Abstract
Nonalcoholic fatty liver disease (NAFLD) is the most common form of liver disease. To elucidate the molecular basis of NAFLD we performed an exome-wide association study of liver fat content. Three variants were associated with increased liver fat at the exome-wide significance level: two in PNPLA3, an established locus for NAFLD, and one (Glu167Lys) in TM6SF2, a gene of unknown function. The Glu167LysTM6SF2 variant was also associated with higher circulating levels of alanine transaminase, a marker of liver injury, and lower levels of LDL-cholesterol, triglycerides and alkaline phosphatase in 3 independent populations (n>80,000). Recombinant Glu167LysTM6SF2 produced 50% less protein than wild-type TM6SF2 when expressed in cultured hepatocytes. Adeno-associated virus-mediated shRNA knockdown of Tm6sf2 in mice increased liver triglyceride content 3-fold and decreased VLDL secretion by 50%. Taken together, these data indicate that TM6SF2 activity is required for normal VLDL secretion, and that impaired TM6SF2 function causally contributes to NAFLD.
Nonalcoholic fatty liver disease (NAFLD) comprises a spectrum of related disorders that stem from the aberrant accumulation of fat in the liver1. Approximately 30% of adults2 have excess liver fat (steatosis), which is stored in the form of triglyceride in cytoplasmic lipid droplets in hepatocytes. Simple steatosis is usually benign but can progress to a chronic inflammatory condition called nonalcoholic steatohepatitis (NASH) and ultimately to cirrhosis, in which the functional cells of the liver are replaced by fibrosis3. The propensity to accumulate hepatic triglyceride varies markedly among individuals2, but the factors underlying this variation have not been fully elucidated.
In 2008, we found a missense variant (I148M) in patatin-like phospholipase domain-containing 3 (PNPLA3) that is strongly associated with hepatic triglyceride content (HTGC) and serum levels of alanine transaminase (ALT)4 Subsequent genome-wide association studies found other common SNPs associated with liver fat content6, and levels of circulating liver enzymes7,8. To identify the functional variants at these loci, we used genotyping arrays (HumanExome BeadChip, Illumina) to perform an exome-wide association study in a multiethnic, population-based study, the Dallas Heart Study (DHS)9. A total of 138,374 sequence variants that were polymorphic and passed our quality control criteria were tested for association with HTGC in 2,736 DHS participants (1,324 non-Hispanic African-Americans, 882 non-Hispanic whites, 467 Hispanic and 61 other ethnicities) with adjustment for age, gender, ancestry and body mass index (BMI) (see Methods). Two sequence variants in PNPLA3 (rs738409 and rs2281135) had the lowest P-values (4.0×10−16 and 6.9×10−12, respectively), followed by a variant (rs58542926) in TM6SF2 (P=5.7×10−08) (Fig. 1a). No other variants exceeded the exome-wide significance threshold. After excluding these 3 SNPs, the quantile-quantile plot of P-values showed no systematic deviation from the expected null distribution (Fig. 1b). The TM6SF2 variant was not associated with other risk factors for hepatic steatosis, including BMI, homeostatic model assessment-insulin resistance (HOMA-IR) or alcohol intake (Supplementary Table 1).
Figure 1.
Exome-wide association with hepatic triglyceride content in the Dallas Heart Study (DHS). (a) Manhattan plot showing the association of 138,374 sequence variants on the HumanExome Array (BeadChip, Illumina) with hepatic triglyceride content in the DHS (n=2,736). The dashed line denotes the Bonferroni corrected significance threshold. (b) Quantile-quantile plot of −log10 P-values. (c) Evolutionary conservation of TM6SF2. Bolded letters denote residues that are conserved among all species shown. Residue 167 is shown in red. (d). Mean hepatic triglyceride content (± s.e.m.) by TM6SF2 genotype (rs58542926) in the DHS. The association was tested using linear regression with adjustment for age, gender, ancestry and BMI. (e) Levels of TM6SF2 mRNA in human tissues. Quantitative Real-time PCR was performed on mRNA extracted from human tissues (Clontech). Each bar represents the average of a triplicate measurement expressed as a fraction of the Ct value obtained from the tissue expressing the highest level (small intestine). The values were normalized to the levels of the 36B4 transcript.
The TM6SF2 variant associated with HTGC is an adenine for guanine substitution in coding nucleotide 499, which replaces glutamate at residue 167 with lysine. Glu167 is highly conserved among mammals and is also an acidic residue (aspartate) in birds (Fig. 1c). The frequency of the Glu167LysTM6SF2 variant was higher in individuals of European ancestry (7.2%), than in African- (3.4%) or Hispanic-Americans (4.7%). Carriers of the Glu167LysTM6SF2 variant had elevated mean and median HTGC in all three ethnic groups, although the difference did not reach statistical significance in Hispanics, likely due to the lower number of Hispanic participants and the lower frequency of the variant in this group (Fig. 1d and source file). The association remained significant after adjusting for ethanol intake, and HOMA-IR (P=5.6×10−7). The effect of the Glu167LysTM6SF2 variant on HTGC was independent of the PNPLA3 rs738409 polymorphism; we found no evidence for statistical interaction between the two risk alleles (P=0.62).
Previously, SNPs near TM6SF2 were found to be associated with NAFLD6,10, 11. The variant at the locus that was most strongly associated with HTGC in the largest GWAS6 (total of 2.4 million imputed or assayed SNPs) was in NCAN (rs2228603). The Glu167LysTM6SF2 variant remained robustly associated (P = 1.3×10−5) with HTGC after conditioning on rs2228603, as well as on other SNPs from the region on the array (Supplementary Table 2). Conversely, conditioning on the Glu167LysTM6SF2 variation abolished the association between the NCAN variant (rs2228603) and HTGC (conditional P=0.8).
