Summary
The microRNA let-7 is a critical regulator of developmental timing events at the larval-to-adult transition in C. elegans. Recently, microRNAs with sequence similarity to let-7 have been identified. We find that doubly mutant animals lacking the let-7 family micro-RNA genes mir-48 and mir-84 exhibit retarded molting behavior and retarded adult gene expression in the hypodermis. Triply mutant animals lacking mir-48, mir-84, and mir-241 exhibit repetition of L2-stage events in addition to retarded adult-stage events. mir-48, mir-84, and mir-241 function together to control the L2-to-L3 transition, likely by base pairing to complementary sites in the hbl-1 3′ UTR and downregulating hbl-1 activity. Genetic analysis indicates that mir-48, mir-84, and mir-241 specify the timing of the L2-to-L3 transition in parallel to the heterochronic genes lin-28 and lin-46. These results indicate that let-7 family microRNAs function in combination to affect both early and late developmental timing decisions.
Introduction
In C. elegans, heterochronic genes control the appropriate temporal execution of stage-specific programs of cell division, cell behavior, and differentiation through the four larval stages, L1–L4 (reviewed in Rougvie, 2001). Mutations in heterochronic genes cause cells in specific lineages to adopt fates normally associated with either earlier or later times in development. Retarded heterochronic mutants reiterate cell fate decisions, and conversely, precocious heterochronic mutants skip cell fate decisions. MicroRNA genes play key roles in this temporal control of larval development (Lee et al., 1993; Reinhart et al., 2000; Rougvie, 2001; Wightman et al., 1993). Progression through the first three larval stages, L1–L3, is controlled by the microRNA lin-4, which downregulates the activities of lin-14 and lin-28 (Lee et al., 1993; Moss et al., 1997; Wightman et al., 1993). Later in larval development, the let-7 micro-RNA controls the L4-to-adult transition (Reinhart et al., 2000) through the downregulation of hbl-1 (Abrahante et al., 2003; Lin et al., 2003), lin-41 (Slack et al., 2000; Vella et al., 2004), and daf-12 (Grosshans et al., 2005). The repression of hbl-1 and lin-41 allows for the activation of the adult-specific transcription factor lin-29. lin-29 mutants, which repeat the L4-stage program indefinitely, display a stronger heterochronic phenotype than let-7 mutants, which repeat the L4-stage program only once. This suggests that other genes in addition to let-7 act upstream of lin-29 in the heterochronic gene pathway to specify the appropriate execution of the adult-stage program. Overexpression of mir-84 results in heterochronic defects (Johnson et al., 2005), further suggesting a role for additional microRNAs in the developmental timing pathway.
Recent cloning and computational efforts have expanded the number of known and predicted micro-RNAs (Bartel, 2004; Berezikov et al., 2005) to include approximately 100 genes in C. elegans (Ambros et al., 2003; Grad et al., 2003; Lau et al., 2001; Lee and Ambros, 2001; Lim et al., 2003; Ohler et al., 2004). As exemplified by let-7, microRNA genes can be extensively conserved across diverse animal taxa, including worms, flies, and humans (Pasquinelli et al., 2000). Many additional microRNAs can be grouped into “families” based primarily on sequence similarity at the 5′ portion of the microRNAs (Ambros et al., 2003; Grad et al., 2003; Lewis et al., 2003; Lim et al., 2003). Three C. elegans genes, mir-48, mir-84, and mir-241, that share complete sequence identity with let-7 for eight consecutive nucleotides at their 5′ ends were identified (Lau et al., 2001; Lim et al., 2003).
To examine the functions of the let-7 family members mir-48, mir-84, and mir-241, we isolated mutants by screening a library of mutagenized worms for deletion mutations. We show that mir-48, mir-84, and mir-241 function together to control developmental timing at the L2-to-L3 transition. Although hbl-1 activity was previously shown to function primarily downstream of let-7 in the L4-to-adult transition, we provide evidence that hbl-1 also functions in the L2-to-L3 transition and that hbl-1, but not lin-28, is a likely downstream target of mir-48, mir-84, and mir-241 activity.
Results
Expression of mir-48, mir-84, and mir-241 and Isolation of Deletion Mutations
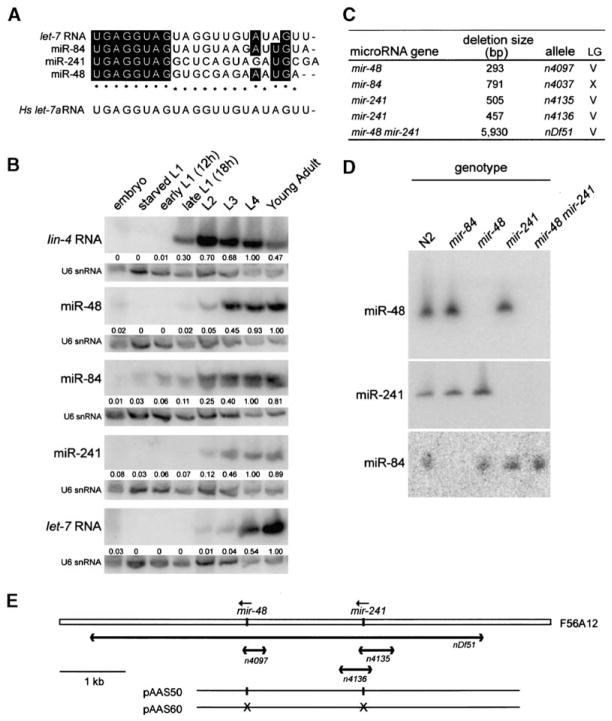
The “let-7 family” is comprised of let-7 and three other C. elegans microRNA genes, mir-48, mir-84, and mir-241 (Lau et al., 2001; Lim et al., 2003), based on the complete conservation of eight nucleotides at their 5′ ends (Figure 1A). To compare temporal expression profiles of the let-7 family microRNAs, Northern blot analysis was performed on RNA isolated from populations of staged worms (Figure 1B). Although all of the let-7 family members reached half-maximal expression after lin-4 RNA, their temporal expression profiles during larval development differed from each other: when normalized to U6 expression, miR-241, miR-48, and miR-84 reached half-maximal expression at about the L3 stage, while let-7 RNA reached half-maximal expression at about the L4 stage (Figure 1B). Previous studies did not detect the four let-7 family members until the L3 stage (Lau et al., 2001; Lim et al., 2003; Reinhart et al., 2000), but we were able to detect earlier expression of all four microRNAs by enhancing the sensitivity of detection (see Experimental Procedures).
Figure 1. Isolation of Deletion Alleles of the let-7 Family MicroRNA Genes mir-48, mir-84, and mir-241.
(A) Alignment of mature ~22 nt microRNA sequences of the four C. elegans let-7 family members and the human let-7 RNA (hs let-7). Shaded boxes and asterisks indicate bases conserved in two or three worm family members, respectively.
(B) Temporal expression patterns of let-7 family microRNA genes, mir-48, mir-84, mir-241, and let-7. Northern blot analysis of RNA isolated from populations of staged worms. As a control for staging of the worms during early larval development, lin-4 expression is observed during the late L1 stage. Northern blots indicate that miR-241, miR-48, and miR-84 reached half-maximal expression at about the L3 stage, while let-7 RNA reached half-maximal expression at about the L4 stage. Each blot was stripped and probed for U6 snRNA expression to standardize loading of RNA samples. The numbers underneath each band indicate the signal of the corresponding microRNA normalized to that of U6 snRNA presented relative to maximal expression for each microRNA.
(C) List of deletion mutations isolated in the mir-48, mir-84, and mir-241 genes.
(D) Northern blot analysis of RNA isolated from wild-type or mutant animals and hybridized with probes to the mature ~22 nt microRNA sequences for miR-48, miR-241, and miR-84. The deletion mutations result in the loss of the corresponding ~22 nt mature microRNA for miR-48, miR-241, and miR-84.
