Abstract
Idiopathic Basal Ganglia Calcification (IBGC) is characterized by bilateral calcification of the basal ganglia associated with a spectrum of neuropsychiatric and motor syndromes. In this study, we set out to determine the frequency of the recently identified IBGC gene SLC20A2 in 27 IBGC cases from the Mayo Clinic Florida Brain Bank using both Sanger sequencing and Taqman copy number analysis to cover the complete spectrum of possible mutations. We identified SLC20A2 pathogenic mutations in 2 of the 27 cases of IBGC (7%). Sequencing analysis identified a p.S113* nonsense mutation in SLC20A2 in one case. Taqman copy-number analysis of SLC20A2 further revealed a genomic deletion in a second case, which was part of a large previously reported Canadian IBGC family with dystonia. Subsequent whole-genome sequencing in this family revealed a 563,256 bp genomic deletion with precise breakpoints on chromosome 8 affecting multiple genes including SLC20A2 and the known dystonia related gene THAP1. The deletion co-segregated with disease in all family members. The deletion of THAP1 in addition to SLC20A2 in the Canadian IBGC family may contribute to the severe and early-onset dystonia in this family. The identification of a SLC20A2 genomic deletion in a familial form of IBGC demonstrates that reduced SLC20A2 in the absence of mutant protein is sufficient to cause neurodegeneration and that previously reported SLC20A2 mutation frequencies may be underestimated.
Keywords: basal ganglia calcification, dystonia, SLC20A2, THAP1, genomic deletion, mutation
INTRODUCTION
Idiopathic Basal Ganglia Calcification (IBGC), also called Fahr’s disease (MIM 213600), can present clinically with extrapyramidal features such as dystonia, parkinsonism, tremor, ataxia or chorea [1–3]. Affected individuals can have a variety of other speech, cognitive and psychiatric abnormalities. The familial form of IBGC is an autosomal dominantly inherited condition that, different from other heredofamilial brain calcinosis syndromes, is not associated with calcium, phosphorus or parathyroid hormone metabolism abnormalities or other causes of secondary calcification such as infections or brain tumors [1]. In IBGC, brain calcification is uniform and most severely affects the basal ganglia. Brain CT scans of IBGC patients show bilateral and symmetric calcifications of the basal ganglia, sometimes associated with additional calcifications in the cerebellum, the white matter and in rare instances the cerebral cortex. However, despite substantial brain calcification, individuals may remain clinically asymptomatic and in our study of a large IBGC family, no difference in the extent and severity of brain calcifications was observed between affected individuals and those individuals with positive CT scans but without clinical symptoms [4]. This variability in clinical penetrance together with the fact that incidental sporadic calcification in the basal ganglia is not uncommon, long complicated genetic studies and hampered identification of genes for familial IBGC [5, 6].
The first evidence of a genetic cause of familial IBGC came from the findings of Geschwind [7]. In their study, a genome-wide search in a three-generation family identified a locus on chromosome 14q (IBGC1). Subsequent studies showed locus heterogeneity with genetic linkage to chromosome 2q37 (IBGC2) and chromosome 8p21.1-8q11.23 (IBGC3) [8, 9]. Our longitudinal clinical, imaging, neuropathologic and genetic studies of a large Canadian family with autosomal dominant dystonia-plus with cerebral calcifications, which we started in 1998, also highlighted the complexity associated with the genetic study of this disorder with no significant genome-wide linkage in this family [3, 4, 10]. Despite these challenges, candidate gene screening in the IBGC3 locus by Wang and colleagues recently led to the identification of the first IBGC gene, SLC20A2, encoding for type III sodium-dependent phosphate transporter 2 (PiT2) [11]. In 7 families of varied ancestry 7 different mutations (including 5 missense mutations, an in-frame single amino acid deletion, and a frame-shift mutation) were identified [11]. Functional studies in a Xenopus model showed impaired phosphate transport activity of PiT2 suggesting a loss-of-function disease mechanism [11]. Subsequent sequencing studies confirmed an important role for SLC20A2 in the etiology of IBGC, explaining up to 41% of familial cases [12–14] and 14% of sporadic cases[15].
In this study we performed mutation analysis of SLC20A2 in a series of pathologically confirmed cases of IBGC from the Mayo Clinic Jacksonville brain bank. In contrast to earlier SLC20A2 screening reports, we performed both sequencing and genomic-copy number analysis. We identified two SLC20A2 mutations, a novel nonsense mutation in one individual and a large genomic deletion encompassing the complete SLC20A2 gene in a large Canadian IBGC family with dystonia-plus.
