Abstract
In the early secretory pathway, opportunistic cleavage of asparagine-linked oligosaccharides by endoplasmic reticulum (ER) mannosidase I targets misfolded glycoproteins for dislocation into the cytosol and destruction by 26 S proteasomes. The low basal concentration of the glycosidase is believed to coordinate the glycan cleavage with prolonged conformation-based ER retention, ensuring that terminally misfolded glycoproteins are preferentially targeted for destruction. Herein the intracellular fate of human ER mannosidase I was monitored to determine whether a post-translational process might contribute to the regulation of its intracellular concentration. The transiently expressed recombinant human glycosidase was subject to rapid intracellular turnover in mouse hepatoma cells, as was the endogenous mouse ortholog. Incubation with either chloroquine or leupeptin, but not lactacystin, led to intracellular stabilization, implicating the involvement of lysosomal acid hydrolases. Inhibition of protein synthesis with cycloheximide led to intracellular depletion of the glycosidase and concomitant ablation of asparagine-linked glycoprotein degradation, confirming the physiologic relevance of the destabilization process. Metabolic incorporation of radiolabeled phosphate, detection by anti-phosphoserine antiserum, and the stabilizing effect of general serine kinase inhibition implied that ER mannosidase I is subjected to regulated proteolysis. Stabilization in response to genetically engineered removal of the amino-terminal cytoplasmic tail, a postulated regulatory domain, and colocalization of green fluorescent protein fusion proteins with Lamp1 provided two additional lines of evidence to support the hypothesis. A model is proposed in which proteolytically driven checkpoint control of ER mannosidase I contributes to the establishment of an equitable glycoprotein quality control standard by which the efficiency of asparagine-linked glycoprotein conformational maturation is measured.
Genetic information is directly transformed into biological activity in response to the correct conformational maturation and deployment of encoded proteins (1, 2). Arguably, this dual intention is best exemplified in the endoplasmic reticulum (ER)2 into which nascent secretory and membrane proteins are translocated. The adoption of native protein structure, facilitated by transient physical engagement with one or more molecular chaperones, precedes productive export from the ER (3). Rather than clogging the secretory pathway, molecules incapable of structural maturation are eliminated by a process coined “ER-associated degradation” (ERAD). The associated multiple requisite steps culminate in the dislocation of misfolded proteins into the cytosol for degradation by 26 S proteasomes (4, 5). Polyubiquitination functions as the signal that mediates both dislocation and proteolysis (Fig. 1) (6, 7).
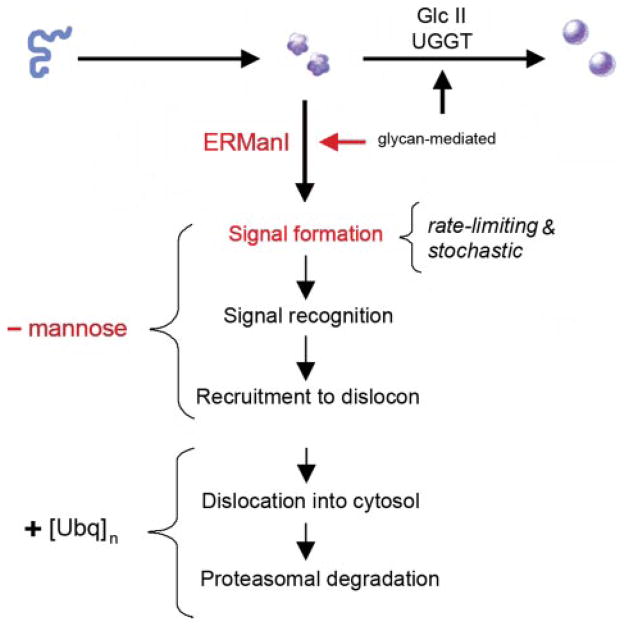
FIGURE 1. Conformational maturation and intracellular disposal of newly synthesized asparagine-linked glycoproteins represent a conformation-based branch point in the early secretory pathway.
Modification of asparagine-linked oligosaccharides by glucosidase II (GlcII) and UDP-Glc:glycoprotein glucosyltransferase (UGGT) facilitates rounds of physical engagement with glycoprotein folding machinery. Conformational maturation (solid sphere) precedes productive transport and deployment. In contrast, nonnative protein structure, in combination with glycans cleaved by ERManI, generates a proposed lumenal bipartite signal for recruitment to the ER dislocon. The low basal ERManI concentration underlies a quantitative trait by which asparagine-linked glycoproteins are selected for destruction on the basis of inefficient conformational. The latter parameter is designated by opportunistic cleavage of asparagine-linked oligosaccharides in response to prolonged conformation-based retention in the ER. Dislocation into the cytosol and elimination by 26 S proteasomes are mediated by polyubiquitination.
As with glycoprotein folding (8), the earliest events of ERAD are orchestrated by the covalent modification of asparagine-linked oligosaccharides (Fig. 1). The capacity of general α-1,2-mannosidase inhibition to abrogate the elimination of numerous misfolded asparagine-linked glycoproteins (2, 9–19) and the effect of gene deletions in yeast (20) implicated ER mannosidase I as a key component in glycoprotein ERAD (GERAD).
Intracellular degradation of terminally misfolded α1-antit-rypsin is accelerated in response to an experimentally elevated concentration of the recombinant glycosidase (18, 21). Its contribution toward a stochastically based substrate discrimination process was strengthened by destabilization of newly synthesized wild type α1-antitrypsin and transferrin under these conditions (18). A luminal bipartite signal consisting of the cleaved glycans and nonnative protein structure is suspected to function as the initial degradation signal. The very low basal concentration of ER mannosidase I (18) is proposed to preferentially target terminally misfolded glycoproteins for ERAD by coordinating the glycan modification with prolonged conformation-based ER retention. The subsequent trajectory of tagged molecules toward the ER dislocon is suspected to result from downstream machinery capable of recognizing the bipartite signal anatomy (Fig. 1).
The complementary nature by which the opposing systems of glycoprotein folding and quality control operate might function as a post-translational checkpoint in eukaryotic genome expression. Despite the fundamental role in which the overall process contributes to normal cell biology and possible involvement in numerous conformational diseases (15, 22), a clear mechanistic understanding of how this branch point operates has not yet emerged. For example, very little is understood about the mechanism by which the intracellular concentration of human ER mannosidase I is regulated, except that no transcriptional control has thus far been detected (23, 24).