(E) The mir-48 and mir-241 locus on chromosome V. The mature microRNA sequences are located within a 1.8 kb genomic region on cosmid F56A12. Deletions at this locus are shown below. The deficiency nDf51 removes both the mir-48 and mir-241 mature microRNA sequences. For rescue experiments, we injected the plasmid pAAS50, which contains a 5073 bp fragment of genomic DNA encompassing both the mir-48 and mir-241 mature microRNA sequences. For control experiments, we injected the plasmid pAAS60, which was generated from pAAS50 but lacked the sequences corresponding to the mir-48 and mir-241 mature microRNA sequences.
To examine the functions of mir-48, mir-84, and mir-241, we isolated worms with deletion mutations in the corresponding genomic loci. These deletions remove the genomic sequence corresponding to the ~22 nt mature microRNA along with flanking regions (Figure 1C). Absence of the ~22 nt mature microRNA in mutant worms was confirmed by Northern blot (Figure 1D). Deletions in single let-7 family member genes did not appreciably affect the expression of the remaining family members. For example, a deletion upstream of mir-48 that removes mir-241 affected the expression of neither mir-48 nor mir-84, as determined by Northern blot analysis of RNA isolated from a population of mixed-stage worms (Figure 1D). Because mir-48 and mir-241 are located within a 2 kb region in the C. elegans genome, we also isolated worms with a deletion, nDf51, that removes both of these microRNA genes (Figure 1E).
mir-48 Single Mutants and mir-48; mir-84 Double Mutants Undergo an Extra Adult Molt
Singly mutant worms with deletions in mir-48, mir-84, or mir-241 displayed an essentially normal phenotype when cultured under standard conditions at 20°C (Table 1). However, at 15°C, adult-stage mir-48 mutants exhibited a weak retarded defect: 69% of young adults inappropriately entered lethargus and executed a partially complete supernumerary molt and sometimes became trapped in the unshed cuticle (Table 1). This phenotype was also observed at a low penetrance (4%, Table 1) when mir-48 animals were cultured at 20°C. mir-84 and mir-241 single mutants had no observable abnormal phenotype at 15°C (data not shown). Thus, like let-7 mutants, mir-48 animals execute a supernumerary molt. However, the characteristics of the extra molt differ: let-7 animals fail to execute the L4-to-adult transition and display an extra larval molt, whereas mir-48 mutants execute the L4-to-adult transition apparently normally and then undergo an extra adult molt.
Table 1.
Phenotypic Consequences of and Genetic Interactions among mir-48, mir-84, and mir-241 Mutations
| Strain | Genotype | Average Number of Seam Cells
|
Percentage of Worms with Alae Formation
|
Percent Lethality at L4 Molt | Percentage of Adult-Stage Lethargus | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Early L4a | L4 Molta
|
||||||||||
| L2 | L3 | L4a | L4 Molta | No Alae | Gapped | Complete | |||||
| N2 | wild-type | 11 (20) | 11 (20) | 11 (20) | 11 (21) | 0 | 0 | 0 | 100 | 0 (104) | 0 (104) |
| MT13650 | mir-48(n4097) | —b | — | — | 11 (22) | 0 | 0 | 5 | 95 | 0 (39) | 4 (135) |
| MT13650 | mir-48(n4097) @ 15°C | — | — | — | 11 (20) | 0 | 0 | 5 | 95 | 0 (66) | 69 (185) |
| MT13651 | mir-84(n4037) | — | — | — | 12 (20) | 0 | 0 | 0 | 100 | 0 (71) | 0 (71) |
| MT13896 | mir-241(n4135) | — | — | — | 11 (13) | 0 | 0 | 0 | 100 | 0 (81) | 0 (81) |
| MT13897 | mir-241(n4136) | — | — | — | 11 (22) | 0 | 0 | 0 | 100 | 0 (35) | 0 (35) |
| MT14118 | mir-241(n4136); mir-84(n4037) | — | — | — | 11 (20) | 0 | 0 | 5 | 95 | 0 (110) | 0 (110) |
| MT13652 | mir-48(n4097); mir-84(n4037) | — | — | — | 11 (20) | 0 | 0 | 0 | 100 | 0 (72) | 79 (72) |
| MT13669 | mir-48 mir-241(nDf51) | 11 (24) | 16 (24)c | 16 (29)c | 16 (27)c | 0 | 8 | 92 | 0 | 37 (81) | 36 (81) |
| VT1066 | mir-48 mir-241(nDf51); mir-84(n4037) | 11 (23) | 19 (31)c | 20 (20)c | 20 (20)c | 0 | 47 | 47 | 6 | 79 (80) | 16 (80) |
| MT14749 | mir-48 mir-241(nDf51); nEx1185d | — | — | — | 12 (32)e,f | — | 0 | 13 | 87 | — | — |
| MT14778 | mir-48 mir-241(nDf51); nEx1192g | — | — | — | 17 (18)e,h | — | 0 | 80 | 20 | — | — |
| VT517 | lin-28(n719) | — | 6 (21)c | 6 (18)c | — | 100 | — | — | — | 0 (108) | — |
| VT1103 | lin-28(n719); mir-48 mir-241(nDf51); mir-84(n4037) | — | 7 (25)c | 6 (19)c | — | 100 | — | — | — | 1 (100) | — |
| VT786 | lin-46(ma164) | — | — | — | 12 (26) | — | 0 | 12 | 88 | 1 (110) | — |
| VT1145 | lin-46(ma164); mir-48 mir-241(nDf51); mir-84(n4037) | — | 21 (17)c | 39 (17)c | 44 (20)c | 0 | 100 | 0 | 0 | 0 (106) | — |
| VT937 | lin-28(n719); lin-46(ma164) | — | — | — | 11 (22) | — | 0 | 0 | 100 | 0 (89) | — |
| VT1102 | lin-28(n719); lin-46(ma164) mir-48 mir-241(nDf51); mir-84(n4037) | 11 (21) | 22 (20)c | 38 (21)c | 39 (20)c | 0 | 100 | 0 | 0 | 22 (27) | — |
| RG559 | hbl-1(ve18) | — | 11 (21) | 13 (24) | 14 (18) | 25 | 0 | 9 | 91 | 9 (34) | — |
| VT1146 | hbl-1(ve18) mir-84(n4037); mir-48 mir-241(nDf51) | — | 11 (17) | 13 (22) | 13 (23) | 0 | 0 | 33 | 67 | 9 (57) | — |
| CT8 | lin-41(ma104) | — | — | 11 (27) | 11 (25) | 19 | 4 | 0 | 96 | 7 (90) | — |
| VT1143 | lin-41(ma104); mir-48 mir-241(nDf51); mir-84(n4037) | 11 (20) | 17 (16)c | 17 (20)c | 18 (20)c | 0 | 0 | 85 | 15 | 3 (97) | — |
Worms scored at the L4 and L4 molt stages for both seam cells and alae formation. Number of worms (n) scored for each group is indicated in parentheses.
—, did not score.
p < 0.01 (comparison with N2).
Extrachromosomal array from injecting 100 ng/μl pAAS50 and 20 ng/μl pTG96 (sur-5::GFP). pAAS50 is a pCR-II TOPO plasmid containing a 5073 bp fragment that contains mir-48 and mir-241.
Worms scored for number of seam cells at the L4 or L4 molt stages. Three lines scored independently for rescue experiments. Data from representative lines are shown.
p < 0.001 (comparison with nDf51).
Extrachromosomal array from injecting 100 ng/μl pAAS60 and 20 ng/μl pTG96 (sur-5::GFP). pAAS60 is the same as pAAS50, except that the sequences corresponding to the mature microRNAs mir-48 and mir-241 have been deleted.
p > 0.05 (comparison with nDf51).
