Abstract
Chemokine CXCL12 is widely expressed in the central nervous system (CNS) and essential for the proper functions of human neural progenitor cells (hNPCs). Although CXCL12 is known to function through its receptor CXCR4, recent data have suggested that CXCL12 binds to chemokine receptor CXCR7 with higher affinity than to CXCR4. However, little is known about the function of CXCR7 in hNPCs. Using a primary hNPC culture system, we demonstrated that CXCL12 promotes hNPC survival in the events of camptothecin-induced apoptosis or growth factor deprivation, and that this effect requires both CXCR7 and CXCR4. Through FACS analysis and immunocytochemistry, we determined that CXCR7 is mainly localized in the early endosome, while CXCR4 is more broadly expressed at the cell surface and on both early and recycling endosomes. Furthermore, we found that endocytosis is required for the pro-survival function of CXCL12. Using dual-color Total Internal Reflection Fluorescence microscopy and immunoprecipitation, we demonstrated that CXCR7 quickly trafficks to plasma membrane in mediating CXCL12 endocytosis and colocalizes with CXCR4 after CXCL12 treatment. Investigating the molecular mechanisms, we found that ERK1/2 endocytotic signaling pathway is essential for hNPC survival upon apoptotic challenges. Consistent with these findings, a significantly higher number of apoptotic NPCs were found in the developing brain of CXCR7 knockout mice. In conclusion, CXCL12 protects hNPCs from apoptotic challenges through CXCR7- and CXCR4-mediated endocytotic signaling. Since survival of hNPCs is important for neurogenesis, CXCR7 may become a new therapeutic target to properly regulate critical processes of brain development.
Introduction
Chemokines play crucial roles in the central nervous system (CNS) during development. There are approximately 50 chemokines interacting with over 20 chemokine receptors 1. Among them CXCL12 has been shown to play an important role in cell migration, proliferation and survival 2. Although CXCR4 has long been believed the unique receptor of CXCL12, recent data has suggested that CXCR7 is also a receptor for CXCL12, with 10-fold higher affinity than CXCR4 3. Exactly what role CXCR7 plays toward the function of CXCL12 remains unclear. CXCR7 by itself does not trigger G-protein dependent signaling but can heterodimerize with CXCR4 and regulate CXCL12-mediated G protein signaling 4. In addition, CXCL12 induces the activation of ERK1/2 and AKT through CXCR7 in astrocytes and Schwann cells 5.
Like CXCL12, CXCR7 is also widely expressed in the CNS. During mouse embryonic development, CXCR7 mRNA is first observed at embryonic day 11.5 (E11.5) and increases strongly between E15 and E18 in the marginal zone/layer I. At postnatal day 1 (P1), CXCR7 decreases rapidly and at P7 only scarce signals can be detected 6. In the cortex, CXCR7 is expressed in GABAergic precursors and in reelin-expressing Cajal-Retzius cells. Also CXCR7 is abundant in neural precursors forming the cortical plate 7. This expression pattern suggests that CXCR7 may have an important role in the development of the CNS. CXCR7 regulation of cell migration has been well documented and has been shown to regulate the migration of primordial germ cells in developing zebrafish 8and interneuron in mouse 9, 10. In addition to the migration, CXCR7 has been shown to promote cancer cell survival 11-13, though little is known about how CXCR7 plays an anti-apoptotic role in the CNS.
To determine the function of CXCR7 in NPCs, we used a primary hNPC culture that we initially characterized in 200414. Since then we have extensively documented the survival 15, differentiation 16 and proliferation of NPCs 17. Here, we further investigated the role of CXCR7 in hNPC survival and the molecular mechanism involved. We found that CXCL12 promotes hNPC survival through the coordination between CXCR7 and CXCR4. Furthermore, CXCR7 and CXCR4 are associated with endosome in hNPCs and the survival of hNPCs is dependent on endocytosis of CXCL12 that activates ERK1/2 signaling in endosomes. The revealing of these molecular events may have strong implications to help us better understand hNPC survival during apoptotic challenges.
Results
CXCL12 enhances hNPC survival during camptothecin induced apoptosis or growth factor deprivation
CXCL12 plays crucial roles in the CNS through inducing NPC migration, proliferation, and neuronal axon projection 18. However, little is known about the effect of CXCL12 on NPC survival. To determine the function of CXCL12 on hNPC survival, we pre-treated hNPCs with CXCL12 for 2 hours and then treated with 10 μM camptothecin for an additional 4 hours. Camptothecin is a cytotoxic chemical that causes DNA damage, thus serving as an apoptosis inducer. Camptothecin dramatically increased the number of TUNEL-positive hNPCs compared with untreated control (Fig. 1A, B) and CXCL12 alleviated camptothecin-induced apoptosis in a dose-dependent manner (10-100 ng/ml) (Fig. 1C-F). To confirm the apoptosis of hNPCs, we also determined cleaved Poly ADP ribose polymerase (PARP, an apoptosis indicator) in the cell lysates at similar experimental settings. Camptothecin increased the levels of cleaved PARP. When treated with increasing concentrations (10-100 ng/ml) of CXCL12, the camptothecin-induced PARP cleavage showed a dose-dependent decrease (Fig. 1G, H), which confirmed that CXCL12 alleviates camptothecin-induced apoptosis in hNPCs.
Figure 1.

CXCL12 protects hNPCs against camptothecin-induced apoptosis. hNPCs were pretreated with CXCL12 for 2 hours and then treated with camptothecin (10 μM) for 4 hours. hNPC apoptosis was determined by TUNEL assay (A-F) or PARP cleavage assay (G, H). A-E) Representative pictures from TUNEL assay were shown. F) Quantification data of TUNEL assay was determined as a percentage of TUNEL positive cells against total cell number. G) Cleaved PARP protein levels were determined by Western blotting. H) Levels of cleaved PARP were normalized as a ratio of cleaved PARP to actin after densimetrical quantification of panel G and shown as fold change relative to control. Results are expressed as the mean ± SEM of triplicate samples and are representative of 3 independent experiments. #, P < 0.05, ##, P < 0.01 compared with control; *, P < 0.05, **, P < 0.01 compared with camptothecin-treated group without CXCL12.
To test the extent of the functional effect of CXCL12 on hNPC survival, we performed CXCL12 treatment with an ongoing apoptosis. CXCL12 was added to the culture 1 hour post camptothecin treatment while hNPCs were treated with camptothecin for 4 hours or 8 hours. Adding CXCL12 after camptothecin treatment decreased the number of TUNEL-positive hNPCs in a dose-dependent manner compared with camptothecin treatment alone (Fig. S1A-F). Similarly, adding CXCL12 after camptothecin treatment decreased the levels of cleaved PARP in a dose-dependent manner compared with camptothecin treatment alone (Fig. S1G, H). Together, these results demonstrated the extent of how CXCL12 enhances hNPC survival with apoptotic challenge or ongoing apoptosis.
Next, we tested whether CXCL12 could promote hNPC survival during growth factor deprivation. hNPCs were deprived of growth factors for 48 hours with or without the treatment of CXCL12. CXCL12 decreased the number of apoptotic hNPCs in a dose-dependent manner (Fig S2A-F). Similarly, growth factor deprivation for 48 hours induced PARP cleavage, whereas treatment with increasing concentrations of CXCL12 reduced PARP cleavage in a dose-dependent manner (Fig. S2G, H). Taken together, these data suggest that CXCL12 blocks apoptosis and helps hNPC survival during DNA damage or growth factor deprivation.
Both CXCR7 and CXCR4 are required for the anti-apoptosis function of CXCL12 in hNPCs
To determine the mechanism involved in CXCL12-induced hNPC survival, we investigated the involvement of CXCR7 and CXCR4, receptors for CXCL12 in hNPCs. We utilized siRNA targeting CXCR7 and CXCR4. Both siRNAs effectively silenced their corresponding receptors, as evaluated by real time RT-PCR and Western blotting of CXCR7 (Fig. S3A, C) and CXCR4 (Fig. S3B, D). Three days after siRNA transfection, hNPCs were pre-treated with CXCL12 for 2 hours and then treated with camptothecin for 4 hours. Apoptotic levels of hNPCs were determined by TUNEL assay and PARP cleavage. In TUNEL assay, CXCR7 or CXCR4 silencing blocked the anti-apoptotic effect of CXCL12 (Fig. 2A-J). Similarly, CXCL12 reduced the levels of cleaved PARP in camptothecin-challenged hNPCs in a dose-dependent manner, whereas silencing of CXCR7 or CXCR4 abolished the effect (Fig. 2K-M). These data suggest that both CXCR7 and CXCR4 are required for the anti-apoptosis function of CXCL12 in hNPCs.