Real time PCR analysis of cDNAs prepared from a panel of human tissues indicated that TM6SF2 is most highly expressed in small intestine, liver and kidney, and is present at lower levels in other tissues (Fig. 1e).
The Glu167LysTM6SF2 variant was also associated with a significant increase in serum alanine transaminase (ALT) activity, consistent with increased hepatic injury (Table 1). These results are similar to what was previously observed with the Ile148MetPNPLA3 variant, which is strongly associated with both hepatic triglyceride content and with elevated ALT activity4,7. To confirm the association with NAFLD, we performed association studies in two additional cohorts: the Dallas Biobank (n=8,585 European-Americans) and a cohort from Copenhagen, Denmark (referred to as the Copenhagen Study in this paper) that included both the Copenhagen City Heart Study and the Copenhagen General Population Study (n=73,532)12 (Supplementary Tables 4 and 5). As in the DHS, the Glu167LysTM6SF2 variant was associated with significantly higher serum activity of ALT in both cohorts (Table 1). Mean serum AST activity was also higher in the Glu167LysTM6SF2 homozygotes from both of these larger cohorts, but the increase only reached significance in the Copenhagen Study. These findings further support the hypothesis that Glu167LysTM6SF2 is associated with NAFLD and are consistent with the notion that the variant compromises hepatic integrity. No genotype-specific differences were observed in plasma levels of bilirubin or gamma-glutamyltransferase (Supplementary Tables 1 and 4), but in all three populations, the Glu167LysTM6SF2 variant was associated with a significant reduction in serum activity of alkaline phosphatase (ALP).
Table 1.
The association between the TM6SF2 (Glu167Lys) variant, liver enzymes and plasma lipid levels in the DHS (n=4,587), the Dallas BioBank (n=8,585), and the Copenhagen Study (n=73,532).
| Trait | EE | EK | KK | P-value |
|---|---|---|---|---|
| DHS | ||||
| n | 4,151 | 423 | 13 | - |
| ALT (U) | 23.5 ± 19.9 | 25.8 ± 18.0 | 29.6 ± 26.8 | 0.014 |
| AST (U) | 24.3 ± 20.6 | 25.1 ± 19.6 | 25.6 ± 16.5 | 0.17 |
| ALP (U) | 71.6 ± 26.4 | 68.0 ± 20.9 | 63.6 ± 16.4 | 0.031 |
| LDL-C (mg/dL) | 109 ± 36 | 105 ± 34 | 94 ± 39 | 0.005 |
| Triglyceride (mg/dL) | 123 ± 102 | 118 ± 91 | 130 ± 66 | 0.037 |
| Dallas Biobank (European-Americans) | ||||
| n | 7,416 | 1,112 | 57 | - |
| ALT (U) | 35.9 ± 16.3 | 36.5 ± 15 | 44.8 ± 23.8 | 0.003 |
| AST (U) | 27 ± 13.8 | 26.9 ± 10.7 | 30.2 ± 13.5 | 0.22 |
| ALP (U) | 69 ± 18.9 | 66.9 ± 17.3 | 65.9 ± 19.1 | 1.2 × 10−4 |
| LDL-C (mg/dL) | 107 ± 34 | 105 ± 32 | 97 ± 32 | 0.029 |
| Triglyceride (mg/dL) | 112 ± 79 | 105.3 ± 67 | 97.5 ± 54 | 6.7 × 10−5 |
| Copenhagen Study | ||||
| n | 61,279 | 11,700 | 553 | - |
| ALT (U) | 22.7 ± 17.2 | 23.8 ± 16.0 | 26.7 ± 20.9 | 7.6 × 10−14 |
| AST (U) (n=8,487) | 22.8 ± 13.2 | 23.9 ± 18.6 | 30.0 ± 36.6 | 1.8 × 10−4 |
| ALP (U) | 91.1 ± 37.5 | 89.7 ± 39.1 | 85.0 ± 34.2 | 4.3 × 10−7 |
| LDL-C (mg/dL) | 128 ± 39 | 127 ± 38 | 112 ± 36 | 4.7 × 10−14 |
| Triglyceride (mg/dL) | 152 ± 106 | 148 ± 96 | 133 ± 90 | 3.7 × 10−9 |
Values are means ± s.d. The association was tested using linear regression with adjustment for age, gender, BMI, and ethnicity (where appropriate). A logarithm transformation was applied to traits with non-normal distributions. AST was only measured in a subset of the Copenhagen Study (n=8,487). Ethnic breakdown for the Dallas Heart Study is provided in Supplementary Table 3. ALT, alanine aminotransferase; AST, aspartate transaminase; ALP, alkaline phosphatase; LDL-C, low density lipoprotein-cholesterol.
The Glu167LysTM6SF2 variant was associated with highly significant reductions in plasma levels of triglyceride and low density lipoprotein-cholesterol (LDL-C) in the DHS, Dallas Biobank, and Copenhagen Study cohorts (Table 1). Other SNPs in the region (rs1040196914, rs1699614815, and rs1721652516) were associated with differences in plasma levels of triglyceride and LDL-C. but TM6SF2 remained associated with HTGC when the genotypes at these loci were controlled for. Conversely, controlling for Glu167LysTM6SF2 abolished the signal for these SNPs (Supplementary Table 2). Plasma levels of HDL-C were not associated with the Glu167LysTM6SF2 variant (Supplementary Table 1). Thus, the Glu167Lys substitution in TM6SF2 results in an increase in HTGC and a decrease in plasma levels of liver-derived triglyceride-rich lipoproteins.
