Abstract
Influenza A viruses cause yearly seasonal epidemics and occasional global pandemics in humans. In the last century, four human influenza A virus pandemics have occured. Ocasionally, influenza A viruses that circulate in other species, cross the species barrier and infect humans. Virus re-assortment (i.e. mixing of gene segments of multiple viruses) and the accumulation of mutations contribute to the emergence of new influenza A virus variants. Fortunately, most of these variants do not have the ability to spread among humans and subsequently cause a pandemic. In this review we focus on the threat of animal influenza A viruses which have shown the ability to infect humans. In addition, genetic factors which could alter the virulence of influenza A viruses are discussed. Identification and characterization of these factors may provide insights into genetic traits which change virulence and help us to understand which genetic determinants are of importance for the pandemic potential of animal influenza A viruses.
Keywords: Influenza A viruses, virulence factors, pandemic threat, transmission
Influenza A virus
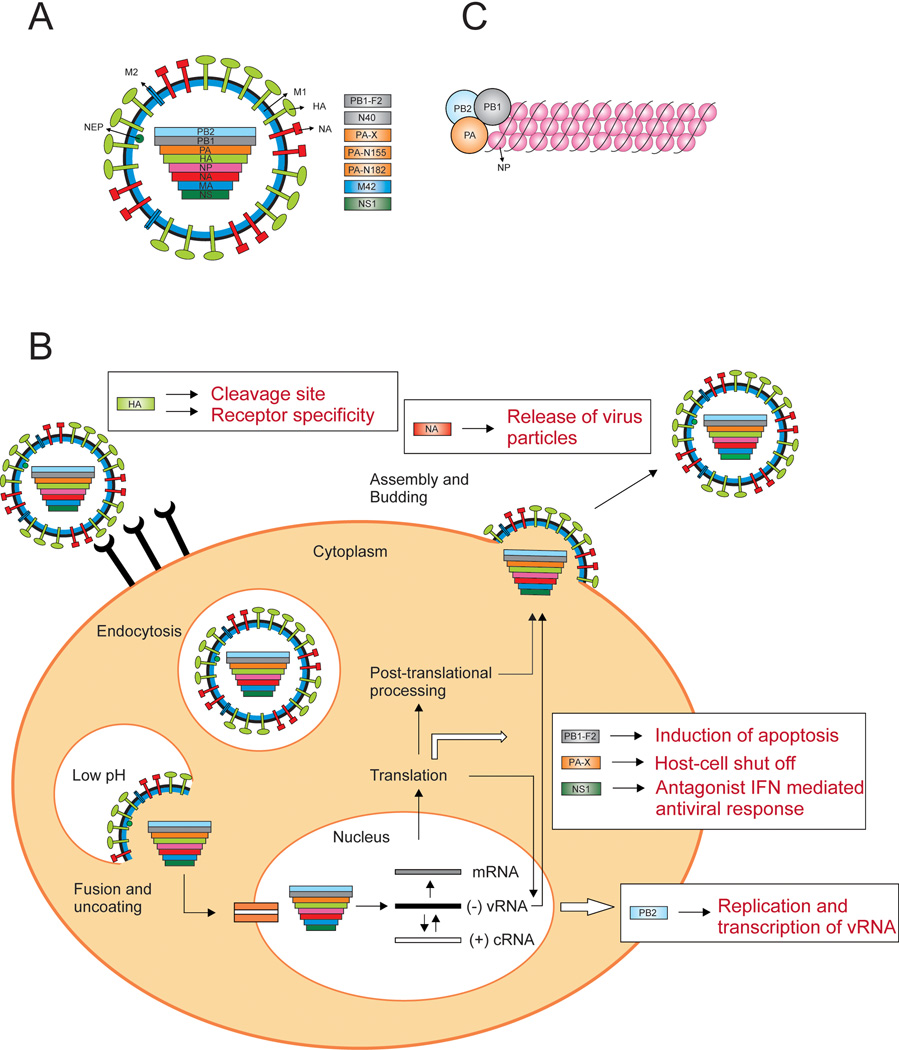
Influenza A virus is a single-stranded negative-sense segmented RNA virus, with a genome consisting of eight gene segments, that can encode up to 16 proteins [1–5] (Fig 1a). Influenza A viruses are divided into subtypes based on the genetic and antigenic properties of their envelope glycoproteins hemagglutinin (HA) and neuraminidase (NA). To date, 17 HA and 10 NA subtypes have been identified. The HA glycoprotein of influenza A viruses is initially synthesized as a single polypeptide precursor (HA0), which needs to be cleaved into HA1 and HA2 subunits by cellular proteases to become biologically active [6]. In the initial stage of virus replication, the HA glycoprotein binds to specific sialic acid (SA) receptors on the surface of susceptible cells (Fig 1b). Human influenza viruses preferentially bind to α2,6-linked SA receptors which are mainly expressed on epithelial cells in the human upper respiratory tract (URT), whereas avian influenza viruses bind to α2,3-linked SA receptors, which are abundantly present on epithelial cells in the intestine of birds and in the lower respiratory tract (LRT) of humans [7]. After binding to these SA receptors, virus particles enter the cell through receptor-mediated endocytosis. Subsequently, a low-pH-triggered conformational change of HA mediates fusion of the viral membrane with the endosomal membrane. The viral ribonucleoproteins (vRNPs) (Fig 1c), are released into the cytoplasm and are translocated tot the nucleus, where transcription and replication takes place by the viral RNA-dependent RNA polymerase complex, consisting of the three polymerase subunits; polymerase basic protein 1 (PB1), polymerase basic protein 2 (PB2), and polymerase acidic protein (PA). M1 and NEP subsequently mediate transport of the newly synthesized vRNPs to the cytoplasm. At the plasma membrane, new virus particles are assembled and released via budding. Efficient release of the viral particles is facilitated by the NA surface glycoprotein which cleaves off SA residues from the cell surface thereby allowing the new virus particle to detach from the cell.
Figure 1. Influenza A virus particle and replication cycle.
A) Schematic representation of influenza A virus particle and gene segments. The influenza genome consists of eight single-stranded RNAs. The non-structural proteins and/or newly identified proteins with unknown function are depicted in rectangles. B) Schematic representation of an influenza A virus replication cycle. The viral HA binds to the appropriate host receptor and the virus enters via receptor-mediated endocytosis. A low-pH-triggered conformational change of HA mediates fusion between the viral en endosomal membrane and the RNPs are released into the cytoplasm. The RNPs trans-locate to the nucleus, where the vRNA transcription and replication takes place by the RNA-dependent RNA polymerases (PB1, PA and PB2). The mRNA is transported out of the nucleus and translated into the viral proteins. Viral proteins that are needed for replication and transcription are trans-located back into the nucleus. Newly synthesized vRNA, along with the polymerase proteins and NP, forms the vRNPs. M1 and NEP subsequently mediate transport of the newly synthesized vRNPs to the cytoplasm. At the plasma membrane, new virus particles are assembled and released via budding. Release from the host cell is mediated by NA, which cleaves off SA from cellular and viral glycoproteins so that the new virus particles can detach from the cell. Several influenza A proteins have been shown to be important determinants of virulence (indicated with boxes). The HA glycoprotein (light green) is important as a tropism factor, for cleavage of HA by cellular proteases, and is a prerequisite for starting viral infection. When some HAs are cleavable by ubiquitous proteases, this can result in systemic virus replication in some hosts (e.g. poultry). HA is also responsible for attachment to the different host cell receptors. The PB2 polymerase protein (light blue), has the ability to increase replication levels of vRNA in the nucleus. The PB1-F2 protein (grey) contributes to viral pathogenicity by inducing apoptosis of infected cells. The PA-X protein (orange) regulates the host-cell shut off. The NS1 protein (dark green) can counteract the innate immune response of the host. The NA protein (red) acts as a virulence factor by allowing efficient release of the virus particle from the cell. C) Schematic illustration of the vRNP structure. The viral RNA is encapsidated by NP, and this structure is bound to the polymerase complex (PB2, PB1 and PA).