The intracellular concentration of ER mannosidase I likely contributes to the establishment of an equitable quality control standard by which the efficiency of glycoprotein conformational maturation can be measured. Therefore, the intracellular fate of the recombinant protein was monitored following biosynthesis. Herein, metabolic pulse-chase radiolabeling demonstrated that the newly synthesized molecules are targeted for lysosomal destruction by a mechanism that involves the amino-terminal cytoplasmic tail. A model is proposed in which proteolysis-driven checkpoint control of ER mannosidase I contributes to the decentralized surveillance of eukaryotic genome expression.
EXPERIMENTAL PROCEDURES
Reagents
Unless stated otherwise, routine chemicals and buffers were purchased from Sigma. Lactacystin was purchased from the E. J. Corey laboratory (Harvard Medical School). Kifunensine was from Toronto Research Chemicals, Inc. Lipofectamine 2000 reagent was from Invitrogen. Brefeldin A was purchased from EPICENTRE (Madison, WI). The murine hepatoma cell line, Hepa 1a, was a kind gift from Dr. Gretchen Darlington (Baylor College of Medicine, Houston, TX). The establishment of Hepa1a (H1A) cells stably transfected with the null(Hong Kong) variant of human α1-antitrypsin (cell line H1A/N13) was previously reported (25).
Antisera
An immunoglobulin fraction of rabbit polyclonal anti-human α1-antitrypsin was purchased from Roche Applied Science. A monoclonal anti-phosphoserine antiserum was purchased from Sigma. Rabbit antiserum ERMI-cc20 (18) was used for the detection of endogenous mouse ERManI in untransfected Hepa1a cells. The transfected human homolog (hER-ManI; accession number NM_007230) was detected following transfection with antiserum (polyclonal anti-ER Man I antibody) under conditions that failed to detect the endogenous murine ortholog. The polyclonal antiserum was generated against the purified recombinant catalytic domain (residues 241–699) expressed in Pichia pastoris (26). Approximately 200 μg (in 0.5 ml of phosphate-buffered saline) of the purified enzyme emulsified with an equal volume of Freund’s complete adjuvant was used for multiple intradermal injections of a male New Zealand White rabbit and included boosting at 3-week intervals. Periodic bleeds from an ear vein were performed following the second boost and allowed to clot at room temperature prior to centrifugation at 2000 × g for 15 min to obtain serum. The aliquots were maintained at −80 °C for long term storage.
Transfection, Metabolic Radiolabeling, and Immunoprecipitation
Transient transfections and cotransfection of H1A cells were performed as previously described, using the Lipofectamine 2000 method (Invitrogen) (18). Changes in the rate of target protein degradation were monitored by pulse-chase metabolic radiolabeling with methionine and immunoprecipitation (15, 18) 72 h post-transfection. In the appropriate experiments, monolayers of semi-confluent cells were subject to preincubation incubated for 1 h at 37 °C in regular growth medium (25) or with medium supplemented with specific inhibitors (lactacystin, 0.025 mM; kifunensine, 0.1–0.2 mM; leupeptin, 0.05 mg/ml) as previously reported (18, 25). Metabolic radiolabeling with [35S]methionine (0.150 mCi/100-mm dish; ICN Pharmaceuticals, Inc.) was performed with methionine-free medium (ICN Pharmaceuticals, Inc.) or with medium containing inhibitors (lactacystin, 0.025 mM; kifunensine, 0.1–0.2 mM; leupeptin, 0.1 mg/ml; chloroquine, 0.2 mM; brefeldin A, 0.005 mg/ml; staurosporine, 100 nM; or genistein, 0.050 mM) for 10 min, followed by a chase in serum-free Dulbecco’s modified Eagle’s medium (Invitrogen) containing 0.2 mM unlabeled methionine with or without inhibitor, as appropriate. Metabolic incorporation and detection of radiolabeled phosphate into cellular proteins, plus digestion with potato acid phosphatase (Sigma), was performed as previously described. At selected intervals, immunoprecipitates were generated from buffered Nonidet P-40 (Nonidet P-40) detergent (Sigma)-generated cell lysates and medium as previously described (27). Following separation by SDS-PAGE, radiolabeled proteins were detected by fluorography and quantified by densitometric scanning using the National Institutes of Health Image Program or by phosphorimaging analysis (18).
Immunoblotting
Approximately 48–72 h post-transfection, 5 × 106 cells were lysed in 1 ml of buffered Nonidet P-40 detergent (Sigma). Equivalent aliquots were resolved by SDS-PAGE and transferred to nitrocellulose prior to blotting with specific antisera. The signals were detected with the recommended ECL Western blotting reagents (Amersham Biosciences) as previously described (14, 15, 18).
Deletion of the hERManI Amino-terminal Cytoplasmic Tail
Programs TMpred from the ExPASy website (us.expasy.org/tools) were used for the identification of amino acids that comprise the amino-terminal cytoplasmic tail of hERManI (accession number NM_007230). PCR was used to modify the hERManI cDNA (accession number NM_007230) to encode a recombinant protein in which the entire amino-terminal cytoplasmic tail was deleted. Priming from nucleotides 234 to 251 was performed with primer HindIIIΔT. (5′-CGAAGCTTCAGCGGAATATGATTCTC) that contained a HindIII site at the 5′ end. Primer ERMITAGRI (5′-GGAATTCTCCACCCTAGGCAGGGGTCC) was used for priming from nucleotides 2106 to 2087 and contained an EcoRI site at the 3′-end. The 1.9-kb PCR fragment was purified and subcloned into pCRII (Invitrogen) for sequence confirmation and then subcloned into the mammalian expression vector pCDNA3.1Zeo(+) (Invitrogen) to generate phERManI(NT)/pCDNA3.1Zeo(+). The methionine residue originally located at position 49 in wild type hER-ManI (accession number NM_007230) was used as the new start codon for expression of the recombinant “no tail” protein, hERManI(NT). Positive bacterial transformants were identified by restriction enzyme digestion. Expression of the encoded protein in mammalian cells was confirmed by immunoprecipitation and Western blotting of the mammalian cell extracts following transfection (14, 15, 18).
Design of Fluorescent Fusion Proteins
For the expression of fluorescent fusion proteins that contain the amino-terminal cytoplasmic tail, transmembrane domain, and a significant portion of the lumenal stem of hERManI (accession number NM_007230), a 500-bp EcoRI (5′-end)-BamHI (nucleotide 569) fragment was purified from DNA construct hERManI/pCDNA3.1Zeo(+) (18) and then subcloned into the corresponding sites in pEGFP-N1 and pDsRed2-N1 (BD Biosciences, Palo Alto, CA) to generate plasmids hERManI(1–158)/pEGFP-N1 and hERManI(1–158)/pDsRED2-N1, respectively. To specifically eliminate the cytoplasmic tail, a 350-bp HindIII-BamHI fragment purified from plasmid hERManI/pCDNA3.1Zeo(+) (18) was sub-cloned into the HindIII-BamHI site of pEGFP-N1 or pDsRed2-N1 to generate constructs hERManI(49–158)/EGFP-N1 and phERManI(49–158)/DsRED2-N1, respectively. Positive bacterial transformants were identified by restriction enzyme digestion, and the encoded recombinant proteins were detected by immunoblotting with antibodies against GFP, RFP (red fluorescent protein) (Invitrogen), and hERManI after transfection into mammalian cells.