Figure 2.
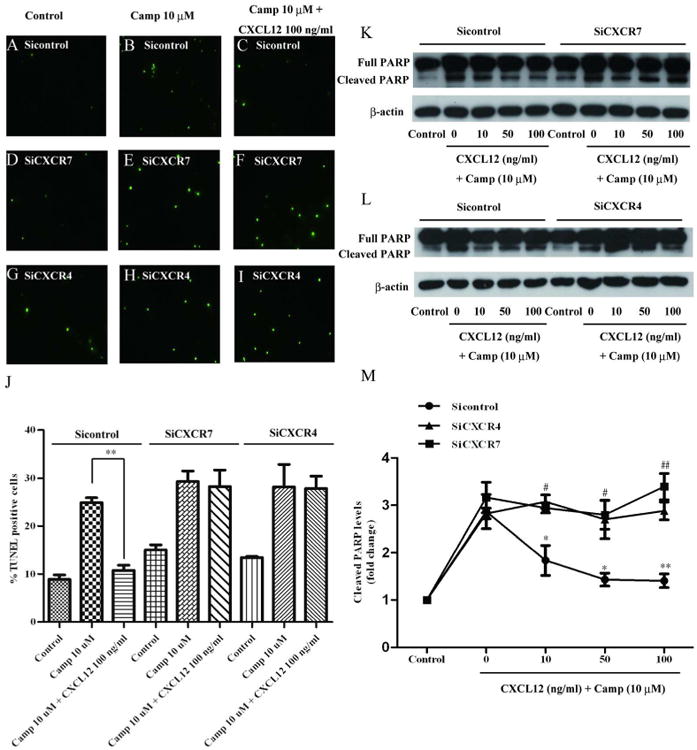
Both CXCR7 and CXCR4 are required for the anti-apoptosis function of CXCL12 in hNPCs. A-I) hNPCs were transfected with CXCR7 (D-F) or CXCR4 (G-I) siRNA and a non-targeting siRNA (A-C) was used as a control. Three days after transfection, hNPCs were pretreated with CXCL12 for 2 hours (C, F, I) and then treated with camptothecin (10 μM, B, C, E, F, H, I) for 4 hours. hNPC apoptosis was determined by TUNEL assay. Representative pictures from TUNEL assay were shown. J) Quantification data of TUNEL assay was determined as a percentage of TUNEL positive cells against total cell number. K, L) hNPC apoptosis was determined by PARP cleavage assay in Western blotting. M) Levels of cleaved PARP were normalized as a ratio of cleaved PARP to actin after densimetrical quantification and shown as fold change relative to control. Results are expressed as the mean ± SEM of triplicate samples from 3 independent experiments. #, P < 0.05, ##, P < 0.01 compared with non-targeting siRNA control with matched concentration of CXCL12 treatment; *, P < 0.05, **, P < 0.01 compared with non-targeting siRNA control with matched concentration of CXCL12 treatment.
CXCR7 and CXCR4 are associated with endosome in hNPCs
To determine the mechanism of how CXCR7 and CXCR4 mediate hNPC survival, we evaluated the expression and subcellular localization of CXCR7 and CXCR4 in hNPCs. Flow cytometry analysis revealed that CXCR7 is mostly expressed in the cytosol with little expression on the cell surface, whereas CXCR4 is strongly expressed both on the cell surface and in the cytosol (Fig. 3A-D). We further labeled hNPCs with antibodies specific to Nestin (green, NPC marker) and either CXCR7 (red) or CXCR4 (red). CXCR7 immunoreactivity was mostly localized to the cytosol, whereas CXCR4 immunoreactivity could be found both on the cell surface and in the cytosol (Fig. 3E, F). To further determine the association between CXCR7, CXCR4, and subcellular organelles, we transfected hNPCs with vector expressing CXCR7-mcherry or CXCR4-EGFP, and stained the cells with EEA1 (early endosome marker), Rab4 (recycling endosome marker), or Rab11 (recycling endosome marker). CXCR7 mostly colocalized with EEA1, with little colocalization with Rab4 or Rab11 (Fig. S4A-C). In contrast, CXCR4 was evenly colocalized with EEA1, Rab4, and Rab11 (Fig. S4D-F). Together, these data suggest that both CXCR4 and CXCR7 are associated with endosome in hNPCs, and CXCR4 has a stronger association with recycling endosome compared with CXCR7.
Figure 3.
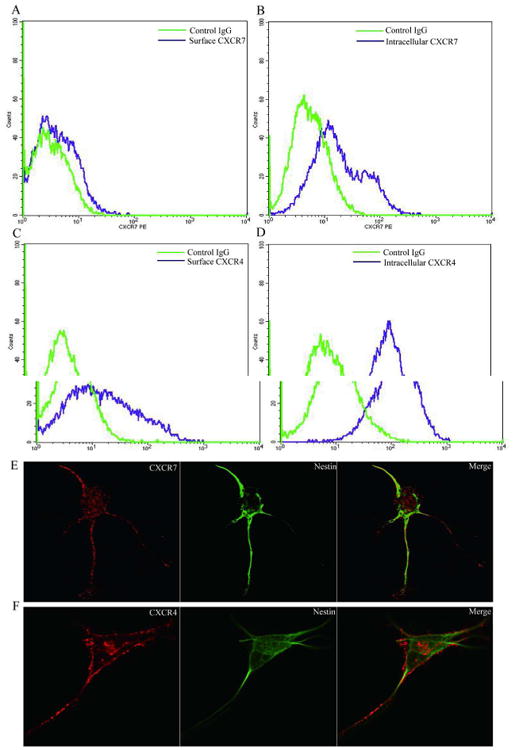
CXCR7 and CXCR4 expression patterns in hNPCs. Using cultured hNPCs, cell surface (A, C) and intracellular (B, D) expression of CXCR7 and CXCR4 was determined by FACS. E, F) Double immunostaining of CXCR7 or CXCR4 (red) with Nestin (green) in hNPCs. Right panels are merged pictures of left and middle panels. Images were acquired from a Zeiss LSM 510 META LASER scanning confocal microscope. Panels are representative of pictures taken from three separate donors.
Endocytosis is necessary for the anti-apoptotic function of CXCL12 in hNPCs
Because little CXCR7 immunoreactivity was found on the cell surface, CXCR7 likely functions through G protein-coupled receptor (GPCR)-independent pathways. Therefore, we tested whether the endocytosis-related signaling pathway is involved in the anti-apoptosis function of CXCR7. We pre-treated hNPCs with MDC (10 μM), an endocytosis inhibitor for 1 hour. MDC completely blocked the endocytosis of fluorescent-labeled CXCL12 (Fig. S5). Treatment of hNPCs with MDC (10 μM) did not significantly change the apoptotic levels of hNPCs (Fig. 4A, D), nor did it change the levels of camptothecin-induced apoptosis (Fig. 4B, E). In contrast, inhibition of endocytosis by MDC abolished the anti-apoptotic effect of CXCL12 on hNPCs (Fig. 4C, F, G). To further test the effect of endocytosis inhibition on the anti-apoptotic effect of CXCL12, we used Western blotting to determine the levels of cleaved PARP. CXCL12 reduced the levels of cleaved PARP in camptothecin-challenged hNPCs in a dose-dependent manner, whereas pretreatment with MDC abolished the effect (Fig. 4H, I). These data suggest that endocytosis is required for the anti-apoptotic function of CXCL12 in hNPCs.
Figure 4.

Endocytosis is necessary for the anti-apoptotic function of CXCL12 in hNPCs. hNPCs were pretreated with the endocytosis inhibitor MDC (10 μM) for 1 hour and then pretreated with CXCL12 for 2 hours, hNPCs were then challenged with camptothecin (10 μM) for 4 hours with or without CXCL12. A-F) TUNEL staining was performed to detect the apoptotic cells. Representative pictures from TUNEL assay were shown. G) Quantification data of TUNEL assay was determined as percentage of TUNEL positive cells against total cell number. H) Cleaved PARP protein levels were determined by Western blotting. I) Levels of cleaved PARP were normalized as a ratio of cleaved PARP to actin after densimetrical quantification of panel H and shown as fold change relative to control. Results are expressed as the mean ± SEM of triplicate samples from 3 independent experiments. *, P < 0.05, compared with camptothecin-treated group without CXCL12.