The influenza A virus genome encodes additional proteins that are either not directly involved in virus replication or for which the function has not yet been elucidated. The polymerase subunit PB1 can encode two more proteins; The PB1-F2 and PB1 N40 proteins. Similarly, the influenza virus PA gene contains a second open reading frame that is accessed via ribosomal frame-shifting, which encodes the PA-X protein. The NS1 protein functions as an antagonist to block the type 1 interferon (IFN) mediated host antiviral response [8]. M42 has only been discovered recently and encodes a novel M2-like protein with a variant extra-cellular domain [2]. Currently, the identification of novel influenza virus proteins is receiving considerable interest and influenza segment PA has now been shown to encode as many as four proteins; PA, PA-X, PA-N155 and PA-N182 [5].
Influenza epidemics and pandemics
Influenza A viruses are the cause of recurrent epidemics and occasional pandemics. The annual epidemics result in about three to five million cases of severe illness, and about 250000 to 500000 deaths worldwide. Infection with influenza A virus results in protective immunity against the viral surface glycoproteins HA and NA. However, the accumulation of point mutations in HA and to a lesser extent NA, allows the virus to escape the host immunity. This phenomenon, known as antigenic drift, explains the occurrence of seasonal influenza epidemics. As a result of this antigenic drift, the vaccine composition has to be updated almost annually [9]. Antigenic shift refers to the introduction of a new influenza A virus subtype in humans. Antigenic shift can be caused by direct introduction of a new influenza A virus from the animal reservoir, or by re-assortment, i.e. the mixing of genes from two (or more) influenza A viruses, between animal and human influenza A viruses. While influenza viruses are continuously changing by antigenic drift, antigenic shift happens only occasionally.
Influenza viruses with pandemic potential may arise in pigs upon re-assortment, as pig cells express both human and avian influenza receptors, therewith providing an opportunity for replication of avian and human influenza viruses in the same cell [10]. In the human population, co-infections with different influenza strains have also been observed [11]. In 2001, an H1N2 virus was identified which was the result of re-assortment between contemporary circulating H1N1 and H3N2 viruses. Fortunately, this virus did not persist in the human population [12]. Re-assortment is also an important factor to increase genetic variation of influenza viruses as multiple re-assortment events have been shown to occur between different lineages of the H3N2 virus [13].
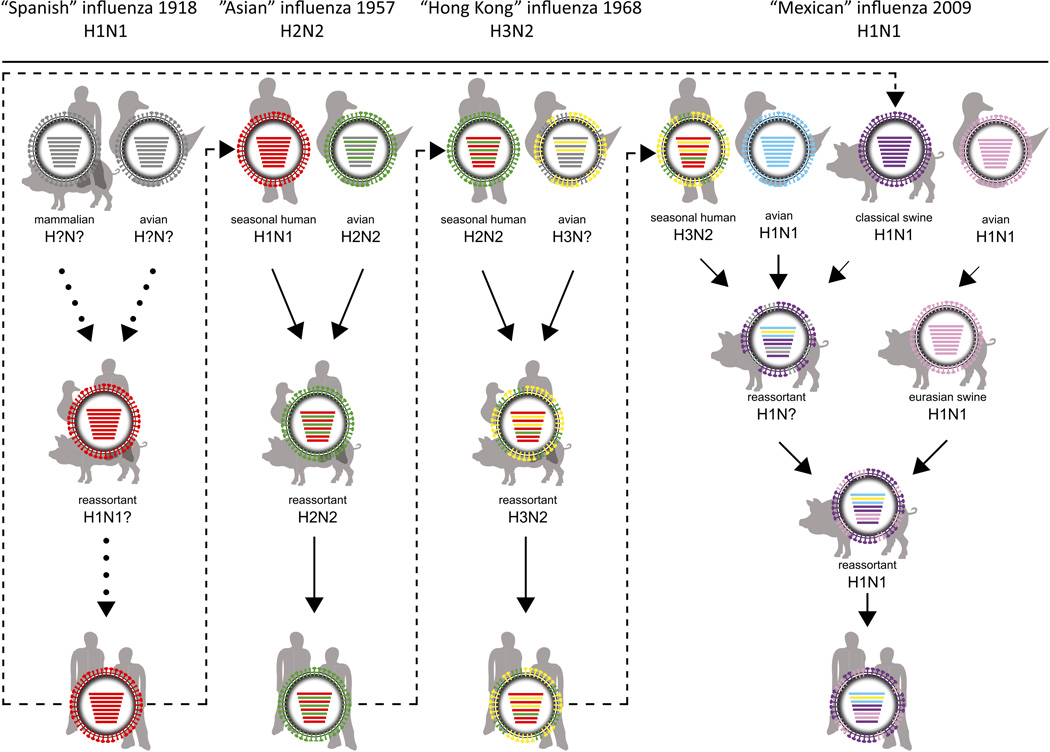
In the last century, four human influenza A virus pandemics have emerged, at least three of which resulted from reassortment of human and animal influenza A viruses [14–16] (Fig 2). Future influenza pandemics are inevitable, but it will be difficult to predict which influenza virus subtype will cause a pandemic, when it will cause a pandemic, and what the severity of the pandemic will be.
Figure 2. Reassortment and adaptation events of pandemic influenza A viruses.
For the 1918 H1N1 “Spanish influenza” pandemic, evidence for two scenarios has been presented: 1) the virus emerged upon reassortment between avian and mammalian influenza viruses and 2) the gradual adaptation of avian influenza genes to the human host. The 1918 H1N1 virus caused seasonal epidemics until 1957, when the H2N2 virus emerged upon re-assortment of the seasonal H1N1 with an avian H2N2 virus, thereby introducing the avian HA, NA, and PB1 genes. The H2N2 virus circulated in humans until 1968, when re-assortment of the H2N2 with an avian H3 virus resulted in the exchange of the H3 HA and PB1 genes and the generation of the H3N2 “Hong Kong” influenza. The pH1N1 virus consists of the NA and M genes of the Eurasian swine lineage, and the other genes of a “triple reassortant” swine influenza virus that had previously acquired its genes upon re-assortment between human, avian, and (classical) swine viruses. Grey colour in virus particles indicates uncertainty of viral gene segment origin or lack of data. Dotted arrows indicate uncertain scenarios and solid arrows indicate events that are supported by scientific evidence. Dashed arrows represent pandemic viruses circulating in subsequent influenza seasons. Partially adapted from [101].
Pandemics threats
Historically, influenza viruses of three HA subtypes (H1, H2 and H3) have acquired the ability to be transmitted efficiently between humans. Currently, influenza viruses of the H1 and H3 subtype co-circulate in humans, however influenza viruses of the H2, H5, H6, H7 and H9 subtype are also considered to represent a pandemic threat.
In 1997, a large outbreak of highly pathogenic avian influenza (HPAI) H5N1 virus in poultry in Hong Kong resulted in the first documented cases of direct transmission of HPAI H5N1 virus from poultry to humans, with a fatal outcome in 6 out of 18 cases [17]. As a result, this outbreak warranted the mass culling of 1.5 million chickens. After 2004, HPAI H5N1 viruses have spread throughout Asia, Europe and Africa since, causing severe disease outbreaks in poultry. Furthermore, HPAI H5N1 viruses have been isolated from mammals on numerous occasions. Since 2003, over 600 cases of human HPAI H5N1 infections have been reported, more than half of which were fatal [18]. Although rare cases of H5N1 virus transmission between humans have been reported, sustained human-to-human transmission of HPAI H5N1 virus has not been detected yet [19]. It is this absence of efficient human-to-human transmission that has prevented an H5N1 pandemic to occur. With the sporadic introduction of HPAI H5N1 virus in the human population, it is feared that these avian H5N1 viruses may mutate or re-assort with contemporary human influenza viruses, possibly resulting in adaptation to humans. Fortunately, at present, re-assortment of HPAI H5 virus with contemporary human influenza viruses has not been detected in nature. However, co-infections of avian and human influenza viruses in humans or pigs may provide new opportunities for re-assortment [20]. Due to the enzootic nature of HPAI H5N1 in poultry, the brood host range (over 20 different mammalian species) and the accumulation of mammalian adaptation mutations, this virus is currently considered represent a significant pandemic threat to humans.
