs possess 4 distinct domains-N-terminal pro-domain, catalytic domain, hinge region, and C-terminal hemopexin-like domain. Transmembrane domain is an additional domain contained by the membrane type MMPs. This additional domain acts like an “anchor” to keep them adhered to the surface of the cell. The catalytic domain is highly conserved among all MMPs with two special features. These features are: S1’ pocket and a loop constituting the outer wall of the S1’ pocket. HEXXHXXGXXH is a conserved active-site sequence motif which contains the glutamic acid residue and coordinates the catalytic zinc (II) ion. This facilitates catalysis. Hence, the active site of MMPs consists of a groove, which is centered on the catalytic zinc ion and an S1’ specificity site, which is significantly variable. A significant interaction between S1’ subsite and P1’ residue of substrates/inhibitors has been observed. The S1'pocket in the case of MMP-2 may be a channel with no bottom while MMP-9 has a small groove-like subsite. The subunit S2’ is a solvent-exposed cleft with hydrophobic P2’ residues in substrates/inhibitors. Subunit S3’ is not well exposed to solvent region.[1] A gelatin binding fibronectin domain is possessed by gelatinases (MMP-2 and MMP-9). This gelatin binding domain consists of three fibronectin-repeats, inserted between the active-site domain and the Zn2+ binding domain. Gelatinase B contains an additional Ser/Thr/Pro-rich collagen type V domain in the hinge region.[2]
Due to their significant role in the extracellular matrix, MMPs are considered as promising targets for cancer therapy. A large number of MMP inhibitors (MMPIs) have been identified as cytostatic and anti-angiogenic agents. Most of the MMPIs have 2 features: (a) Hydrophobic extension protruding from the site of catalysis into the large and hydrophobic S1’ pocket (P1’ group) and (b) a chelating moiety that interacts with the catalytic zinc ion. In P1’ region of a molecule (the region that interacts with S1’ site of MMPs), a longer-chain aliphatic substituent tends to decrease the potency for MMP-1 and increase for MMP-3 while an aromatic substituent is expected to generate broad-spectrum inhibition.
A large number of plant extracts/compounds have been found capable of inhibiting MMPs. The procyanidin-rich maritime pine bark extract Pycnogenol has well-documented inhibitory effect on MMPs.[3] Inhibition of MMP-2 and MMP-9 by Flavonoids (primuletin/5-hydroxyflavone, luteolin 7-O-glucoside, luteolin) has also been reported.[4] A study demonstrated that Magnolia obovatal leaves extract exerted its anti-cancer effects through blocking migration and invasion by inhibition of MMP-2 expression and activity.[5] A strong inhibition, of > 90% at a concentration of 100 μg/ml, was found in the butanol fractions of Cinnamomum cassia, Magnolia obovata, Magnolia officinalis and Euonymus alatus.[6] Green tea polyphenol epigallocatechin-3-O-gallate has been shown to inhibit the MMP-2/-9 activity as well as the invasiveness of tumor cells. Hydrolysable tannins (plant polyphenols) have also been found to suppress tumor cell invasiveness via inhibiting the activity of MMP-2/-9.[7]
Ageratum houstonianum, a member of Asteraceae (compositae), is a short-lived (i.e. annual or biennial) herbaceous plant growing 0.3-1 m tall. It is native to Mexico, Central America and the Caribbean, but has naturalized other parts of the world as weed such as the coastal districts of eastern Australia, the tropical regions of the world, including south-eastern USA (i.e. Florida, Alabama, Georgia, South Carolina and North Carolina) and some Pacific islands (e.g. Hawaii, Fiji and French Polynesia).[8] In India, it is found in plains and hills and in forests as under growth.[9] Formerly, it has been found that A. houstonianum contains flavones (agehoustin A, B, C, D, eupalestin, agecorynin C) and pyrrolizidine alkaloids (heliohoustine, lycopsamine and retrohoustine) in aerial parts of the plant; oxygen heterocyclesageratone, Ageratum benzofuran in the roots; precocenes in the leaf, stem, root, flowers especially in leaves; triterpenesfriedlin, friedelanol and steroids sit sterol, Beta, stigmasterol in the entire plant.[10] A number of compounds from Ageratum species have been reported for its medicinal and insecticidal uses.[11] Traditionally, it has been used to treat tumors. Precocenes (7-methoxy-2, 2-dimethylchromene [precocene I] and ageratochromene [precocene II]) are responsible for destruction of parenchymal cells of corpus allatum leading to decrease in juvenile hormone. This leads in induction of precocious metamorphosis, sterilization, diapause induction, elimination of sex pheromone production and anti-feeding.[12,13] Due to this reason these have been used to develop insecticides.
Osteosarcoma is an aggressive malignant neoplasm, which exhibits osteoblastic differentiation and produces malignant osteoid, which arise from primitive transformed cells of mesenchymal origin. In osteosarcoma tissue, both MMP-2 and MMP-9 are over expressed in comparison with their expression in non-affected stromal tissue. Indeed, MMP-9 was suggested as a prognostic factor for the development of metastasis in high-grade osteosarcoma.[14,15] MG-63 is an osteosarcoma derived cell line. These cells can also be transfected.
It is pertinent to mention that the natural compounds mentioned herein were identified by Gas Chromatography-Mass Spectrometry (GC-MS) analysis in a previous study.[16] These compounds were present in the (AB-2) active band isolated from methanolic crude extract of leaves of A. houstonianum.[16] In the present computational study, we have tested six of these compounds, namely (1,2-benzenedicarboxylic acid-bis (2-ethylhexyl) ester; squalene; 3,5-bis (1,1-dimethylethyl) phenol; pentamethyl tetrahydro-5H-chromene; (1,4-cyclohexylphenyl) ethanone and 6-vinyl-7-methoxy-2,2-dimethylchromene for their ability to inhibit MMP-2 and MMP-9.