Indirect Immunofluorescence and Confocal Microscopy
24 h-post transfection, the cells were grown on coverslips in 6-well plates for an additional 24 h, then washed in phosphate-buffered saline, air-dried at room temperature, and fixed in methanol for 5 min. For indirect immunofluorescent staining, fixed cells were incubated in phosphate-buffered saline blocking solution containing 1% bovine serum albumin and 0.5% Tween 20 detergent at room temperature for 1 h and then subjected to the primary antibody reactions at 4 °C overnight using a goat polyclonal antibody against lysosomal-associated membrane protein 1 (Lamp1) (1:200 dilution) and the appropriate rabbit polyclonal ERManI antibody (1:200 dilution). Secondary antibody reactions were performed at room temperature, for 1 h, using anti-goat or anti-rabbit IgG conjugated with fluorescein isothiocyanate or Texas Red. For high resolution studies, after mounting in GEL/MOUNT TM (Biomeda Corp., Foster City, CA), fluorescent images were obtained using a Zeiss LSM 510 confocal microscope system and then analyzed with LSM 510 software and Adobe Photoshop v7.
RESULTS
Rapid Intracellular Turnover of Transfected Recombinant Human ER Mannosidase I
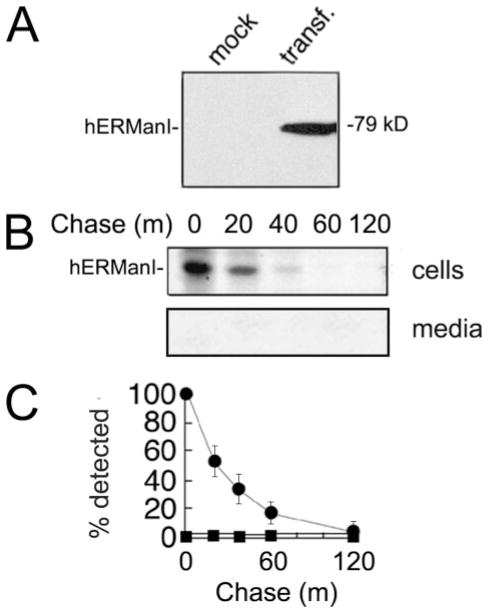
Modification of asparagine-linked oligosaccharides by ER mannosidase I is the rate-limiting step for GERAD in H1A cells (14, 15, 28). Moreover, its experimentally elevated concentration can inappropriately target newly synthesized wild type glycoproteins into GERAD (18). Our immediate objective was to test the prediction that the human homolog is subject to tight intracellular regulation. According to ECL Western blotting of cell lysates, the transiently expressed recombinant human ER mannosidase I was absent from mock transfected cells but detected as a single immunoreactive 79-kDa band following transient expression (Fig. 2A). A combination of metabolic pulse-chase radiolabeling and immunoprecipitation allowed us to monitor the fate of the protein following a 10-min pulse with [35S]methionine. The entire radiolabeled cohort rapidly disappeared from cells (T½ = 20 min) and was absent from the medium (Fig. 2, B and C), indicating that its loss was not caused by secretion.
FIGURE 2. Intracellular turnover of transfected (transf.) recombinant human ER mannosidase I.
A, detection by ECL Western blotting of transiently expressed recombinant human ER mannosidase I. B, pulse-chase metabolic radiolabeling with [35S]Met and selective immunoprecipitation of recombinant human ER mannosidase I. C, results from B are depicted as a graph. All of the experiments were initiated ~48–72 h post-transfection. Specific antiserum (“Experimental Procedures”) resulted in the detection of the recombinant protein in Nonidet P-40 cell lysates (cells) and tissue culture medium (media). Duration of the metabolic pulse in B was 10 min.
Involvement of Lysosomal Hydrolases
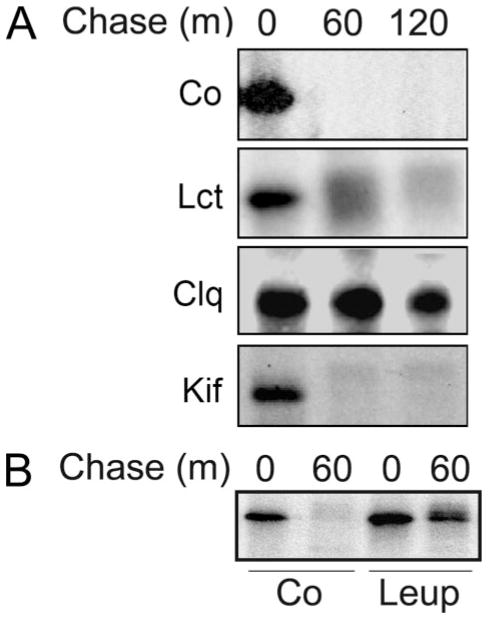
Because immunoreactive recombinant human ER mannosidase I was not detected in the insoluble fraction of the Nonidet P-40 cell lysates (data not shown), we concluded that intracellular degradation, rather than insolubility, was responsible for our observation. In mammalian cells, 26 S proteasomes and lysosomes are the two major systems by which proteins are degraded (29). To determine whether either might be responsible for the rapid turnover of transfected human recombinant ER mannosidase I, metabolic pulse-chase radiolabeling and immunoprecipitation were repeated in cells that had been incubated with specific inhibitors of either system. Incubation with lactacystin (0.025–0.1 mM), an irreversible inhibitor of multicatalytic proteasomes (30), had only a minor effect on the disappearance of radiolabeled human ER mannosidase I (Fig. 3A, Lct) as compared with control (Fig. 3A, Co) during a 120-min chase. Elevated concentrations of the inhibitor did not prove to be more effective (data not shown). In contrast, the entire cohort was detectable following the same period in which the medium was supplemented with chloroquine (Fig. 3A, Clq), a lysosomotrophic amine capable of raising the pH of acidic intracellular compartments. To verify this observation, the cells were incubated in medium containing ammonium chloride, another lysosomotrophic amine (31). Although the treatment exerted a stabilizing effect (data not shown), the apparent cytotoxicity precluded any accurate interpretation of the results. Therefore, incubation with medium supplemented with the lysosomal protease inhibitor leupeptin (32) was performed as an alternative method to test our hypothesis. Under these conditions, intracellular turnover of pulse-radiolabeled recombinant human ER mannosidase I was substantially inhibited (Fig. 3B).