PATIENTS AND METHODS
Patients
A total of 27 patients (16 males, 11 females) were selected from the Mayo Clinic Florida (MCF) Brain Bank based on the presence of significant basal ganglia calcification upon pathological examination, in excess of what would be expected due to aging alone. Pathologic assignment was made without bias for clinical presentation with the recognition that IBGC can be asymptomatic. Most cases were sporadic or had an unknown family history, but one individual was part of a Canadian IBGC family with dystonia-plus described previously [4, 10]. All patients were white except one African-American and one American-Asian case. In the majority, basal ganglia calcification was accompanied by primary pathological findings characteristic of other neurodegenerative diseases, in particular Alzheimer’s disease (AD), Lewy body disease (LBD), progressive supranuclear palsy (PSP) or frontotemporal lobar degeneration (FTLD) due to the fact that they were derived from a brain bank for neurodegenerative disorders. The average age at death was 81 years (range 62–94 years). Clinical diagnoses were also variable including AD, frontotemporal dementia (FTD), Parkinson’s disease (PD), PSP and dementia with Lewy bodies (DLB). Two individuals presented with dystonia as the predominant clinical manifestation. Patients provided written consent and the ethics review board at Mayo Clinic approved the study.
Clinical and pathological assessment of mutation carriers
The patient with familial IBGC with dystonia-plus who carried the SLC20A2 deletion was investigated prospectively and examined multiple times during the course of his illness from 1998 to the time of death in 2008. As part of a large Canadian family, he and several family members were investigated by us and others since 1985. Prospective head CT studies were conducted on this man and were previously reported[4]. For the second mutation carrier (p.S113*), a retrospective review of the available medical records was conducted, which included a head CT study. A standardized macroscopic and microscopic neuropathologic evaluation was conducted on all 27 brains that included histologic studies with H&E as well as thioflavin S fluorescent microscopy for assessing Alzheimer type pathology. Immunohistochemistry was routinely used to screen for α-synuclein and TDP-43 pathology and additional stains were used as indicated by pathologic findings. In all cases the presence and extent of basal ganglia calcification was noted, with attention to its association with calcification in periventricular white matter, hippocampal fissure and cerebellar deep nuclei and white matter. When basal ganglia calcification was excessive, which indicated both parenchymal calcospherites as well as small and large vessel calcific vasculopathy, Fahr’s disease was noted in the neuropathologic report and recorded in a relational database (Microsoft Access) used for tracking specimens and linking them to clinical and genetic data.
Molecular genetics and analysis
Genomic DNA was extracted from brain (MCF brain bank cases) or blood (affected and unaffected relatives of Canadian IBGC family) using the relevant Gentra Puregene kit (Qiagen). All coding exons of SLC20A2 (exons 2–11) were amplified by PCR using primers designed to flanking intronic sequence and labeled with one of two unique M13 sequences to facilitate sequencing (primer sequences available upon request). PCR products were purified using AMPure (Agencourt Biosciences) and sequenced in both directions using Big Dye Cycle Sequencing chemistry (Life Technologies). Sequence reactions were purified with CleanSEQ (Agencourt Biosciences) and analyzed on an ABI3730xl Genetic Analyzer. Sequence analysis was performed using Sequencher 4.5 (Gene Codes).
Duplex real-time quantitative PCR assays were performed on an ABI7900 to detect copy number variations of SLC20A2 and THAP1 using TaqMan copy number assays Hs01067215 (SLC20A2, exon 6), Hs06217808 (SLC20A2, 4245bp downstream of the SLC20A2 stopcodon) and Hs03061373 (THAP1, exon 3). For each assay, RNaseP was used as the reference (Life Technologies).
To confirm the presence of a genomic deletion on chromosome 8 and confirm the deletion breakpoint, a PCR based assay was developed. Primers were designed on either side of the deletion breakpoint based on exact deletion coordinates obtained from whole-genome sequencing analysis of one case by Complete Genomics (Mountain View, CA, USA) (F primer chr8: ACTACCTTGCTTATGACATTGAC; R primer chr8: AGCTACAACTGCCTATATGCTAC). Amplification of a 361-bp product indicated the presence of the deletion. A primer set amplifying an arbitrarily selected region on chromosome 5 was included in the PCR assay to confirm DNA quality (F primer chr5: AACTGTTGTTCACTCCAGC; R primer chr5: CCATTCCTGCACTCTCAC)
Whole genome sequencing
Whole genome DNA paired-end sequencing was performed by “unchained combinatorial probe anchor ligation sequencing” as described previously[16]. The genome was sequenced to 169 Gb (gigabase), which is equivalent coverage of 60X. The single nucleotide variation (SNV) calls for the sample with respect to the human reference genome (NCBI Build37) were made as described previously[16]. Detection of structural variation (SV) and copy number variation are described in Complete Genomics Data File Format document (http://www.completegenomics.com/customer-support/documentation/100357139.html).