METHODS
To determine the inhibition potential of 1,2-benzenedicarboxylic acid-bis (2-ethylhexyl) ester, squalene, 3,5-bis (1,1-dimethylethyl) phenol, pentamethyl tetrahydro-5H-chromene, (1,4-cyclohexylphenyl) ethanone and 6-vinyl-7-methoxy-2,2-dimethylchromene, virtual docking was performed at the “catalytic domain” of MMP-2 and MMP-9. The crystal structures of MMP-2 and MMP-9, solved at 2.3 Å and 2.0 Å resolutions respectively, were retrieved from the Protein Data Bank (PDB ID: 1HOV and 1GKC, respectively). PDB structure of natural compounds 1,2-benzenedicarboxylic acid-bis (2-ethylhexyl) ester (Chemspider ID-21106505), squalene (Pubchem CID-638072), 3,5-bis (1,1-dimethylethyl) phenol (Pubchem CID-70825), pentamethyl tetrahydro-5H-chromene (Pubchem CID-605742), (1,4-cyclohexylphenyl) ethanone (Pubchem CID-87715) and 6-vinyl-7-methoxy-2,2-dimethylchromene) (Pubchem CID-188454) were retrieved from either Pubchem or Chemspider databases. Thereafter, each of these ligands (natural compounds) was docked to the enzymes (MMP-2 and MMP-9) using “Autodock4.2”, separately. For energy minimization of the ligand molecule, we used the MMFF94 force field. After merging the Non-polar hydrogen atoms, rotatable bonds were defined. With the aid of AutoDock tools, essential hydrogen atoms, Kollman united atom type charges, and solvation parameters were added. Accordingly, with the help of “Autogrid program,” affinity (grid) maps of 40 × 40 × 40 Å grid points and having spacing of 0.375 Å were generated. We aim to target grid coordinates in proximity with the S1’ pockets of the active site of MMP-2 and MMP-9. The values of x, y and z coordinates used for targeting the “S1’ pocket” was 21.64, 55.63, 47.74 for MMP-2 and 65.185, 30.533, 117.013 for MMP-9. We used “AutoDock parameter set,” and “distance-dependent dielectric functions” for the calculation of the Van der Waals and the electrostatic terms, respectively. We performed docking simulations using the “Lamarckian Genetic Algorithm” and the “Solis and Wets local search method.” Orientation and torsions of the ligand molecules were set randomly at the initial position. Each docking experiment was derived from 100 different runs. These runs were set to terminate after a maximum of 2,500,000 energy evaluations. We set the population size to 150 for the docking experiments. A translational step of 0.2 Å, and quaternion and torsion steps of 6 were applied during the search. We used Discovery Studio 2.5 (Accelrys) for generating the final figures.
RESULTS AND DISCUSSION
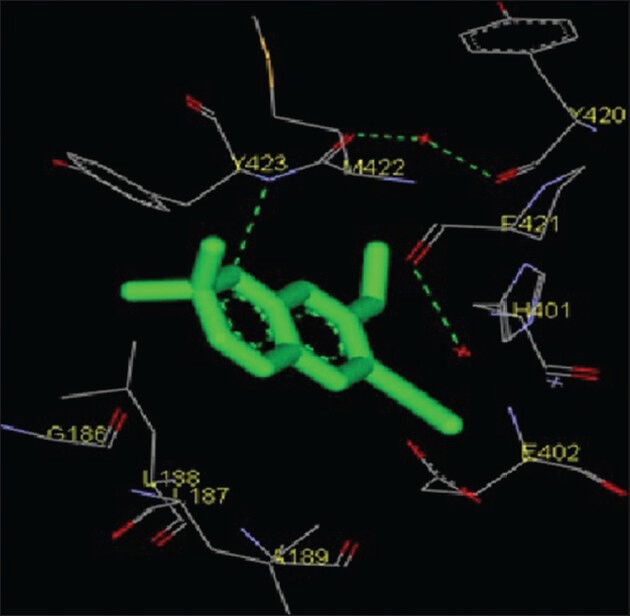
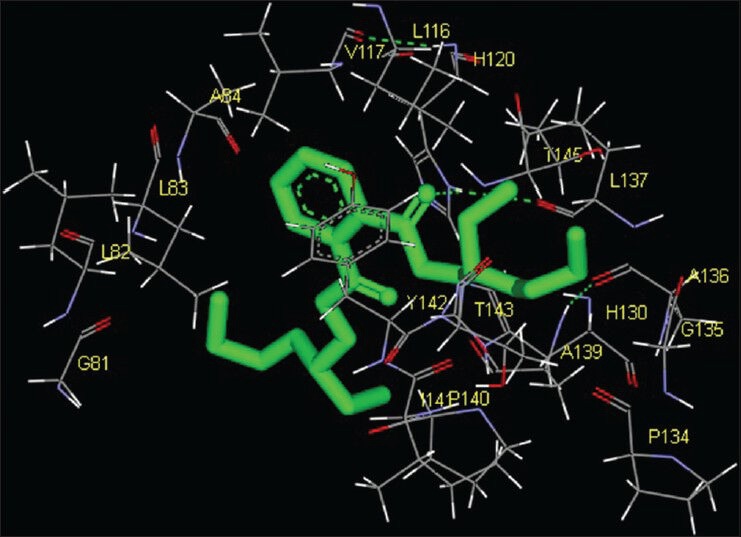
Molecular interactions of the six natural ligands, namely: (1,4-Cyclohexylphenyl) ethanone; 3,5-bis (1,1-dimethylethyl) phenol; pentamethyl tetrahydro-5H-chromene; 6-vinyl-7-methoxy-2,2-dimethylchromene; 1,2-benzenedicarboxylic acid-bis (2-ethylhexyl) ester and squalene, docked to MMP-2 active site are shown in Figures 1–6. Figures 7–12 represent molecular interactions of the same ligands (respectively) with MMP-9 active site. In the present study, the catalytic domain of both MMPs were found to interact with (1,4-cyclohexylphenyl) ethanone through the amino acid residues L83, Y112, V117, L116, H120, T145, Y142, T143, I141, A139, L137, P134, G135, A136 for MMP-2 and L418, H411, Y420, P421, M422, R424, Y423, L397, H401, V398, E402, A189, L188, L187 for MMP-9 [Figures 1 and 7; Table 1]. The free energy of binding and estimated inhibition constant (Ki) for the “(1,4-cyclohexylphenyl) ethanone-MMP catalytic domain-interaction” were determined to be-7.95 kcal/mol and 1.48 μM, respectively for MMP-2 and −8.2 kcal/mol and 0.98 μM, respectively for MMP-9. Four carbon atoms, namely C3, C8, C13, C14 were determined to be involved in hydrophobic interactions via CD1, CD2, CZ, CG1 with amino residues LEU83, TYR112, LEU116, VAL117 and LEU137 of MMP-2. Eight carbon atoms, namely C1, C3, C5, C6, C8, C10, C13, C14, were found to be involved in hydrophobic interactions via CB, CD1, CG1, CG2 with amino acid residues LEU188, LEU397, VAL398 and LEU418 of MMP-9. Four carbon atoms, namely C7, C8, C10, C11, were found to be involved in pi-pi interactions via CE1 and CD1 with amino acid residues HIS120 and TYR142 of MMP-2. Similarly, four carbon atoms, namely C8, C9, C10, C11, were found to be involved in pi-pi interaction via CD1, CD2, CE1, CG with amino acid residues HIS401 and TYR423 of MMP-9. Total intermolecular energy of docking for (1,4-cyclohexylphenyl) ethanone-MMP catalytic domain-interaction was found to be −8.55 kcal/mol for MMP-2 and −9.21 kcal/mol for MMP-9. “Van der Waals,” “hydrogen bond” and “desolvation” energy components together contributed −8.59 kcal/mol for MMP-2 and −9.14 kcal/mol for MMP-9 while the “Electrostatic” energy component was found to be +0.04 kcal/mol for MMP-2 and −0.07 kcal/mol for MMP-9. Total interacting surface area for (1,4-cyclohexylphenyl) ethanone-MMP catalytic domain-interaction was found to be 572.163 Å2 for MMP-2 and 576.424 Å2 for MMP-9 while hydrogen bonds, polar interactions and cation-pi interactions were absent.