Several outbreaks of HPAI H7 viruses in poultry have resulted in transmission to humans. In 2003, a large outbreak of an HPAI H7N7 virus in poultry in the Netherlands resulted in 89 cases of human infections, one of which was fatal [21]. HPAI H7N7 virus displayed an unusual tissue tropism; the virus targeted the conjunctiva, resulting in conjunctivitis, a symptom rarely reported for other influenza virus subtypes [22]. Recently, a novel reassortant influenza H7N9 virus of avian origin emerged in China. This LPAI H7N9 virus is associated with severe and fatal respiratory disease. As of July 2013, 135 confirmed cases of human infection with H7N9 virus have been reported, 44 of which have resulted in deaths [23]. The emergence of this new H7N9 virus in humans emphasizes the pandemic potential of influenza A viruses of the H7 subtype.
Since the mid-1990's, H9N2 viruses have become endemic in poultry populations throughout Eurasia. The first human case of infection with an avian H9N2 virus was documented in 1999 in Hong Kong [24] and sporadically human infections have been described since [25]. Furthermore, this subtype has been isolated from pigs and numerous reassortment events between H9N2 virus and other influenza virus subtypes (i.e. H7N9, H5N1 and, H6N2) [26] have been reported. H9N2 viruses with either avian and human or human receptor specificity [27] are now prevalent in many Eurasian countries, thereby increasing the possibility of this virus to infect humans. This year, the first human infected case of avian H6N1 was documented [28]. Avian influenza A viruses of the H6 subtype were found to replicate in mice and ferrets without prior adaptation [29]. Given the first human case, the high prevalence and, frequent re-assortment of H6 viruses in birds, concerns have risen about the possible emergence of a pandemic H6 virus [30, 31].
Influenza viruses of the H2 subtype have not circulated in humans since 1968 and therefore a large proportion of the current world population is likely to be susceptible to infection with H2 viruses if they would re-emerge. As H2 viruses continue to circulate in swine and several avian species [32], they again pose a potential pandemic threat.
The pH1N1 virus in 2009 illustrated that a new pandemic does not necessarily require the introduction of a virus with an HA subtype that is new to the human population. A novel influenza A (H3N2) variant virus (H3N2v) containing seven gene segments of swine influenza origin and the pH1N1 M segment was isolated from 12 humans in 2011 [33]. Although there is a low level of cross-reactive antibodies with a human H3N2 virus that circulated in the 1990’s [34], the H3N2v is antigenically different from the currently circulating seasonal H3N2 viruses and can thus potentially infect a large proportion of the human population.
Our understanding of avian influenza virus infections of humans is still rather limited. The H7N9 outbreak that started in may 2013 highlights the continuous threat of avian influenza viruses and underlines the unpredictability of which viruses are likely to cross the species barrier.
Insight into mechanisms by which influenza viruses cross the species barrier is therefore crucial to interpret surveillance data and to allow the early detection of influenza viruses with pandemic potential. Since the pH1N1 virus emerged after extensive re-assortment in swine, it is important to continue surveillance activities not only in birds, but also in swine to monitor novel swine influenza viruses which may have the ability to infect humans. Recently, re-assortment studies demonstrated that pH1N1 virus was found to preferentially incorporate the NA and PB1 gene segments of a seasonal H3N2 virus [35, 36]. In addition, a recent study showed that co-circulating H1N1, H1N2 and H3N2 viruses can re-assort rapidly in swine [37]. Thus, it is possible that additional re-assortment events occur between currently circulating swine or human influenza viruses.
Influenza A virus virulence factors
HA
Amino acid substitutions in HA that affect the receptor binding preference can influence the cellular host range and tissue tropism which may alter virulence. Specific amino acid residues in HA determine the receptor binding specificity of human and avian influenza viruses and these specific residues differ among virus subtypes. Binding patterns of pandemic influenza A viruses of the H1 subtype are determined by amino acids at position 190 and 225 in HA (H3 numbering) [38, 39]. For influenza viruses of the H2 and H3 subtypes, positions 226 and 228 are important for receptor binding specificity [40]. Since the HA proteins of the 1918, 1957, and 1968 pandemic strains were derived from avian influenza viruses, adaptation of avian HA proteins to the human receptor is considered to be a prerequisite for efficient human-to-human transmission [40]. Numerous studies have described amino acid substitutions in HA of HPAI H5N1 viruses that change and/or increase binding to human α2,6-linked SA receptors that are present in the human URT [41, 42]. The majority of these amino acid substitutions are located in or near the receptor binding site of HA. However, a potential N-glycosylation motif at amino acid position 154–156 of HA, which is proximal but not immediately adjacent to the receptor-binding site, may also affect binding preference and virulence [43, 44].
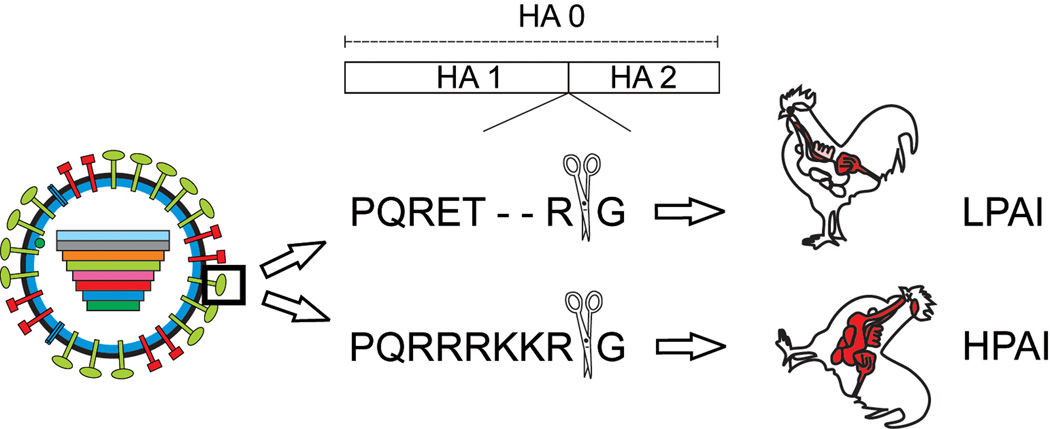
One of the best-known virulence determinants of HPAI viruses is the multi basic cleavage site (MBCS) in HA. The sequence of this cleavage site determines by which cellular proteases HA0 is cleaved and consequently determines viral tropism and virulence (Fig 3). Cleavage of HA by cellular proteases is a crucial step in the replication cycle; virus particles with un-cleaved HAs are not infectious. The HA of low pathogenic avian influenza (LPAI) viruses and human influenza viruses harbors a monobasic cleavage site that is cleaved by trypsin-like proteases. These trypsin-like proteases are only present in the respiratory tract of humans and the respiratory and/or intestinal tract of birds, thereby restricting virus replication to these tissues [45]. Cleavage of the human influenza virus HA is thought to occur by trypsin-like serine proteases of the type II trans-membrane serine protease (TTSP) family that are present at the plasma membrane, or by extra-cellular proteases present in the respiratory tract [6].
Figure 3. Cleavage site as an important virulence factor.
The HA0 is cleaved into two subunits HA1 and HA2 by cellular proteases which recognize either a mono basic or multi basic cleavage site. The HA0 of LPAI viruses harbors a mono basic cleavage site and is cleaved by trypsin-like proteases only, thereby limiting replication of these viruses to sites where these enzymes are expressed; i.e. respiratory and intestinal tract. The HA0 of HPAI viruses of the H5 and H7 subtype can be cleaved by ubiquitously expressed furin-like proteases, facilitating systemic replication in chickens.
Influenza viruses of the H5 and H7 subtype may become highly pathogenic after circulation in poultry. The switch from a low pathogenic to a high pathogenic phenotype of these H5 and H7 influenza viruses is caused by the introduction of basic amino acid residues into the HA0 cleavage site. This HA0 can subsequently be cleaved by ubiquitously expressed pro-protein convertases of the subtilisin family, like furin or PC5/6, thereby facilitating systemic replication in chickens [6]. Recent studies demonstrated that non-H5 and -H7 subtypes could support a highly pathogenic phenotype which demonstrates that introduction of an MBCS in a non-H5 or - H7 avian influenza virus is not deleterious [46, 47].