Different endocytotic properties of CXCR7 and CXCR4 in mediating CXCL12 endocytosis
Next, we investigated the molecular events associated with CXCR7- and CXCR4-mediated CXCL12 endocytosis. To selectively visualize events within 100 nm of the plasma membrane, where CXCR7 and CXCR4 initiates CXCL12 endocytosis, we used the Total Internal Reflection Fluorescence microscope (TIRFm) 19. We transfected hNPCs with CXCR7-mcherry and CXCR4-EGFP, and then examined endocytosis of CXCL12 using a dual-color TIRFm system. Time-lapse images were captured every 3 seconds for 20 minutes. We identified CXCR7-mcherry fluorescent puncta under TIRFm illumination, which suggests that CXCR7-positive vesicles are in proximity to the plasma membrane. In addition, we observed that CXCR7 puncta were much more mobile than CXCR4 puncta (Fig. 5 video 1). The addition of CXCL12 (100 ng/ml) at 2.5 minutes appeared to enhance the movement of CXCR7 to the plasma membrane (Fig. 5A, B; Fig. 5 video 1 and 2). CXCR7 fluorescence intensity of the whole cell measured within ∼100 nm of the membrane by TIRF microscopy increased significantly after CXCL12 treatment (Fig. 5C). In contrast, the fluorescence intensity of CXCR4 did not have a significant change after CXCL12 treatment in the duration of 17.5 minutes under recording (Fig. 5D). These data indicate that CXCR7 may play a more active role in mediating CXCL12 endocytosis, at least at the early moments of endocytosis.
Figure 5.
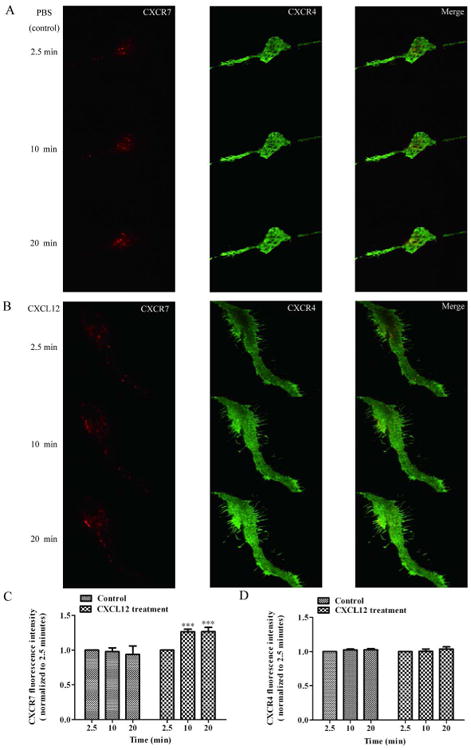
CXCR7 quickly responds to CXCL12 and trafficks to the plasma membrane to mediate CXCL12 endocytosis. hNPCs were transfected with CXCR7-mcherry and CXCR4-EGFP for 24 hours. The trafficking of CXCR7 and CXCR4 were acquired by time-lapse dual-color TIRFm every 3 seconds. PBS (control) or CXCL12 (100 ng/ml) was added to hNPCs after recording for 2.5 minutes and then the recording continued for another 17.5 minutes. A, B) Representative TIRFm images of CXCR7-mcherry (left panels) and CXCR4-EGFP (middle panels) at the time points 2.5 minutes, 10 minutes and 20 minutes with PBS (A) or CXCL12 (B) treatment. Right panels are merged pictures of left and middle panels. C, D) Levels of CXCR7 (C) and CXCR4 (D) in close proximity to the plasma membrane in cells treated with PBS or CXCL12. The results were normalized to the fluorescent intensity at the time point 2.5 minutes. The average normalized intensity of nine cells are expressed as mean ± SEM. ***, P < 0.001 compared with the PBS-treated group at the matched time point.
Increased interaction between CXCR7 and CXCR4 after CXCL12 endocytosis
To further study CXCL12 endocytosis, we utilized Alex647-labeled CXCL12 and used confocal microscopy to monitor the localization of CXCL12 during endocytosis. hNPCs were also co-transfected with CXCR7-mcherry and CXCR4-EGFP to study their association with CXCL12 at different time points of endocytosis. CXCL12 colocalized with CXCR7 as early as 5 minutes, consistent with the TIRFm data. At 2 hours after CXCL12 treatment, colocalization among CXCL12, CXCR7 and CXCR4 was evident, and at 6 hours more CXCL12, CXCR7 and CXCR4 colocalization was found (Fig. S6A). The close colocalization indicates a close association, and possibly protein-protein interaction between CXCR7 and CXCR4. To test whether CXCR7 and CXCR4 have physical interaction, we used immunoprecipitation with anti-CXCR4 antibody followed by Western blotting for CXCR7. Little CXCR7 was detected in the precipitated protein mix of the untreated hNPCs (Fig. S6B). However, after CXCL12 treatment, CXCR7 increased in a time-dependent manner (Fig. S6B), suggesting increased protein-protein interactions between CXCR7 and CXCR4 upon CXCL12 treatment. Furthermore, to test if transfected CXCR7-mcherry or CXCR4-EGFP colocalized with endogenous proteins, we transfected hNPCs with either CXCR7-mcherry or CXCR4-EGFP and used immunocytochemistry to determine the endogenous CXCR7 or CXCR4 levels. Cells transfected with CXCR7-mcherry were immunostained with endogenous CXCR4, and cells transfected with CXCR4-GFP were immunostained with endogenous CXCR7. The confocal imaging showed after CXCL12 treatment, CXCR7-mcherry and CXCR4-EGFP colocalized with endogenous CXCR4 and CXCR7, respectively (Fig. S6C).
CXCL12 mediates hNPC survival through the ERK1/2 signaling pathway
Activation of ERK1/2 by CXCL12 promotes cell survival on Retinal Ganglion cells 20. Furthermore, recent data has suggested that CXCR7 recruits β-arrestin2 and increase ERK1/2 phosphorylation in cytoplasmic vesicles in HEK293 cells 21. To further investigate the molecular mechanisms by which the CXCL12-induced endocytosis promotes cell survival, we tested whether CXCL12 mediates hNPC survival through ERK1/2 signaling pathway. We pretreated hNPCs with ERK1/2 inhibitor PD98059 (20 μM). Inhibition of ERK1/2 significantly reduced the anti-apoptotic effect of CXCL12 on hNPCs (Fig. 6A, B), suggesting a critical role of ERK1/2 in CXCL12-mediated hNPC survival.
Figure 6.
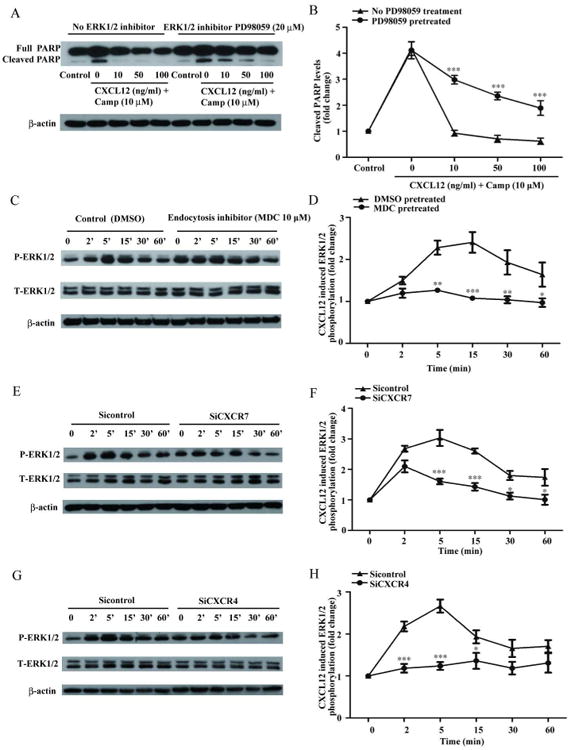
CXCL12 mediates hNPC survival through the ERK1/2 signaling pathway. A) hNPCs were pretreated with ERK1/2 inhibitor PD98059 (20 μM) for 2 hours and then treated with camptothecin (10 μM). Cleaved PARP protein levels were determined by Western blotting. B) Levels of cleaved PARP were normalized as a ratio of cleaved PARP to actin after densimetrical quantification of panel A and shown as fold change relative to control. C) hNPCs were pretreated with MDC or DMSO (control) for 1 hour and CXCL12-mediated ERK1/2 activation were detected from 2 minutes to 60 minutes through Western blotting. D) Levels of phosphorylated ERK were normalized as a ratio to total ERK1/2 after densimetrical quantification of panel C and shown as fold change relative to control. E-H) ERK1/2 activation after CXCR7 (E) or CXCR4 (G) siRNA transfection. Quantification of ERK1/2 phosphorylation in panel E (F) and panel G (H) were shown. All results are expressed as mean ± SEM of triplicate samples and are representative of 3 independent experiments ***, P < 0.001; **, P < 0.01; *, P < 0.05 compared with control.