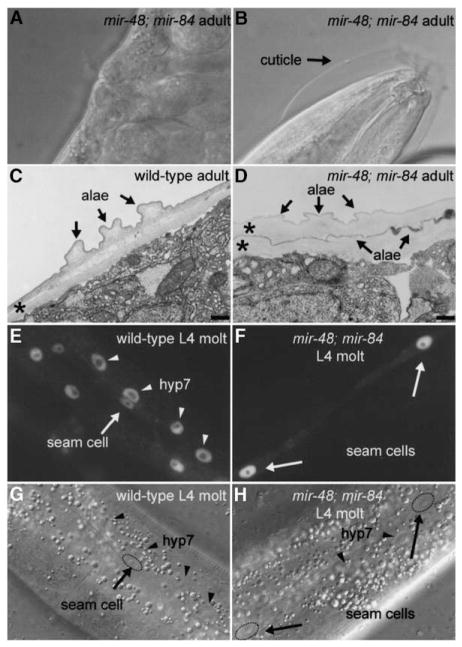
To test whether mir-48, mir-84, and mir-241 function redundantly, double and triple mutants were generated. mir-84; mir-241 animals displayed no detectable abnormalities (Table 1). mir-48; mir-84 adult-stage animals exhibited a strongly penetrant extra molting phenotype at 20°C (Figures 2A and 2B; Table 1), similar to that of mir-48 mutant animals at 15°C. By electron microscopy, two cuticles, both with adult lateral alae, were apparent on mir-48; mir-84 adults (Figure 2D), in contrast to the single cuticle on wild-type adults (Figure 2C). At the L4 molt, mir-48; mir-84 animals displayed no abnormal phenotype in the lateral hypodermal seam cells; worms had the normal number of seam cells generated from the V lineage, complete alae formation, and normal expression of the adult stage-specific transgene col-19::gfp (Abrahante et al., 1998; Liu et al., 1995). In contrast, in the main body hypodermal syncytial cell, hyp7, col-19::gfp expression was reduced or absent in mir-48; mir-84 worms at the L4 molt (Figure 2F). Thus, in mir-48; mir-84 double mutants at the L4 molt, hyp7 displayed larval characteristics and likely failed to make the larval-to-adult transition appropriately. The supernumerary molting phenotype of mir-48; mir-84 animals may be a consequence of the retarded state of hyp7. In contrast, the hypodermal seam cells in mir-48; mir-84 double mutants at the L4 molt primarily displayed adult characteristics: seam cells stopped dividing, formed alae, and expressed col-19::gfp. This is a weaker heterochronic phenotype than that of let-7(lf) worms. Whereas mir-48; mir-84 worms exhibited a larval-to-adult delay only in hyp7, let-7(lf) worms exhibit a larval-to-adult delay in both hyp7 and in the seam cells.
Figure 2. mir-48; mir-84 Double Mutants Display Supernumerary Molting Behavior in Adult Worms.
(A and B) Nomarski DIC images of a mir-48; mir-84 animal with a (A) fully formed vulva and with embryos visible inside of the worm to show that the worm is in the adult stage and (B) unshed cuticle surrounding the anterior region of the worm.
(C and D) Electron micrographs of adult-stage cuticle in a (C) wild-type animal and a (D) mir-48; mir-84 adult. Asterisks indicate cuticles. The scale bar is 300 nm. (C) Normal single cuticle with alae in a wild-type adult. (D) Two cuticles, both with alae structures (arrows), are visible in a mir-48; mir-84 adult.
(E and F) Fluorescence micrographs of a (E) wild-type and a (F) mir-48; mir-84 worm at the L4 molt stage that carry the col-19::gfp transgene maIs105. (E) Fluorescence micrograph of a wild-type L4 molt-stage worm. Expression of col-19::gfp is observed in nuclei of the hyp7 syncytium (arrowheads) and hypodermal seam cells (arrows) in a wild-type L4 molt-stage-worm. (F) Expression of col-19::gfp is observed in hypodermal seam cells (arrows) in a mir-48; mir-84 L4 molt-stage worm. col-19::gfp expression is reduced or absent in hyp7.
(G and H) Nomarski DIC images of the hypodermis of the animals in (E) and (F), respectively, showing seam cell (arrows) and hyp7 (arrowheads) nuclei.
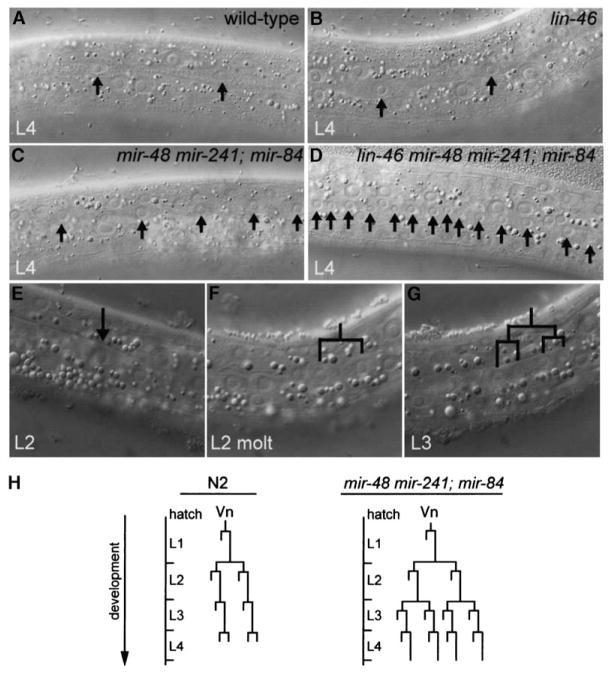
Seam Cells of mir-48 mir-241 Double Mutants and mir-48 mir-241; mir-84 Triple Mutants Repeat the L2-Stage Developmental Program
mir-48 mir-241 double mutants and mir-48 mir-241; mir-84 triple mutants displayed a retarded cell lineage phenotype in the lateral hypodermis. Extra seam cells were observed at the L3 and L4 stages (Table 1, Figure 3C) compared to wild-type worms (Table 1, Figure 3A). In the L2-stage of wild-type worms, five of the six V lineage seam cells on each side of the animal execute a “proliferative” program with two rounds of cell division, which results in an increase in the number of seam cells from six in the L1 stage to 11 in the L2 stage. In the L3 and L4 stages, a single stem cell-like division occurs, with only the posterior daughter retaining seam cell identity. Therefore, the number of V lineage seam cells on each side of wild-type animals remains constant at 11 after the L2 stage. However, in mir-48 mir-241 and in mir-48 mir-241; mir-84 L3 larvae, the average number of V lineage seam cells was 16 (range = 11–18) and 19 (range = 14–23), respectively. No further increase in the number of seam cells was detected in L4 larvae. In addition, mir-48 mir-241; mir-84 triple mutants failed to generate complete alae at the L4 molt (Table 1). The retarded seam cell and alae phenotypes were not observed in mutants transformed with a 5 kb genomic fragment containing the mir-48 and mir-241 loci (Table 1, Figure 1E), indicating that the defect was caused by mutations within the 5 kb region corresponding to the transgene. Further, when the sequences for mir-48 and mir-241 mature ~22 nt microRNAs were deleted from this genomic fragment, no rescue was observed, indicating that the phenotype was caused specifically by loss of the microRNA sequences (Table 1, Figure 1E).
Figure 3. Extra Seam Cells Generated as a Consequence of Reiteration of the L2-Stage Developmental Program in Animals Lacking mir-48, mir-84, and mir-241 MicroRNA Activity.
Nomarski DIC images of the hypodermis. (A and B) Arrows indicate seam cells. Normal seam cell number is observed in a (A) wild-type and (B) lin-46 mutant animal at the L4 stage.
(C) Extra seam cells in a mir-48 mir-241; mir-84 triple mutant at the L4 stage.
(D) Enhanced extra-seam-cell phenotype of a lin-46 mir-48 mir-241; mir-84 mutant animal at the L4 stage.
(E–G) Sequential Nomarski DIC images of a mir-48 mir-241; mir-84 mutant animal (E) during the late L2 stage, (F) during the L2 molt, and (G) during the early L3 stage. The same V seam cell lineage in a single animal is shown in all three images. (E) A single seam cell prior to cell division (arrow). (F) The seam cell in (E) divided during the L2 molt, resulting in two daughters. (G) Each of the daughters from (F) executed another round of cell divisions, resulting in four daughter nuclei in the early-L3 stage. The posterior daughter of each pair of daughter nuclei retains the seam cell identity, and the anterior daughter fuses with the hyp7 syncytium (Sulston and Horvitz, 1977).
(H) Diagram of Vn (V1–V4, V6) lineage of a normal N2 wild-type animal (adapted from Sulston and Horvitz, 1977) and a mir-48 mir-241; mir-84 triple mutant animal. Reiteration of the L2-stage proliferative program is observed in V lineage seam cells of mir-48 mir-241; mir-84 triple mutants.