In mammals, the association between the presence of an MBCS and systemic spread is less obvious. In the ferret model, incorporation of an MBCS in the HA of a human influenza H3N2 virus did not resulted in increased virulence or a change in tissue tropism. These findings suggest that, in addition to an MBCS, other factors are involved to result in systemic spread in ferrets. The presence of an MBCS in H5 HA also does not always result in systemic spread in mammals since the inoculation of cynomolgus macaques with HPAI H5N1 resulted in respiratory tract infection only [48]. However, in mice, deletion of the MBCS of an HPAI H5N1 virus results in a virus that only causes respiratory tract infection in contrast to the systemic spread of the wild-type HPAI H5N1 virus, indicating that the MBCS is a major virulence factor in mice [49]. Although the extra-respiratory tractinfection of HPAI H5N1 virus in mammals is likely caused by multiple factors [50, 51], the MBCS has been shown to be essential [52].
Polymerase proteins
The influenza virus polymerase proteins, and in particular PB2, have been shown to be important determinants of virulence. Amino acid substitution lysine (K) to glutamic acid (E) at position 627 in PB2 has been studied extensively in the context of mammalian adaptation [53]. This substitution is suggested to occur in order to adapt to physiological constraints, e.g. differences in body temperature. Most avian influenza A viruses, which preferentially replicate at a relatively high temperature of around 41 degrees Celsius in the digestive tract of birds, have a E residue at position 627. In contrast, human influenza A viruses replicate at the lower temperature of around 33 degrees Celsius, which is the temperature in the human URT. These viruses typically have a K residue at this position. The E627K substitution was acquired rapidly when avian viruses were pass-aged in mice [54]. Moreover, this mutation has been implicated with increased virulence of human HPAI H5N1 virus isolates and was found in a fatal human case of infection with HPAI H7N7 [44, 49]. In the absence of the E627K mutation, an aspartate (D) to asparagine (N) substitution at position 701 of H5N1 PB2 was found to increase virulence and to expand the host range of avian H5N1 virus to mammalian hosts [54–56]. The adaptive mutation D701N caused enhancement of binding of PB2 to importin alpha1 in mammalian cells resulting in increased transport of PB2 into the nucleus [57]. Noteworthy, unlike other human influenza viruses, pH1N1 does not contain the mammalian adaptation residues 627K and/or 701N. When the mammalian adaptation substitutions E627K or D701N were introduced in pH1N1, no increase in virulence was observed [58]. However, the absence of these mammalian adaptation markers in PB2 have been compensated to some extent by a serine (S) and arginine (R) substitution at position 590 and 591 that may affect the protein's interaction with viral and/or cellular factors and hence its ability to support virus replication in mammals [59]. Recently, numerous other mutations in PB1, PA, NP and NEP have been described, that can overcome the poor polymerase activity of avian influenza viruses in human cells [60].
PB1-F2
PB1-F2 contributes to the virulence of influenza A viruses by inducing apoptosis of infected cells [61]. Moreover, PB1-F2 promotes and increases severity of secondary pneumonia [62]. The PB1-F2 protein contributed to the virulence of HPAI H5N1 and the 1918 H1N1, 1957 H2N2 and, 1968 H3N2 pandemic strains [63, 64]. In particular, an N to S substitution at position 66 (N66S) of HPAI H5N1 and 1918 pandemic influenzaPB1-F2, is partly responsible for the high virulence of these viruses [63]. The PB1-F2 N66S variant reduces the production of interferon (IFN), which is part of the innate immune response [65]. In contrast to previous pandemic influenza viruses, pH1N1 does not encode a PB1-F2, because of three premature stop co-dons. Surprisingly, when the pH1N1 PB1-F2’s coding capacity was restored, the virulence was only modestly affected in mice and ferrets [66, 67].
PA-X
The PA gene also encodes a newly identified protein PA-X [4]. This protein modulates the host response by repressing cellular gene expression, i.e. host-cell shut off. PA-X deficient influenza viruses cause more severe disease in mice, as a result of an accelerated host response. Moreover, influenza viruses lacking PA-X differ in host-cell shutoff compared to wild-type virus. Truncation of PA-X protein appears to be associated with influenza virus lineages circulating in particular hosts, indicating that there may be some species specificity to the evolution of PA-X [68].
NS1
In response to the presence of pathogens in the host, IFNs are secreted by cells and ‘interfere’ with viral replication. To establish productive infection, influenza viruses have mechanisms to evade host immune responses, including the type-I IFN response. The influenza virus NS1 protein has several ways to act as an IFN antagonist [69].
NS1 has been studied extensively as a molecular determinant of virulence. Influenza viruses lacking the IFN antagonist NS1 are only able to replicate in cells or mice that have a compromised IFN response [8]. H5N1 viruses, unlike other human, avian and swine influenza viruses, are relatively resistant to the antiviral effects of INFs, which result in increased levels of pro-inflammatory cytokines [70]. This effect requires an E at amino acid position 92 of the NS1 molecule and allows virus replication in the presence of IFN; this mutation was a determinant of virulence in pigs [71]. Furthermore, an E instead of a D residue at this position increased virulence of HPAI H5N1 in mice.
Large-scale genome sequence analysis of avian influenza virus isolates indicated that four carboxyterminal residues of the NS1 protein form a PDZ (postsynaptic density protein 95, Drosophila disc large tumor suppressor, and zonula occludens 1 protein) ligand domain of the X-S/T-X-V type [72]. This PDZ ligand domain of NS1 has also been shown to influence influenza virus virulence. Pandemic 1918 H1N1 and H5N1 HPAIviruses contain a PDZ ligand domain motif which increases virulence when introduced into a mouse-adapted influenza strain [73]. This demonstrates that NS1 can modulate virulence through different mechanisms. pH1N1 lacks the ability to block host gene expression in both human and swine cell lines [74]. Additionally, pH1N1 has a truncated NS1 protein with a 11 amino acid deletion at its C-terminus, and therefore lacks the PDZ-binding domain [16]. However, even when these functions are restored for pH1N1, they do not appear to have a significant effect on replication, virulence or transmission of pH1N1 in various animal models [74].
NA
Considering that HA binds to SA receptors and NA cleaves SA from cellular receptors, the balance between HA and NA activity is critical for virus replication and transmission [75].
Two influenza viruses are known that have developed additional mechanisms that promote cleavage of HA. The NA of the neurovirulent laboratory H1N1 strain A/WSN/33 recruits plasminogen which, when converted to plasmin, cleaves HA in the absence of trypsin [76]. On the other hand, the 1918 H1N1 NA gene enables the virus to replicate in the absence of trypsin. Additionally, this NA protein was shown to play a critical role in the high virulence of the 1918 pandemic H1N1 in mice [77].
In 2003, an outbreak of HPAI H7N7 virus in poultry in the Netherlands resulted in the death of one person and 89 human cases of conjunctivitis. When the sequence of the virus obtained from the fatal case was compared to the sequence of a virus isolated from a patient with conjunctivitis, four amino acid substitutions in the NA gene were identified [21]. These mutations all contributed to an increased NA activity, resulting in more efficient replication in mammalian cells most likely by preventing the formation of virus aggregates [44].
When avian influenza viruses are transmitted from wild birds to poultry, genetic changes as a result of adaptation to the new host frequently occur. One example of such a change is a deletion in the stalk region of the NA that has been reported in several viruses isolated from unrelated poultry outbreaks [78]. This shortened NA stalk region is frequently detected upon transmission of avian influenza viruses from waterfowl to domestic poultry and is associated with increased virulence [79, 80]. It is not yet clear how this shortened NA stalk region influences virulence, however, deletion in the NA stalk does not enhance the release of progeny viruses since the active site in the head cannot efficiently access its substrate [81].