To determine whether endocytotic signaling is involved in ERK1/2 activation, we pretreated hNPCs with MDC (10 μM) for 1 hour and determined ERK1/2 phosphorylation upon CXCL12 treatment through Western blotting. Quantification of Western blotting suggested that MDC blocks ERK1/2 activation at the time points between 5 and 60 minutes but not at earlier time points (Fig. 6C, D). This result is consistent with a previous report that showed the CXCL12-mediated ERK1/2 activation at early time points is mainly through GPCR signaling and the sustained activation of ERK1/2 at later time points is through endocytotic signaling 22, 23. To further test whether ERK1/2 activation is dependent on CXCR7- or CXCR4-mediated endocytotic signaling, we transfected hNPCs with specific siRNA targeting either CXCR7 or CXCR4, and determined CXCL12-induced ERK1/2 activation through Western blotting. CXCL12 induced ERK1/2 phosphorylation in hNPCs. CXCR7 silencing significantly reduced CXCL12-mediated ERK1/2 phosphorylation at later time points (5 minutes to 60 minutes) (Fig. 6E, F). In contrast, CXCR4 silencing blocked both early (2 minutes) and late (5 minutes to 15 minutes) ERK1/2 activation (Fig 6G, H). These data suggest that sustained ERK1/2 activation by CXCL12 is dependent on both CXCR4 and CXCR7. Furthermore, hNPCs were transfected with CXCR7-mcherry and treated with CXCL12 (100 ng/ml) for 30 minutes. Then we detected the ERK1/2 activation in hNPCs by immunocytochemistry. The confocal imaging showed after treatment with CXCL12 for 30 minutes, the ERK1/2 were activated at CXCR7-positive endosomes (Fig. S7). Taken together, we demonstrated that CXCL12 enhances hNPC survival through CXCR7- and CXCR4-mediated endocytotic signaling and ERK1/2 activation.
Increased apoptosis of NPCs in the developing brain of CXCR7 deficient mice
To further determine the relevance of CXCR7 in NPC survival in vivo, we obtained an established CXCR7 knockout mouse model 24. The migration defects of interneuron in CNS development of CXCR7 knockout mice have been well documented9. However, the survival of NPCs during the developmental stage of CXCR7 knockout mice is unclear. To determine whether there is any defects of NPC survival in the CXCR7 knockout mice, we obtained floating brains sections from E16.5 embryos. Immunohistochemical analysis of Nestin and TUNEL positive cells in the lateral and medial ganglionic eminences revealed a significantly higher percentage of TUNEL positive NPCs in the CXCR7 knockout mice compared with the wild-type control mice (Fig 7). These in vivo data suggest that CXCR7 plays a crucial role in NPC survival during the development of mouse brain.
Figure 7.

Increased apoptosis of NPCs in the developing brain of CXCR7 deficient mice. Floating coronal sections from E16.5 CXCR7 wild-type and knockout mouse brain were stained with Nestin (green), TUNEL (red), and DAPI (blue) (A-H). Representative confocal images of wild-type and CXCR7-/- sections from lateral and medial ganglionic eminences in the developing cerebrum were shown along with high-magnification images of the corresponding small box area (scale bar, 20 μm). (I), Quantification data of TUNEL-positive NPCs in the lateral and medial ganglionic eminences of CXCR7 knockout mice compared to wild-type mice. The results are expressed as mean ± SEM (n=11), ***, P < 0.001 compared with wild-type mice.
Discussion
CXCL12 and its receptor CXCR4 are constitutively expressed during the development of the central nervous system (CNS) and play important roles in establishing and maintaining CNS homeostasis2. Recent data has suggested that CXCL12 binds to chemokine receptor CXCR7 with higher affinity than to CXCR4. However, little is known about the function of CXCR7 in hNPCs. Here we demonstrated that CXCL12 promotes hNPC survival during chemical-induced DNA damage or growth factor deprivation (Figs. 1, S1, S2). More importantly, CXCR7 and CXCR4, two cognate receptors for CXCL12, are both required for the anti-apoptotic function of CXCL12, because siRNA silencing of either receptor abrogated the protective effect of CXCL12 on hNPCs against chemical-induced DNA damage (Figs. 2, S3). Interestingly, CXCR7 mediates NPC survival through interaction with CXCR4 (Fig. S6) and an endocytotic signaling that involves ERK1/2 activation (Figs. 4, 6, S4, S5, and S7). The relevance of CXCR7 to NPC survival is further demonstrated by in vivo data showing defects of NPC survival in the developing brain of CXCR7 knockout mice (Fig. 7). To our knowledge, this is the first report demonstrating that CXCR7 is required for hNPC survival.
Using a hypoxia model, Bakondi et al demonstrated that CXCL12 promotes NPC survival through CXCR7 25. In light of the study, we have expanded the apoptotic stimuli to chemical-induced DNA damage and growth factor deprivation and found that CXCL12 also promotes the survival of hNPCs (Figs. 1, S1, S2). Furthermore, we have found that CXCR7 and CXCR4 are both required for the anti-apoptotic function of CXCL12. These results are consistent with the original report 25 in that both studies have found that CXCR7 is required for NPC survival. However, the results on CXCR4 indicate that the CXCR4 functionality is more variable in different NPC models. The difference in the requirement of CXCR4 in NPC survival may be due to the specificity of the siRNA versus inhibitors used in the studies, different apoptotic challenges, or there is a species-specific effect for CXCR4.
CXCR7 is highly conserved between mammalian species and widely expressed in a variety of cell types 26. Interestingly, the expression pattern of CXCR7 varies among different cell types. In MCF-7 cells, CXCR7 is expressed both on the cell surface and in the cytosol 27, but in human T lymphocytes, there is little CXCR7 expression on the cell surface and most expression localized to early endosome 28. In the CNS, high levels of CXCR7 expression levels have been found on the cell surface of astrocytes 5. On the contrary, CXCR7 is mostly expressed in recycling vesicles (Rab4-positive cells), but not on the cell surface in neurons 10. Our study finds that CXCR7 expression is unique in hNPCs in that CXCR7 is mostly expressed in cytosol whereas CXCR4 is expressed both on the cell surface and in cytosol (Fig. 3). Furthermore, the subcellular location of CXCR7 is different from CXCR4. CXCR7 localized in early endosome (EEA1 positive) with little expression in recycling vesicles (Rab11 and Rab4 positive), whereas CXCR4 is expressed in early endosome as well as recycling vesicles (Fig. S4). These unique expression patterns of CXCR7 and CXCR4 in hNPCs suggest that they may have similar downstream signaling in early endosome but CXCR4 may be more prone to traffick to the plasma membrane through the recycling vesicles.
The expression patterns of CXCR7 and CXCR4 in early endosome also indicate their role in CXCL12 endocytosis. Indeed we have determined that endocytosis is necessary for the anti-apoptotic function of CXCL12 in hNPCs (Fig. 4). We visualized the molecular events associated with CXCR7- and CXCR4-mediated CXCL12 endocytosis using TIRFm and confocal microscopy and found that CXCR7 fluorescent puncta moved more quickly and trafficked faster to the cell membrane in response to CXCL12 treatment than CXCR4 puncta. In contrast, CXCR4 stimulates CXCL12 endocytosis in a slower manner, at least in the first 17.5 minutes after CXCL12 treatment there is no significant decrease of surface CXCR4 receptors (Figs. 5, video 1 and 2). In contrast, the confocal microscopy (Fig. S6) indicates that the endocytosis of CXCL12 by CXCR4 does occur, likely in a delayed manner after CXCL12 treatment. These data suggest that the CXCR7 receptor have a more rapid dynamic of internalization, which are consistent with previous studies in CXCR7 mutant mice suggesting that interneurons uptake CXCL12 through CXCR7 and prevent the rapid desensitization of CXCR4 receptors9, 10. Together, these data illustrate a CXCR7- and CXCR4-mediated endocytotic signaling pathway that guards hNPCs against apoptosis.