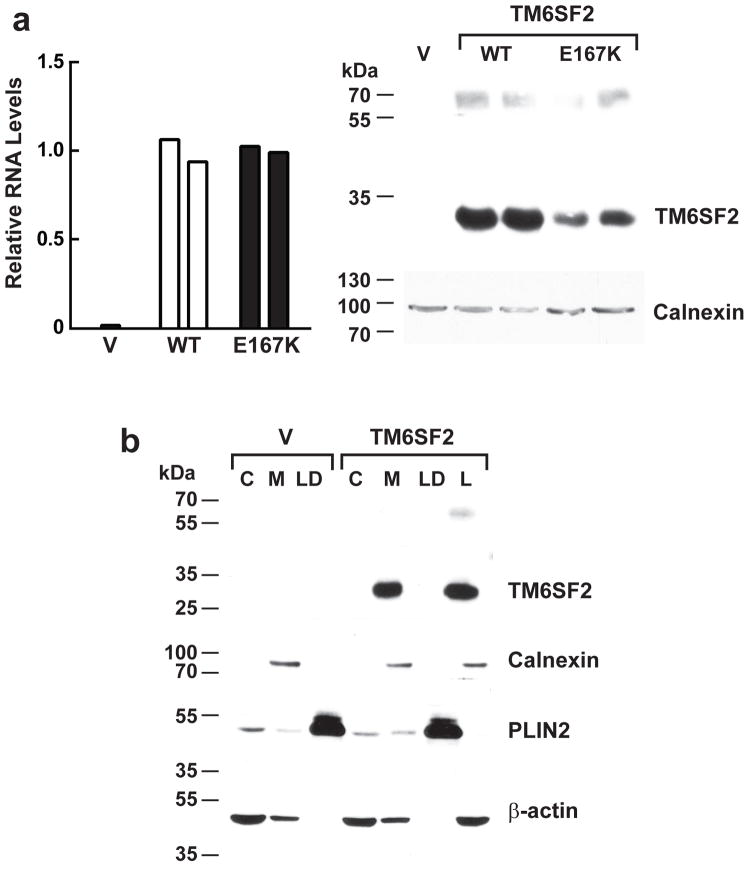
The specific biological role of TM6SF2 is not known. The protein is predicted to have 7 transmembrane domains (TMHMM 2.0)17, but does not contain any known functional domains18. To assess the effect of the Glu167Lys substitution on the expression and localization of TM6SF2 we expressed the human wild-type and mutant proteins in a human hepatoma cell line, HuH-7. Levels of TM6SF2 mRNA were comparable in cells expressing wild-type and mutant human TM6SF2 (Fig. 2a, left), but levels of the mutant protein was reduced 46% (Fig. 2a, right). Similar results were obtained in cells solubilized in 3% SDS and 8M urea. When lysates from Hepa1c1c7 cells expressing human TM6SF2-V5 were fractionated on sucrose density gradients, TM6SF2 was recovered exclusively in the membrane fraction (Fig. 2b). These findings suggest that TM6SF2 is a polytopic membrane protein and that the 167Lys isoform is misfolded and undergoes accelerated intracellular degradation.
Figure 2.
Expression of TM6SF2 in cultured hepatocytes. (a) Plasmids encoding wild-type and mutant human TM6SF2 were expressed in HuH7 cells. Two days after transfection, the TM6SF2 mRNA levels were measured using Real-Time PCR (left). The cells were harvested and solubilized in RIPA buffer (150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris, pH=8). Quantitative immunoblotting was performed using a LI-COR Odyssey infrared imaging system as described in the Methods (right). The experiment was performed twice and the results were similar. The blots shown are representative of two independent experiments. V, vector. (b) Recombinant wild-type hTM6SF2 was expressed in Hepa1c1c7 cells. After two days, the cells were fractionated and subjected to immunoblotting as described in the Methods. C, cytosol; M, membranes; LD, lipid droplets; L, whole cell lysate. The experiment was performed twice and the results were similar. The blots shown are representative of two independent experiments.
To directly assess the effect of loss of TM6SF2 function on HTGC, we used recombinant adeno-associated viral (AAV) vectors expressing shRNAs to selectively knockdown (KD) the gene in livers of mice. Expression of two different shRNAs targeting mouse Tm6sf2 resulted in >90% KD of TM6SF2 mRNA (Fig. 3a, left), without any significant changes in TM6SF2 mRNA in adipose tissue or small intestine (Supplementary Fig. 1). Empty AAV or AAV expressing nonfunctional shRNAs were used as controls (Supplementary Fig. 1b). Hepatic inhibition of Tm6sf2 increased HTGC 3-fold (Fig. 3a, middle) and significantly decreased plasma levels of cholesterol, in animals fed a chow diet ad lib (Fig. 3a, right). Plasma triglyceride levels tended to be lower in KD mice fed ad lib (Fig. 3a, right) and were consistently lower after a 4 hour fast (Fig 3c). ). FPLC fractionation of plasma lipoproteins isolated after a 4 h fast revealed that Tm6sf2 KD reduced the cholesterol content of both the LDL and HDL fractions and the triglyceride content of the VLDL fraction (Fig. 3b). Mean serum ALT levels were unchanged in KD mice when compared to controls (mean±s.d.: 136±8 U and 131±4 U vs. 132±8 U; P=0.33 and P=0.68, respectively
Figure 3.