RESULTS
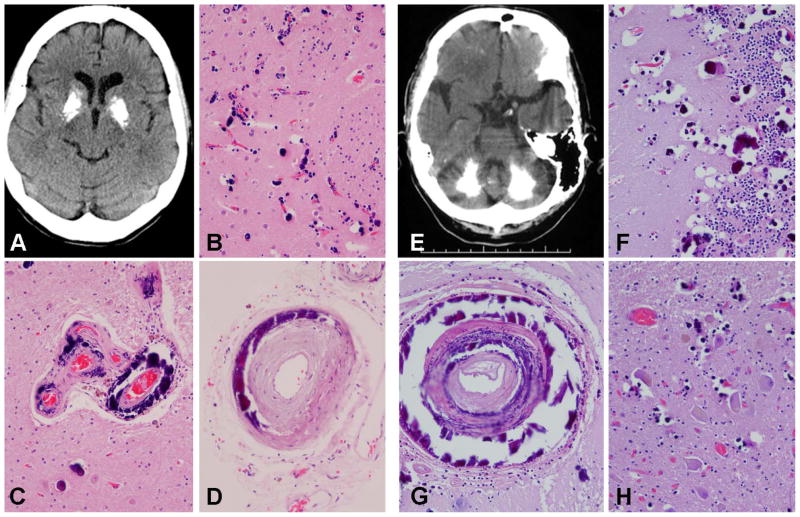
Sequencing analysis of SLC20A2 in 27 unrelated pathologically confirmed IBGC cases led to the identification of one case with a novel coding variant predicted to induce a nonsense mutation, c.338C>G (p.S113*), in exon 3 and one case carrying the known variant c.1422G>T (p.E474D) in exon 8. The latter variant, changing one negatively-charged polar amino acid for another, is not well conserved, is predicted to be tolerated by both SIFT and PolyPhen, and has a low population frequency (1/4,300 European American individuals) in the NHLBI GO Exome Sequencing Project, suggesting it is likely not pathogenic [17]. In contrast, p.S113*, induces a protein truncation similar to several of the previously reported SLC20A2 mutations and is therefore predicted to be pathogenic [11, 12]. The patient with this mutation presented with asymmetric parkinsonism with poor response to Levodopa at the age of 68 years. She also had mild ataxic tremor in both upper extremities and dyskinetic/dystonic facial movements. She was diagnosed clinically as Parkinsonism, possible corticobasal degeneration. Her family history was negative for PD or tremor; however, medical history of her ancestors was not well known. Head CT demonstrated the presence of bilateral basal ganglia calcifications (Figure 1A). She came to autopsy at the age of 73 and on coronal sections of the cerebrum there was gritty, yellow-brown discoloration of the globus pallidus and the internal portion of the putamen, which corresponded to extensive basophilic mineral deposits in small and large vessels as well as many small parenchymal calcospherites (Figure 1B–D). DNA samples of relatives were not available for study.
Figure 1. Novel pathogenic SLC20A2 mutations: clinical and pathological manifestations.
A-D (p.S113*): Brain axial unenhanced computed tomography image of the p.S113* carrier shows marked symmetric calcification of the basal ganglia (A). Histologic studies showed extensive small vessel calcific vasculopathy and calcospherites in the putamen (B); large and small vessel deposits in the globus pallidus (C) and deposits in arteries in subarachnoid space (D). E–H (SLC20A2 genomic deletion): Brain axial unenhanced computed tomography image of pedigree member II-3 shows marked extensive bilateral cerebellar calcifications (E). Histologic studies showed extensive small vessel calcific vasculopathy and many calcospherites in the cerebellar molecular and internal granular cell layer (F); intramural deposits in large vessels in the cerebellar deep white matter (G); and calcific deposits in the deep cerebellar nuclei (H).