Transmission
Human-to-human transmission of influenza viruses can occur through direct contact, indirect contact via fomites (contaminated environmental surfaces), and/or airborne transmission via small aerosols or large respiratory droplets. Efficient and sustained human-to-human transmission is critical for the circulation of seasonal and pandemic influenza viruses in the human population. Transmission has been studied extensively in mammalian models, in particular the ferret and guinea pig [82]. Ferrets are naturally susceptible to both human and avian influenza viruses and upon infection develop similar symptoms and pulmonary pathology as humans. In addition, cells of the ferret respiratory tract express predominantly α2,6-linked SA receptors in the URT and α2,3-linked SA receptors in the LRT, similar to humans [83, 84]. Avian influenza viruses do not transmit via the airborne route in the ferret model [85, 86]. Therefore, the ferret model is a valuable tool to study viral traits for influenza virus transmission in mammals. This is highly relevant as it is currently unclear what exactly determines transmission of influenza viruses in mammals via aerosols or respiratory droplets. For this reason, the ferret model was used extensively to compare the transmissibility of pH1N1 with the contemporary seasonal H1N1 virus, when it first emerged in humans [86]. In addition, transmission of the 1918 H1N1 virus was studied in ferrets. These studies showed that changes in the HA receptor-binding domain and PB2 were critical to initiate transmission of an avian-derived influenza virus [39, 87]. Similar genetic changes were required for the Asian H2N2 virus. An early H2N2 virus (1957) failed to transmit to naïve ferrets. However a Q to L at position 226 in HA was sufficient to change its binding preference from avian to human receptors, subsequently resulting in transmission between ferrets [88]. Overall, amino acid substitutions in HA and polymerase proteins can affect host range and transmission of influenza viruses [56, 59, 87].
As described above, avian influenza viruses of the H5, H7, and H9 subtypes have infected humans on several occasions and are therefore considered a potential pandemic threat. However, the requirements for a virus to become pandemic (i.e. transmissible between humans) are poorly understood. In order to study the determinants that could lead to a pandemic virus, an avian H9N2 virus that harbored the internal genes from a human H3N2 virus was adapted to replication in mammals by serial pass-aging in ferrets. This ‘adapted’ virus was found to be transmitted efficiently between ferrets via respiratory droplets [89]. This indicates that avian H9N2 viruses may acquire the ability to be transmitted between humans.
The lack of sustained transmission of HPAI H5N1 virus between humans has been confirmed in guinea pigs and ferrets. Early attempts to create airborne-transmissible H5 viruses by generating reassortant viruses between H5N1 and human influenza viruses did not result in H5 viruses that could be transmitted between mammals via the airborne route [90–92]. Based on evidence from previous influenza pandemics, it has been hypothesized that a switch of receptor binding preference from avian α2,3-linked to human α2,6-linked SA receptors is required for an avian virus to become transmissible between humans. Nevertheless, changing the receptor binding preference alone was not sufficient to confer airborne transmission of H5N1 virus, indicating that additional adaptive changes are required for H5N1 viruses to become transmissible [90, 93]. Herfst et al. recently demonstrated airborne transmissibility of a fully avian, ferret-adapted H5N1 virus. This research proved that avian-origin H5N1 viruses with a human SA receptor binding preference can become airborne-transmissible, but that indeed additional mutations were required for this phenotype [93]. Recent transmission studies in guinea pigs demonstrate that reassortants between H5N1 and pH1N1 virus that harbor the H5N1 HA with a dual SA receptor preference, are airborne-transmissible between guinea pigs as well [94]. In addition, phenotypical analysis of airborne H5 virus demonstrated that, the stability of HA in an acidic environment is important for airborne transmission [95].
For H7N9 virus, only one case of non-sustainable transmission between humans has been reported to date [96]. However, transmission experiments in ferrets indicated that this virus has a limited ability to be transmitted via the airborne route is limited [97–100].
The recent H7N9 outbreak again accentuates that increased understanding of the mechanisms and molecular determinants that facilitate avian influenza viruses to cross the species barrier and become airborne-transmissible in humans, is urgently needed. It is still impossible to predict when a new influenza virus will emerge in humans to cause the next pandemic, and what the subtype of this virus will be [101]. Therefore, surveillance of bird and swine influenza viruses should specifically target particular mutations that render viruses more virulent or airborne-transmissible in humans, as described above. Detection of such genetic traits should trigger more aggressive control programs than those employed currently. Improving pandemic preparedness by developing new vaccines that induce broader and stronger immune responses than the current influenza vaccines is another research priority. The ultimate goal of influenza vaccine research should be to design a universal vaccine that would induce protection against all influenza virus subtypes.
Table 1.
Important determinants of viral pathogenicity
| Protein | Function | Position | Reference |
|---|---|---|---|
| HA | Alter cellular host range and tissue tropism | Receptor binding site | [40, 102] |
| Determines by which cellular proteases HA is cleaved | Cleavage site | [6] | |
| Potential N-glycosylation motif which affects binding | 154–156 | [43, 44] | |
| PB2 | Replication advantage in mammalian species | 627 | [53] |
| 701 | [54, 57] | ||
| 590/591 | [59] | ||
| PA-PB1-NP- | Increased polymerase activity in mammalian cells | N/A | [60] |
| NEP | |||
| PB1-F2 | Induction of apoptosis; Antagonize the IFN response | 66 | [61, 65] |
| PA-X | Regulates host-cell shutoff | N/A | [4] |
| NS-1 | Evasion of host immune response | C-term | [72, 73] |
| 92 | [71] | ||
| NA | Release of virus particles | N/A | [75, 103] |
| Additional mechanisms that promote cleavage of HA | unknown | [76, 77] | |
| Adaptation and increased virulence upon deletion | Stalk region | [79, 80] |
Acknowledgements
The research of the authors is sponsored under contract HHSN266200700010C from National Institutes of Health/National Institute of Allergy and Infectious Diseases (NIH/NIAID) and FP7 Program ANTIGONE of the European Union.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Palese P, Shaw ML. Fields Virology Lippincott. Philadelphia, USA: Williams & Wilkins; 2007. pp. p 1647–p 1690. [Google Scholar]
- 2.Wise HM, Hutchinson EC, Jagger BW, Stuart AD, Kang ZH, Robb N, Schwartzman LM, Kash JC, Fodor E, Firth AE, Gog JR, Taubenberger JK, Digard P. Identification of a novel splice variant form of the influenza a virus m2 ion channel with an antigenically distinct ectodomain. PLoS Pathog. 2012;8(11):e1002998. doi: 10.1371/journal.ppat.1002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wise HM, Foeglein A, Sun J, Dalton RM, Patel S, Howard W, Anderson EC, Barclay WS, Digard P. A complicated message: Identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. Journal of virology. 2009;83(16):8021–8031. doi: 10.1128/JVI.00826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science. 2012;337(6091):199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muramoto Y, Noda T, Kawakami E, Akkina R, Kawaoka Y. Identification of novel influenza A virus proteins translated from PA mRNA. Journal of virology. 2012 doi: 10.1128/JVI.02656-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertram S, Glowacka I, Steffen I, Kuhl A, Pohlmann S. Novel insights into proteolytic cleavage of influenza virus hemagglutinin. Reviews in medical virology. 2010;20(5):298–310. doi: 10.1002/rmv.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. H5N1 Virus Attachment to Lower Respiratory Tract. Science. 2006;312(5772):399. doi: 10.1126/science.1125548. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252(2):324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 9.Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305(5682):371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 10.Ito T, Couceiro JN, Kelm S, Baum LG, Krauss S, Castrucci MR, Donatelli I, Kida H, Paulson JC, Webster RG, Kawaoka Y. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. Journal of virology. 1998;72(9):7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falchi A, Arena C, Andreoletti L, Jacques J, Leveque N, Blanchon T, Lina B, Turbelin C, Dorleans Y, Flahault A, Amoros JP, Spadoni G, Agostini F, Varesi L. Dual infections by influenza A/H3N2 and B viruses and by influenza A/H3N2 and A/H1N1 viruses during winter 2007, Corsica Island, France. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2008;41(2):148–151. doi: 10.1016/j.jcv.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Gregory V, Bennett M, Orkhan MH, Al Hajjar S, Varsano N, Mendelson E, Zambon M, Ellis J, Hay A, Lin YP. Emergence of influenza A H1N2 reassortant viruses in the human population during 2001. Virology. 2002;300(1):1–7. doi: 10.1006/viro.2002.1513. [DOI] [PubMed] [Google Scholar]
- 13.Holmes EC, Ghedin E, Miller N, Taylor J, Bao Y, St George K, Grenfell BT, Salzberg SL, Fraser CM, Lipman DJ, Taubenberger JK. Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and re-assortment among recent H3N2 viruses. PLoS Biol. 2005;3(9):e300. doi: 10.1371/journal.pbio.0030300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taubenberger JK, Morens DM. 1918 Influenza: the mother of all pandemics. Emerging infectious diseases. 2006;12(1):15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scholtissek C, Rohde W, Von Hoyningen V, Rott R. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology. 1978;87(1):13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- 16.Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459(7249):931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Jong JC, Claas EC, Osterhaus AD, Webster RG, Lim WL. A pandemic warning? Nature. 1997;389(6651):554. doi: 10.1038/39218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.WHO. Confirmed Human Cases of Avian Influenza A(H5N1) [Cited 05 July 2013];2013 http://www.who.int/influenza/human_animal_interface/EN_GIP_20130705CumulativeNumberH5N1cases_2.pdf.