Knockdown of Tm6sf2 expression in the liver of mice is associated with increased hepatic triglyceride content. (a) Adeno-associated virus vectors (AAV) expressing two different anti-Tm6sf2 shRNAs or vector alone were administered intravenously into the tail veins of 8-week old chow-fed C47BL/6J male mice (n=8/group). After two weeks, the livers were harvested and the levels of TM6SF2 mRNA were measured using Real-Time PCR (left). Lipids were extracted from the livers of the AAV-treated mice and triglyceride levels were quantified using enzymatic assays (middle). The plasma levels of cholesterol and triglyceride were measured in ad-lib fed AAV-treated mice at the end of the dark cycle (right). The experiment was performed three times and the results were similar. (b) Plasma lipoproteins from chow-fed C47BL/6J male mice (n=6/group) after a 4-h fast were fractionated using FPLC as described in the Methods. Cholesterol and triglyceride were measured enzymatically in each column fraction. The experiment was performed twice and the results were similar. (c) Hepatic VLDL secretion in Tm6sf2 KD mice. Plasma triglyceride levels after 4 hour fasting (left), and triglyceride accumulation after Triton WR 1339 injection (right) were measured in chow-fed male mice (8 week old, 4/group) treated with the AAV Tm6sf2-shRNA8 or control mice as described in the Methods. Two weeks after infection, mice were fasted for 4 h and injected via the tail vein with Triton WR1339 (500 μg/g body weight). Blood was sampled at the indicated times and plasma triglyceride levels were measured. Mean triglyceride levels at each time point are shown. The slopes of the lines were calculated by least squares regression and compared using a t-test. The experiment was performed twice and the results were similar. Error bars indicate standard deviations.
An increase in HTGC together with a decrease in plasma cholesterol and triglycerides is consistent with a defect in VLDL secretion. To determine the effect of Tm6sf2 KD on VLDL secretion, we inhibited intravascular lipoprotein lipase, which hydrolyzes the triglyceride in VLDL, and measured the rate of accumulation of triglyceride in plasma. Triglyceride accumulation was markedly reduced in the Tm6sf2 KD mice (mean±s.d.: 4.5±1.6 mg/dL/min vs. 11.1±0.6 mg/dL/min, Fig. 3c). These data indicate that TM6SF2 normally acts to promote VLDL secretion, and that the increased HTGC associated with the TM6SF2-167Lys variant in humans results from a reduction in TM6SF2 function.
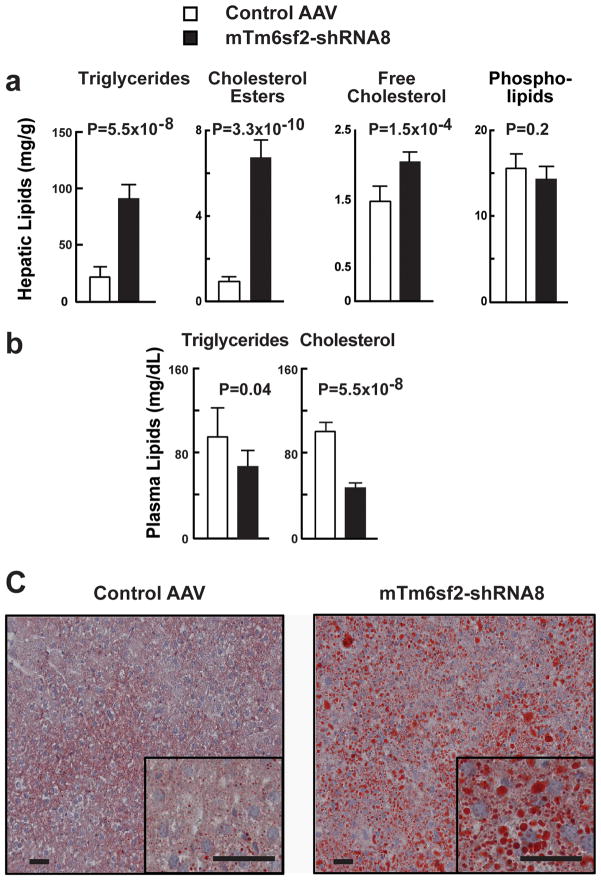
High sucrose diets, which increase hepatic triglyceride synthesis, exacerbated the effects of Tm6sf2 KD on HTGC (Fig. 4). In sucrose-fed mice, levels of triglyceride and cholesterol esters were increased in liver (Fig. 4a), and decreased in plasma (Fig. 4b). Oil Red O staining, which stains neutral lipids, showed an increase in the number and size of lipid droplets in the KD mice (Fig. 4c). Therefore, KD of Tm6sf2 selectively in mouse liver recapitulated the effects on HTGC and plasma lipids of the TM6SF2-167Lys variant observed in humans.
Figure 4.
Sucrose feeding in TM6SF2-KD mice. AAV expressing anti-Tm6sf2 shRNAs or vector alone were administered intravenously into the tail veins of 8-week old C47BL/6J male mice (n=6/group) and the mice were fed a high-sucrose diet for 4 weeks. (a) Lipids were extracted from the livers of the AAV-treated mice and hepatic triglyceride levels were quantified using enzymatic assays. (b) Plasma levels of cholesterol and triglyceride in TM6SF2-KD mice were measured enzymatically. The differences in mean hepatic triglyceride concentrations were compared using two-sample t-tests. (c) Sections from liver tissue of the same mice stained with Oil Red O, as described in Methods. Magnification: main image ×20, inset ×64. Bar represents 40 μm. The experiment was performed twice and the results were similar. Error bars show standard deviations.