Our data suggested that CXCL12 protects hNPCs from apoptosis and both CXCR7 and CXCR4 are required for the anti-apoptotic functions of CXCL12. CXCR4 is known to regulate survival of CNS cell types including glioma cells, neural and oligodendrocyte progenitors 29, 30. Recent evidence indicates that the receptor CXCR7 is also involved in survival signaling 31, 32. In line with these reports, we found that CXCR7 deletion has a detrimental effect on NPCs, as there was significantly higher number of apoptotic NPCs in the developing brains of CXCR7 deficient mice compared to those of wild-type mice (Fig. 7). CXCR4 enhances survival through GPCR, However, whether CXCR7 function through GPCR is more debatable. Recent evidence has suggested that CXCR7 and CXCR4 are able to form CXCR7/CXCR4 heterodimers33, which is likely to boost the signal transduction of each other. Through confocal live imaging, we show the colocalization of CXCR7 and CXCR4 intracellular after CXCL12 endocytosis (Fig. S6A). However, the exact functions of CXCL12 endocytosis as well as CXCR7/CXCR4 heterodimerization remain to be determined.
Previous studies suggested that ERK is an effector in the endocytotic signaling pathways and treatment with endocytosis inhibitor blocks ERK activation 34. Our study has extended the endocytotic signaling pathways and ERK1/2 activation to the anti-apoptotic function of CXCL12 through ERK1/2. It is unclear whether CXCL12 activated ERK phosphorylation is through a classical GPCR pathway or an endocytotic signaling pathway. Previous study suggests that GPCR activation that leads to ERK phosphorylation occurs in the early stages of endocytosis signaling, mostly in the first 2 minutes, while ERK activation mediated by β-arresin2 takes place in a delayed manner, from 5 minutes onward 23. In our study, CXCL12 induces sustained activation of ERK1/2, which is dependent on CXCR7 or CXCR4 based on the time point indicated. Silencing of CXCR7 may inhibit endocytotic ERK activation while silencing CXCR4 may inhibit both GPCR-dependent ERK activation and endocytotic ERK activation. This is in agreement with the confocal microscopy that showed ERK activation colocalized with CXCR7 positive vesicles after CXCL12 treatment (Fig. S7). Interestingly, after CXCL12 endocytosis, colocalization between CXCR7 and CXCR4 increased in the endosome, suggesting that CXCR7 and CXCR4 may act in concert in mediating downstream endocytotic signaling. The physical interactions between CXCR7 and CXCR4 were also confirmed by immunoprecipitation. However, the exact mechanism how CXCR4 interacts with CXCR7 that triggers endocytotic signaling remains unclear. Biologically, the endocytosis signaling pathways are slower than the G-protein pathways 35, but more constant and stable. These characteristics of endocytosis may be more beneficial for cells in response to apoptotic challenges.
In summary, our studies demonstrate that CXCL12 protects hNPCs from apoptotic challenges, such as camptothecin or growth factor deprivation. The anti-apoptotic function of CXCL12 is dependent on CXCR7- and CXCR4-mediated endocytosis, ERK1/2 activation, and may involve interaction between CXCR7 and CXCR4. These molecular events may be used to develop potential therapeutic targets for promoting hNPC survival during apoptotic challenges.
Materials and methods
Reagents
Antibodies for Western blotting: CXCR7, CXCR4, cleaved PARP, phospho-ERK1/2, total ERK1/2, and β-actin protein levels were detected using Anti-CXCR7 (Abcam 1:1000), anti-CXCR4 (Abcam 1:1000), anti-PARP, anti-phospho-ERK1/2, anti-total-ERK1/2 (Cell Signaling Technology, 1:1000), and anti-β-actin (Sigma-Aldrich, 1:100000), respectively. Recombinant human CXCL12 was obtained from R&D Systems. CXCL12-Alex647 was purchased from ALMAC Sciences.
Cell culture
Human NPCs were isolated and cultured from human brain tissue. Briefly, hNPCs were cultured in 75 mm flasks and grown in suspension in neurosphere initiation medium (NPIM), which consisted of X-Vivo 15 (LONZA) with N2 supplement (Invitrogen), neural cell survival factor-1 (BioWhittaker), bFGF (20 ng/mL, Sigma-Aldrich), epidermal growth factor (EGF, 20 ng/mL, Sigma-Aldrich), and leukemia inhibitory factor (10 ng/mL, Millipore Bioscience Research Reagents). All studies utilizing human subjects were performed in full compliance with the University of Nebraska Medical Center and National Institutes of Health's ethical guidelines.
Animal
The CXCR7 knockout mice were kindly provided by Dr. Liang Zhou (Northwestern, Chicago IL) 24. All mice were housed and bred at University of Nebraska Medical Center, in accordance with ethical guidelines for care and use of laboratory animals set forth by the National Institutes of Health. All procedures were approved by the Institutional Animal Care and Use Committee of the University of Nebraska Medical Center.
siRNA silencing of CXCR7 and CXCR4
siRNA silencing in hNPCs was performed as previously described 36. Briefly, pre-designed siRNA duplex targeted against CXCR7 and CXCR4 was purchased from Applied Biosystems Inc. hNPCs were transfected with 100 nM siRNA duplex for 72 hours in the presence of siIMPORTER (Upstate Cell Signaling Solutions) according to the manufacturer's instructions. Non-specific control siRNA from Applied Biosystems Inc. were also transfected at the same concentration as controls to CXCR7 and CXCR4 siRNA.
RNA extraction and TaqMan real-time RT-PCR
Total RNA was isolated with TRIzol Reagent (Invitrogen) and RNeasy Kit according to the manufacture's protocol (Qiagen). Primers used for real-time reverse-transcription polymerase chain reaction (real-time RT-PCR) include CXCR7 and CXCR4 and GAPDH from Applied Biosystems Inc. Real-time RT-PCR was carried out using the one-step quantitative TaqMan assay in a StepOne™ Real-Time PCR system (Applied Biosystems Inc.). Relative CXCR7 and CXCR4 mRNA levels were determined and standardized with a GAPDH internal control using comparative ΔΔCT method. All primers used in the study were tested for amplification efficiencies and the results were similar.
Immunoprecipitation (IP) and Western blotting
Cell lysates were collected using IP buffer and precleared using protein G (Thermo Scientific). Then, precleared cell lysates were incubated with monoclonal antibody of CXCR4 (R&D Systems) overnight on a rotator. Rabbit antibody of CXCR7 (Abcam) was used to detect the CXCR7-CXCR4 interaction using Western blotting.
Immunocytochemistry and confocal microscopy
For immunofluorescence staining, hNPCs were fixed using 4% Paraformaldehyde (PFA), and permeabilized with 0.5% triton-X and 3% BSA in PBS. After washing, hNPCs were incubated with primary antibodies overnight. Antibodies used for immunostaining include monoclonal anti-CXCR7 (R&D Systems, 1:100), anti-rab11 (BD Biosciences, 1:100), anti-rab4 (BD Biosciences, 1:100); anti-EEA1 (BD Biosciences, 1:150). Cultures were washed and then secondary antibodies, anti-mouse IgG (coupled with Alexa Flour 488, Invitrogen) or anti-rabbit IgG (coupled with Alexa Fluor 568, Invitrogen), were added for 1 hour at room temperature. Nuclear DNA was labeled with DAPI (Invitrogen) for 10 minutes after the secondary antibody at room temperature. Cover slips were mounted on glass slides with mounting medium (Sigma-Aldrich). Triple immunostaining was examined by a Zeiss META 510 confocal microscope (Carl Zeiss MicroImaging, LLC). For CXCR7 and CXCR4 trafficking experiments, full-length human CXCR7 and CXCR4 were subcloned into pmcherry-N1 (Clontech) and pEGFP-N1 (Clontech), respectively. hNPCs were seeded into 35-mm glass-bottom dishes (MatTek) and transfected with CXCR7-mcherry and CXCR4-EGFP using Lipofectamine LTX reagent (Invitrogen). Twenty-four hours after transfection, hNPCs were serum-starved overnight and then treated with CXCL12-Alex647 (100 ng/ml). The colocalization of CXCR7, CXCR4, and CXCL12 was detected using a Zeiss Meta 510 confocal microscope (Carl Zeiss MicroImaging, LLC).