- 19.Wang H, Feng Z, Shu Y, Yu H, Zhou L, Zu R, Huai Y, Dong J, Bao C, Wen L, Wang H, Yang P, Zhao W, Dong L, Zhou M, Liao Q, Yang H, Wang M, Lu X, Shi Z, Wang W, Gu L, Zhu F, Li Q, Yin W, Yang W, Li D, Uyeki TM, Wang Y. Probable limited person-to-person transmission of highly pathogenic avian influenza A (H5N1) virus in China. Lancet. 2008;371(9622):1427–1434. doi: 10.1016/S0140-6736(08)60493-6. [DOI] [PubMed] [Google Scholar]
- 20.Nidom CA, Takano R, Yamada S, Sakai-Tagawa Y, Daulay S, Aswadi D, Suzuki T, Suzuki Y, Shinya K, Iwatsuki-Horimoto K, Muramoto Y, Kawaoka Y. Influenza A (H5N1) viruses from pigs, Indonesia. Emerging infectious diseases. 2010;16(10):1515–1523. doi: 10.3201/eid1610.100508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rimmelzwaan GF, Schutten M, Van Doornum GJ, Koch G, Bosman A, Koopmans M, Osterhaus AD. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A. 2004;101(5):1356–1361. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Wit E, Fouchier RA. Emerging influenza. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2008;(1):1–6. doi: 10.1016/j.jcv.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.WHO. Human infection with avian influenza A(H7N9) virus – update. 2013 http://www.who.int/csr/don/2013_08_11/en/index.html.
- 24.Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, Lai RW, Orr WK, Shortridge KF. Human infection with influenza H9N2. Lancet. 1999;354(9182):916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 25.Guo Y, Dong J, Wang M, Zhang Y, Guo J, Wu K. Characterization of hemagglutinin gene of influenza A virus subtype H9N2. Chinese medical journal. 2001;114(1):76–79. [PubMed] [Google Scholar]
- 26.Yu H, Zhou YJ, Li GX, Ma JH, Yan LP, Wang B, Yang FR, Huang M, Tong GZ. Genetic diversity of H9N2 influenza viruses from pigs in China: a potential threat to human health? Veterinary microbiology. 2011;149(1–2):254–261. doi: 10.1016/j.vetmic.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Matrosovich MN, Krauss S, Webster RG. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology. 2001;281(2):156–162. doi: 10.1006/viro.2000.0799. [DOI] [PubMed] [Google Scholar]
- 28.Yuan J, Zhang L, Kan X, Jiang L, Yang J, Guo Z, Ren Q. Origin and molecular characteristics of a novel 2013 avian influenza A H6N1 virus causing human infection in Taiwan. Clin Infect Dis. 2013 doi: 10.1093/cid/cit479. [DOI] [PubMed] [Google Scholar]
- 29.Gillim-Ross L, Santos C, Chen Z, Aspelund A, Yang CF, Ye D, Jin H, Kemble G, Subbarao K. Avian influenza h6 viruses productively infect and cause illness in mice and ferrets. Journal of virology. 2008;82(21):10854–10863. doi: 10.1128/JVI.01206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffmann E, Stech J, Leneva I, Krauss S, Scholtissek C, Chin PS, Peiris M, Shortridge KF, Webster RG. Characterization of the influenza A virus gene pool in avian species in southern China: was H6N1 a derivative or a precursor of H5N1? Journal of virology. 2000;74(14):6309–6315. doi: 10.1128/jvi.74.14.6309-6315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munster VJ, Baas C, Lexmond P, Waldenstrom J, Wallensten A, Fransson T, Rimmelzwaan GF, Beyer WE, Schutten M, Olsen B, Osterhaus AD, Fouchier RA. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 2007;3(5):e61. doi: 10.1371/journal.ppat.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krauss S, Obert CA, Franks J, Walker D, Jones K, Seiler P, Niles L, Pryor SP, Obenauer JC, Naeve CW, Widjaja L, Webby RJ, Webster RG. Influenza in migratory birds and evidence of limited intercontinental virus exchange. PLoS Pathog. 2007;3(11):e167. doi: 10.1371/journal.ppat.0030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearce MB, Jayaraman A, Pappas C, Belser JA, Zeng H, Gustin KM, Maines TR, Sun X, Raman R, Cox NJ, Sasisekharan R, Katz JM, Tumpey TM. Pathogenesis and transmission of swine origin A(H3N2)v influenza viruses in ferrets. Proc Natl Acad Sci U S A. 2012;109(10):3944–3949. doi: 10.1073/pnas.1119945109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shu B, Garten R, Emery S, Balish A, Cooper L, Sessions W, Deyde V, Smith C, Berman L, Klimov A, Lindstrom S, Xu X. Genetic analysis and antigenic characterization of swine origin influenza viruses isolated from humans in the United States, 1990–2010. Virology. 2012;422(1):151–160. doi: 10.1016/j.virol.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 35.Schrauwen EJ, Herfst S, Chutinimitkul S, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Kuiken T, Fouchier RA. Possible Increased Pathogenicity of Pandemic (H1N1) 2009 Influenza Virus upon Reassortment. Emerging infectious diseases. 2011;17(2):200–208. doi: 10.3201/eid1702.101268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Angel M, Kimble JB, Pena L, Wan H, Perez DR. In vivo selection of H1N2 influenza virus reassortants in the ferret model. Journal of virology. 2013;87(6):3277–3283. doi: 10.1128/JVI.02591-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lycett SJ, Baillie G, Coulter E, Bhatt S, Kellam P, McCauley JW, Wood JL, Brown IH, Pybus OG, Leigh Brown AJ. Estimating re-assortment rates in co-circulating Eurasian swine influenza viruses. The Journal of general virology. 2012;93(Pt 11):2326–2336. doi: 10.1099/vir.0.044503-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glaser L, Stevens J, Zamarin D, Wilson IA, Garcia-Sastre A, Tumpey TM, Basler CF, Taubenberger JK, Palese P. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. Journal of virology. 2005;79(17):11533–11536. doi: 10.1128/JVI.79.17.11533-11536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tumpey TM, Maines TR, Van Hoeven N, Glaser L, Solorzano A, Pappas C, Cox NJ, Swayne DE, Palese P, Katz JM, Garcia-Sastre A. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science. 2007;315(5812):655–659. doi: 10.1126/science.1136212. [DOI] [PubMed] [Google Scholar]
- 40.Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, Kawaoka Y. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. Journal of virology. 2000;74(18):8502–8512. doi: 10.1128/jvi.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chutinimitkul S, van Riel D, Munster VJ, van den Brand JM, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Fouchier RA, de Wit E. In vitro assessment of attachment pattern and replication efficiency of H5N1 influenza A viruses with altered receptor specificity. Journal of virology. 2010;84(13):6825–6833. doi: 10.1128/JVI.02737-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312(5772):404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 43.Chen H, Bright RA, Subbarao K, Smith C, Cox NJ, Katz JM, Matsuoka Y. Polygenic virulence factors involved in pathogenesis of 1997 Hong Kong H5N1 influenza viruses in mice. Virus research. 2007;128(1–2):159–163. doi: 10.1016/j.virusres.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 44.de Wit E, Munster VJ, van Riel D, Beyer WE, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Fouchier RA. Molecular determinants of adaptation of highly pathogenic avian influenza H7N7 viruses to efficient replication in the human host. Journal of virology. 2010;84(3):1597–1606. doi: 10.1128/JVI.01783-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garten W, Bosch FX, Linder D, Rott R, Klenk HD. Proteolytic activation of the influenza virus hemagglutinin: The structure of the cleavage site and the enzymes involved in cleavage. Virology. 1981;115(2):361–374. doi: 10.1016/0042-6822(81)90117-3. [DOI] [PubMed] [Google Scholar]