TM6SF2 is expressed at highest levels in human intestine (Fig. 1e), which is the source of dietary-derived triglyceride-rich lipoprotein (i.e. chylomicrons). Alkaline phosphatase is also expressed in the intestine. Reduction in alkaline phosphatase activity in Glu167LysTM6SF2 carriers may be due to intestinal action of TM6SF2. Kaliannan et al. found that deletion of intestinal alkaline phosphatase produces features of metabolic syndrome in mice.19 We are currently testing the hypothesis that TM6SF2 plays a role in lipoprotein synthesis and alkaline phosphatase activity in the intestine, and that reduced intestinal alkaline phosphatase activity contributes to the increased HTGC associated with the Glu167LysTM6SF2 variant.
ONLINE METHODS
Study populations
The Dallas Heart Study (DHS) is a multiethnic population-based probability sample of Dallas County residents, weighted to include 50% African-American and 50% non-African-American participants1. Sampling design and recruitment procedures were previously described in detail20. The study was initiated in 2000, and converted from a cross-sectional to a longitudinal study in 2007, when all participants were invited for a repeat evaluation (DHS-2). The sampling process was designed to obtain a probability sample of residents that was representative of the target population, and to identify subjects to be recruited later for mechanistic substudies of hypertension and dyslipidemia. On the basis of power calculations for several such studies, the recruitment target was set at 3,000 subjects completing all phases of initial data collection. The DHS-2 cohort was augmented by voluntary participation of friends and spouses of the original participants. The DHS was approved by the Institutional Review Board of the University of Texas Southwestern Medical Center, and all subjects provided written informed consent. Each participant completed a detailed staff-administered survey, including questions about socioeconomic status, medical history, and medication use. Race/ethnicity was self-reported. A total of 4,708 DHS participants (52% non-Hispanic African-Americans, 29% non-Hispanic whites, 16% Hispanics, and 3% other ethnicities) who underwent clinical examination during DHS-1 or DHS-2 were included in the present analysis.
Findings in the DHS were replicated in two independent cohorts: the Dallas Biobank and the Copenhagen Study. The Dallas Biobank is a repository of DNA and plasma samples from individuals ascertained at various locations in North–Central Texas. All participants were over 18 years of age and gave written informed consent for inclusion in the database. Individuals for this study were European-Americans who completed a preventive medicine examination at the Cooper Clinic in Dallas, Texas between 2008 and 2012 after signing an informed consent that was approved by the Cooper Research Institute IRB. The Copenhagen Study combines participants in two studies of the Danish population: the Copenhagen General Population Study (CGPS)12 and the Copenhagen City Heart Study (CCHS)12. The CGPS and CCHS are prospective studies of the Danish general population initiated in, respectively, 2003 and 1976–1978, with ongoing enrollment. Individuals were selected based on the National Danish Civil Registration System to reflect the adult Danish population aged 20 to 100+ years. Data were obtained from a self-administered questionnaire reviewed together with an investigator on the day of attendance, a physical examination, and from blood samples including DNA extraction. Blood samples for DNA extraction and biochemical analyses were drawn on the day of enrollment in the CGPS (2003–2011), and on the day of enrollment at the 1991–1994 and 2001–2003 examinations of the CCHS. Studies were approved by Institutional Review Boards and Danish ethical committees, and were conducted according to the Declaration of Helsinki. Written informed consent was obtained from participants. All participants were white and of Danish descent, as determined by the National Danish Person Registration System. There was no overlap of individuals between the studies. We included 64,000 consecutive participants from the CGPS, and 9,532 participants from the CCHS, yielding a total of 73,532.
Clinical measurements
Body-mass index (BMI) was calculated as weight measured in kilograms divided by squared height in meters. Homeostatic model assessment of insulin resistance (HOMA-IR) was calculated from fasting concentrations of glucose and insulin. Plasma levels of high-density lipoprotein cholesterol (HDL-C) and triglycerides were determined by beta-quantification, and low-density lipoprotein cholesterol (LDL-C) concentrations were estimated using the Friedewald equation21. Liver enzyme levels (alanine transaminase (ALT), aspartate transaminase (AST), and alkaline phosphatase (ALP) were determined by enzymatic assays. Hepatic triglyceride content was measured with 1H-MRS as previously described22,23. Hepatic triglyceride measurements were available for a total of n=2,815 participants (including n=2,287 of the DHS-1 and n=528 DHS-2 participants).
Genotyping and quality control
Genomic DNA was extracted from circulating leukocytes. A total of 4,625 DHS-1 and DHS-2 participants were genotyped using Illumina Infinium HumanExome BeadChip (Illumina, San Diego, CA, USA), which captured a total of 247,870 markers, including functional exonic variants (>90%), disease-associated tag markers from recently published GWAS, ancestry-informative markers, and other markers. Genotypes were called using Illumina GenomeStudio software. Several quality-control filters were applied to samples and variants prior to analysis. Individuals were excluded for the following reasons: a call rate <99% (n=25) or duplicate discordance (n=1). Variants were excluded based on a call rate <99% (n=1,795) or a deviation from Hardy-Weinberg equilibrium (HWE) in African-Americans with p<0.0001 (n=221). In addition, variants that were monomorphic in our study population (61,947) or had a single heterozygote (n=27,705) were removed from analysis. We did not further filter variants based on minor allele frequency. After exclusions, a total of 156,202 variants and 4,591 individuals were available for analysis (including 138,374 variants in 2,735 individuals with measures of HTGC). Genotyping of the Dallas Biobank Sample and the Copenhagen Study was performed using a Taqman assay (dbSNPrs58542926; Applied Biosystems ID: C_89463510_10). Genotyping of the Copenhagen Study was performed by Taqman assay using primers 1 and 2 (Supplementary Table 6).