FIGURE 3. Pharmacologic inhibition of recombinant human ER mannosidase I turnover.
A, pulse-chase metabolic radiolabeling with [35S]Met and selective immunoprecipitation of recombinant human ER mannosidase I. Co, normal medium; Lct, medium with the addition of lactacystin; Clq, medium with the addition of chloroquine; Kif, medium with the addition of kifunensine (“Experimental Procedures”). B, same as above, but with the addition of leupeptin (Leup) (“Experimental Procedures”). Duration of the metabolic pulse in B was 10 min.
To determine whether the observations reflected a cellular response to the elevation of total ER mannosidase I activity, the fate of the recombinant protein was monitored in transfected cells incubated with kifunensine, a general membrane-permeable inhibitor of α-mannosidase activity (33). However, the manipulation exerted no apparent stabilizing effect (Fig. 3A, Kif). Furthermore, enzymatic inactivation of recombinant human ER mannosidase I in response to the addition of a Myc tag to the carboxyl terminus, as previously reported (18), did not hinder the rate of intracellular turnover (data not shown), negating the notion that the glycosidase instability reflected a cellular response to the elevated mannosidase activity.
Lysosome-mediated Destabilization of Endogenous Mouse ER Mannosidase I
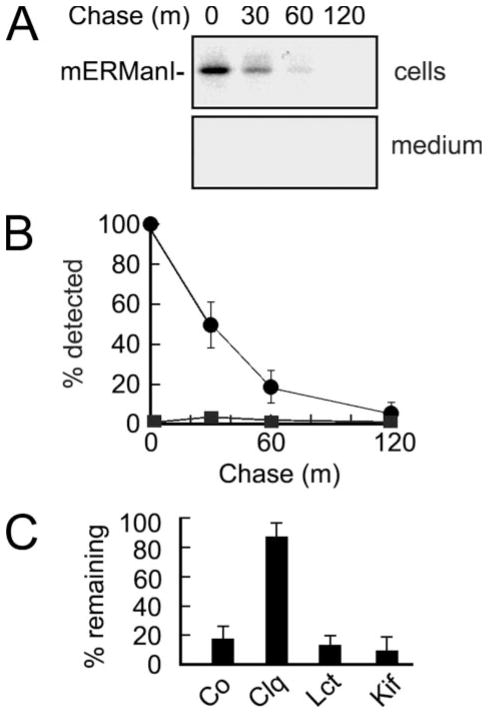
We asked whether the endogenous mouse ER mannosidase I is subject to rapid intracellular turnover, because this would be predicted if instability of the human homolog reflected a physiologically relevant process. For this, a peptide-specific antiserum was generated to monitor the fate of newly synthesized endogenous mouse ER mannosidase I in mock transfected H1A cells (see “Experimental Procedures”). A slight cross-reaction with the human homolog (not shown) was not problematic because the analyses were performed in nontransfected cells. As previously observed for the recombinant human homolog, the entire cohort of newly synthesized endogenous mouse ER mannosidase I was rapidly depleted from Nonidet P-40 cell lysates following a 10-min pulse without detection in the chase medium (Fig. 4, A and B). Moreover, the rate of disappearance (Fig. 4B) and pattern of stabilization with cell-permeable inhibitors (Fig. 4C) was comparable with that of the transfected human homolog (Fig. 3A).
FIGURE 4. Endogenous mouse ER mannosidase I is subject to rapid intracellular turnover.
A, pulse-chase metabolic radiolabeling with [35S]Met and selective immunoprecipitation of endogenous mouse ER mannosidase I from Nonidet P-40 cell lysates (cells) and tissue culture medium (medium). Duration of the metabolic pulse was 10 min. B, results from A are depicted as a graph. C, the effects of chloroquine (Clq), lactacystin (Lct), and kifunensine (Kif) on the intracellular fate of endogenous mouse ER mannosidase I, relative to control (Co), are depicted as a bar graph. All of the experiments were initiated ~48–72 h post-transfection. Specific antiserum (“Experimental Procedures”) resulted in the detection of the recombinant protein in Nonidet P-40 cell lysates and tissue culture medium. Duration of the metabolic pulse in B was 10 min.
Physiologic Relevance of ER Mannosidase I Destabilization
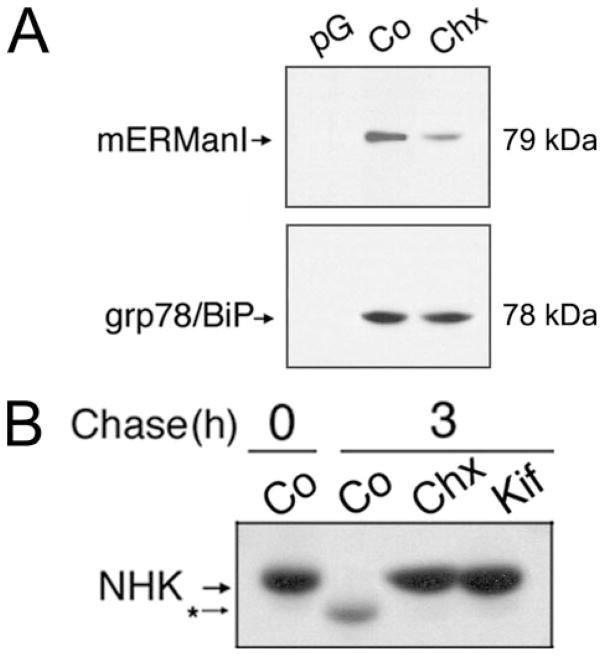
Because short-lived proteins undergo intracellular depletion in response to the inhibition of protein synthesis, the intracellular concentration of endogenous mouse ER mannosidase I was examined following a 1-h incubation in regular growth medium supplemented with cycloheximide. ECL Western blotting of Nonidet P-40 lysates demonstrated that the inhibition of translation resulted in a reduction in the level of the endogenous mouse homolog (Fig. 5, compare A and B). The absence of any demonstrable change in the intracellular concentration of the long-lived ER resident molecular chaperone grp78/BiP (34), as compared with control (Fig. 5A, lower panel), provided confirmatory evidence for the short intracellular half-life of ER mannosidase I under normal conditions.
FIGURE 5. Intracellular depletion of ER mannosidase I and concomitant inhibition of GERAD in response to a 1-h incubation in medium containing cycloheximide.