Figure 1.
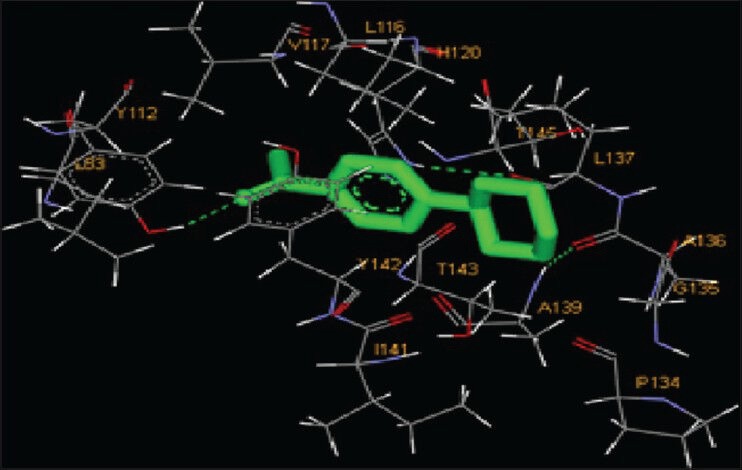
Interaction of (1,4-cyclohexylphenyl) ethanone docked to the “catalytic site” of the matrix metalloproteinase-2. The ligand, (1,4-cyclohexylphenyl) ethanone, is shown in “stick” representation
Figure 6.
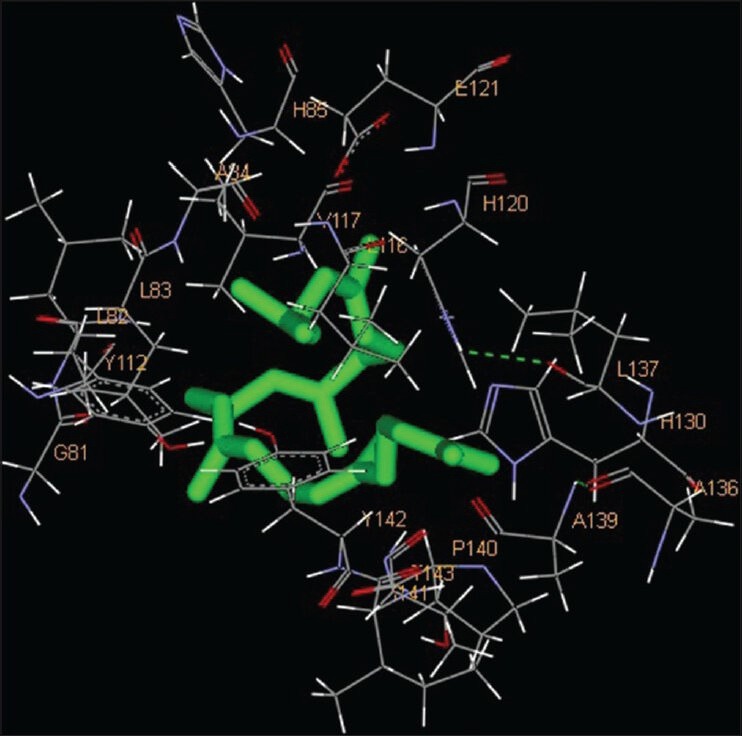
Interaction of squalene docked to the “catalytic site” of the matrix metalloproteinase-2. The ligand, squalene, is shown in “stick” representation
Figure 7.
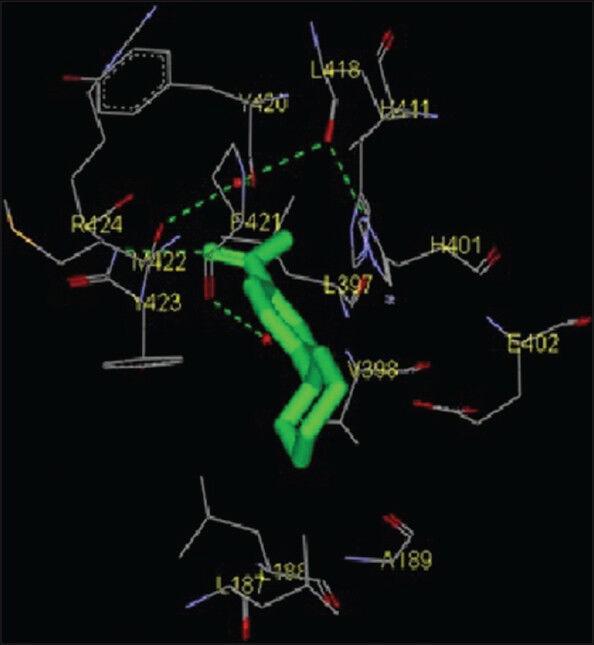
Interaction of (1,4-cyclohexylphenyl) ethanone docked to the “catalytic site” of the matrix metalloproteinase-9. The ligand, (1,4-cyclohexylphenyl) ethanone, is shown in “stick” representation
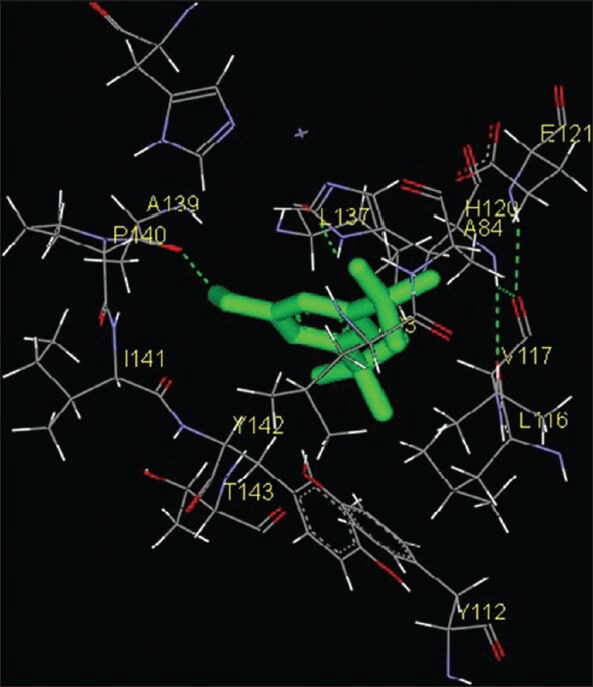
Figure 12.