We hypothesized that the extra seam cells observed in mir-48 mir-241 and mir-48 mir-241; mir-84 mutants arose from inappropriate repetition of the L2-stage seam cell proliferative program during the L3 stage. To investigate this possibility, we observed seam cell division patterns in individual mir-48 mir-241 and mir-48 mir-241; mir-84 mutant worms from the late-L2 stage through the L3 stage. In all animals examined, two rounds of cell division were observed in V lineage seam cells during the L3 stage (Figures 3E–3G). The number of seam cells observed by lineage analysis to undergo an extra round of cell division varied among individual worms (range = 3–10 of 11 V lineage seam cells, n = 6). This variability was consistent with the range of seam cell numbers counted in L3-stage worms (see above). These data demonstrate that mir-48 mir-241 doubly mutant and mir-48 mir-241; mir-84 triply mutant worms repeat the L2-stage developmental program in the lateral hypodermal seam cells during the L3 stage and indicate that mir-48, mir-84, and mir-241 act redundantly to specify the appropriate timing of stage-specific seam cell behavior.
Do mir-48, mir-84, and mir-241 Act through lin-28 to Control the L2-to-L3 Transition?
The heterochronic gene lin-28 is a key regulator of the timing of L2-stage events in the hypodermis. In lin-28(lf) mutants, the L2-stage seam cell proliferative program is skipped (Ambros and Horvitz, 1984). In contrast, seam cells of mir-48 mir-241; mir-84 animals repeat the L2-stage program. Thus, lin-28 and mir-48 mir-241; mir-84 have opposing activities. To determine the relationship between mir-48, mir-84, mir-241, and lin-28, we performed genetic epistasis analysis. We found that lin-28(lf); mir-48 mir-241; mir-84 worms displayed a precocious phenotype like that of lin-28(lf), including reduced numbers of seam cells and precocious adult alae during the L4 stage (Table 1). Thus, the lin-28 precocious phenotype in the hypodermis was epistatic to the mir-48 mir-241; mir-84 retarded phenotype. Because the lin-28 3′ UTR contains a putative let-7 binding site (Reinhart et al., 2000), mir-48, mir-84, and mir-241 could act to directly repress the activity of lin-28; alternatively, mir-48, mir-84, and mir-241 could act in an independent parallel pathway to control the L2-to-L3 transition.
If lin-28 is a target of mir-48, mir-84, and mir-241, then the level of LIN-28 protein should be elevated in mir-48 mir-241; mir-84 triple mutants. To test this, we used a lin-28::gfp::lin-28 fusion construct to monitor the LIN-28 protein level (Moss et al., 1997). In wild-type worms, lin-28::gfp::lin-28 is expressed most abundantly in the L1 stage, is downregulated beginning at the L2 stage, and is essentially absent by the L3 and L4 stages (Moss et al., 1997). We observed no abnormalities in the downregulation of lin-28::gfp::lin-28 expression in mir-48 mir-241; mir-84 triple mutants (Figure S1; see the Supplemental Data available with this article online). In contrast, lin-28::gfp::lin-28 expression remained elevated in lin-4(lf) worms (Figure S1, Moss et al., 1997). Western blot analysis also indicated little, if any, increase in LIN-28 protein levels in the absence of mir-48, mir-84, and mir-241 (data not shown). The apparently normal downregulation of LIN-28 in the absence of mir-48, mir-84, and mir-241 suggests that lin-28 is not a direct target of these microRNAs.
If lin-28 is not a target of the mir-48, mir-84, and mir-241 microRNAs, but rather acts in a parallel pathway, then what accounts for the epistasis of lin-28 over mir-48, mir-84, and mir-241? This epistasis could be explained by the function of lin-46, a downstream effector of lin-28. lin-46 acts downstream of lin-28 to specify L3-specific cell fates, and mutations in this gene completely suppress the precocious phenotype of lin-28(lf) mutants (Pepper et al., 2004). If mir-48, mir-84, and mir-241 function in a pathway parallel to both lin-28 and lin-46 to control the L2-to-L3 transition, then retarded development like that in mir-48 mir-241; mir-84 triple mutants is predicted for lin-28(lf); lin-46(lf) mir-48 mir-241; mir-84 worms. Indeed, despite the complete absence of lin-28 activity, lin-28(lf); lin-46(lf) mir-48 mir-241; mir-84 quintuply mutant worms displayed a strong retarded phenotype with extra seam cells and retarded adult alae formation (Table 1). This observation provides additional evidence that the L2 reiteration phenotype is not caused primarily by elevated levels of lin-28 activity. Therefore, we propose that mir-48, mir-84, and mir-241 function in a pathway parallel to lin-28 and lin-46, and that these pathways converge on a shared downstream effector to control the L2-to-L3 transition.
Interestingly, the lin-28(lf); lin-46(lf) mir-48 mir-241; mir-84 quintuply mutant animals displayed a stronger retarded phenotype than that of mir-48 mir-241; mir-84 triple mutants. Whereas mir-48 mir-241; mir-84 worms reiterated the L2-stage program only once in the L3 stage, lin-28(lf); lin-46(lf) mir-48 mir-241; mir-84 worms had a greater number of seam cells in L4-stage worms, consistent with an additional execution of the L2-stage developmental program in the L4 stage (Table 1). Moreover, vulva formation was retarded in lin-28(lf); lin-46(lf) mir-48 mir-241; mir-84 worms, while vulva formation in mir-48 mir-241; mir-84 triple mutants occurred with no obvious timing defects. The retarded phenotype observed in lin-28(lf); lin-46(lf) mir-48 mir-241; mir-84 worms was independent of lin-28 activity, in that the same phenotype was observed in lin-46(lf) mir-48 mir-241; mir-84 worms (Table 1, Figure 3D). lin-46(lf) single mutants display an L2 reiteration phenotype at 15°C, but not, however, at 20°C (Pepper et al., 2004) (Figure 3C). Our data indicate that lin-46 likely acts in parallel with mir-48, mir-84, and mir-241 to specify L3-stage events.
Depletion of hbl-1 Activity, but Not lin-41 Activity, Suppresses mir-48 mir-241; mir-84 Defects at the L2-to-L3 Transition
One candidate downstream effector of mir-48, mir-84, and mir-241 activities is the C. elegans hunchback homolog hbl-1 (Fay et al., 1999). hbl-1 regulates developmental timing and also contains eight let-7 complementary sites in its 3′ UTR (Abrahante et al., 2003; Lin et al., 2003). If the L2 reiteration phenotype of mir-48 mir-241; mir-84 animals is caused by failure to repress hbl-1, then it is expected that a decrease in hbl-1 activity, either genetically with the partial loss-of-function hbl-1 allele, ve18 (Abrahante et al., 2003; Lin et al., 2003), or by RNAi, should suppress this phenotype. mir-48 mir-241; hbl-1(ve18) mir-84 mutant worms did not display extra seam cells at the L3 stage (Table 1), indicating that hbl-1(ve18) suppressed the L2 reiteration phenotype. The retarded alae phenotype caused by the loss of mir-48, mir-84, and mir-241 was also suppressed; at the L4 molt, 85% of mir-48 mir-241; hbl-1(ve18) mir-84 quadruply mutant worms exhibited complete alae formation, compared to only 6% of mir-48 mir-241; mir-84 triple mutants (Table 1). Similar results were obtained when hbl-1 activity was depleted by RNAi in mir-48 mir-241; mir-84 animals. hbl-1(RNAi) administered postembryonically resulted in strong suppression of the L2 reiteration phenotype of L3-stage mir-48 mir-241; mir-84 worms and suppression of the retarded alae phenotype at the L4 molt (Table 2). hbl-1(RNAi) also suppressed the enhanced L2 reiteration phenotype of lin-46(lf) mir-48 mir-241; mir-84 animals (Table 2), suggesting that both the lin-46 and the mir-48, mir-84, and mir-241 pathways converge on hbl-1 to control the L2-to-L3 transition.