A, detection of endogenous mERManI and grp78/BiP by ECL Western blotting from Nonidet P-40 cell lysates under control conditions (Co) or following incubation with cycloheximide (Chx). Protein G used without antiserum (pG). B, pulse-chase metabolic radiolabeling with [35S]Met and immunoprecipitation of α1-antitrypsin variant null(Hong Kong) (NHK) in cell line H1A/N13 (“Experimental Procedures”) under control conditions (Co) or following treatment with cycloheximide (Chx) or kifunensine (Kif). Duration of the metabolic pulse in B was 10 min.
The data were then exploited to investigate the effect of cycloheximide treatment on the intracellular fate of a terminally misfolded genetic variant of α1-antitrypsin, designated null(Hong Kong), which is a well established substrate for glycoprotein ERAD (14, 18, 21, 25, 28, 35, 36). Intracellular degradation of the radiolabeled molecules was ablated, as compared with control, in response to cycloheximide treatment (Fig. 5B, compare Co and CHX). Moreover, the inhibitory effect mimicked that of kifunensine treatment (Fig. 5B, Kif). Importantly, both cycloheximide and kifunensine were able to independently ablate the discrete electrophoretic mobility shift of radiolabeled null(Hong Kong) in SDS-PAGE (Fig. 5B, compare CHX, Kif, and Co), implying that both treatments had blocked the removal of mannose from asparagine-linked oligosaccharides (15–18). Taken together, the results strengthened the conclusion that endogenous mouse ER mannosidase I is a short-lived intracellular protein.
Evidence for Proteolytic Down-regulation
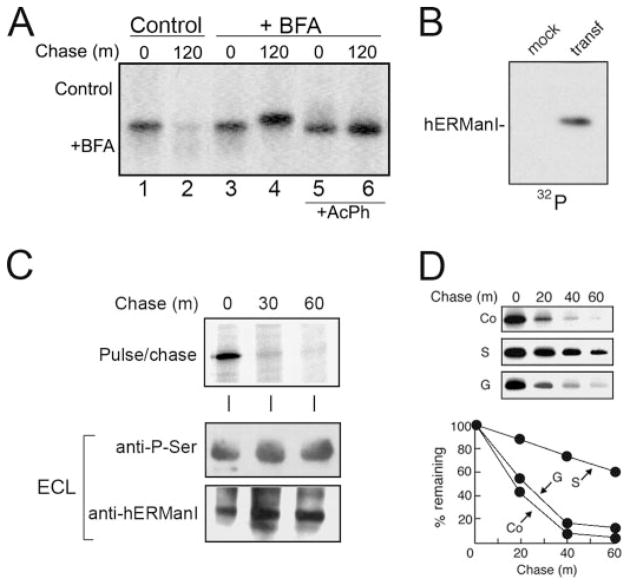
Intracellular turnover of ER mannosidase I is a predictable aspect of the stochastic model for substrate selection. However additional data were needed to determine whether the molecules are inherently labile or might be subject to regulated proteolysis. In the next set of experiments, we tested the hypothesis that vesicular transport contributes to the delivery of ER mannosidase I to lysosomes. The human homolog was chosen for our investigation because of the available corresponding recombinant DNA that allowed for genetic manipulation. Incubation with medium supplemented with brefeldin A, a known inhibitor of vesicle transport (20, 37), ablated intracellular turnover of the radiolabeled molecules as compared with control (Fig. 6A, compare lanes 2 and 4). An additional important observation was that the entire population of undegraded molecules exhibited a slower rate of electrophoretic migration in SDS-PAGE as compared with their newly synthesized counterparts (Fig. 6A, compare lanes 3 and 4). Restoration in response to digestion of Nonidet P-40 cell lysates with potato acid phosphatase (Fig. 6A, compare lanes 5 and 6) implied that phosphorylation of ER mannosidase I was responsible for the altered electrophoretic mobility.
FIGURE 6. Detectable phosphorylation of recombinant human ER man-nosidase I and arrested intracellular turnover in response to selective inhibition of serine kinase activity.
A, pulse-chase metabolic radiolabeling with [35S]Met and selective immunoprecipitation of recombinant human ER mannosidase I from Nonidet P-40 cell lysates under normal conditions (Control) or with the addition of brefeldin A (+BFA). Samples treated with potato acid phosphatase (+AchP) are shown. B, selective immunoprecipitation of recombinant human ER mannosidase I from mock transfected (mock) and transfected (transf) Hepa1a Nonidet P-40 cell lysates followed a 3-h incubation with phosphate-free growth medium supplemented with radiolabeled inorganic phosphate (32P). C, metabolic pulse-chase radiolabeling with [35S]methionine (top panel), ECL Western blotting with an anti-phospho-Ser antibody of human ER mannosidase I immunoprecipitates from mock transfected (mock) and transfected (transf) Hepa1a cells (middle panel), and ECL Western blotting for human ERManI (bottom panel). D, pulse-chase metabolic radiolabeling with [35S]Met and selective immunoprecipitation of recombinant human ER mannosidase I under control (Co) conditions or following treatment with staurosporine (S) or genistein (G). The quantified results are shown (graph). Duration of the metabolic pulses in A and D was 10 min.
Under steady-state conditions, radiolabeled phosphate was metabolically incorporated into transfected recombinant human ER mannosidase I even in the absence of the vesicular transport inhibitor (Fig. 6B), indicating that the covalent modification had not represented an artifact of the treatment. Moreover, the modification was recognized by anti-phosphoserine antiserum under steady-state conditions (Fig. 6C, bottom panel). Finally, in metabolic pulse-chase radiolabeling experiments the rate of intracellular turnover was significantly diminished as compared with control in cells incubated with medium containing staurosporine (Fig. 6D), a membrane-permeable inhibitor of general Ser/Thr kinase activity. In contrast, genistein (Fig. 6D), a general protein-tyrosine kinase inhibitor (38), had no apparent effect. Taken together, the data argue against the notion that human ER mannosidase I is merely labile.