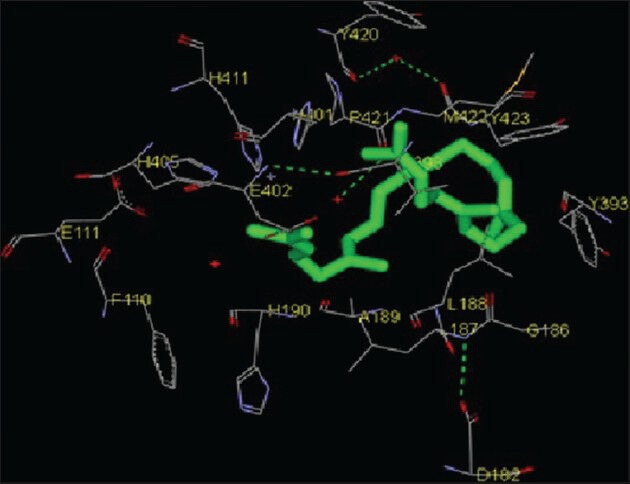
Interaction of squalene docked to the “catalytic site” of the matrix metalloproteinase-9. The ligand, squalene, is shown in “stick” representation
Table 1.
Amino acid residues involved in “natural compounds and MMPs (MMP-2 and MMP-9)-interactions”
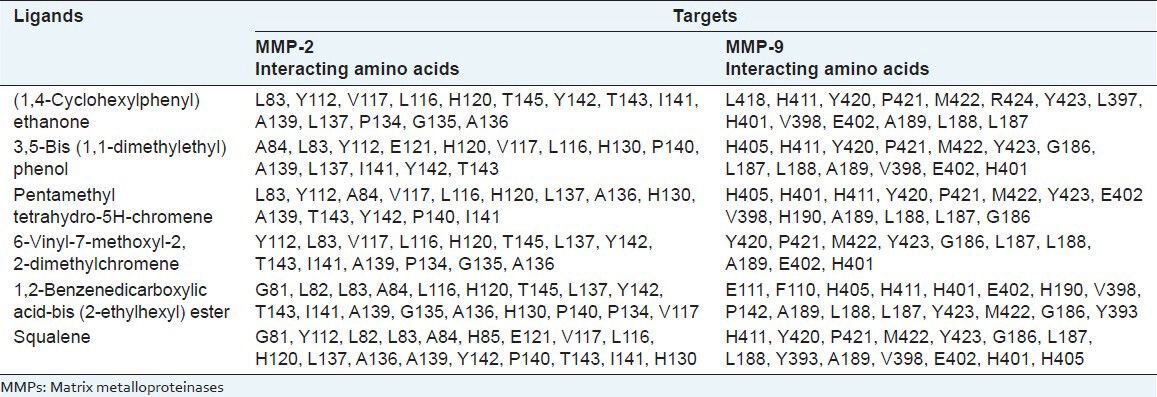
3,5-bis (1,1-dimethylethyl) phenol showed the second highest inhibition against both MMPs. The catalytic domain of MMP-2 was found to interact with 3,5-bis (1,1-dimethylethyl) phenol via 14 amino acid residues, namely A84, L83, Y112, E121, H120, V117, L116, H130, P140, A139, L137, I141, Y142 and T143, whereas MMP-9 interacted with thirteen amino acid residues, namely H405, H411, Y420, P421, M422, Y423, G186, L187, L188, A189, V398, E402 and H401 [Figures 2 and 8 and Table 1]. The free energy of binding and estimated inhibition constant (Ki) for the “3,5-bis (1,1-dimethylethyl) phenol-MMP catalytic domain-interaction” were determined to be −7.43 kcal/mol and 55.22 μM, respectively for MMP-2; while the ∆ G and Ki values for similar interaction involving MMP-9 were found to be −6.78 kcal/mol and 10.69 μM, respectively. Six carbon atoms of MMP-2, namely C3, C4, C5, C6, C7 and C10 were found to be involved in hydrophobic interactions with amino acid residues LEU116 and LEU137. Similarly, six carbon atoms of MMP-9, namely C2, C7, C8, C9, C10, C11 were found to be involved in hydrophobic interactions with amino acid residues LEU187, LEU188, VAL398, HIS401 and TYR423. Two carbon atoms, namely C4 and C13, were found to be involved in π – π interaction with amino acid residue HIS401 of MMP-9. One oxygen atom, O1 and one hydrogen atom, H1 were found to make polar contacts with amino acid residue GLU402 of MMP-9. H1 was also found to make a cation-pi contact with HIS401. Total intermolecular energy of docking for 3,5-bis (1,1-dimethylethyl) phenol-MMP catalytic domain-interaction was found to be −6.70 kcal/mol and −6.12 kcal/mol for MMP-2 and MMP-9, respectively. Van der Waals, hydrogen bond and desolvation energy components together contributed −6.22 kcal/mol for MMP-2 and −5.90 kcal/mol for MMP-9 while the “Electrostatic” energy component was found to be −0.09 kcal/mol for MMP-2 and −0.21 kcal/mol for MMP-9. Total interacting surface area for 3,5-bis (1,1-dimethylethyl) phenol-MMP catalytic domain-interaction was found to be 565.113 Å2 for MMP-2 and 647.901 Å2 for MMP-9. No hydrogen bonds were present.