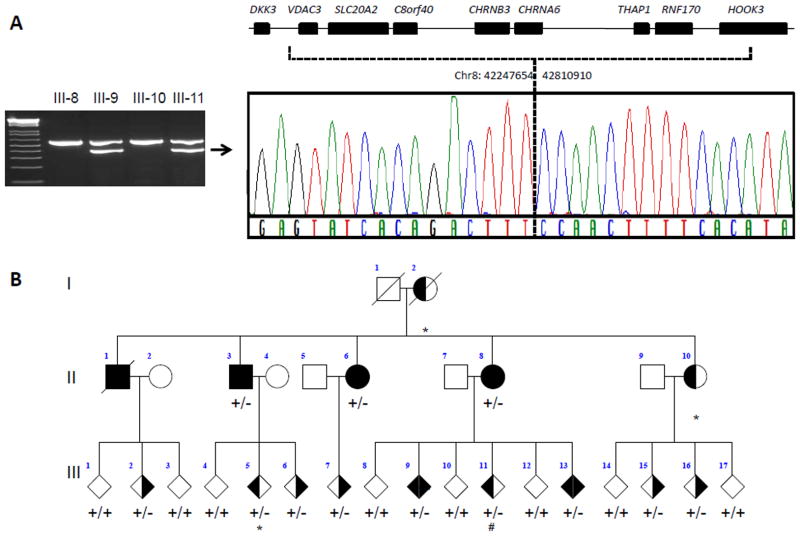
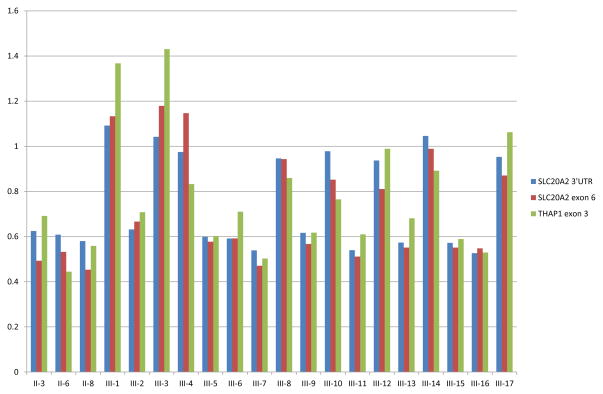
In anticipation of the proposed loss-of-function disease mechanism associated with SLC20A2 mutations [11, 14] we also performed copy-number analysis of SLC20A2. Two Taqman copy-number assays were used, one targeting SLC20A2 exon 6 and one assay targeting a sequence just downstream of the 3′untranslated region (3′UTR) of SLC20A2. In one case, previously found negative for SLC20A2 by sequencing, both assays suggested a decrease in copy number. Interestingly, this was the proband of a Canadian family affected by familial IBGC that had been followed since 1985 whose genetic basis had remained elusive [3, 4, 10, 18] (abbreviated pedigree in Figure 3B). In 2006, we reported a detailed genealogic, clinical, imaging, and neuropathologic study of this family describing 11 patients with clinical symptoms and brain calcinosis, 8 individuals with brain calcinosis without symptoms, and 15 at-risk individuals [4]. Since most patients presented with dystonia with or without additional symptoms such as chorea or postural tremor, the family was defined as having autosomal dominant dystonia-plus with cerebral calcifications. Prospective head CT studies were conducted on this family demonstrating the presence of bilateral basal ganglia and cerebellar calcifications (Figure 1E)[4]. Sections from the cerebellum showed calcific vasculopathy as well as parenchymal calcospherites in the molecular and granular cell layers, as well as in the deep cerebellar nuclei and in large arteries in the deep white matter (Figure F–H). Since the two Taqman probes that we used for SLC20A2 were ~32kb apart, we hypothesized that a large genomic region partially or completely encompassing SLC20A2 and potentially other genes was deleted in this family. From the genes located in the proximity of SLC20A2, THAP1 is particularly important because of its direct association with hereditary dystonia [19]. To explore the extent of the deletion, and to establish if THAP1 was also affected, we performed a third Taqman copy number assay located in exon 3 of THAP1. This assay indicated that the deletion extended into THAP1. Importantly, when we analyzed all 3 probes in DNA from the additional 11 affected and 8 unaffected family members, a clear segregation of the deletion with disease was observed (Figure 2). To precisely identify the deletion breakpoint, we next used whole-genome sequencing and determined that the deleted region of chromosome 8 was located in p11.21, spanning 563,256 bp (42,247,654 – 42,810,910) and including complete deletion of 7 genes (VDAC3, SLC20A2, c8ORF40, CHRNB3, CHRNA6, THAP1 and RNF170) and partial deletion of one other (HOOK3) (Figure 3). To determine whether the deletion breakpoint was identical in all affected family members, we implemented a PCR-based method using primers that align to the non-deleted genomic sequence surrounding the deleted region. No PCR product was detected in any of the individuals without basal ganglia calcification whereas the same 361-bp PCR product was observed in all individuals with basal ganglia calcification (either symptomatic or asymptomatic), further confirming co-segregation of disease with the chromosome 8 deletion in this family (Figure 3).
Figure 3. Segregation of chromosome 8 deletion with disease in IBGC family.