Contribution by the Amino-terminal Cytosolic Tail
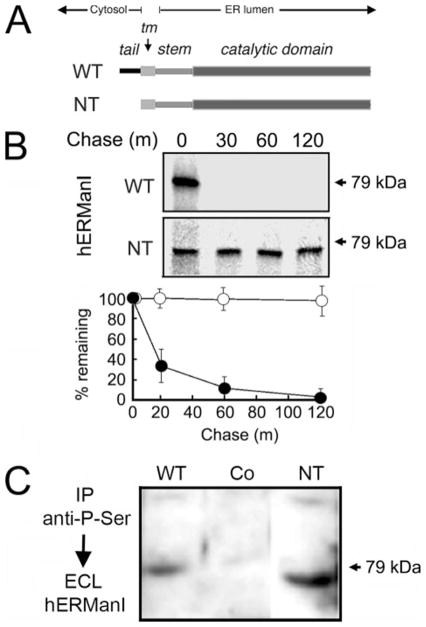
One interpretation of the aforementioned data is that human ER mannosidase I is subject to regulated proteolysis. The modular architecture of the glycosidase was exploited to identify a domain through which its down-regulation might be mediated. The prediction was that removal of a candidate regulatory domain would ablate rapid intracellular turnover. Therefore, the fate of the protein was monitored following genetically engineered removal of the amino-terminal cytoplasmic tail. Removal of the tail does not influence the remaining structure of the molecule, including the original transmembrane domain, noncleavable ER sorting signal, and proposed stem and catalytic domains (Fig. 7A). The fates of wild type (WT) and tail-less (NT) human ER mannosidase I were monitored by metabolic pulse-chase radiolabeling following transient transfection. Both proteins were synthesized as single radiolabeled bands, although of slightly different molecular weights, as predicted (Fig. 7B). Importantly, its absence did not diminish the efficiency of protein translation. As shown in Fig. 7B, the radiolabeled wild type molecules were rapidly degraded during a 1-h chase, whereas the tail-less counterpart remained stable during this period. Stabilization of the tail-less molecule implied that it had not misfolded, otherwise the molecules would have been subject to intracellular destruction by ERAD.
FIGURE 7. Arrested intracellular turnover in response to genetically engineered removal of the amino-terminal cytoplasmic tail.
A, relative topology in the ER membrane of the domains for wild type (WT) human ER mannosidase I, including the amino-terminal cytoplasmic tail (tail), transmembrane domain (tm), and lumenal domains (stem and catalytic domain) before (WT) and after genetic-engineered removal of the amino-terminal cytoplasmic tail (NT). B, pulse-chase metabolic radiolabeling with [35S]Met (10-min pulse) and selective immunoprecipitation from Nonidet P-40 cell lysates following transient expression of the wild type recombinant human ER mannosidase (WT) and following genetic-engineered removal of the amino-terminal cytoplasmic tail (NT). In the lower panel, the results are presented as a graph. C, Nonidet P-40 lysates from untransfected cells (Co), and cells transfected with either WT and NT were immunoprecipitated (IP) with anti-phospho-antiserum and fractionated by SDS-PAGE, and human ERManI was detected by ECL Western blotting.
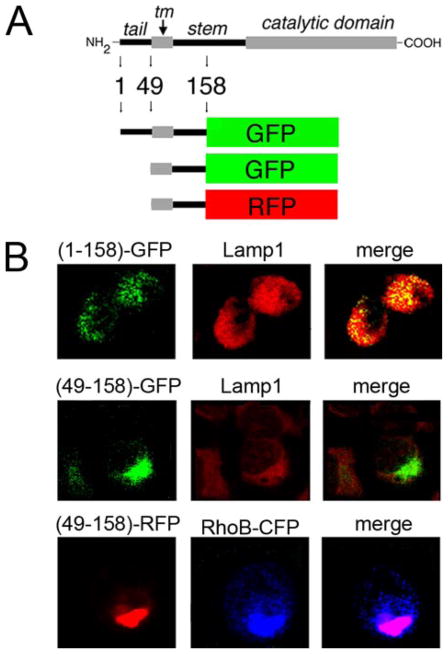
In a subsequent set of experiments, ECL Western blotting demonstrated that recombinant NT was phosphorylated under steady state conditions (Fig. 7C). The observation precluded any conclusion of the involvement of the tail in targeting ER mannosidase I to lysosomes. Therefore, to further investigate the involvement of the tail, fluorescent fusion proteins were generated in which the lumenal stem of human ER mannosidase I was fused in-frame to GFP (Fig. 8A). The stem was chosen as the site for fusion because the putative signal might extend into the transmembrane domain. Also, the absence of distinct structural features in the stem would prevent random misfolding of the chimeric molecules.
FIGURE 8. Colocalization with lysosomes and contribution of the amino-terminal cytoplasmic tail.
A, schematic diagram of hERManI domain structure and design of fluorescent fusion proteins (“Experimental Procedures”). B, confocal microscopy 72 h post-transfection of Hepa1a cells was used to detect and localize the fluorescent fusion proteins shown in A. Endogenous Lamp1 was used as a marker for lysosomes. Cotransfected RhoB-CFP served a fluorescent marker for endosomes.
Our initial experiments demonstrated that incubation with chloroquine disrupted the intracellular architecture of H1A cells (not shown), precluding our original strategy to test for colocalization with lysosomes. Therefore, the experimental design was performed under steady-state conditions. A considerable fraction of recombinant hERMI (1–158)-GFP colocalized with the endogenous lysosomal marker, Lamp1 (Fig. 8B, top set of panels). To substantiate the involvement of the ER mannosidase I tail, the encoding nucleotides were selectively removed (Fig. 8B). Consistent with our hypothesis, the manipulation ablated any detectable colocalization with Lamp1 (Fig. 8B, middle panels). Importantly, the tail-less chimeric molecule had apparently sequestered within cells (Fig. 8B, middle panels). To identify the apparent site of accumulation, GFP was replaced with red fluorescent protein (RFP) (Fig. 8A) to allow for analyses following coexpression with the cyan-colored RhoB-CFP. Colocalization of the two molecules (Fig. 8B, lower panels) indicated that removal of the tail had led to accumulation of the recombinant fusion proteins in an expanded endosomal compartment (39). In no set of experiments were any of the fluorescent fusion proteins detected at the cell surface (not shown). These findings imply that sequestration in endosomes likely represents mistargeting of the tail-less fusion protein rather than identification of a compartment through which the molecules are normally transported to lysosomes.
DISCUSSION
Successful gene expression is rooted in the correct conformational maturation of encoded proteins and selective removal of those molecules unable to attain this structural milestone. The earliest events by which cells manage the numerous proteins translocated into the ER is central to the branch point composed of protein folding and degradation. Considering the stochastic role played by the low basal ER mannosidase I concentration and its potential contribution to a post-translational checkpoint, we asked whether the molecule is subject to regulated proteolysis.
In our present study, an analysis of the endogenous mouse ortholog was incorporated into the experimental design to ensure that no observations were artifactual. Rapid intracellular turnover of the transfected molecules confirmed the existence of a short intracellular half-life. The conclusion was strengthened by the capacity of cycloheximide treatment to deplete cells of ER mannosidase I. The physiological relevance of ER mannosidase I destabilization was verified by the short half-life of the endogenous mouse ER mannosidase I, plus its depletion in response to cycloheximide treatment and ability to mimic the effect of kifunensine on the fate of a GERAD substrate. Importantly, our findings begin to address the long-standing question as to how cycloheximide is capable, at least in some situations, to stabilize multiple GERAD substrates (40). However, it should be noted that cycloheximide is capable of influencing numerous intracellular systems. Therefore, it is premature to propose its use as a diagnostic tool for the identification of GERAD clients.