Figure 2.
Interaction of 3,5-bis(1,1-dimethylethyl) phenol docked to the “catalytic site” of the matrix metalloproteinase-2. The ligand, 3,5-bis (1,1-dimethylethyl) phenol, is shown in “stick” representation
Figure 8.
Interaction of 3,5-bis(1,1-dimethylethyl) phenol docked to the “catalytic site” of the matrix metalloproteinase-9. The ligand, 3,5-bis (1,1-dimethylethyl) phenol, is shown in “stick” representation
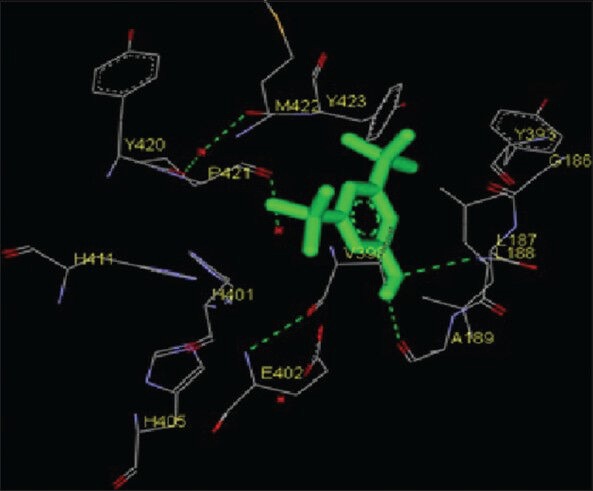
Chromenes are structurally simple compounds belonging to a large class of molecules known as benzopyranes and chroman-4-one moiety is an integral part of many natural products. These compounds and its derivatives possess diverse biological activities which include antitumor, leishmanicidal and bacteriostatic activities as well. In the present study, two chromenes, pentamethyl tetrahydro-5H-chromene and 6-vinyl-7-methoxy-2,2-dimethylchromene were considered. The catalytic domain of both MMPs were found to interact with pentamethyl tetrahydro-5H-chromene through the amino acid residues L83, Y112, A84, V117, L116, H120, L137, A136, H130, A139, T143, Y142, P140, I141 for MMP-2 and H405, H401, H411, Y420, P421, M422, Y423, E402, V398, H190, A189, L188, L187, G186 for MMP-9 [Figures 3 and 9; Table 1]. The free energy of binding and estimated inhibition constant (Ki) for the “pentamethyl tetrahydro-5H-chromene-MMP catalytic domain-interaction” were determined to be −7.02 kcal/mol and 7.15 μM for MMP-2 while −6.65 kcal/mol and 13.46 μM for MMP-9. Six carbon atoms, namely C4, C5, C8, C11, C12, C14, were found to be involved in hydrophobic interactions with amino residues LEU83, ALA84, VAL117 and HIS120 of MMP-2. Accordingly, seven carbon atoms, namely C4, C5, C6, C7, C9, C11, C13, were found to be involved in hydrophobic interactions with amino acid residues LEU187, LEU188, ALA189, VAL398, HIS401 and HIS411 of MMP-9. One of the oxygen atom O1 of pentamethyl tetrahydro-5H-chromene was observed to make polar bonds with amino acid residue HIS120 of MMP-2. Total intermolecular energy of docking for “pentamethyl tetrahydro-5H-chromene-MMP catalytic domain-interaction” was found to be −7.02 kcal/mol for MMP-2 and −6.27 kcal/mol for MMP-9. “Van der Waals”, “hydrogen bond” and “desolvation” energy components together contributed −6.99 kcal/mol for MMP-2 and −6.29 kcal/mol for MMP-9 while the “Electrostatic” energy component was found to be −0.03 kcal/mol for MMP-2 and + 0.02 kcal/mol for MMP-9. Total interacting surface area for pentamethyl tetrahydro-5H-chromene-MMP catalytic domain-interaction was found to be 558.465 Å2 for MMP-2 and 611.974 Å2 for MMP-9 while hydrogen bonds, pi-pi interactions and cation-pi interactions were absent. However, 6-vinyl-7-methoxy-2,2-dimethylchromene interacted with the catalytic domain of MMPs via Y112, L83, V117, L116, H120, T145, L137, Y142, T143, I141, A139, P134, G135, A136 amino acid residues of MMP-2 and Y420, P421, M422, Y423, G186, L187, L188, A189, E402, H401 amino acid residues of MMP-9 [Figures 4 and 10; Table 1]. The free energy of binding and estimated inhibition constant (Ki) for the “6-vinyl-7-methoxy-2,2-dimethylchromene-MMP catalytic domain-interaction” were determined to be −6.59 kcal/mol and 14.70 μM for MMP-2 and −6.08 kcal/mol and 34.8 μM for MMP-9. Six carbon atoms, namely C3, C9, C11, C12, C13, C14, were found to be involved in hydrophobic interactions with amino residues LEU116, HIS120, LEU137, TYR142 of MMP-2. Two carbon atoms, namely C6 and C13, were found to be involved in hydrophobic interactions with amino acid residues HIS401 and TYR420 of MMP-9. Two carbon atoms, namely C10, C11, were found to be involved in pi-pi interaction with amino acid residue TYR142 of MMP-2. Three carbon atoms, namely C3, C4, C8, were found to be involved in pi-pi interaction with amino acid residues HIS401 and TYR423 of MMP-9. One of the oxygen atom O1 of 6-vinyl-7-methoxy-2,2-dimethylchromene was observed to make polar bonds with amino acid residue HIS401 of MMP-9. Total intermolecular energy of docking for 6-vinyl-7-methoxy-2,2-dimethylchromene-MMP catalytic domain-interaction was found to be −7.20 kcal/mol for MMP-2 and −8.39 kcal/mol for MMP-9. Van der Waals, hydrogen bond and desolvation energy components together contributed −7.21 kcal/mol for MMP-2 and −8.28 kcal/mol for MMP-9 while the “Electrostatic” energy component was found to be + 0.01 kcal/mol for MMP-2 and −0.10 kcal/mol for MMP-9. Total interacting surface area for 6-vinyl-7-methoxy-2,2-dimethylchromene-MMP catalytic domain-interaction was found to be 602.848 Å2 for MMP-2 and 631.14 Å2 for MMP-9 while hydrogen bonds and cation-pi interactions were absent.
Figure 3.