A: Agarose gel image shows duplex PCR products that confirm the presence of the deletion in a representative set of family members. The upper band (557bp) corresponds to a control fragment confirming the quality and presence of the DNA template. The lower band (361bp) is generated using primers located on each side of the deletion breakpoint based on coordinates defined by the whole-genome sequencing analysis. Sequencing analysis of the lower band using primers flanking the deletion breakpoint was performed and confirmed deletion of chr8:42247654–42810910, which includes 7 complete and one partial gene from the chromosome 8p11.21 region. B: Pedigree drawing shows segregation of the genomic deletion on chromosome 8 with disease as determined by the duplex PCR. Square represents male, circle represents female, diamond indicates disguised gender, diagonal line indicates subject is deceased, dystonia phenotype is represented by left side shading, calcifications by right sided shading, subjects with both dystonia and calcifications appear as solid, deletion shown by +/− and wild type by +/+, subject with * denote that no CT data was available. # denotes that this individual showed mild calcification in the basal ganglia which was not considered abnormal at time of CT scan.
Figure 2. SLC20A2 and THAP1 copy-number analyses in Canadian IBGC family.
Copy number analysis of SLC20A2 and THAP1 probes in all family members of the Canadian IBGC family with DNA available. For each individual, their pedigree number (see Figure 3B) appears along the x-axis, copy number along the y-axis, where a value of 1 represents two alleles. Probes correspond to Taqman assays Hs06217808 (SLC20A2 3′UTR), Hs01067215 (SLC20A2 exon 6) and Hs03061373 (THAP1 exon 3).
DISCUSSION
We performed SLC20A2 mutation analysis in a pathologically defined cohort of cases with severe basal ganglia calcification in excess of what would be expected due to aging alone. In two patients (7% of the cohort) SLC20A2 mutations were identified. One patient carried a novel p.S113* nonsense mutation and the other patient was shown to carry a ~563kb genomic deletion on chromosome 8, which included the entire SLC20A2 locus. To define the precise boundaries of the genomic deletion in this patient, we performed whole genome sequencing. This approach allowed us to identify the exact deletion breakpoint as well as the transition sequence of the breakpoint. This allowed design of a PCR assay to confirm the deletion and to perform segregation analysis in the family. This new technology provides considerable advantage over existing methods in precisely detecting the location of the breakpoint and its sequence compared with SNP microarrays or fluorescent in-situ hybridization which would otherwise have been needed to confirm the presence of the deletion. The SLC20A2 mutation frequency observed in our cohort is low compared to the 41% previously reported by Hsu et al.[12] in a large study of 218 subjects from 29 families with IBGC of various ancestries and the study by Chen et al [15], reporting 2 mutations in 14 sporadic IBGC patients. This difference can be explained by the unique nature of our autopsy sample, which was selected based on pathological evidence of excessive basal ganglia calcification often in the setting of another neurodegenerative disease process, and not enriched in cases with a family history of basal ganglia calcification. In fact, family history (of IBGC or any neurodegenerative disease) was negative or unknown in 25 of the 27 cases in our series.
This is the first study to use copy-number analysis in addition to traditional sequencing to determine the full spectrum of mutations in SLC20A2. The combined use of different techniques increased the power of mutational analysis in our study and allowed us to detect a genomic deletion of SLC20A2 in our large Canadian IBGC family. This finding is important and shows that the loss of 50% SLC20A2 expression is sufficient to cause disease in the absence of truncated SLC20A2 protein, which confirms the earlier proposed haploinsufficiency disease mechanism [11]. Our findings also underscore the importance of covering the complete mutational spectrum when performing SLC20A2 mutation screening, and we suggest that genomic copy-number analysis for SLC20A2 should be included in the screening of patients with IBGC, especially in the setting of clinical genetic testing.
IBGC patients can present with a wide spectrum of neuropsychiatric symptoms, including parkinsonism, tremor, dystonia and dementia, among others. The typical age at onset for IBGC is between 30 and 50 years, however onset ages vary widely. Our patient with the p.S113* mutation presented with parkinsonian symptoms at the age of 68. In contrast, the mean age at symptom onset in our large Canadian family carrying the SLC20A2 deletion was only 19 years4. Moreover dystonia was extremely common in this family, affecting 8 out of 11 patients, and some also had chorea, intellectual decline, postural tremor, or dysarthria. Interestingly, the chromosome 8 deletion in this family affects 7 other genes in addition to SLC20A2, including THAP1, a gene associated with hereditary dystonia. Loss-of-function mutations in THAP1 are the genetic cause of DYT6 or idiopathic torsion dystonia of mixed type (MIM602629), a progressive dystonia that involves mostly cranio-cervical (spasmodic dysphonia, oromandibular dystonia, torticollis) and upper limb muscles [20, 21]. While the brain calcinosis in our family is likely related to the deletion of SLC20A2, the THAP1 deletion may contribute to the dystonia phenotype, especially the oromandibular type, and the early-onset of dystonia in several family members (range 8–25 years), which is in line with THAP1 mutations and different from the typical onset of 30–50 years in cases with IBGC4. This is the first complete THAP1 deletion reported in dystonia patients; however, mutations affecting the translation initiation methionine start codon were found that are predicted to result in a similar loss of THAP1 protein [22, 23]. Of the other known genes in the deletion region, RNF170 is also of interest because a specific missense mutation in this gene was found in an autosomal dominant form of sensory ataxia; however, the disease mechanism associated with this particular mutation was thought to be a toxic gain-of-function, and the effect of a loss of RNF170 is not known [24]. Given that this is the first family in which a genomic deletion of SLC20A2 has been reported, genotype-phenotype correlations are premature.