Table 2.
hb1-1 Depletion by RNAi Suppresses the Phenotype of Extra Seam Cells and Retarded Alae of a mir-48 mir-241; mir-84 Triple Mutant and of a lin-46 mir-48 mir-241; mir-84 Mutant
| Strain | Genotype | dsRNA Construct | Number of Seam Cells (n)
|
Percentage of Worms with Precocious Alae during L4
|
Percentage of Worms with Alae at L4 Molt/Young Adult
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L3 | L4 | No Alae | Gapped | Complete | n | No Alae | Gapped | Complete | n | |||
| N2 | wild-type | control | 11 (15) | 11 (20) | 100 | 0 | 0 | 20 | 0 | 0 | 100 | 26 |
| hbl-1 | 9 (33) | 10 (33) | 0 | 24 | 76 | 33 | 0 | 14 | 86 | 7 | ||
| lin-41 | 11 (16) | 11 (40) | 60 | 40 | 0 | 40 | 0 | 29 | 71 | 24 | ||
| hbl-1 and lin-41 | 10 (27) | 11 (35) | 3 | 57 | 40 | 35 | 0 | 46 | 54 | 13 | ||
| VT1066 | mir-48 mir-241; mir-84 | control | 17 (21) | 19 (23) | 100 | 0 | 0 | 23 | 43 | 54 | 4 | 28 |
| hbl-1 | 9 (33) | 14 (48) | 89 | 11 | 0 | 36 | 0 | 32 | 68 | 25 | ||
| lin-41 | 16 (20) | 17 (32) | 93 | 7 | 0 | 29 | 0 | 47 | 53 | 30 | ||
| hbl-1 and lin-41 | 10 (20) | 11 (30) | 0 | 60 | 40 | 25 | 0 | 29 | 71 | 21 | ||
| VT786 | lin-46 | control | 12 (9) | 12 (20) | 100 | 0 | 0 | 20 | 0 | 19 | 81 | 26 |
| hbl-1 | 9 (6) | 10 (20) | 0 | 20 | 80 | 20 | 0 | 0 | 100 | 15 | ||
| lin-41 | 12 (1) | 12 (21) | 81 | 19 | 0 | 21 | 0 | 12 | 88 | 17 | ||
| hbl-1 and lin-41 | 8 (3) | 10 (20) | 11 | 37 | 53 | 19 | 0 | 8 | 92 | 24 | ||
| VT1145 | lin-46 mir-48 mir-241; mir-84 | control | 22 (17) | 40 (20) | 100 | 0 | 0 | 20 | 100 | 0 | 0 | 18 |
| hbl-1 | 10 (24) | 14 (36) | 100 | 0 | 0 | 36 | 0 | 80 | 20 | 20 | ||
| lin-41 | 22 (20) | 40 (31) | 100 | 0 | 0 | 31 | 100 | 0 | 0 | 36 | ||
| hbl-1 and lin-41 | 11 (22) | 11 (36) | 0 | 50 | 50 | 36 | 0 | 29 | 71 | 17 | ||
Because lin-41 acts redundantly with hbl-1 to regulate the L4-to-adult transition (Abrahante et al., 2003; Lin et al., 2003), we tested whether lin-41 might also act with hbl-1 to regulate the L2-to-L3 transition. Unlike reduction of hbl-1 activity, reduction of lin-41 activity had little effect on the L2 reiteration phenotype (Tables 1 and 2), but did partially suppress the retarded alae phenotype (Table 2). These data indicate that mir-48, mir-84, and mir-241 control the L2-to-L3 transition by repressing hbl-1 and not by repressing lin-41.
mir-48, mir-84, and mir-241 Are Necessary for Temporal Downregulation of hbl-1 Expression
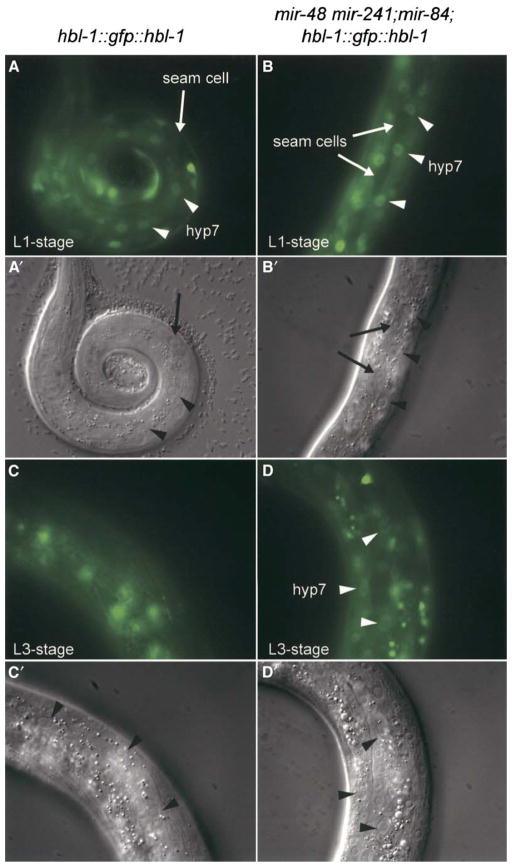
If mir-48, mir-84, and mir-241 regulate hbl-1 expression during wild-type development, then mir-48 mir-241; mir-84 triple mutants should exhibit an elevated level of HBL-1 protein. hbl-1 is expressed in the hypodermis during embryogenesis, and this expression decreases during early larval development, becoming undetectable by the L3 stage (Abrahante et al., 2003; Fay et al., 1999; Lin et al., 2003). The repression of hbl-1 in the hypodermal syncytium hyp7 during early larval development requires the hbl-1 3′ UTR (Abrahante et al., 2003; Lin et al., 2003). To test whether hbl-1 activity is upregulated in mir-48 mir-241; mir-84 triple mutants, we used an hbl-1::gfp::hbl-1 fusion construct as a reporter for the level of HBL-1 protein (Fay et al., 1999). In 80% of mir-48 mir-241; mir-84 triple mutants at the L3 stage, hbl-1::gfp::hbl-1 expression was detected in hyp7 (Figure 4D). In contrast, hbl-1::gfp::hbl-1 expression was detected in hyp7 in only 11% of wild-type worms at the L3 stage (Figure 4C). Our data are consistent with a model that mir-48, mir-84, and mir-241 act together to regulate the level of HBL-1 protein in hyp7. However, because single and double mutants that do not display the L2 reiteration phenotype were not examined for elevated hbl-1::gfp::hbl-1 activity, it remains possible that the individual family members, mir-48, mir-84, and mir-241, differ in their capacity to repress hbl-1 activity.
Figure 4. mir-48, mir-84, and mir-241 Are Necessary for the Downregulation of hbl-1::gfp::hbl-1 in the hyp7 Syncytium.
(A–D) Larval stage and genotype are indicated for each panel. All fluorescent images were taken with identical exposure times. Fluorescent micrographs of a (A and C) wild-type and a (B and D) mir-48 mir-241; mir-84 mutant animal carrying the hbl-1::gfp::hbl-1 containing transgene ctIs39 (Fay et al., 1999). (A and B) hbl-1::gfp::hbl-1 is expressed at the L1 stage in hyp7 in a (A) wild-type and (B) mir-48 mir-241; mir-84 mutant animal. Little or no expression is visible in the seam cells. Arrows indicate examples of seam cells, and arrowheads indicate examples of hyp7 nuclei. (C) hbl-1::gfp::hbl-1 expression is undetectable in hyp7 in a L3-stage wild-type animal. (D) hbl-1::gfp::hbl-1 expression is elevated in hyp7 in a L3-stage mir-48 mir-241; mir-84 animal.
(A′–D′) Corresponding Nomarski DIC images for images shown in (A)–(D). Arrows indicate examples of seam cells, and arrowheads indicate examples of hyp7 nuclei.
Discussion
Functional Cooperation among MicroRNA Family Members
Our data demonstrate that the let-7 family microRNA genes mir-48, mir-84, and mir-241 function together to control developmental timing in C. elegans. Synthetic heterochronic phenotypes were observed in double and triple mutants, indicating functional cooperation or redundancy among these let-7 family microRNAs. mir-48 single mutants displayed a supernumerary adult-stage lethargus. The penetrance of this phenotype was greatly enhanced in mir-48; mir-84 double mutants. mir-48 mir-241 double mutants also displayed extra seam cell divisions during the L3 stage, the formation of incomplete adult-stage alae, and lethality associated with vulva bursting at the L4-to-adult transition. All of these retarded heterochronic defects occurred with higher frequency in triply mutant worms that lack mir-48, mir-84, and mir-241.