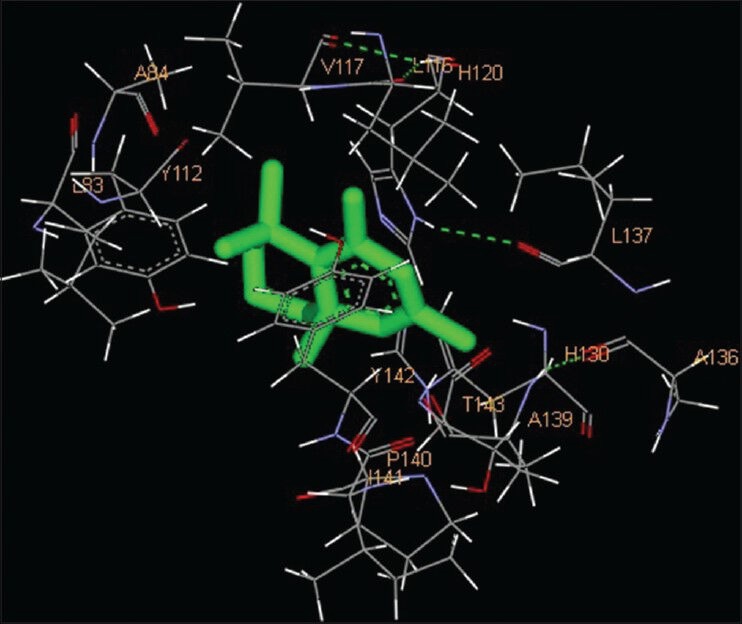
Interaction of pentamethyl tetrahydro-5H-chromene docked to the “catalytic site” of the matrix metalloproteinase-2. The ligand, pentamethyl tetrahydro-5H-chromene, is shown in “stick” representation
Figure 9.
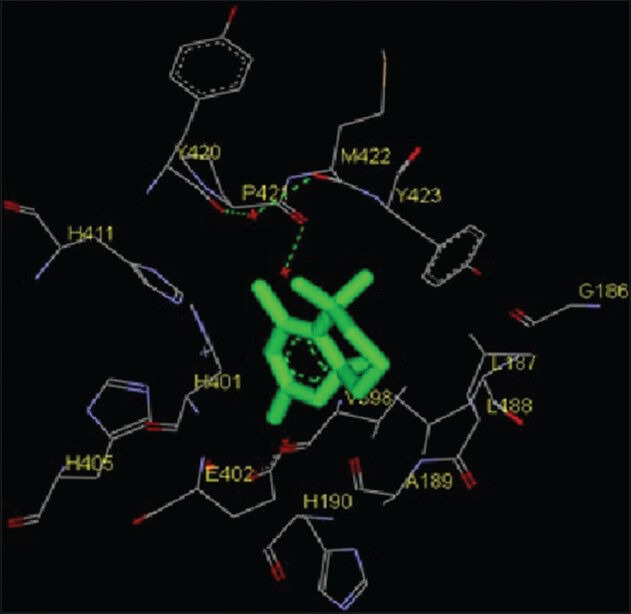
Interaction of pentamethyl tetrahydro-5H-chromene docked to the “catalytic site” of the matrix metalloproteinase-9. The ligand, pentamethyl tetrahydro-5H-chromene, is shown in “stick” representation
Figure 4.
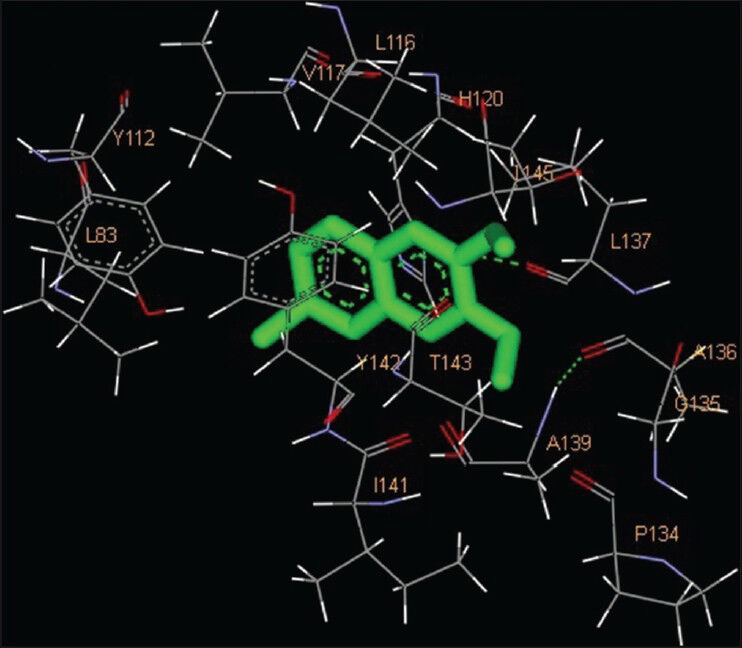
Interaction of 6-vinyl-7-methoxyl-2,2-dimethylchromene docked to the “catalytic site” of the matrix metalloproteinase-2. The ligand, 6-vinyl-7-methoxyl-2,2-dimethylchromene, is shown in “stick” representation
Figure 10.
Interaction of 6-vinyl-7-methoxyl-2,2-dimethylchromene docked to the “catalytic site” of the matrix metalloproteinase-9. The ligand, 6-vinyl-7-methoxyl-2,2-dimethylchromene, is shown in “stick” representation
The catalytic domain of both MMPs interacted with 1,2-benzenedicarboxylic acid-bis (2-ethylhexyl) ester via amino acid residues G81, L82, L83, A84, L116, H120, T145, L137, Y142, T143, I141, A139, G135, A136, H130, P140, P134, V117 of MMP-2 and E111, F110, H405, H411, H401, E402, H190, V398, P142, A189, L188, L187, Y423, M422, G186, Y393 of MMP-9 [Figures 5 and 11; Table 1]. The free energy of binding and estimated inhibition constant (Ki) for the “1,2-benzenedicarboxylic acid-bis (2-ethylhexyl) ester-MMP catalytic domain-interaction” were determined to be −5.27 kcal/mol and 136.31 μM, respectively for MMP-2 while these values were −5.73 kcal/mol and 62.59 μM, respectively for MMP-9. Total intermolecular energy of docking for 1,2-benzenedicarboxylic acid-bis (2-ethylhexyl) ester-MMP catalytic domain-interaction was found to be −9.18 kcal/mol for MMP-2 and −7.00 kcal/mol for MMP-9. Van der Waals, hydrogen bond and desolvation energy components together contributed −9.26 kcal/mol for MMP-2 and −6.96 kcal/mol for MMP-9 while the “Electrostatic” energy component was found to be + 0.08 kcal/mol for MMP-2 and −0.04 kcal/mol for MMP-9. Total interacting surface area for 1,2-benzenedicarboxylic acid-bis (2-ethylhexyl) ester-MMP catalytic domain-interaction was found to be 877.259 Å2 for MMP-2 and 701.168 Å2 for MMP-9.