SLC20A2 was identified only recently as the first IBGC disease gene and is responsible for the IBGC3 locus on chromosome 8 [11]. The large clinical variability associated with IBGC together with the relatively high frequency of incidental sporadic calcification in the basal ganglia in the general elderly population provides an ongoing challenge for the genetic study of IBGC. Even though a 10 cM density genome-wide linkage study had previously found no significant genome-wide linkage in our Canadian IBGC family with dystonia-plus, the region on chromosome 8 close to SLC20A2 was reported as a region of interest, with suggestive LOD-scores between 1 and 1.5 [4]. The lack of significant linkage likely resulted from sparse marker density in this region, especially close to the centromere. Interestingly, a pathogenic SLC20A2 mutation was also reported in the family that defined the IBGC2 locus on chromosome 14 [12]. In this family, the presence of two phenocopies led to the inaccurate genetic mapping of the disease gene [12]. While the identification of SLC20A2 mutations in both families further underlines the importance of this gene in the pathogenesis of IBGC, genetic heterogeneity of IBGC clearly exists. In fact, recently, exome sequencing identified another gene (PDGFRB) in a large French IBGC family, followed by the identification of mutations in its main ligand PDGFB in at least 6 other families [25]. Moreover, despite these advances the genetic cause of at least half of the familial IBGC patients currently remains unexplained [26].
In summary, we identified two novel mutations in SLC20A2, a nonsense mutation in one patient and a unique genomic deletion encompassing both SLC20A2 and THAP1 gene in a large Canadian IBGC family with dystonia-plus. To cover the complete mutational spectrum, we suggest that genomic copy-number analysis for SLC20A2 should be included when analyzing IBGC patients. Moreover, THAP1 mutation screening in dystonia patients should also be expanded to include genomic copy-number analysis.
Acknowledgments
We would like to thank all those who have contributed to our research, particularly the patients and families who donated DNA samples for this work. This work is supported by a Morris K. Udall Parkinson’s Disease Research Center of Excellence (National Institute of Neurological Disorders and Stroke P50NS072187), Mayo Clinic Florida Research Committee, and the Mayo Clinic Center for Individualized Medicine.
References
- 1.Baba Y, Broderick DF, Uitti RJ, Hutton ML, Wszolek ZK. Heredofamilial brain calcinosis syndrome. Mayo Clinic proceedings Mayo Clinic. 2005;80 (5):641–651. doi: 10.4065/80.5.641. [DOI] [PubMed] [Google Scholar]
- 2.Brodaty H, Mitchell P, Luscombe G, Kwok JJ, Badenhop RF, McKenzie R, Schofield PR. Familial idiopathic basal ganglia calcification (Fahr’s disease) without neurological, cognitive and psychiatric symptoms is not linked to the IBGC1 locus on chromosome 14q. Human genetics. 2002;110 (1):8–14. doi: 10.1007/s00439-001-0650-x. [DOI] [PubMed] [Google Scholar]
- 3.Larsen TA, Dunn HG, Jan JE, Calne DB. Dystonia and calcification of the basal ganglia. Neurology. 1985;35 (4):533–537. doi: 10.1212/wnl.35.4.533. [DOI] [PubMed] [Google Scholar]
- 4.Wszolek ZK, Baba Y, Mackenzie IR, Uitti RJ, Strongosky AJ, Broderick DF, Baker MC, Melquist S, Hutton ML, Tsuboi Y, Allanson JE, Carr J, Kumar A, Calne SM, Miklossy J, McGeer PL, Calne DB, Stoessl AJ. Autosomal dominant dystonia-plus with cerebral calcifications. Neurology. 2006;67 (4):620–625. doi: 10.1212/01.wnl.0000230141.40784.09. [DOI] [PubMed] [Google Scholar]
- 5.Forstl H, Krumm B, Eden S, Kohlmeyer K. What is the psychiatric significance of bilateral basal ganglia mineralization? Biological psychiatry. 1991;29 (8):827–833. doi: 10.1016/0006-3223(91)90201-v. [DOI] [PubMed] [Google Scholar]