Pharmacological inhibition studies, plus intracellular colocalization of GFP fusion proteins with Lamp1, provided separate lines of evidence that proteolytic regulation of human ER mannosidase I involves lysosomal proteases. The simplest explanation for using lysosomes, rather than proteasomes, to down-regulate ER mannosidase I is to avoid the potential complications that might arise from controlling the glycosidase concentration by the system for which it is intended to function. Involvement of the amino-terminal cytoplasmic tail is reasonable, especially when one considers the difference in length between that of the human and yeast orthologs (9, 41).
Prior to this study, the underlying clockwork mechanics by which the critical glycan-based signal determinant is generated had remained poorly defined (2, 18). Our observations challenge the prior notion that the poor catalytic activity of ER mannosidase I (9, 10) functions as the sole mechanism by which the rate at which glycoproteins are targeted for GERAD. Of greater importance, the data imply that long-lived cellular factors, such as alternative mannosidases, are unable to compensate for the role played by ER mannosidase I in GERAD. After the initiation of our study, Nagata and co-workers (21) reported the rapid turnover of recombinant human ER mannosidase I by a nonproteasomal pathway when transiently expressed in a cultured human embryonic kidney cell line. However, no explanation about the possible physiologic relevance of their observation was provided.
Based on our findings, we conclude that ER mannosidase I is subject to regulated proteolysis as a means to govern the performance-based measurement of inefficient glycoprotein conformational maturation. The intracellular ER mannosidase I concentration represents a quantitative trait that underlies the critical decision of how long a nonnative glycoprotein is allowed to remain in the ER. Although functioning at distinct organizational levels, the impaired progression of damaged gene products (i.e. encoded proteins) through the exocytic pathway is broadly reminiscent of the capacity by which damaged DNA impedes progression through the cell cycle. Also, it is our contention that use of the novel regulatory system is consistent with a role for ER mannosidase I in mediating the first step in GERAD. Moreover, we take note of the operational resemblance between down-regulation of ER mannosidase I and the proteolytic regulation of several nuclear-situated cell cycle checkpoint proteins (42). Taken together, our findings lend support to a model in which ER mannosidase I contributes to an important tempo-spatial decision by which the surveillance of eukaryotic genome expression is decentralized (2, 43). The notion is certainly consistent with the fact that the nucleus and ER are signature organelles in eukaryotic cells and is similar to a conclusion recently made by Lennarz and co-workers (44) concerning coevolution of cell cycle checkpoint regulation and ER quality control.
The role by which phosphorylation might contribute to ER mannosidase I instability remains elusive. The same can be said for the stabilizing effect of brefeldin A. Presently, we do not know how the glycosidase is transported to lysosomes. Also, because the proteolytic down-regulation of human ER mannosidase I is operative under basal conditions, it is not yet known whether some natural cellular strategy might exist to adjust the glycosidase concentration under conditions of ER stress, possibly at a post-translational level. Moreover, despite our conclusions it should be noted that Lederkremer and Glickman (45) have proposed a model in which vesicle recycling rather than strict ER retention contributes to the “timed” degradation (46) of secretion-incompetent glycoproteins. In that model nonnative protein structure diverts glycoproteins into a specific targeting pathway, and a window of opportunity in which glycoproteins are exposed to ER mannosidase I represents the emergent clock. Additional investigation of the model is warranted, although it is difficult to reconcile how the rapid intracellular destruction of the glycosidase might contribute to this process.
Finally, it should be mentioned that the secretion of structurally aberrant proteins from cultured mammalian cells has been reported (47), bringing into question the actual existence of quality control. Our findings indicate that the system does, in fact, exist and plays a vital role in the selection of substrates for GERAD. At least four explanations might explain the unexpected phenomenon in which structurally aberrant proteins are secreted. These are: (i) the failure of heterologous systems to adequately monitor foreign proteins, (ii) the saturation of endogenous folding, retention, and/or degradation systems, (iii) aberrant cell adaptation in response to prolonged cell culture that results in the loss of retention and/or ERAD elements, and/or (iv) a previously unanticipated aspect of the overall system.
Footnotes
This work was supported in part by National Institutes of Health Grants HL62553 and DK 064232, plus Grants 1Y102-6 and F03-12 from the Fernandez Liver Research Initiative, Grants RO2-5 and RO6-06 from the Alpha1-Foundation (to R. N. S.), and National Institutes of Health Grants GM47533 and RR005351 (to K. W. M.).
The abbreviations used are: ER, endoplasmic reticulum; ERManI, endoplasmic reticulum mannosidase I; ERAD, ER-associated degradation; GERAD, glycoprotein ERAD; GFP, green fluorescent protein; h, human; WT, wild type; NT, tail-less.