Figure 5.
Interaction of 1,2-benzenedicarboxylic acid-bis(2-ethylhexyl) ester docked to the “catalytic site” of the matrix metalloproteinase-2. The ligand, 1,2-benzenedicarboxylic acid-bis(2-ethylhexyl) ester, is shown in “stick” representation
Figure 11.
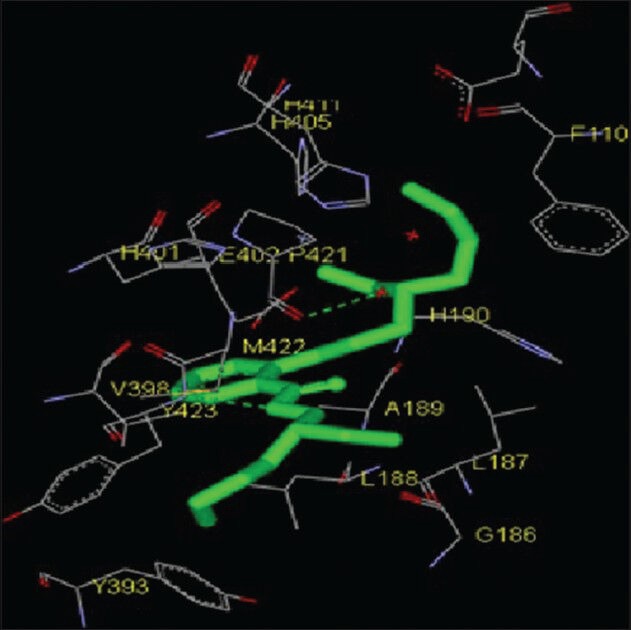
Interaction of 1,2-benzenedicarboxylic acid-bis(2-ethylhexyl) ester docked to the “catalytic site” of the matrix metalloproteinase-9. The ligand, 1,2-benzenedicarboxylic acid-bis(2-ethylhexyl) ester, is shown in “stick” representation
Squalene is a hydrocarbon and a triterpene and is a natural and vital part of the synthesis of cholesterol, steroid hormones, and vitamin D in the human body. Experimental studies have shown that squalene can effectively inhibit chemically induced skin, colon, and lung tumorigenesis in rodents.[17,18] Squalene showed weakest inhibition among all compounds. Eighteen amino acids were found to be interacting with the catalytic domain of MMP-2, namely G81, Y112, L82, L83, A84, H85, E121, V117, L116, H120, L137, A136, A139, Y142, P140, T143, I141 and H130 whereas 14 amino acid residues were found to be interacting with the catalytic domain of MMP-9. These amino acids were H411, Y420, P421, M422, Y423, G186, L187, L188, Y393, A189, V398, E402, H401 and H405 [Figures 6 and 12; Table 1]. The free energy of binding and estimated inhibition constant (Ki) for the “squalene-MMP catalytic domain-interaction” were determined to be −4.09 kcal/mol and 1.01 μM, respectively for MMP-2 while these values were −5.04 kcal/mol and 0.20 μM, respectively for MMP-9. Significant hydrophobic interactions were found in case of MMP-9. Nine carbon atoms were found to be interacting, namely C1, C5, C9, C11, C13, C15, C17, C21, C27 with amino acid residues LEU188, HIS401 and TYR423. Total intermolecular energy of docking for squalene-MMP catalytic domain-interaction was found to be −7.80 kcal/mol for MMP-2 and −9.52 kcal/mol for MMP-9. Van der Waals, hydrogen bond and desolvation energy components together contributed −7.83 kcal/mol for MMP-2 and −9.53 kcal/mol for MMP-9 while the “Electrostatic” energy component was found to be + 0.03 kcal/mol for MMP-2 and + 0.00 kcal/mol for MMP-9. Total interacting surface area for squalene-MMP catalytic domain-interaction was found to be 832.981 Å2 for MMP-2 and 997.269 Å2 for MMP-9.
Hence, the order of compounds on the basis of their inhibition potential is (1,4-cyclohexylphenyl) ethanone > 3,5-bis (1,1-dimethylethyl) phenol > pentamethyl tetrahydro-5H-chromene > 6-vinyl-7-methoxyl-2,2-dimethylchromene > 1,2-benzenedicarboxylic acid-bis (2-ethylhexyl) ester > squalene.
Among all compounds docked, (1,4-cyclohexylphenyl) ethanone exhibited strongest binding to the catalytic domain of MMP-2 and MMP-9 as it displayed a lower Ki and a higher negative ∆ G value compared to other compounds. Furthermore, many derivatives of (1,4-cyclohexylphenyl) ethanone have been prepared and were found effective against various cancerous cell lines.[19,20,21,22] It is known that Tyr 145 in the prime side of the substrate-binding cleft of MMP-2 is responsible for the selective inhibition while the catalytic center of MMP-9 is composed of the active-site zinc ion, coordinated by three histidine residues (H401, H405 and H411) and the essential glutamic acid residue (E402). Our interacting amino acid residues, docking poses and the active pocket are coherent with those observed in the studies quoted above. This affirms the accuracy of our docking experiments. Hence, this study is expected to aid future design of more specific pharmacological compounds. It has been observed that in silico results often correlate well with the results obtained in wet lab experiments. However, we find it pertinent to state that further in vitro and in vivo studies are required to validate the findings mentioned herein.
CONCLUSION
This study explores molecular interactions between human MMPs (MMP-2 and MMP-9) and six natural compounds. Moreover, we have provided a comparative account of the interactions of different natural compounds. Hydrophobic interactions play an important role in the correct positioning of these natural compounds within the catalytic site of MMP enzymes to permit docking with the enzymes. Accordingly, docking of (1,4-cyclohexylphenyl) ethanone to MMP-2 and MMP-9 is largely dominated by hydrophobic interactions. Such information may aid in the design of versatile MMPIs. Further in vitro and in vivo studies are warranted to validate the anti-cancer potential of (1,4-cyclohexylphenyl) ethanone. This study predicts that (1,4-cyclohexylphenyl) ethanone is a more efficient inhibitor of human MMP-2 and MMP-9 enzymes compared to the other natural compounds used in this study with reference to Ki and ∆ G values.