Flow cytometry assay
hNPCs were cultured in 6 well plates at the density of 0.5 million/well. Cells were resuspended in 3% FBS-PBS and the cell surface expression of CXCR7 and CXCR4 were detected using PE-conjugated CXCR7 monoclonal antibody and APC-conjugated CXCR4 monoclonal antibody from R&D, respectively. A matching mouse IgG isotype control was used. For intracellular expression of CXCR7 and CXCR4, cells were permeabilized with a permeabilization buffer before antibody incubation. Data was acquired on a FACSCalibur flowcytometer (BD Biosciences) using CellQuest Software (BD Biosciences). At least 10,000 cells were analyzed per sample.
In vitro TUNEL assay
hNPCs were seeded on poly-D-lysine coated coverslips at the density of 0.1million/well. After treatment, cells were fixed and permeablized with 0.5% triton-X, and the apoptotic cells were determined by in situ cell death detection kit with Fluorescein (Roche) according to the manufacture's protocol. Images were taken using a Nikon Eclipse E800 fluorescent microscope. All obtained images were imported into Image-ProPlus, version 7.0 (Media Cybernetics) for quantifying number of apoptotic cells. The assessors were blinded during image acquisition or quantification.
Free-floating immunohistochemistry and image analysis
The brains from E16.5 wild-type or CXCR7 knockout embryos were rapidly removed and immersed in freshly depolymerized 4% paraformaldehyde for 48 h and then cryoprotected by 30% sucrose for 48 hours before sectioning. Fixed, cryoprotected brains were frozen and sectioned in the horizontal plane at 30 μm using a Cryostat (Leica Microsystems Inc., Bannockburn, IL), with sections collected serially in PBS. Antibody to Nestin was used for the detection of NPCs. Apoptotic cells were determined by in situ cell death detection kit with TMR red (Roche) according to the manufacture's protocol. Images were taken using a Zeiss Meta 710 confocal microscope (Carl Zeiss MicroImaging, LLC) (20× object, tile scan 2×2 mode). Eleven brain section images from three litters of CXCR7 wild-type or knockout mice were imported into Image-ProPlus, version 7.0 (Media Cybernetics, Sliver Spring, MD) for quantifying levels of TUNEL/Nestin double positive staining. Apoptotic NPCs in the lateral and medial ganglionic eminences in the developing cerebrum were analyzed.
Dual-color TIRFm
An objective-type TIRF microscope was used to observe CXCR7-and CXCR4-mediated CXCL12 endocytosis. The setup consisted of two solid state lasers (488 nm and 561 nm), a high-numerical-aperture (NA) oil immersion objective (60×, 1.45 NA), an inverted microscope (IX71; Olympus, Tokyo, Japan), an EMCCD (Hamamatsu, Japan) and temperature controller (Live Cell Instrument, Korea). Images were acquired every 3 seconds, CXCL12 (100 ng/ml) was added to hNPCs after recording for 2.5 minutes and then the recording continued for another 17.5 minutes. All images and movies were acquired and analyzed using Metamorph software (Universal Imaging).
Statistical analysis
Data were analyzed by one-way ANOVA followed by the Tukey's test for pairwise comparisons or by two-way ANOVA followed by post-tests using GraphPad Prism software. Significance was considered with a p value less than 0.05. All experiments were performed with at least three donors to account for any donor specific differences. Assays were performed at least three times in triplicate or quadruplicate.
Supplementary Material
Figure S1. CXCL12 protects hNPCs against ongoing camptothecin-induced apoptosis. hNPCs were treated with camptothecin (10 μM) for 1 hour and then CXCL12 (50 ng/ml) for 4 or 8 hours. Cultures were fixed or lysed and hNPC apoptosis was determined by TUNEL (A-F) or PARP cleavage assay (G, H). A-E) Representative pictures from TUNEL assay were shown. F) Quantification of TUNEL assay was determined as percentage of TUNEL positive cells against total cell number. Results are expressed as the mean ± SEM and are representative of 3 independent experiments using 3 different donors. *, P < 0.05, compared with camptothecin-treated group without CXCL12. G) Cleaved PARP protein levels were determined by Western blotting. H) Levels of cleaved PARP were normalized as a ratio of cleaved PARP to actin after densimetrical quantification of panel G and shown as fold change relative to control. Representative data were shown from 3 independent experiments using 3 different donors.
Figure S2. CXCL12 protects hNPCs from apoptosis induced by growth factor deprivation. A) hNPCs were deprived of growth factors for 48 hours with or without the treatment of CXCL12. Cultures were fixed or lysed and hNPC apoptosis was determined by TUNEL (A-F) or PARP cleavage assay (G, H). A-E) Representative pictures from TUNEL assay were shown. F) Quantification of TUNEL assay was determined as a percentage of TUNEL positive cells against total cell number. G) Cleaved PARP protein levels were determined by Western blotting. Actin was used as loading control. H) Levels of cleaved PARP were normalized as a ratio of cleaved PARP to actin after densimetrical quantification of panel G and shown as fold change relative to control. All results are expressed as mean ± SEM of triplicate samples and are representative of 3 independent experiments. ##, P < 0.01 compared with control; *, P < 0.05, **, P < 0.01, ***, P < 0.001 compared with growth factor deprivation group without CXCL12.
Figure S3. SiRNA silencing efficiency for CXCR7 and CXCR4. A, B) Forty eight hours after transfection, RNA was collected and expression of CXCR7 (A) and CXCR4 (B) were analyzed using Real-time RT PCR. Data were normalized to GAPDH and presented as fold change compared to control. C, D) Seventy-two hours after transfection, protein levels of CXCR7 (C) or CXCR4 (D) were determined by Western blotting.
Figure S4. Endosomal localization pattern of CXCR7 and CXCR4 in hNPCs. Co-localization of CXCR7 (A-C) and CXCR4 (D-F) with endosomal markers were determined by confocal microscopy. hNPCs were transfected with CXCR7-mcherry and CXCR4-EGFP for 24 hours, then cells were fixed and immunostained with monoclonal antibody against EEA1 (early endosome marker, middle panels A, D), Rab4 (recycling marker, middle panels B, E) or Rab11 (recycling marker, middle panels C, F). Right panels are merged pictures of left and middle panels. Images were acquired from a Zeiss LSM 510 META LASER scanning confocal microscope. Panels are representative of pictures taken from three separate donors.
Figure S5. MDC blocks endocytosis of fluorescent-labeled CXCL12. hNPCs were pretreated with DMSO, vehicle control (A) or MDC (B) for 1 hour and then were treated with CXCL12-Alex647 for 2 hours. A) Endocytosis of CXCL12 without MDC treatment. B) Lack of CXCL12 endocytosis after MDC treatment.
Figure S6. CXCL12 induces CXCR4 and CXCR7 interaction after endocytosis. A) hNPCs were transfected with CXCR7-mcherry and CXCR4-EGFP. 24 hours later, cells were treated with CXCL12-ALEX647 (100 ng/ml) and triple-color confocal images were acquired in real time. Right panels are merged pictures of left and middle panels. B) hNPCs were treated with CXCL12 (100 ng/ml) for 2 and 6 hours. Cell lysates were collected and immunoprecipitated (IP) with monoclonal-antibody against CXCR4. CXCR7 protein levels in the lysates were determined by immunoblotting with rabbit-anti-CXCR7 antibody. Mouse IgG was used as control for immunoprecipitation. Immunoblotting of cell extracts before immunoprecipitation was used as control (Input). C) hNPCs were transfected with either CXCR7-mcherry or CXCR4-EGFP. 48 hours later, cells transfected with CXCR7-mcherry were immunostained with CXCR4, and cells transfected with CXCR4-GFP were immunostained with CXCR7. Images were acquired from a Zeiss LSM 510 META LASER scanning confocal microscope. Panels are representative of pictures taken from three separate donors.