- 46.Munster VJ, Schrauwen EJ, de Wit E, van den Brand JM, Bestebroer TM, Herfst S, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Insertion of a multi-basic cleavage motif into the hemagglutinin of a low-pathogenic avian influenza H6N1 virus induces a highly pathogenic phenotype. Journal of virology. 2010;84(16):7953–7960. doi: 10.1128/JVI.00449-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veits J, Weber S, Stech O, Breithaupt A, Graber M, Gohrbandt S, Bogs J, Hundt J, Teifke JP, Mettenleiter TC, Stech J. Avian influenza virus hemagglutinins H2, H4, H8, and H14 support a highly pathogenic phenotype. Proc Natl Acad Sci U S A. 2012;109(7):2579–2584. doi: 10.1073/pnas.1109397109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rimmelzwaan GF, Kuiken T, van Amerongen G, Bestebroer TM, Fouchier RA, Osterhaus AD. Pathogenesis of influenza A (H5N1) virus infection in a primate model. Journal of virology. 2001;75(14):6687–6691. doi: 10.1128/JVI.75.14.6687-6691.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293(5536):1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- 50.Bogs J, Veits J, Gohrbandt S, Hundt J, Stech O, Breithaupt A, Teifke JP, Mettenleiter TC, Stech J. Highly pathogenic H5N1 influenza viruses carry virulence determinants beyond the polybasic hemagglutinin cleavage site. PLoS One. 2010;5(7):e11826. doi: 10.1371/journal.pone.0011826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schrauwen EJ, Bestebroer TM, Munster VJ, de Wit E, Herfst S, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Insertion of a multi-basic cleavage site in the haemagglutinin of human influenza H3N2 virus does not increase pathogenicity in ferrets. The Journal of general virology. 2011;92(Pt 6):1410–1415. doi: 10.1099/vir.0.030379-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schrauwen EJ, Herfst S, Leijten LM, van Run P, Bestebroer TM, Linster M, Bodewes R, Kreijtz JH, Rimmelzwaan GF, Osterhaus AD, Fouchier RA, Kuiken T, van Riel D. The Multi-basic Cleavage Site in H5N1 Virus Is Critical for Systemic Spread along the Olfactory and Hematogenous Routes in Ferrets. Journal of virology. 2012;86(7):3975–3984. doi: 10.1128/JVI.06828-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subbarao EK, London W, Murphy BR. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. Journal of virology. 1993;67(4):1761–1764. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Z, Chen H, Jiao P, Deng G, Tian G, Li Y, Hoffmann E, Webster RG, Matsuoka Y, Yu K. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. Journal of virology. 2005;79(18):12058–12064. doi: 10.1128/JVI.79.18.12058-12064.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, Qui PT, Cam BV, Ha do Q, Guan Y, Peiris JS, Chinh NT, Hien TT, Farrar J. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nature medicine. 2006;12(10):1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steel J, Lowen AC, Mubareka S, Palese P. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 2009;5(1):e1000252. doi: 10.1371/journal.ppat.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gabriel G, Herwig A, Klenk HD. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog. 2008;4(2):e11. doi: 10.1371/journal.ppat.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Herfst S, Chutinimitkul S, Ye J, de Wit E, Munster VJ, Schrauwen EJ, Bestebroer TM, Jonges M, Meijer A, Koopmans M, Rimmelzwaan GF, Osterhaus AD, Perez DR, Fouchier RA. Introduction of virulence markers in PB2 of pandemic swine-origin influenza virus does not result in enhanced virulence or transmission. Journal of virology. 2010;84(8):3752–3758. doi: 10.1128/JVI.02634-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mehle A, Doudna JA. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc Natl Acad Sci U S A. 2009;106(50):21312–21316. doi: 10.1073/pnas.0911915106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manz B, Schwemmle M, Brunotte L. Adaptation of avian influenza a virus polymerase in mammals to overcome the host species barrier. Journal of virology. 2013;87(13):7200–7209. doi: 10.1128/JVI.00980-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O'Neill R, Schickli J, Palese P, Henklein P, Bennink JR, Yewdell JW. A novel influenza A virus mitochondrial protein that induces cell death. Nature medicine. 2001;7(12):1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- 62.McAuley JL, Hornung F, Boyd KL, Smith AM, McKeon R, Bennink J, Yewdell JW, McCullers JA. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell host & microbe. 2007;2(4):240–249. doi: 10.1016/j.chom.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 2007;3(10):1414–1421. doi: 10.1371/journal.ppat.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McAuley JL, Chipuk JE, Boyd KL, Van De Velde N, Green DR, McCullers JA. PB1-F2 proteins from H5N1 and 20 century pandemic influenza viruses cause immunopathology. PLoS Pathog. 2010;6(7):e1001014. doi: 10.1371/journal.ppat.1001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Varga ZT, Ramos I, Hai R, Schmolke M, Garcia-Sastre A, Fernandez-Sesma A, Palese P. The influenza virus protein PB1-F2 inhibits the induction of type I interferon at the level of the MAVS adaptor protein. PLoS Pathog. 2011;7(6):e1002067. doi: 10.1371/journal.ppat.1002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hai R, Schmolke M, Varga ZT, Manicassamy B, Wang TT, Belser JA, Pearce MB, Garcia-Sastre A, Tumpey TM, Palese P. PB1-F2 expression by the 2009 pandemic H1N1 influenza virus has minimal impact on virulence in animal models. Journal of virology. 2010;84(9):4442–4450. doi: 10.1128/JVI.02717-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ozawa M, Basnet S, Burley LM, Neumann G, Hatta M, Kawaoka Y. Impact of amino acid mutations in PB2, PB1-F2, and NS1 on the replication and pathogenicity of pandemic (H1N1) 2009 influenza viruses. Journal of virology. 2011;85(9):4596–4601. doi: 10.1128/JVI.00029-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shi M, Jagger BW, Wise HM, Digard P, Holmes EC, Taubenberger JK. Evolutionary conservation of the PA-X open reading frame in segment 3 of influenza A virus. Journal of virology. 2012;86(22):12411–12413. doi: 10.1128/JVI.01677-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hale BG, Randall RE, Ortin J, Jackson D. The multifunctional NS1 protein of influenza A viruses. The Journal of general virology. 2008;89(Pt 10):2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 70.Peiris JS, Yu WC, Leung CW, Cheung CY, Ng WF, Nicholls JM, Ng TK, Chan KH, Lai ST, Lim WL, Yuen KY, Guan Y. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet. 2004;363(9409):617–619. doi: 10.1016/S0140-6736(04)15595-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seo SH, Hoffmann E, Webster RG. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nature medicine. 2002;8(9):950–954. doi: 10.1038/nm757. [DOI] [PubMed] [Google Scholar]
- 72.Obenauer JC, Denson J, Mehta PK, Su X, Mukatira S, Finkelstein DB, Xu X, Wang J, Ma J, Fan Y, Rakestraw KM, Webster RG, Hoffmann E, Krauss S, Zheng J, Zhang Z, Naeve CW. Large-scale sequence analysis of avian influenza isolates. Science. 2006;311:1576–1580. doi: 10.1126/science.1121586. [DOI] [PubMed] [Google Scholar]