Association Analysis
To account for possible population stratification, we computed principal components of ancestry based on markers with a minor allele frequency >0.1% in the combined sample using the EIGENSTRAT software version 4.224. Each sequence variant was tested for association with hepatic triglyceride content using linear regression, assuming an additive genetic model (with genotypes coded as 0, 1, 2), and adjusted for age, gender, BMI, and the four leading principal components of ancestry. For very rare variants (<8 carriers) the analysis was performed using Wilcoxon rank-sum test, without covariate adjustment. A power transformation (1/4) was applied to liver fat measurements to achieve approximate normality of the residuals. Based on the number of tests performed, we set a significance threshold at 10−07 for each variant to maintain a family-wise type-I error rate of 5%. A quantile-quantile (Q-Q) plot of log10 p-values did not show a systematic deviation from the expected distribution.
In the follow-up and replication analyses, the association between TM6SF2-167Lys variant and clinical traits was tested using linear regression with adjustment for age, gender, BMI, and ethnicity (where appropriate). For hepatic fat content, we tested an additional model adjusted for HOMA-IR and alcohol consumption. For variables with skewed distributions a logarithm (BMI, ALT, AST, ALP, triglycerides) or a square-root (LDL) transformation was applied prior to analysis, to ensure that the residuals were approximately normal and had constant variance. All reported p-values are two-sided.
Mouse Studies
All experimental protocols were approved by the University of Texas Southwestern Medical Center Institutional Animal Care and Research Committee. Mice were housed in a vivarium and maintained under a standard 12 h light:12 h dark cycle and fed a chow diet (Teklad Mouse/Rat Diet 7001, Harlan Teklad) ad libitum unless otherwise stated. Experiments were performed in eight week old male C57BL/6J mice (Jackson Laboratory). For AAV experiments, half of the mice were selected at random and injected via the tail vein with 1×1011 genome copies (GC) of AAV8 control or anti-Tm6sf2 shRNA, respectively. Initial studies were performed in 8 mice per group to provide 80% power to detect a 1.5 s.d. in liver triglyceride content. The investigator was not blinded to the group allocation during the experiment. After 14 days the mice were fasted for 4 h at the end of the dark cycle, then sacrificed. Tissues and plasma were harvested and frozen at −80°C. prior to analysis. Hepatic and plasma lipids were measured as described below. The experiment was repeated except the mice were fed were fed a high sucrose diet (MP Biomedicals; 901683) for 4 weeks. In each experiment, data from all mice in the experiment were included the analysis.
Quantitative Real-Time PCR assays of mRNA abundance
Total RNA was isolated from livers of mice using RNA STAT-60 (Tel-Test Inc.), and Real-Time PCR measurements were performed. Expression levels of mouse TM6SF2 mRNA were measured using oppositely-oriented primers (Primers 3 and 4, Supplementary Table 6). Mouse 36B4 mRNA was used as an internal control.
Oligonucleotides specific for each gene that was analyzed were used to amplify by PCR in 2×SYBRGreen PCR Master Mix (Applied Biosystems) in a volume of 66 μL according to manufacturer’s instructions.
The relative expression levels of TM6SF2 in human tissues were assayed in cDNA samples prepared from 22 human tissue RNA (Human Total RNA Master Panel II, Clontech) by Real-Time PCR using two oppositely-oriented primers (primers 5 and 6, Supplementary Table 6). Human 36B4 mRNA was used as an internal control.
Plasma fast performance liquid chromatography
Eight week old C57BL/6J male mice (Jackson Laboratory) were injected via the tail vein with 1 × 1011 genome copies (GC) of AAV8 control or anti-Tm6sf2 shRNA, respectively (4 mice per group) and fed with regular chow for 14 days. After a 4 h fast, plasma was isolated from blood samples collected from the tail veins of four mice. The plasma samples were pooled (total volume = 400 μl/group) Lipoproteins were separated by FPLC using a Superose 6 column (GE Healthcare). A total of 42 fractions (300 μl each) were collected. The cholesterol and triglyceride content of each fraction was measured using enzymatic assays (Infinity, Thermo Scientific).
Liver and plasma chemistries
Lipids were extracted from liver tissue (~100 mg) using the method of Folch and Lees26. Hepatic triglyceride, cholesterol, and phosphatidylcholine were measured using enzymatic assays (Infinity, Thermo Electron Corp., and Wako Inc.) and normalized to the sample weight. Serum levels of ALT, AST, alk phos, triglyceride and cholesterol were measured using the Vitros 250 system (GMI, Inc.).
Histological studies
Lipid droplet size and distribution were determined in mice liver fed with high sucrose diet for 4 weeks after mTm6sf2 KD by AAV-shRNA. The slice of liver was fixed in 4% paraformaldehyde for 48 hours and equilibrated in 10% sucrose for 24 hours and then in 18% sucrose for 24 hours at 4°C, prior to cryosectioning by Molecular Pathology Core facility of University of Texas Southwestern Medical Center. Cryosections of livers stored at −80 °C were brought to room temperature and air-dried for at least 2 h and then fixed in methanol-free 4% paraformaldehyde. Slides were washed with distilled water three times and then incubated for 10 min in 0.18% Oil Red O (Sigma) prepared in 60% isopropyl alcohol. The slides were washed in distilled water five times. Nuclei were counterstained with hematoxylin, and coverslips were affixed with aqueous mounting medium (Vector Laboratories, Inc., Burlingame, CA). Slides were viewed using a Leica microscope (DM2000; original magnification, ×20 and ×63).