Figure S7. CXCL12 induced ERK1/2 activation in CXCR7-positive endosome. hNPCs were transfected with plasmid expressing CXCR7-mcherry and then treated with CXCL12 for 30 minutes. ERK1/2 phosphorylation and CXCR7 were visualized by immunocytochemistry. A) Phosphorylated ERK1/2 in CXCR7-positive endosome without CXCL12 treatment. B) Phosphorylated ERK1/2 in CXCR7-positive endosome with CXCL12 treatment.
Figure 5 video 1. CXCR7 and CXCR4 endocytotic trafficking with PBS treatment in TIRFm.
Figure 5 video 2. CXCR7 and CXCR4 endocytotic trafficking with CXCL12 treatment in TIRFm.
Acknowledgments
We thank Dr. Changhai Tian, Dr. Xiqian Lan, Lijun Sun and Hamilton Vernon, who provided technical support for this work. We thank Dr. Liang Zhou at Northwestern University who provided the CXCR7 knockout mice. We thank Janice A. Taylor and James R. Talaska of the Confocal Laser Scanning Microscope Core Facility at the University of Nebraska Medical Center for providing assistance with confocal microscopy and the Nebraska Research Initiative and the Eppley Cancer Center for their support of the Core Facility. This work was supported in part by research grants by the National Institutes of Health: R01 NS 41858-01, R01 NS 061642-01, 3R01NS61642-2S1, R21 MH 083525-01, P01 NS043985, and P20 RR15635-01 (JZ) and National Natural Science Foundation of China (NSFC) # 81028007.
Footnotes
Author Contribution: Bing Zhu: Conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing
Dongsheng Xu: Conception and design, collection and/or assembly of data, data analysis and interpretation,
Xiaobei Deng: Collection and/or assembly of data, data analysis and interpretation,
Qiang Chen: Collection and/or assembly of data, Data analysis and interpretation
Yunlong Huang: Data analysis and interpretation, manuscript writing
Hui Peng: Collection and/or assembly of data, data analysis and interpretation
Yuju Li: Collection and/or assembly of data
Beibei Jia: Collection and/or assembly of data
Wallace B. Thoreson: Provision of study material, data analysis and interpretation
Wenjun Ding: Data analysis and interpretation
Jianqing Ding: Provision of study material, data analysis and interpretation
Lixia Zhao: Collection and/or assembly of data
Yi Wang: Collection and/or assembly of data
Kristin Leland Wavrin: Collection and/or assembly of data
Shumin Duan: Conception and design, financial support, data analysis and interpretation, manuscript writing, final approval of manuscript
Jialin Zheng: Conception and design, financial support, data analysis and interpretation, manuscript writing, final approval of manuscript
References
- 1.Tiveron MC, Cremer H. CXCL12/CXCR4 signalling in neuronal cell migration. Curr Opin Neurobiol. 2008;18:237–244. doi: 10.1016/j.conb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Li M, Ransohoff RM. Multiple roles of chemokine CXCL12 in the central nervous system: a migration from immunology to neurobiology. Prog Neurobiol. 2008;84:116–131. doi: 10.1016/j.pneurobio.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burns JM, Summers BC, Wang Y, et al. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J Exp Med. 2006;203:2201–2213. doi: 10.1084/jem.20052144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levoye A, Balabanian K, Baleux F, et al. CXCR7 heterodimerizes with CXCR4 and regulates CXCL12-mediated G protein signaling. Blood. 2009;113:6085–6093. doi: 10.1182/blood-2008-12-196618. [DOI] [PubMed] [Google Scholar]
- 5.Odemis V, Boosmann K, Heinen A, et al. CXCR7 is an active component of SDF-1 signalling in astrocytes and Schwann cells. J Cell Sci. 2010;123:1081–1088. doi: 10.1242/jcs.062810. [DOI] [PubMed] [Google Scholar]
- 6.Tiveron MC, Boutin C, Daou P, et al. Expression and function of CXCR7 in the mouse forebrain. J Neuroimmunol. 2010 doi: 10.1016/j.jneuroim.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 7.Schonemeier B, Kolodziej A, Schulz S, et al. Regional and cellular localization of the CXCl12/SDF-1 chemokine receptor CXCR7 in the developing and adult rat brain. J Comp Neurol. 2008;510:207–220. doi: 10.1002/cne.21780. [DOI] [PubMed] [Google Scholar]
- 8.Sasado T, Yasuoka A, Abe K, et al. Distinct contributions of CXCR4b and CXCR7/RDC1 receptor systems in regulation of PGC migration revealed by medaka mutants kazura and yanagi. Dev Biol. 2008;320:328–339. doi: 10.1016/j.ydbio.2008.05.544. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Li G, Stanco A, et al. CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron. 2011;69:61–76. doi: 10.1016/j.neuron.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez-Alcaniz JA, Haege S, Mueller W, et al. Cxcr7 controls neuronal migration by regulating chemokine responsiveness. Neuron. 2011;69:77–90. doi: 10.1016/j.neuron.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 11.Yao X, Zhou L, Han S, et al. High expression of CXCR4 and CXCR7 predicts poor survival in gallbladder cancer. J Int Med Res. 2011;39:1253–1264. doi: 10.1177/147323001103900413. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Shiozawa Y, Wang Y, et al. The role of CXCR7/RDC1 as a chemokine receptor for CXCL12/SDF-1 in prostate cancer. J Biol Chem. 2008;283:4283–4294. doi: 10.1074/jbc.M707465200. [DOI] [PubMed] [Google Scholar]
- 13.Maksym RB, Tarnowski M, Grymula K, et al. The role of stromal-derived factor-1--CXCR7 axis in development and cancer. Eur J Pharmacol. 2009;625:31–40. doi: 10.1016/j.ejphar.2009.04.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng H, Huang Y, Rose J, et al. Stromal cell-derived factor 1-mediated CXCR4 signaling in rat and human cortical neural progenitor cells. J Neurosci Res. 2004;76:35–50. doi: 10.1002/jnr.20045. [DOI] [PubMed] [Google Scholar]
- 15.Peng H, Huang Y, Duan Z, et al. Cellular IAP1 regulates TRAIL-induced apoptosis in human fetal cortical neural progenitor cells. J Neurosci Res. 2005;82:295–305. doi: 10.1002/jnr.20629. [DOI] [PubMed] [Google Scholar]
- 16.Peng H, Kolb R, Kennedy JE, et al. Differential expression of CXCL12 and CXCR4 during human fetal neural progenitor cell differentiation. J Neuroimmune Pharmacol. 2007;2:251–258. doi: 10.1007/s11481-007-9081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, Peng H, Cui M, et al. CXCL12 increases human neural progenitor cell proliferation through Akt-1/FOXO3a signaling pathway. J Neurochem. 2009;109:1157–1167. doi: 10.1111/j.1471-4159.2009.06043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carbajal KS, Schaumburg C, Strieter R, et al. Migration of engrafted neural stem cells is mediated by CXCL12 signaling through CXCR4 in a viral model of multiple sclerosis. Proc Natl Acad Sci U S A. 2010;107:11068–11073. doi: 10.1073/pnas.1006375107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keyel PA, Watkins SC, Traub LM. Endocytic adaptor molecules reveal an endosomal population of clathrin by total internal reflection fluorescence microscopy. J Biol Chem. 2004;279:13190–13204. doi: 10.1074/jbc.M312717200. [DOI] [PubMed] [Google Scholar]
- 20.Chalasani SH, Baribaud F, Coughlan CM, et al. The chemokine stromal cell-derived factor-1 promotes the survival of embryonic retinal ganglion cells. J Neurosci. 2003;23:4601–4612. doi: 10.1523/JNEUROSCI.23-11-04601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajagopal S, Kim J, Ahn S, et al. Beta-arrestin- but not G protein-mediated signaling by the “decoy” receptor CXCR7. Proc Natl Acad Sci U S A. 2010;107:628–632. doi: 10.1073/pnas.0912852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shenoy SK, Drake MT, Nelson CD, et al. beta-arrestin-dependent, G protein-independent ERK1/2 activation by the beta2 adrenergic receptor. J Biol Chem. 2006;281:1261–1273. doi: 10.1074/jbc.M506576200. [DOI] [PubMed] [Google Scholar]