The C. elegans genome encodes at least 19 micro-RNA gene families containing from 2 to 8 members with significant sequence conservation within the ~22 nt microRNA sequence (Ambros et al., 2003; Grad et al., 2003; Lim et al., 2003). Sequence conservation among family members is strongest near the 5′ end of the microRNA in the region known as the “seed,” which has been proposed to reflect a potential for family members to direct the repression of shared target genes (Bartel, 2004; Brennecke et al., 2005; Doench and Sharp, 2004; Kloosterman et al., 2004; Lai, 2002; Lewis et al., 2003, 2005; Lim et al., 2005; Mallory et al., 2004; Stark et al., 2003). Because mir-48, mir-84, and mir-241 display complete sequence conservation in the seed region at the 5′ end, it is possible that they could repress a common set of targets and hence could be functionally equivalent. Our findings suggest that let-7, mir-48, mir-84, and mir-241 may all act to repress a shared target, hbl-1 (see below). let-60 RAS also has been proposed to be a target of mir-84 based on overexpression experiments (Johnson et al., 2005). Elevated levels of let-60 RAS expression lead to a multivulva phenotype (Beitel et al., 1990; Han and Sternberg, 1990); however, we did not observe a multivulva phenotype in mir-84 single mutants nor in mir-48 mir-241; mir-84 triple mutants.
Our results leave open the possibility that mir-48, mir-84, and mir-241 are not functionally equivalent in all respects. Sequence differences in the 3′ end of the let-7 family microRNAs may direct the repression of some distinct sets of targets, the repression of which could function coordinately to regulate developmental timing. Target sites that lack strong complementarity at the microRNA 5′ end can direct repression if there is extensive compensatory pairing at the 3′ end, thus allowing for distinct activities of microRNA family members (Brennecke et al., 2005; Doench and Sharp, 2004). Indeed, let-7 complementary sites in the lin-41 mRNA have extensive complementarity to the let-7 3′ region, along with imperfect pairing to the let-7 5′ seed region (Slack et al., 2000). The specificity imparted by compensatory 3′ pairing may function to enable repression of lin-41 by let-7 and not allow for the repression of lin-41 by mir-48, mir-84, or mir-241 (Brennecke et al., 2005; Lewis et al., 2005). Similarly, extensive 3′ pairing to one of the other three let-7 family members might compensate for a lack of strong 5′ pairing and therefore could restrict the repression of specific targets to individual let-7 family members.
hbl-1 Is Likely a Direct Target of the let-7 Family of MicroRNAs
Our findings suggest that the four let-7 family micro-RNAs may all act to repress hbl-1. Reduction of hbl-1 activity can suppress the heterochronic defects observed in both mir-48 mir-241; mir-84 (this study) and let-7 (Abrahante et al., 2003; Lin et al., 2003) mutant animals, indicating that hbl-1 functions downstream of the let-7 family microRNAs. Moreover, the failure to appropriately downregulate hbl-1 can be detected in the hypodermis of mir-48 mir-241; mir-84 mutants and in neuronal cells of let-7 mutants (Abrahante et al., 2003; Lin et al., 2003). The hbl-1 3′ UTR contains eight let-7 complementary sites (Abrahante et al., 2003; Lin et al., 2003). Because these potential binding sites differ in sequence, each may be able to bind the individual let-7 family microRNAs with differing efficacies. The relative contribution of individual let-7 family microRNAs to the repression of hbl-1 activity remains to be tested.
Previous studies showed a role for hbl-1 activity in controlling the L4-to-adult transition (Abrahante et al., 2003; Lin et al., 2003). Our findings indicate that hbl-1 also controls the L2-to-L3 transition in the hypodermis. This early role for hbl-1 is consistent with the observation that reduction of hbl-1 activity by RNAi results in a decreased number of seam cells in L2-stage animals (Abrahante et al., 2003). A reduced number of seam cells likely reflects a partial omission of the L2-stage proliferative program. This precocious phenotype is relatively weak in comparison to that of lin-28(lf) mutants, in which all seam cells generated from the V lineage fail to execute the L2-stage program (Ambros, 1989; Ambros and Horvitz, 1984; Moss et al., 1997). This weak phenotype may be a consequence of residual hbl-1 activity of the partial loss-of-function allele, ve18. It is possible that complete loss of hbl-1 activity would result in a stronger precocious L2-omission phenotype similar to that seen in lin-28(lf) mutants.
mir-48, mir-84, and mir-241 Function in Parallel to the lin-28 and lin-46 Pathway
An important regulator of the L2-to-L3 transition is lin-28 (Ambros, 1989; Ambros and Horvitz, 1984; Moss et al., 1997), yet multiple lines of evidence suggest that the control of the L2-to-L3 transition by mir-48, mir-84, and mir-241 does not occur through regulation of lin-28 activity. First, a lin-28::gfp::lin-28 reporter transgene that recapitulates the wild-type temporal regulation of LIN-28 protein and that rescues the phenotype of lin-28(lf) worms (Moss et al., 1997) was not derepressed in mir-48 mir-241; mir-84 triple mutants. Second, we found that the level of endogenous LIN-28 protein was not significantly elevated in mir-48 mir-241; mir-84 triple mutants, whereas, in lin-4 retarded mutants, LIN-28 protein is abnormally abundant at later larval stages (Seggerson et al., 2002). Third, two alleles of lin-58 that contain mutations upstream of mir-48, and hence lead to the misexpression of mir-48, enhance the precocious phenotype of a lin-28 null mutant (Abrahante et al., 1998; Li et al., 2005), indicating that mir-48 does not act exclusively through lin-28. Lastly, we found that the L2 reiteration phenotype of mir-48 mir-241; mir-84 triple mutants could occur independently of lin-28 activity. These data together indicate that mir-48, mir-84, and mir-241 control the L2-to-L3 transition primarily through downstream effectors other than lin-28, even though the lin-28 3′ UTR contains a let-7 complementary site. It is possible that the let-7 family microRNAs may contribute to the repression of lin-28 expression, but to a degree undetectable by our assays.
Our genetic epistasis analysis indicates that mir-48, mir-84, and mir-241 function in parallel with the lin-28 and lin-46 pathway to downregulate hbl-1 activity and hence control the L2-to-L3 transition (Table 1; Figure 5). One model to account for this convergence of pathways on hbl-1 would be that LIN-46, in its putative role as a scaffolding protein (Pepper et al., 2004), could control assembly of a protein complex that directly interacts with HBL-1 protein to inhibit its activity in parallel with the repression of hbl-1 mRNA translation exerted by mir-48, mir-84, and mir-241. Alternatively, LIN-46 could interact with RNA binding protein(s) and directly potentiate the activity of the mir-48, mir-84, and mir-241 microRNAs.
Figure 5. Model for the Early Role of mir-48, mir-84, and mir-241 in the Heterochronic Gene Pathway at the L2-to-L3 Transition and the Late Role for let-7 in the L4-to-Adult Transition.