- 6.Harrington MG, Macpherson P, McIntosh WB, Allam BF, Bone I. The significance of the incidental finding of basal ganglia calcification on computed tomography. Journal of neurology, neurosurgery, and psychiatry. 1981;44 (12):1168–1170. doi: 10.1136/jnnp.44.12.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geschwind DH, Loginov M, Stern JM. Identification of a locus on chromosome 14q for idiopathic basal ganglia calcification (Fahr disease) American journal of human genetics. 1999;65 (3):764–772. doi: 10.1086/302558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai X, Gao Y, Xu Z, Cui X, Liu J, Li Y, Xu H, Liu M, Wang QK, Liu JY. Identification of a novel genetic locus on chromosome 8p21.1–q11.23 for idiopathic basal ganglia calcification. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2010;153B (7):1305–1310. doi: 10.1002/ajmg.b.31102. [DOI] [PubMed] [Google Scholar]
- 9.Volpato CB, De Grandi A, Buffone E, Facheris M, Gebert U, Schifferle G, Schonhuber R, Hicks A, Pramstaller PP. 2q37 as a susceptibility locus for idiopathic basal ganglia calcification (IBGC) in a large South Tyrolean family. Journal of molecular neuroscience : MN. 2009;39 (3):346–353. doi: 10.1007/s12031-009-9287-3. [DOI] [PubMed] [Google Scholar]
- 10.Wider C, Dickson DW, Schweitzer KJ, Broderick DF, Wszolek ZK. Familial idiopathic basal ganglia calcification: a challenging clinical-pathological correlation. Journal of neurology. 2009;256 (5):839–842. doi: 10.1007/s00415-009-5025-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C, Li Y, Shi L, Ren J, Patti M, Wang T, de Oliveira JR, Sobrido MJ, Quintans B, Baquero M, Cui X, Zhang XY, Wang L, Xu H, Wang J, Yao J, Dai X, Liu J, Zhang L, Ma H, Gao Y, Ma X, Feng S, Liu M, Wang QK, Forster IC, Zhang X, Liu JY. Mutations in SLC20A2 link familial idiopathic basal ganglia calcification with phosphate homeostasis. Nature genetics. 2012;44 (3):254–256. doi: 10.1038/ng.1077. [DOI] [PubMed] [Google Scholar]
- 12.Hsu SC, Sears RL, Lemos RR, Quintans B, Huang A, Spiteri E, Nevarez L, Mamah C, Zatz M, Pierce KD, Fullerton JM, Adair JC, Berner JE, Bower M, Brodaty H, Carmona O, Dobricic V, Fogel BL, Garcia-Estevez D, Goldman J, Goudreau JL, Hopfer S, Jankovic M, Jauma S, Jen JC, Kirdlarp S, Klepper J, Kostic V, Lang AE, Linglart A, Maisenbacher MK, Manyam BV, Mazzoni P, Miedzybrodzka Z, Mitarnun W, Mitchell PB, Mueller J, Novakovic I, Paucar M, Paulson H, Simpson SA, Svenningsson P, Tuite P, Vitek J, Wetchaphanphesat S, Williams C, Yang M, Schofield PR, de Oliveira JR, Sobrido MJ, Geschwind DH, Coppola G. Mutations in SLC20A2 are a major cause of familial idiopathic basal ganglia calcification. Neurogenetics. 2013;14 (1):11–22. doi: 10.1007/s10048-012-0349-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemos RR, Oliveira MF, Oliveira JR. Reporting a new mutation at the SLC20A2 gene in familial idiopathic basal ganglia calcification. European journal of neurology : the official journal of the European Federation of Neurological Societies. 2013;20 (3):e43–44. doi: 10.1111/ene.12044. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Guo X, Wu A. Association between a novel mutation in SLC20A2 and familial idiopathic basal ganglia calcification. PloS one. 2013;8 (2):e57060. doi: 10.1371/journal.pone.0057060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen WJ, Yao XP, Zhang QJ, Ni W, He J, Li HF, Liu XY, Zhao GX, Murong SX, Wang N, Wu ZY. Novel SLC20A2 mutations identified in southern Chinese patients with idiopathic basal ganglia calcification. Gene. 2013;529 (1):159–162. doi: 10.1016/j.gene.2013.07.071. [DOI] [PubMed] [Google Scholar]