Expression of TM6SF2 in cultured hepatocytes
The cDNA for human TM6SF2 was cloned downstream of the cytomegalovirus promoter/enhancer elements in pCMV6-XL5 plasmid (OriGene Technologies). Single nucleotide changes were introduced using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and confirmed by Sanger sequencing. A V5-epitope tag (GKPIPNPLLGLDST) was placed at the C-terminus of each plasmid construct.
Human hepatoma (HuH-7) cells, which tested negative for mycoplasma, were obtained from the Department of Molecular Genetics (UT Southwestern) were transfected with plasmids expressing wild-type or mutant human TM6SF2-V5 using Lipofectamine 2000 (Life Technologies). HuH-7 cells were grown to 90% confluence in DMEM with 10% FCS plus 100 IU/ml penicillin and 100 μg/ml streptomycin for 48 h. Cells were then washed 3 times with ice cold PBS, harvested and divided into two aliquots. One aliquot was used determine TM6SF2 mRNA levels by RT-PCR. The other aliquot was suspended in RIPA buffer (150 mM NaCl, 1.0% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris, pH 8.0) supplemented with protease inhibitors (Protease Inhibitor Cocktail, Roche) and disrupted by 20 passages through a 25-gauge needle. The homogenates were centrifuged at 12,000 × g for 10 min at 4 °C to pellet nuclei and unbroken cells. Post-nuclear supernatant protein concentrations were determined by BCA Protein Assay (Thermo Scientific). Identical amounts (25 μg) of protein were size-fractionated on 10% SDS-PAGE gels, and immunoblotted using anti-V5 antibody (Invitrogen). Proteins were visualized using Super-Signal ECL (Pierce Biotechnology).
Subcellular fractionation of cultured cells
Hepa1c1c7 cells (obtained from the Department of Molecular Genetics, UT Southwestern) were transfected with PCMV6-XL5 (OriGene Technologies) expressing V5-tagged wild-type human TM6SF2 using Lipofectamine 2000 (Life Technologies). The cells, which tested negative for mycoplasma, were grown in DMEM plus 10% FCS and 400 μM oleate conjugated to albumin. After 48 h, cells were washed three times in iced-cold PBS and disrupted in Buffer C (20 mm Tris, 1 mm EDTA, pH 7.4) plus protease inhibitors (Protease Inhibitor Cocktail, Roche), the homogenates were passaged through a 25-gauge needle × 20 and then were centrifuged at 4,000 × g for 10 min at 4 °C to pellet nuclei and unbroken cells. Post-nuclear supernatants were adjusted to a concentration of Buffer C plus 20% sucrose and added to the bottom of an ultracentrifuge tube. A step gradient was constructed by overlaying the post-nuclear supernatant with 5 ml of Buffer C plus 5% sucrose followed by Buffer C alone. The tube was clamped and centrifuged at 28,000 × g for 40 min at 4 °C. After centrifugation, the lipid layer was isolated by slicing the tube, combined with 20 volumes of −80 °C acetone, and stored overnight at −20 °C. The precipitated proteins were pelleted by centrifugation at 4,000 × g for 1 h at 4 °C and then resuspended in Buffer C plus 1% SDS. The lipid droplet, postnuclear supernatant (cytosol), and membrane fractions were subjected to immunoblotting using an anti-V5 antibody (R960-25, Invitrogen), polyclonal anti-ADRP antibody (20R-AP002, Fitzgerald Industries International). Antibodies against calnexin (SPA860F, StressGen) and β-actin (122625, Cell Signaling) were used as internal controls.
Adeno-associated viral knockdown of Tm6sf2 in mice
Ten short hairpin RNA (shRNA 1–10) sequences were designed to specifically target mouse Tm6sf2 using a proprietary algorithm. Nineteen-nucleotide (nt) sequences starting from nt 604, 508, 470, 616, 855, 899, 247, 210, 977, 323, 334, 946 of Tm6sf2, respectively, were synthesized as complementary antiparallel oligonucleotides with a loop sequence (ttcaagaga) and BbsI overhang. The forward and reverse oligonucleotides were annealed and ligated into the BbsI site of AAV cis plasmid downstream of the human H1 promoter (pAAV-shRNA). To evaluate the potency of these shRNAs, the ten pAAV-shRNAs were transfected into a CHO cell line that stably expresses mouse Tm6sf2 using Nucleofector (Lonza) per manufacturer’s instructions. AAV cis plasmid containing the H1 promoter and termination sequence was used as a negative control. After 48 h, cells were harvested and RNA was isolated and prepared for real time PCR analysis25. The relative mRNA expression levels were normalized to the expression of β-actin. Primers and probe sets for mouse Tm6sf2 and β-actin were purchased from Applied Biosystems (Foster City, CA). The two most potent shRNAs (sh5 and sh8) were selected for AAV vector production.
The AAV8-Tm6sf2 shRNAs and the negative control AAV vector containing H1 promoter and termination sequence were produced by the helper-free triple plasmid transfection method as described7. The pAAV cis plasmid, an adenoviral helper plasmid, and a chimeric packaging plasmid containing AAV2 rep gene and AAV8 cap gene were co-transfected into HEK293 cells. The AAV vectors were subsequently purified by two rounds of cesium chloride density gradient ultracentrifugation, and titers were determined via a real-time PCR analysis.