See text for details. Our data indicate that mir-48, mir-84, and mir-241 act in parallel with lin-28 and lin-46 and upstream of hbl-1 to control specification of L3-stage events. Both the lin-28 and lin-46 pathway and the mir-48, mir-84, mir-241 pathway converge on hbl-1 to specify the L2-to-L3 transition. let-7 functions later to control specification of adult-stage events through its downstream effectors, hbl-1 and lin-41. mir-48, mir-84, and mir-241 control the exit from the molting cycle and may contribute to the repression of hbl-1 and lin-41 at the L4-to-adult transition.
mir-48, mir-84, and mir-241 Coordinate Developmental Timing among Hypodermal Cell Types
Our data suggest that mir-48, mir-84, and mir-241 control developmental timing in two physically associated but distinct cell types in the hypodermis: the postmitotic main body hypodermal syncytial cell, hyp7, and the proliferative seam cells. Two lines of evidence point to a role in hyp7 for mir-48, mir-84, and mir-241 to repress hbl-1 and control hyp7 temporal behavior. First, mir-48; mir-84 mutant worms displayed heterochronic defects in hyp7; the expression of the adult-specific transgene col-19::gfp was retarded in hyp7 but was regulated normally in the seam cells. Thus, the supernumerary molt observed in mir-48; mir-84 double mutants may be a consequence of a heterochronic defect in hyp7. Second, our data indicate that mir-48, mir-84, and mir-241 act in hyp7 to repress hbl-1 activity. In mir-48 mir-241; mir-84 worms, hbl-1::gfp::hbl-1 was misregulated in hyp7. Thus, the 3′ UTR-dependent downregulation of hbl-1::gfp::hbl-1 in hyp7 that occurs in wild-type animals (Abrahante et al., 2003; Lin et al., 2003) can be accounted for largely by the regulation of hbl-1 by mir-48, mir-84, and mir-241.
mir-48, mir-84, and mir-241 may also function in the hypodermal seam cells to control developmental timing. Reduction of hbl-1 activity genetically or by hbl-1 RNAi affected stage-specific behavior of seam cells, resulting in suppression of the retarded seam cell and alae phenotypes of mir-48 mir-241; mir-84 worms. This could be a consequence of the repression of hbl-1 by mir-48, mir-84, and mir-241 in the seam cells. Interestingly, hbl-1::gfp::hbl-1 cannot be detected in the seam cells after the L1 stage (Abrahante et al., 2003; Fay et al., 1999; Lin et al., 2003), suggesting that, at the time of the L2-to-L3 transition, the amount of hbl-1 expression in seam cells is relatively low. Thus, mir-48, mir-84, and mir-241 may function cell autonomously in the seam cells at the L3 stage to downregulate hbl-1, albeit beginning from a level already below the threshold of detection by our assays. Alternatively, since we could easily observe repression of hbl-1::gfp::hbl-1 at the L2-to-L3 transition in hyp7, it is conceivable that the stage-specific behavior of seam cells may be controlled non-cell autonomously by a hbl-1-regulated signal from hyp7. Non-cell autonomous signaling from hyp7 to neighboring cells has been proposed in the pathway to specify the fates of vulval precursor cells (VPCs). Mosaic analyses suggest that the sites of action of the multivulva (Muv) gene locus lin-15 (Herman and Hedgecock, 1990) and of the synthetic Muv genes lin-37 (Hedgecock and Herman, 1995) and lin-35 (Myers and Greenwald, 2005) are in hyp7. One model is that hyp7 generates a signal to neighboring VPCs to inhibit vulval cell fate specification. Similarly, a signal from hyp7 to the lateral hypodermal seam cells may regulate the temporal behavior of seam cells and thereby help coordinate developmental timing throughout the hypodermis.
In summary, the results presented here demonstrate a role for the let-7 family microRNA genes mir-48, mir-84, and mir-241 in the heterochronic pathway to control the L2-to-L3 cell fate transitions in the hypodermis. Proper progression through the L1 and L2 larval stages requires downregulation of lin-14 and lin-28, primarily through the action of the microRNA lin-4. Our findings extend the involvement of microRNAs in the regulation of C. elegans developmental timing to include a requirement for the downregulation of hbl-1 by the combined action of the three let-7 family microRNAs, mir-48, mir-84, and mir-241 (Figure 5), in the hypodermis. We find that the L2-to-L3 transition is controlled by complex genetic mechanisms involving two microRNA-regulated pathways that converge on hbl-1: the lin-4, lin-28, lin-46 pathway and the mir-48, mir-84, mir-241 pathway (Figure 5). These parallel inputs to hbl-1 may serve to couple hbl-1 downregulation to distinct upstream temporal signals. Further, the functional redundancy among mir-48, mir-84, and mir-241 could reflect alternative mechanisms for triggering the L2-to-L3 transition throughout the hypodermis. mir-48, mir-84, and mir-241 seem to have more minor roles, compared to let-7, at the L4-to-adult transition in the hypodermis, indicating that different microRNA family members can be deployed for distinct roles, perhaps through differences in temporal or spatial expression patterns and/or differences in target specificity. These findings suggest that we may expect analogous forms of genetic redundancy and regulatory complexity in pathways involving other families of related microRNAs.
Experimental Procedures
Nematode Methods
C. elegans strains were grown under standard conditions as described (Wood et al., 1988). Strains used are listed in Table 1. The wild-type strain used was var. Bristol N2 (Brenner, 1974). Strains were grown at 20°C, except where otherwise indicated. For building multiply mutant strains, mutant alleles of mir-48, mir-84, and mir-241 were identified by performing PCR reactions that amplified the genomic region flanking the deletion mutations. For the sequences of primers used to identify mutant alleles of mir-48, mir-84, and mir-241, see Table S1.
Identification, Isolation, and Rescue of MicroRNA Deletion Strains
Deletion mutants of mir-48, mir-84, and mir-241 were isolated from a frozen library of EMS or UV-TMP mutagenized worms (Jansen et al., 1997; Liu et al., 1999). To enhance the detection of relatively small deletions, a “poison” primer was included in the first round of nested PCR reactions (Edgley et al., 2002). Additional detail is available in the Supplemental Data. For the sequences of primers used in identification of microRNA deletion strains, see Table S1.
Mutant alleles were backcrossed at least three times before further analysis. For rescue experiments, we injected pAAS50, a pCR-II TOPO plasmid containing a 5073 bp fragment that was amplified from genomic DNA from wild-type N2 worms. This fragment contains mir-48 and mir-241, but no other experimentally defined or predicted gene. The sequences of the primers used to amplify the genomic DNA were 5′-CCAATTCTCTGTCTCCCCTTC-3′ and 5′-CAGAATTGTGTCGTCGTGTTC-3′. As a control, we used pAAS60, a plasmid generated from pAAS50 that lacked the sequences corresponding to the mature miR-48 and miR-241. The deletions were confirmed by sequence analysis of PCR-amplified genomic DNA sequences. To generate transgenic animals, germline transformation was performed as described (Mello and Fire, 1995). Plasmid DNA for pAAS50 or pAAS60 was injected (100 ng/μl) into mir-48 mir-241(nDf51) animals with pTG96 (sur-5::gfp at 20 ng/μl) as a coinjection marker. Seam cells were counted, alae were scored at the L4 molt, and worms were subsequently scored for GFP expression.
Northern Blot Analysis
Northern blots were performed as described (Lau et al., 2001; Lee and Ambros, 2001). Total RNA was isolated from a mixed population of animals for each strain, and 10 μg total RNA was loaded in each lane. To maximize the sensitivity of microRNA detection for developmental Northern blots, oligonucleotide probes were labeled with the Starfire Oligos Kit (IDT, Coralville, IA) and 32P ATP (Amersham). Starfire oligonucleotide probes have approximately 10-fold greater specific activity as compared to traditional end-labeled oligonucleotide probes (Behlke et al., 2000). Probes used were the following 32P end-labeled (Figure 1D) or Starfire-labeled (Figure 1B) oligonucleotides: 5′-TCGCATCTACTGAGCCTAC-3′ (miR-48), 5′-TACAATATTACATACTACC-3′ (miR-84), 5′-TCATTTCTCGCACCTA CCT-3′ (miR-241), 5′-TCACACTTGAGGTCTCAGG-3′ (lin-4 RNA), 5′-AACTATACAACCTACTACC-3′ (let-7 RNA), and 5′-TGTCATCCTTGCGCAGG-3′ (U6 snRNA). For developmental Northern blots, RNA from staged animals was prepared as described in Lee and Ambros (2001). RNA samples were run on acrylamide denaturing gels and then transferred to GeneScreen Plus membranes (Perkin Elmer, Boston, MA) by electrophoresis. After transfer, blots were crosslinked with UV and baked at 80°C for 1 hr. Probes were hybridized to the membranes at 34°C in 7% SDS, 0.2 M Na2PO4 (pH 7.0) overnight. Membranes were washed at 34°C, twice with 2× SSPE, 0.1% SDS and twice with 0.5× SSPE, 0.1% SDS. The blots were exposed on Molecular Dynamics Phosphorimager screens, and signals were quantified by using ImageQuant (Molecular Dynamics).