- 23.Kim J, Ahn S, Rajagopal K, et al. Independent beta-arrestin2 and Gq/protein kinase Czeta pathways for ERK stimulated by angiotensin type 1A receptors in vascular smooth muscle cells converge on transactivation of the epidermal growth factor receptor. J Biol Chem. 2009;284:11953–11962. doi: 10.1074/jbc.M808176200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cruz-Orengo L, Holman DW, Dorsey D, et al. CXCR7 influences leukocyte entry into the CNS parenchyma by controlling abluminal CXCL12 abundance during autoimmunity. J Exp Med. 2011;208:327–339. doi: 10.1084/jem.20102010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bakondi B, Shimada IS, Peterson BM, et al. SDF-1alpha secreted by human CD133-derived multipotent stromal cells promotes neural progenitor cell survival through CXCR7. Stem Cells Dev. 2011;20:1021–1029. doi: 10.1089/scd.2010.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thelen M, Thelen S. CXCR7, CXCR4 and CXCL12: an eccentric trio? J Neuroimmunol. 2008;198:9–13. doi: 10.1016/j.jneuroim.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 27.Luker KE, Gupta M, Steele JM, et al. Imaging ligand-dependent activation of CXCR7. Neoplasia. 2009;11:1022–1035. doi: 10.1593/neo.09724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartmann TN, Grabovsky V, Pasvolsky R, et al. A crosstalk between intracellular CXCR7 and CXCR4 involved in rapid CXCL12-triggered integrin activation but not in chemokine-triggered motility of human T lymphocytes and CD34+ cells. J Leukoc Biol. 2008;84:1130–1140. doi: 10.1189/jlb.0208088. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Y, Larsen PH, Hao C, et al. CXCR4 is a major chemokine receptor on glioma cells and mediates their survival. J Biol Chem. 2002;277:49481–49487. doi: 10.1074/jbc.M206222200. [DOI] [PubMed] [Google Scholar]
- 30.Dziembowska M, Tham TN, Lau P, et al. A role for CXCR4 signaling in survival and migration of neural and oligodendrocyte precursors. Glia. 2005;50:258–269. doi: 10.1002/glia.20170. [DOI] [PubMed] [Google Scholar]
- 31.Calatozzolo C, Canazza A, Pollo B, et al. Expression of the new CXCL12 receptor, CXCR7, in gliomas. Cancer Biol Ther. 2011;11:242–253. doi: 10.4161/cbt.11.2.13951. [DOI] [PubMed] [Google Scholar]
- 32.Hou KL, Hao MG, Bo JJ, et al. CXCR7 in tumorigenesis and progression. Chin J Cancer. 2010;29:456–459. doi: 10.5732/cjc.009.10404. [DOI] [PubMed] [Google Scholar]
- 33.Decaillot FM, Kazmi MA, Lin Y, et al. CXCR7/CXCR4 heterodimer constitutively recruits beta-arrestin to enhance cell migration. J Biol Chem. 2011;286:32188–32197. doi: 10.1074/jbc.M111.277038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pierce KL, Maudsley S, Daaka Y, et al. Role of endocytosis in the activation of the extracellular signal-regulated kinase cascade by sequestering and nonsequestering G protein-coupled receptors. Proc Natl Acad Sci U S A. 2000;97:1489–1494. doi: 10.1073/pnas.97.4.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorkin A, von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol. 2009;10:609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng H, Erdmann N, Whitney N, et al. HIV-1-infected and/or immune activated macrophages regulate astrocyte SDF-1 production through IL-1beta. Glia. 2006;54:619–629. doi: 10.1002/glia.20409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. CXCL12 protects hNPCs against ongoing camptothecin-induced apoptosis. hNPCs were treated with camptothecin (10 μM) for 1 hour and then CXCL12 (50 ng/ml) for 4 or 8 hours. Cultures were fixed or lysed and hNPC apoptosis was determined by TUNEL (A-F) or PARP cleavage assay (G, H). A-E) Representative pictures from TUNEL assay were shown. F) Quantification of TUNEL assay was determined as percentage of TUNEL positive cells against total cell number. Results are expressed as the mean ± SEM and are representative of 3 independent experiments using 3 different donors. *, P < 0.05, compared with camptothecin-treated group without CXCL12. G) Cleaved PARP protein levels were determined by Western blotting. H) Levels of cleaved PARP were normalized as a ratio of cleaved PARP to actin after densimetrical quantification of panel G and shown as fold change relative to control. Representative data were shown from 3 independent experiments using 3 different donors.
Figure S2. CXCL12 protects hNPCs from apoptosis induced by growth factor deprivation. A) hNPCs were deprived of growth factors for 48 hours with or without the treatment of CXCL12. Cultures were fixed or lysed and hNPC apoptosis was determined by TUNEL (A-F) or PARP cleavage assay (G, H). A-E) Representative pictures from TUNEL assay were shown. F) Quantification of TUNEL assay was determined as a percentage of TUNEL positive cells against total cell number. G) Cleaved PARP protein levels were determined by Western blotting. Actin was used as loading control. H) Levels of cleaved PARP were normalized as a ratio of cleaved PARP to actin after densimetrical quantification of panel G and shown as fold change relative to control. All results are expressed as mean ± SEM of triplicate samples and are representative of 3 independent experiments. ##, P < 0.01 compared with control; *, P < 0.05, **, P < 0.01, ***, P < 0.001 compared with growth factor deprivation group without CXCL12.
Figure S3. SiRNA silencing efficiency for CXCR7 and CXCR4. A, B) Forty eight hours after transfection, RNA was collected and expression of CXCR7 (A) and CXCR4 (B) were analyzed using Real-time RT PCR. Data were normalized to GAPDH and presented as fold change compared to control. C, D) Seventy-two hours after transfection, protein levels of CXCR7 (C) or CXCR4 (D) were determined by Western blotting.
Figure S4. Endosomal localization pattern of CXCR7 and CXCR4 in hNPCs. Co-localization of CXCR7 (A-C) and CXCR4 (D-F) with endosomal markers were determined by confocal microscopy. hNPCs were transfected with CXCR7-mcherry and CXCR4-EGFP for 24 hours, then cells were fixed and immunostained with monoclonal antibody against EEA1 (early endosome marker, middle panels A, D), Rab4 (recycling marker, middle panels B, E) or Rab11 (recycling marker, middle panels C, F). Right panels are merged pictures of left and middle panels. Images were acquired from a Zeiss LSM 510 META LASER scanning confocal microscope. Panels are representative of pictures taken from three separate donors.
Figure S5. MDC blocks endocytosis of fluorescent-labeled CXCL12. hNPCs were pretreated with DMSO, vehicle control (A) or MDC (B) for 1 hour and then were treated with CXCL12-Alex647 for 2 hours. A) Endocytosis of CXCL12 without MDC treatment. B) Lack of CXCL12 endocytosis after MDC treatment.
Figure S6. CXCL12 induces CXCR4 and CXCR7 interaction after endocytosis. A) hNPCs were transfected with CXCR7-mcherry and CXCR4-EGFP. 24 hours later, cells were treated with CXCL12-ALEX647 (100 ng/ml) and triple-color confocal images were acquired in real time. Right panels are merged pictures of left and middle panels. B) hNPCs were treated with CXCL12 (100 ng/ml) for 2 and 6 hours. Cell lysates were collected and immunoprecipitated (IP) with monoclonal-antibody against CXCR4. CXCR7 protein levels in the lysates were determined by immunoblotting with rabbit-anti-CXCR7 antibody. Mouse IgG was used as control for immunoprecipitation. Immunoblotting of cell extracts before immunoprecipitation was used as control (Input). C) hNPCs were transfected with either CXCR7-mcherry or CXCR4-EGFP. 48 hours later, cells transfected with CXCR7-mcherry were immunostained with CXCR4, and cells transfected with CXCR4-GFP were immunostained with CXCR7. Images were acquired from a Zeiss LSM 510 META LASER scanning confocal microscope. Panels are representative of pictures taken from three separate donors.
Figure S7. CXCL12 induced ERK1/2 activation in CXCR7-positive endosome. hNPCs were transfected with plasmid expressing CXCR7-mcherry and then treated with CXCL12 for 30 minutes. ERK1/2 phosphorylation and CXCR7 were visualized by immunocytochemistry. A) Phosphorylated ERK1/2 in CXCR7-positive endosome without CXCL12 treatment. B) Phosphorylated ERK1/2 in CXCR7-positive endosome with CXCL12 treatment.
Figure 5 video 1. CXCR7 and CXCR4 endocytotic trafficking with PBS treatment in TIRFm.
Figure 5 video 2. CXCR7 and CXCR4 endocytotic trafficking with CXCL12 treatment in TIRFm.