- 16.Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, Dahl F, Fernandez A, Staker B, Pant KP, Baccash J, Borcherding AP, Brownley A, Cedeno R, Chen L, Chernikoff D, Cheung A, Chirita R, Curson B, Ebert JC, Hacker CR, Hartlage R, Hauser B, Huang S, Jiang Y, Karpinchyk V, Koenig M, Kong C, Landers T, Le C, Liu J, McBride CE, Morenzoni M, Morey RE, Mutch K, Perazich H, Perry K, Peters BA, Peterson J, Pethiyagoda CL, Pothuraju K, Richter C, Rosenbaum AM, Roy S, Shafto J, Sharanhovich U, Shannon KW, Sheppy CG, Sun M, Thakuria JV, Tran A, Vu D, Zaranek AW, Wu X, Drmanac S, Oliphant AR, Banyai WC, Martin B, Ballinger DG, Church GM, Reid CA. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327 (5961):78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 17.Exome Variant Server. [ http://evs.gs.washington.edu/EVS/]
- 18.Miklossy J, Mackenzie IR, Dorovini-Zis K, Calne DB, Wszolek ZK, Klegeris A, McGeer PL. Severe vascular disturbance in a case of familial brain calcinosis. Acta neuropathologica. 2005;109 (6):643–653. doi: 10.1007/s00401-005-1007-7. [DOI] [PubMed] [Google Scholar]
- 19.Fuchs T, Gavarini S, Saunders-Pullman R, Raymond D, Ehrlich ME, Bressman SB, Ozelius LJ. Mutations in the THAP1 gene are responsible for DYT6 primary torsion dystonia. Nature genetics. 2009;41 (3):286–288. doi: 10.1038/ng.304. [DOI] [PubMed] [Google Scholar]
- 20.Muller U. The monogenic primary dystonias. Brain : a journal of neurology. 2009;132 (Pt 8):2005–2025. doi: 10.1093/brain/awp172. [DOI] [PubMed] [Google Scholar]
- 21.Spatola M, Wider C. Overview of primary monogenic dystonia. Parkinsonism & related disorders. 2012;81(Suppl 1):S158–161. doi: 10.1016/S1353-8020(11)70049-9. [DOI] [PubMed] [Google Scholar]
- 22.Blanchard A, Ea V, Roubertie A, Martin M, Coquart C, Claustres M, Beroud C, Collod-Beroud G. DYT6 dystonia: review of the literature and creation of the UMD Locus-Specific Database (LSDB) for mutations in the THAP1 gene. Human mutation. 2011;32 (11):1213–1224. doi: 10.1002/humu.21564. [DOI] [PubMed] [Google Scholar]
- 23.De Carvalho Aguiar P, Fuchs T, Borges V, Lamar KM, Silva SM, Ferraz HB, Ozelius L. Screening of Brazilian families with primary dystonia reveals a novel THAP1 mutation and a de novo TOR1A GAG deletion. Movement disorders : official journal of the Movement Disorder Society. 2010;25 (16):2854–2857. doi: 10.1002/mds.23133. [DOI] [PubMed] [Google Scholar]
- 24.Valdmanis PN, Dupre N, Lachance M, Stochmanski SJ, Belzil VV, Dion PA, Thiffault I, Brais B, Weston L, Saint-Amant L, Samuels ME, Rouleau GA. A mutation in the RNF170 gene causes autosomal dominant sensory ataxia. Brain : a journal of neurology. 2011;134 (Pt 2):602–607. doi: 10.1093/brain/awq329. [DOI] [PubMed] [Google Scholar]
- 25.Keller A, Westenberger A, Sobrido MJ, Garcia-Murias M, Domingo A, Sears RL, Lemos RR, Ordonez-Ugalde A, Nicolas G, da Cunha JE, Rushing EJ, Hugelshofer M, Wurnig MC, Kaech A, Reimann R, Lohmann K, Dobricic V, Carracedo A, Petrovic I, Miyasaki JM, Abakumova I, Mae MA, Raschperger E, Zatz M, Zschiedrich K, Klepper J, Spiteri E, Prieto JM, Navas I, Preuss M, Dering C, Jankovic M, Paucar M, Svenningsson P, Saliminejad K, Khorshid HR, Novakovic I, Aguzzi A, Boss A, Le Ber I, Defer G, Hannequin D, Kostic VS, Campion D, Geschwind DH, Coppola G, Betsholtz C, Klein C, Oliveira JR. Mutations in the gene encoding PDGF-B cause brain calcifications in humans and mice. Nature genetics. 2013;45 (9):1077–1082. doi: 10.1038/ng.2723. [DOI] [PubMed] [Google Scholar]
- 26.Nicolas G, Pottier C, Maltete D, Coutant S, Rovelet-Lecrux A, Legallic S, Rousseau S, Vaschalde Y, Guyant-Marechal L, Augustin J, Martinaud O, Defebvre L, Krystkowiak P, Pariente J, Clanet M, Labauge P, Ayrignac X, Lefaucheur R, Le Ber I, Frebourg T, Hannequin D, Campion D. Mutation of the PDGFRB gene as a cause of idiopathic basal ganglia calcification. Neurology. 2013;80 (2):181–187. doi: 10.1212/WNL.0b013e31827ccf34. [DOI] [PubMed] [Google Scholar]