Microscopy and Phenotype Analysis
The number of lateral hypodermal seam cells was counted at specific larval stages; staging was assessed by gonad morphology. Nomarski DIC microscopy was used to score seam cells and alae formation. Seam cells were identified according to their characteristic morphology and position along the lateral midline of the worm. Worms were anesthetized with 1 mM levamisole. Seam cells derived from the V lineage between the pharynx and the anus were scored on one side of individual animals. Cell lineage analysis was performed by picking individual L2-stage worms and monitoring seam cells derived from the V lineage as described by Sulston and Horvitz (1977), except that movies of worms were taken by using iVeZeen software (Boinx, Germering, Germany) and were analyzed with Quicktime software (Apple, Cupertino, CA). Three mir-48 mir-241 and three mir-48 mir-241; mir-84 animals were analyzed. Electron microscopy of adult animals was performed as described (Bargmann et al., 1993).
RNAi Assays
The plasmid pAA26, which expresses hbl-1 dsRNA, was constructed by amplifying a 1.5 kb region from genomic DNA, digesting with NotI, and inserting into the NotI site of the cloning vector pPD129.36 (Timmons et al., 2001) with convergent T7 promoters. Oligonucleotides used for hbl-1 amplification were AA141, 5′-ATC TATGCGGCCGCGACGGTGCTCAATCAGATAGC-3′, and AA142, 5′-ATCTATGCGGCCGCGACGGTGCTCAATCAGATAGC-3′. pAA26 was transformed into bacterial strain HT115 (Timmons et al., 2001). RNAi of lin-41 was performed by using a bacterial strain obtained from the MRC geneservice (Cambridge, UK), gene pair name C12C8.3 (Fraser et al., 2000; Kamath and Ahringer, 2003). dsRNA-expressing bacteria were grown from single colonies overnight in LB with 200 μg/ml ampicillin and 12.5 μg/ml tetracycline and were then plated on NGM plates containing 1 mM IPTG, 200 μg/ml ampicillin, and 12.5 μg/ml tetracycline. For suppression of hbl-1 and lin-41, overnight cultures of each bacterial strain were mixed 1:1 and plated. Embryos were hatched on plates with dsRNA-expressing bacteria, and larvae were scored by using Nomarski DIC microscopy at indicated stages for seam cell and alae phenotypes.
Supplementary Material
Acknowledgments
We thank Rosalind Lee and Chris Hammell for RNA preparation and Northern blot analysis, Andrew Hellman for performing PCR screening for deletion mutations, Beth Castor for DNA sequence determinations, Na An for strain management, Erika Hartwieg for performing EM analyses, and the Caenorhabditis Genetics Center for strains. The Caenorhabditis Genetics Center is supported by the National Institutes of Health (NIH) National Center for Research Resources. A.L.A. was supported by a Ruth L. Kirschstein National Research Service Award postdoctoral fellowship (5F32GM065721-02). E.A.M. was a Wellcome Trust Prize Traveling Research Fellow (#061641). D.P.B. and V.A. were supported by grants from the NIH (GM067031 to D.P.B and GM34028 to V.A.). H.R.H. is the David H. Koch Professor of Biology at Massachusetts Institute of Technology and was supported by and is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental Data including Supplemental Experimental Procedures, a figure, and a table are available at http://www.developmentalcell.com/cgi/content/full/9/3/403/DC1/.
References
- Abrahante JE, Miller EA, Rougvie AE. Identification of heterochronic mutants in Caenorhabditis elegans. Temporal mis-expression of a collagen:green fluorescent protein fusion gene. Genetics. 1998;149:1335–1351. doi: 10.1093/genetics/149.3.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- Ambros V. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell. 1989;57:49–57. doi: 10.1016/0092-8674(89)90171-2. [DOI] [PubMed] [Google Scholar]
- Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Behlke MA, Dames SA, McDonald WH, Gould KL, Devor EJ, Walder JA. Use of high specific activity StarFire oligonucleotide probes to visualize low-abundance pre-mRNA splicing intermediates in S. pombe. Biotechniques. 2000;29:892–897. doi: 10.2144/00294pf01. [DOI] [PubMed] [Google Scholar]
- Beitel GJ, Clark SG, Horvitz HR. Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature. 1990;348:503–509. doi: 10.1038/348503a0. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgley M, D’Souza A, Moulder G, McKay S, Shen B, Gilchrist E, Moerman D, Barstead R. Improved detection of small deletions in complex pools of DNA. Nucleic Acids Res. 2002;30:e52. doi: 10.1093/nar/gnf051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay DS, Stanley HM, Han M, Wood WB. A Caenorhabditis elegans homologue of hunchback is required for late stages of development but not early embryonic patterning. Dev Biol. 1999;205:240–253. doi: 10.1006/dbio.1998.9096. [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Grad Y, Aach J, Hayes GD, Reinhart BJ, Church GM, Ruvkun G, Kim J. Computational and experimental identification of C. elegans microRNAs. Mol. Cell. 2003;11:1253–1263. doi: 10.1016/s1097-2765(03)00153-9. [DOI] [PubMed] [Google Scholar]
- Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev. Cell. 2005;8:321–330. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Han M, Sternberg PW. let-60, a gene that specifies cell fates during C. elegans vulval induction, encodes a ras protein. Cell. 1990;63:921–931. doi: 10.1016/0092-8674(90)90495-z. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Herman RK. The ncl-1 gene and genetic mosaics of Caenorhabditis elegans. Genetics. 1995;141:989–1006. doi: 10.1093/genetics/141.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman RK, Hedgecock EM. Limitation of the size of the vulval primordium of Caenorhabditis elegans by lin-15 expression in surrounding hypodermis. Nature. 1990;348:169–171. doi: 10.1038/348169a0. [DOI] [PubMed] [Google Scholar]
- Jansen G, Hazendonk E, Thijssen KL, Plasterk RH. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nat Genet. 1997;17:119–121. doi: 10.1038/ng0997-119. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 2004;32:6284–6291. doi: 10.1093/nar/gkh968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li M, Jones-Rhoades MW, Lau NC, Bartel DP, Rougvie AE. Regulatory mutations of mir-48, a C. elegans let-7 family microRNA, cause developmental timing defects. Dev Cell. 2005;9:415–422. doi: 10.1016/j.devcel.2005.08.002. this issue. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The micro-RNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ. The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev. Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- Liu LX, Spoerke JM, Mulligan EL, Chen J, Reardon B, Westlund B, Sun L, Abel K, Armstrong B, Hardiman G, et al. High-throughput isolation of Caenorhabditis elegans deletion mutants. Genome Res. 1999;9:859–867. doi: 10.1101/gr.9.9.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Kirch S, Ambros V. The Caenorhabditis elegans heterochronic gene pathway controls stage-specific transcription of collagen genes. Development. 1995;121:2471–2478. doi: 10.1242/dev.121.8.2471. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J. 2004;23:3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Fire A. DNA transformation. In: Epstein HF, Shakes DC, editors. Methods in Cell Biology: Caenorhabditis elegans: Modern Biological Analysis of an Organism. San Diego, CA: Academic Press; 1995. pp. 451–482. [Google Scholar]
- Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Myers TR, Greenwald I. lin-35 Rb Acts in the major hypodermis to oppose Ras-mediated vulval induction in C. elegans. Dev. Cell. 2005;8:117–123. doi: 10.1016/j.devcel.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Ohler U, Yekta S, Lim LP, Bartel DP, Burge CB. Patterns of flanking sequence conservation and a characteristic upstream motif for microRNA gene identification. RNA. 2004;10:1309–1322. doi: 10.1261/rna.5206304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Pepper AS, McCane JE, Kemper K, Yeung DA, Lee RC, Ambros V, Moss EG. The C. elegans heterochronic gene lin-46 affects developmental timing at two larval stages and encodes a relative of the scaffolding protein gephyrin. Development. 2004;131:2049–2059. doi: 10.1242/dev.01098. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rougvie AE. Control of developmental timing in animals. Nat Rev Genet. 2001;2:690–701. doi: 10.1038/35088566. [DOI] [PubMed] [Google Scholar]
- Seggerson K, Tang L, Moss EG. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev Biol. 2002;243:215–225. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans hetero-chronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wood WB the Community of C. elegans Researchers. The Nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.