Eight week old male C57BL/6J mice (Jackson Laboratory) were injected via the tail vein with 1 × 1011 genome copies (GC) of AAV8 control or AAV-shRNA, respectively (8 mice/group). The mice were maintained on a 12-hour-light/12-hour-dark cycle and fed the Teklad Mouse/Rat Diet 7001 chow diet Harlan Teklad) ad libitum. After 14 days the mice were fasted for 4 h at the end of the dark cycle, then sacrificed. Tissues and plasma were harvested and frozen at −80°C. Hepatic and plasma lipids were measured as described above. The experiment was repeated except the mice were fed were fed a high sucrose diet (MP Biomedicals; 901683) for 4 weeks. Mice were sacrificed 4 h after food withdrawal.
VLDL synthesis
To measure the effect of Tm6sf2 KD on VLDL secretion, 8 week old C57BL/6J male mice (4/group) were transduced with 1 × 1011 GC of control AAV or AAV-shRNA8 in the tail vein (n=4). After 14 d, mice were fasted 4 h and injected with 500 μg/g body weight of TRITON WR 1339 (Sigma), which blocks intravascular lipoprotein lipase activity. Blood was obtained from the tail vein prior to injection and then 30, 60, 90, and 120 min post injection.
Supplementary Material
Acknowledgments
We thank Fang Xu for excellent technical assistance. This work was supported by grants from the NIH: HL20948, 1HL092550, DK090066 and the National Center for Advancing Translational Sciences (UL1TR001105). TFV acknowledges the contributions of Dr Guanping Gao from the University of Massachusetts for advice and assistance with AAV vector production.
Footnotes
Author Contributions
The manuscript was prepared by all of the authors. The experiments were performed by E.S, and H.H.Z. The genetic analysis and association studies were performed by J.K. and S.S. T.F.V., A. T.-H., B.G.N. J.C.C. and H.H.H. provided experimental design and coordination.
Accession numbers: TM6SF2: NM_001001524
Competing Financial Interests
The authors do not have any competing financial interests.
References
- 1.Tiniakos DG, Vos MB, Brunt EM. Nonalcoholic fatty liver disease: pathology and pathogenesis. Annu Rev Pathol. 2010;5:145–71. doi: 10.1146/annurev-pathol-121808-102132. [DOI] [PubMed] [Google Scholar]
- 2.Browning JD, et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387–95. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 3.McCullough AJ. The clinical features, diagnosis and natural history of nonalcoholic fatty liver disease. Clin Liver Dis. 2004;8:521–33. viii. doi: 10.1016/j.cld.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Romeo S, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–5. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li JZ, et al. Chronic overexpression of PNPLA3I148M in mouse liver causes hepatic steatosis. J Clin Invest. 2012;122:4130–44. doi: 10.1172/JCI65179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Speliotes EK, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan X, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet. 2008;83:520–8. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambers JC, et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43:1131–8. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Victor RG, et al. The Dallas Heart Study: a population-based probability sample for the multidisciplinary study of ethnic differences in cardiovascular health. Am J Cardiol. 2004;93:1473–80. doi: 10.1016/j.amjcard.2004.02.058. [DOI] [PubMed] [Google Scholar]
- 10.Gorden A, et al. Genetic variation at NCAN locus is associated with inflammation and fibrosis in non-alcoholic fatty liver disease in morbid obesity. Hum Hered. 2013;75:34–43. doi: 10.1159/000346195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernaez R, et al. Association between variants in or near PNPLA3, GCKR, and PPP1R3B with ultrasound-defined steatosis based on data from the Third National Health and Nutrition Examination Survey. Clin Gastroenterol Hepatol. 2013;11:1183–1190. e2. doi: 10.1016/j.cgh.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stender S, Frikke-Schmidt R, Nordestgaard BG, Tybjaerg-Hansen A. Extreme bilirubin levels as a causal risk factor for symptomatic gallstone disease. JAMA Intern Med. 2013;173:1222–8. doi: 10.1001/jamainternmed.2013.6465. [DOI] [PubMed] [Google Scholar]
- 13.Schonfeld G. Familial hypobetalipoproteinemia: a review. J Lipid Res. 2003;44:878–83. doi: 10.1194/jlr.R300002-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–13. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kathiresan S, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–97. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kathiresan S, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 18.Marchler-Bauer A, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–9. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaliannan K, et al. Intestinal alkaline phosphatase prevents metabolic syndrome in mice. Proc Natl Acad Sci U S A. 2013;110:7003–8. doi: 10.1073/pnas.1220180110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Victor RG, et al. The Dallas Heart Study: a population-based probability sample for the multidisciplinary study of ethnic differences in cardiovascular health. Am J Cardiol. 2004;93:1473–80. doi: 10.1016/j.amjcard.2004.02.058. [DOI] [PubMed] [Google Scholar]
- 21.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative centrifuge. Clin Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- 22.Szczepaniak LS, et al. Magnetic resonance spectroscopy to measure hepatic triglyceride content: prevalence of hepatic steatosis in the general population. Am J Physiol Endocrinol Metab. 2005;288:462–468. doi: 10.1152/ajpendo.00064.2004. [DOI] [PubMed] [Google Scholar]
- 23.Browning JD, et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387–95. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 24.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 25.Chen Z, et al. AAV8-mediated long-term expression of human LCAT significantly improves lipid profiles in hCETP;Ldlr(+/−) mice. J Cardiovasc Transl Res. 2011;4:801–10. doi: 10.1007/s12265-011-9309-8. [DOI] [PubMed] [Google Scholar]
- 26.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.