References
- 1.Gerstein M, Lan N, Jansen R. Science. 2002;295:284–287. doi: 10.1126/science.1068664. [DOI] [PubMed] [Google Scholar]
- 2.Cabral CM, Liu Y, Sifers RN. Trends Biochem Sci. 2001;26:619–624. doi: 10.1016/s0968-0004(01)01942-9. [DOI] [PubMed] [Google Scholar]
- 3.Ellgaard L, Helenius A. Curr Opin Cell Biol. 2001;13:431–437. doi: 10.1016/s0955-0674(00)00233-7. [DOI] [PubMed] [Google Scholar]
- 4.McCracken AA, Brodsky JL. Bioessays. 2003;25:868–877. doi: 10.1002/bies.10320. [DOI] [PubMed] [Google Scholar]
- 5.Jarosch E, Taxis C, Volkwein C, Bordallo J, Finley D, Wolf DH, Sommer T. Nat Cell Biol. 2002;4:134–139. doi: 10.1038/ncb746. [DOI] [PubMed] [Google Scholar]
- 6.Fewell SW, Travers KJ, Weissman JS, Brodsky JL. Annu Rev Genet. 2001;35:149–191. doi: 10.1146/annurev.genet.35.102401.090313. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida Y, Tokunaga F, Chiba T, Iwai K, Tanaka K, Tai T. J Biol Chem. 2003;278:43877–43884. doi: 10.1074/jbc.M304157200. [DOI] [PubMed] [Google Scholar]
- 8.Parodi AJ. Biochem J. 2000;348:1–13. [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez DS, Karaveg K, Vandersall-Nairn AS, Lal A, Moremen KW. J Biol Chem. 1999;274:21375–21386. doi: 10.1074/jbc.274.30.21375. [DOI] [PubMed] [Google Scholar]
- 10.Helenius A. Mol Biol Cell. 1994;5:253–265. doi: 10.1091/mbc.5.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karaveg K, Moremen KW. J Biol Chem. 2005;280:29837–29848. doi: 10.1074/jbc.M505130200. [DOI] [PubMed] [Google Scholar]
- 12.Karaveg K, Siriwardena A, Tempel W, Liu ZJ, Glushka J, Wang BC, Moremen KW. J Biol Chem. 2005;280:16197–16207. doi: 10.1074/jbc.M500119200. [DOI] [PubMed] [Google Scholar]
- 13.Tremblay LO, Herscovics A. Glycobiology. 1999;9:1073–1078. doi: 10.1093/glycob/9.10.1073. [DOI] [PubMed] [Google Scholar]
- 14.Cabral CM, Choudhury P, Liu Y, Sifers RN. J Biol Chem. 2000;275:25015–25022. doi: 10.1074/jbc.M910172199. [DOI] [PubMed] [Google Scholar]
- 15.Cabral CM, Liu Y, Moremen KW, Sifers RN. Mol Biol Cell. 2002;13:2639–2650. doi: 10.1091/mbc.E02-02-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Choudhury P, Cabral CM, Sifers RN. J Biol Chem. 1999;274:5861–5867. doi: 10.1074/jbc.274.9.5861. [DOI] [PubMed] [Google Scholar]
- 17.Termine D, Wu Y, Liu Y, Sifers RN. Methods. 2005;35:348–353. doi: 10.1016/j.ymeth.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y, Swulius MT, Moremen KW, Sifers RN. Proc Natl Acad Sci U S A. 2003;100:8229–8234. doi: 10.1073/pnas.1430537100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oda Y, Hosokawa N, Wada I, Nagata K. Science. 2003;299:1394–1397. doi: 10.1126/science.1079181. [DOI] [PubMed] [Google Scholar]
- 20.Jakob CA, Burda P, Roth J, Aebi M. J Cell Biol. 1998;142:1223–1233. doi: 10.1083/jcb.142.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hosokawa N, Tremblay LO, You Z, Herscovics A, Wada I, Nagata K. J Biol Chem. 2003;278:26287–26294. doi: 10.1074/jbc.M303395200. [DOI] [PubMed] [Google Scholar]
- 22.Sifers RN. Nat Struct Biol. 1995;2:355–357. doi: 10.1038/nsb0595-355. [DOI] [PubMed] [Google Scholar]
- 23.Travers KJ, Patil CK, Weissman JS. Adv Protein Chem. 2001;59:345–390. doi: 10.1016/s0065-3233(01)59011-7. [DOI] [PubMed] [Google Scholar]
- 24.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 25.Sifers RN, Brashears-Macatee S, Kidd VJ, Muensch H, Woo SL. J Biol Chem. 1988;263:7330–7335. [PubMed] [Google Scholar]
- 26.Vallee F, Karaveg K, Herscovics A, Moremen KW, Howell PL. J Biol Chem. 2000;275:41287–41298. doi: 10.1074/jbc.M006927200. [DOI] [PubMed] [Google Scholar]
- 27.Choudhury P, Liu Y, Bick RJ, Sifers RN. J Biol Chem. 1997;272:13446–13451. doi: 10.1074/jbc.272.20.13446. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Choudhury P, Cabral CM, Sifers RN. J Biol Chem. 1997;272:7946–7951. doi: 10.1074/jbc.272.12.7946. [DOI] [PubMed] [Google Scholar]
- 29.Trombetta ES, Parodi AJ. Annu Rev Cell Dev Biol. 2003;19:649–676. doi: 10.1146/annurev.cellbio.19.110701.153949. [DOI] [PubMed] [Google Scholar]
- 30.Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber SL. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 31.Seglen PO. Methods Enzymol. 1983;96:737–764. doi: 10.1016/s0076-6879(83)96063-9. [DOI] [PubMed] [Google Scholar]
- 32.Gordon SR, DeMoss J. Exp Cell Res. 1999;246:233–242. doi: 10.1006/excr.1998.4298. [DOI] [PubMed] [Google Scholar]
- 33.Weng S, Spiro RG. Arch Biochem Biophys. 1996;325:113–123. doi: 10.1006/abbi.1996.0014. [DOI] [PubMed] [Google Scholar]
- 34.Wooden SK, Li LJ, Navarro D, Qadri I, Pereira L, Lee AS. Mol Cell Biol. 1991;11:5612–5623. doi: 10.1128/mcb.11.11.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hosokawa N, Wada I, Hasegawa K, Yorihuzi T, Tremblay LO, Herscovics A, Nagata K. EMBO Rep. 2001;2:415– 422. doi: 10.1093/embo-reports/kve084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshida H, Matsui T, Hosokawa N, Kaufman RJ, Nagata K, Mori K. Dev Cell. 2003;4:265–271. doi: 10.1016/s1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 37.Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akiyama T, Ishida J, Nakagawa S, Ogawara H, Watanabe S, Itoh N, Shibuya M, Fukami Y. J Biol Chem. 1987;262:5592–5595. [PubMed] [Google Scholar]
- 39.Hynes TR, Mervine SM, Yost EA, Sabo JL, Berlot CH. J Biol Chem. 2004;279:44101–44112. doi: 10.1074/jbc.M405151200. [DOI] [PubMed] [Google Scholar]
- 40.Amshoff C, Jack HM, Haas IG. Biol Chem. 1999;380:669– 677. doi: 10.1515/BC.1999.083. [DOI] [PubMed] [Google Scholar]
- 41.Herscovics A. Biochimie (Paris) 2001;83:757–762. doi: 10.1016/s0300-9084(01)01319-0. [DOI] [PubMed] [Google Scholar]
- 42.Zhou BB, Elledge SJ. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 43.Ellgaard L, Molinari M, Helenius A. Science. 1999;286:1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- 44.Zhao G, Zhou X, Wang L, Li G, Kisker C, Lennarz WJ, Schindelin H. J Biol Chem. 2006;281:13751–13761. doi: 10.1074/jbc.M600137200. [DOI] [PubMed] [Google Scholar]
- 45.Lederkremer GZ, Glickman MH. Trends Biochem Sci. 2005;30:297–303. doi: 10.1016/j.tibs.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 46.Fagioli C, Sitia R. J Biol Chem. 2001;276:12885–12892. doi: 10.1074/jbc.M009603200. [DOI] [PubMed] [Google Scholar]
- 47.Sekijima Y, Wiseman RL, Matteson J, Hammarstrom P, Miller SR, Sawkar AR, Balch WE, Kelly JW. Cell. 2005;121:73–85. doi: 10.1016/j.cell.2005.01.018. [DOI] [PubMed] [Google Scholar]