- 73.Jackson D, Hossain MJ, Hickman D, Perez DR, Lamb RA. A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc Natl Acad Sci U S A. 2008;105(11):4381–4386. doi: 10.1073/pnas.0800482105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hale BG, Steel J, Medina RA, Manicassamy B, Ye J, Hickman D, Hai R, Schmolke M, Lowen AC, Perez DR, Garcia-Sastre A. Inefficient control of host gene expression by the 2009 pandemic H1N1 influenza A virus NS1 protein. Journal of virology. 2010;84(14):6909–6922. doi: 10.1128/JVI.00081-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wagner R, Matrosovich M, Klenk HD. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Reviews in medical virology. 2002;12(3):159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 76.Goto H, Kawaoka Y. A novel mechanism for the acquisition of virulence by a human influenza A virus. Proc Natl Acad Sci U S A. 1998;95(17):10224–10228. doi: 10.1073/pnas.95.17.10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solorzano A, Swayne DE, Cox NJ, Katz JM, Taubenberger JK, Palese P, Garcia-Sastre A. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science. 2005;310(5745):77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 78.Li J, zu Dohna H, Anchell NL, Adams SC, Dao NT, Xing Z, Cardona CJ. Adaptation and transmission of a duck-origin avian influenza virus in poultry species. Virus research. 2010;147(1):40–46. doi: 10.1016/j.virusres.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 79.Munier S, Larcher T, Cormier-Aline F, Soubieux D, Su B, Guigand L, Labrosse B, Cherel Y, Quere P, Marc D, Naffakh N. A genetically engineered waterfowl influenza virus with a deletion in the stalk of the neuraminidase has increased virulence for chickens. Journal of virology. 2010;84(2):940–952. doi: 10.1128/JVI.01581-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sorrell EM, Song H, Pena L, Perez DR. A 27-amino-acid deletion in the neuraminidase stalk supports replication of an avian H2N2 influenza A virus in the respiratory tract of chickens. Journal of virology. 2010;84(22):11831–11840. doi: 10.1128/JVI.01460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Els MC, Air GM, Murti KG, Webster RG, Laver WG. An 18-amino acid deletion in an influenza neuraminidase. Virology. 1985;142(2):241–247. doi: 10.1016/0042-6822(85)90332-0. [DOI] [PubMed] [Google Scholar]
- 82.Bouvier NM, Lowen AC. Animal Models for Influenza Virus Pathogenesis and Transmission. Viruses. 2010;2(8):1530–1563. doi: 10.3390/v20801530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. The American journal of pathology. 2007;171(4):1215–1223. doi: 10.2353/ajpath.2007.070248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu Q, Wang W, Cheng X, Zengel J, Jin H. Influenza H1N1 A/Solomon Island/3/06 virus receptor binding specificity correlates with virus pathogenicity, antigenicity, and immunogenicity in ferrets. Journal of virology. 2010;84(10):4936–4945. doi: 10.1128/JVI.02489-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Herlocher ML, Elias S, Truscon R, Harrison S, Mindell D, Simon C, Monto AS. Ferrets as a transmission model for influenza: sequence changes in HA1 of type A (H3N2) virus. The Journal of infectious diseases. 2001;184(5):542–546. doi: 10.1086/322801. [DOI] [PubMed] [Google Scholar]
- 86.Munster VJ, de Wit E, van den Brand JM, Herfst S, Schrauwen EJ, Bestebroer TM, van de Vijver D, Boucher CA, Koopmans M, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Fouchier RA. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science. 2009;325(5939):481–483. doi: 10.1126/science.1177127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Van Hoeven N, Pappas C, Belser JA, Maines TR, Zeng H, Garcia-Sastre A, Sasisekharan R, Katz JM, Tumpey TM. Human HA and polymerase subunit PB2 proteins confer transmission of an avian influenza virus through the air. Proc Natl Acad Sci U S A. 2009;106(9):3366–3371. doi: 10.1073/pnas.0813172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pappas C, Viswanathan K, Chandrasekaran A, Raman R, Katz JM, Sasisekharan R, Tumpey TM. Receptor specificity and transmission of H2N2 subtype viruses isolated from the pandemic of 1957. PLoS One. 2010;5(6):e11158. doi: 10.1371/journal.pone.0011158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sorrell EM, Wan H, Araya Y, Song H, Perez DR. Minimal molecular constraints for respiratory droplet transmission of an avian-human H9N2 influenza A virus. Proc Natl Acad Sci U S A. 2009;106(18):7565–7570. doi: 10.1073/pnas.0900877106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maines TR, Chen LM, Van Hoeven N, Tumpey TM, Blixt O, Belser JA, Gustin KM, Pearce MB, Pappas C, Stevens J, Cox NJ, Paulson JC, Raman R, Sasisekharan R, Katz JM, Donis RO. Effect of receptor binding domain mutations on receptor binding and transmissibility of avian influenza H5N1 viruses. Virology. 2011;413(1):139–147. doi: 10.1016/j.virol.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jackson S, Van Hoeven N, Chen LM, Maines TR, Cox NJ, Katz JM, Donis RO. Re-assortment between avian H5N1 and human H3N2 influenza viruses in ferrets: a public health risk assessment. Journal of virology. 2009;83(16):8131–8140. doi: 10.1128/JVI.00534-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schrauwen EJ, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA, Herfst S. Re-assortment between Avian H5N1 and human influenza viruses is mainly restricted to the matrix and neuraminidase gene segments. PLoS One. 2013;8(3):e59889. doi: 10.1371/journal.pone.0059889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336(6088):1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang Y, Zhang Q, Kong H, Jiang Y, Gao Y, Deng G, Shi J, Tian G, Liu L, Liu J, Guan Y, Bu Z, Chen H. H5N1 hybrid viruses bearing 2009/H1N1 virus genes transmit in guinea pigs by respiratory droplet. Science. 2013;340(6139):1459–1463. doi: 10.1126/science.1229455. [DOI] [PubMed] [Google Scholar]
- 95.Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486(7403):420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Qi X, Qian YH, Bao CJ, Guo XL, Cui LB, Tang FY, Ji H, Huang Y, Cai PQ, Lu B, Xu K, Shi C, Zhu FC, Zhou MH, Wang H. Probable person to person transmission of novel avian influenza A (H7N9) virus in Eastern China, 2013: epidemiological investigation. BMJ. 2013;347:f4752. doi: 10.1136/bmj.f4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhu H, Wang D, Kelvin DJ, Li L, Zheng Z, Yoon SW, Wong SS, Farooqui A, Wang J, Banner D, Chen R, Zheng R, Zhou J, Zhang Y, Hong W, Dong W, Cai Q, Roehrl MH, Huang SS, Kelvin AA, Yao T, Zhou B, Chen X, Leung GM, Poon LL, Webster RG, Webby RJ, Peiris JS, Guan Y, Shu Y. Infectivity, Transmission, and Pathology of Human H7N9 Influenza in Ferrets and Pigs. Science. 2013 doi: 10.1126/science.1239844. [DOI] [PubMed] [Google Scholar]
- 98.Watanabe T, Kiso M, Fukuyama S, Nakajima N, Imai M, Yamada S, Murakami S, Yamayoshi S, Iwatsuki-Horimoto K, Sakoda Y, Takashita E, McBride R, Noda T, Hatta M, Imai H, Zhao D, Kishida N, Shirakura M, de Vries RP, Shichinohe S, Okamatsu M, Tamura T, Tomita Y, Fujimoto N, Goto K, Katsura H, Kawakami E, Ishikawa I, Watanabe S, Ito M, Sakai-Tagawa Y, Sugita Y, Uraki R, Yamaji R, Eisfeld AJ, Zhong G, Fan S, Ping J, Maher EA, Hanson A, Uchida Y, Saito T, Ozawa M, Neumann G, Kida H, Odagiri T, Paulson JC, Hasegawa H, Tashiro M, Kawaoka Y. Characterization of H7N9 influenza A viruses isolated from humans. Nature. 2013 doi: 10.1038/nature12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Belser JA, Gustin KM, Pearce MB, Maines TR, Zeng H, Pappas C, Sun X, Carney PJ, Villanueva JM, Stevens J, Katz JM, Tumpey TM. Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature. 2013 doi: 10.1038/nature12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Richard M, Schrauwen EJ, de Graaf M, Bestebroer TM, Spronken MI, van Boheemen S, de Meulder D, Lexmond P, Linster M, Herfst S, Smith DJ, van den Brand JM, Burke DF, Kuiken T, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Limited airborne transmission of H7N9 influenza A virus between ferrets. Nature. 2013 doi: 10.1038/nature12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sorrell E, Schrauwen E, Linster M, De Graaf M, Herfst S, Fouchier R. Predicting 'airborne' influenza viruses: (trans-) mission impossible? Current opinion in virology. 2011;1(6):635–642. doi: 10.1016/j.coviro.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Imai M, Kawaoka Y. The role of receptor binding specificity in interspecies transmission of influenza viruses. Current opinion in virology. 2012;2(2):160–167. doi: 10.1016/j.coviro.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xu R, Zhu X, McBride R, Nycholat CM, Yu W, Paulson JC, Wilson IA. Functional balance of the hemagglutinin and neuraminidase activities accompanies the emergence of the 2009 H1N1 influenza pandemic. Journal of virology. 2012;86(17):9221–9232. doi: 10.1128/JVI.00697-12. [DOI] [PMC free article] [PubMed] [Google Scholar]