ACKNOWLEDGMENT
The authors extend sincere thanks to all of the staff of Integral University, Lucknow, India for co-operation.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Verma RP, Hansch C. Matrix metalloproteinases (MMPs): Chemical-biological functions and (Q) SARs. Bioorg Med Chem. 2007;15:2223–68. doi: 10.1016/j.bmc.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 2.Van den Steen PE, Dubois B, Nelissen I, Rudd PM, Dwek RA, Opdenakker G. Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9) Crit Rev Biochem Mol Biol. 2002;37:375–536. doi: 10.1080/10409230290771546. [DOI] [PubMed] [Google Scholar]
- 3.Grimm T, Schäfer A, Högger P. Antioxidant activity and inhibition of matrix metalloproteinases by metabolites of maritime pine bark extract (pycnogenol) Free Radic Biol Med. 2004;36:811–22. doi: 10.1016/j.freeradbiomed.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 4.Ende C, Gebhardt R. Inhibition of matrix metalloproteinase-2 and-9 activities by selected flavonoids. Planta Med. 2004;70:1006–8. doi: 10.1055/s-2004-832630. [DOI] [PubMed] [Google Scholar]
- 5.Lee SK, Chun HK, Yang JY, Han DC, Son KH, Kwon BM. Inhibitory effect of obovatal on the migration and invasion of HT1080 cells via the inhibition of MMP-2. Bioorg Med Chem. 2007;15:4085–90. doi: 10.1016/j.bmc.2007.03.081. [DOI] [PubMed] [Google Scholar]
- 6.Seo UK, Lee YJ, Kim JK, Cha BY, Kim DW, Nam KS, et al. Large-scale and effective screening of Korean medicinal plants for inhibitory activity on matrix metalloproteinase-9. J Ethnopharmacol. 2005;97:101–6. doi: 10.1016/j.jep.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 7.Tanimura S, Kadomoto R, Tanaka T, Zhang YJ, Kouno I, Kohno M. Suppression of tumor cell invasiveness by hydrolyzable tannins (plant polyphenols) via the inhibition of matrix metalloproteinase-2/-9 activity. Biochem Biophys Res Commun. 2005;330:1306–13. doi: 10.1016/j.bbrc.2005.03.116. [DOI] [PubMed] [Google Scholar]
- 8.Weeds of Australia – Biosecurity Queensland. [Last accesed on 2013 Dec 03]. Available from: http://www.keyserver.lucidcentral.org/weeds/data/03030800-0b07-490a-8d04-0605030c0f01/media/Html/Ageratum_houstonianum.htm .
- 9.Pullaiah T, Moulali DA. Vol. 2. Jodhpur: Scientific Publishers; 1997. Flora of Andhra Pradesh. [Google Scholar]
- 10.Srinivas RK, Sanjeeva KA, Ganapaty S. Evaluation of Ageratum houstonianum whole plant for its anti-diabetic activity. J Adv Scit Res. 2012;3:67–70. [Google Scholar]
- 11.Sharma PD, Sharma OP. Natural products chemistry and biological properties of the Ageratum plant. Toxicol Environ Chem. 1995;50:1–4. [Google Scholar]
- 12.Menut C, Lamaty G, Zollo PH, Kuiate JR, Bessière JM. Aromatic plants of tropical central Africa. Part X chemical composition of the essential oils of Ageratum houstonianum Mill and Ageratum conyzoides L. from Cameroon. Flavour Fragr J. 1993;8:1–4. [Google Scholar]
- 13.Bowers WS, Areguillin M. Discovery and identification of an antijuvenile hormone from Chrysanthemum cornarium. Mem Inst Oswaldo Cruz. 1987;82:51–4. [Google Scholar]
- 14.Felx M, Guyot MC, Isler M, Turcotte RE, Doyon J, Khatib AM, et al. Endothelin-1 (ET-1) promotes MMP-2 and MMP-9 induction involving the transcription factor NF-kappaB in human osteosarcoma. Clin Sci (Lond) 2006;110:645–54. doi: 10.1042/CS20050286. [DOI] [PubMed] [Google Scholar]
- 15.Himelstein BP, Asada N, Carlton MR, Collins MH. Matrix metalloproteinase-9 (MMP-9) expression in childhood osseous osteosarcoma. Med Pediatr Oncol. 1998;31:471–4. doi: 10.1002/(sici)1096-911x(199812)31:6<471::aid-mpo2>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 16.Zeeshan M, Rizvi SM, Khan MS, Kumar A. Isolation, partial purification and evaluation of bioactive compounds from leaves of Ageratum houstonianum. EXCLI J. 2012;11:78–88. [PMC free article] [PubMed] [Google Scholar]
- 17.Smith TJ. Squalene: Potential chemopreventive agent. Expert Opin Investig Drugs. 2000;9:1841–8. doi: 10.1517/13543784.9.8.1841. [DOI] [PubMed] [Google Scholar]
- 18.Auffray B. Protection against singlet oxygen, the main actor of sebum squalene peroxidation during sun exposure, using Commiphora myrrha essential oil. Int J Cosmet Sci. 2007;29:23–9. doi: 10.1111/j.1467-2494.2007.00360.x. [DOI] [PubMed] [Google Scholar]
- 19.Hitesh P, Saavani S. Synthesis and anti-cancer activity of new thiosemicarbazones of 1-(5-chloro-1H-benzimidazol-2-yl) ethanone. Pelagia Res Libr Der Pharm Sin. 2012;3:199–210. [Google Scholar]
- 20.İlhan I, Yusuf O, Zerrin I. Synthesis and anticancer activity of some bisquinoxaline derivatives. Turk J Pharm Sci. 2011;8:179–88. [Google Scholar]
- 21.Al-Said MS, Bashandy MS, Al-Qasoumi SI, Ghorab MM. Anti-breast cancer activity of some novel 1,2-dihydropyridine, thiophene and thiazole derivatives. Eur J Med Chem. 2011;46:137–41. doi: 10.1016/j.ejmech.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 22.Magdy A, Atef M. Synthesis and cellular cytotoxicities of new N-substituted indole-3-carbaldehyde and their indolylchalcones. J Chem Sci. 2009;121:455–62. [Google Scholar]


