Abstract
In inflammatory bowel diseases (IBD), particularly ulcerative colitis (UC), intestinal macrophages (MΦs), eosinophils and the eosinophil-selective chemokine CCL11 have been associated with disease pathogenesis. MΦs, a source of CCL11, have been reported to be of a mixed classical (NF-κB-mediated) and alternatively activated (STAT-6-mediated) phenotype. The importance of NF-κB and STAT-6 pathways to the intestinal MΦ/CCL11 response and eosinophilic inflammation in the histopathology of experimental colitis is not yet understood. Our gene array analyses demonstrated elevated STAT-6- and NF-κB-dependent genes in pediatric UC colonic biopsies. Dextran sodium sulphate (DSS) exposure induced STAT-6 and NF-κB activation in mouse intestinal F4/80+CD11b+Ly6Chi (inflammatory) MΦs. DSS-induced CCL11 expression, eosinophilic inflammation and histopathology were attenuated in RelA/p65Δmye mice but not in the absence of STAT-6. Deletion of p65 in myeloid cells did not affect inflammatory MΦ recruitment or alter apoptosis, but did attenuate lipopolysaccharide-induced cytokine production (IL-6) and Ccl11 expression in purified F4/80+CD11b+Ly6Chi inflammatory MΦs. Molecular and cellular analyses revealed a link between expression of calprotectin (S100a8/S100a9), Ccl11 expression and eosinophil numbers in the DSS-treated colon. In vitro studies of bone marrow-derived MΦs showed calprotectin-induced CCL11 production via a p65-dependent mechanism. Our results indicate that myeloid cell-specific NF-κB-dependent pathways play an unexpected role in CCL11 expression and maintenance of eosinophilic inflammation in experimental colitis. These data indicate that targeting myeloid cells and NF-κB-dependent pathways may be of therapeutic benefit for the treatment of eosinophilic inflammation and histopathology in IBD.
Introduction
Inflammatory bowel diseases (IBD) are chronic, relapsing, remitting diseases of the gastrointestinal (GI) tract. While the precise etiologies of IBD (ulcerative colitis [UC] and Crohn’s disease [CD]) remain unclear, experimental and clinical studies indicate that activation of innate immune pathways trigger macrophage (MΦ) and dendritic cell (DC) activation and subsequent cytokine production (interleukin [IL]-1β, IL-6 and tumor necrosis factor [TNF]-α), driving IL-23/Th17 development and granulocyte (neutrophils and eosinophils) recruitment and activation leading to pathophysiological features of disease (1, 2). Monocytes/MΦs are elevated in colonic biopsy samples from IBD patients, and these cells produce large amounts of pro-inflammatory cytokines (IL-6, TNF-α and IL-23), as well as various chemokines, and retain respiratory burst activity (3–5). Corroborative experimental studies employing chemical (dextran sodium sulphate [DSS]) and spontaneous models of IBD (IL-10−/−) have identified a role for MΦs in augmentation and exacerbation of the intestinal inflammatory responses and pathogenesis in IBD (6–8).
Clinical and experimental evidence indicates a pathogenic role for eosinophils in both chemical (DSS) and spontaneous murine models (SAMP1/Yit and IL-10−/−) of colitis and in human IBD (9–12). Recent studies have reported that inflammatory MΦs express the eosinophil-specific chemokine CCL11 and have implicated this pathway in the regulation of eosinophilic inflammation in experimental colitis (13). Moreover, intestinal CD68+ MΦs in colonic biopsy samples from pediatric UC patients are positive for CCL11 and CCL11 mRNA levels positively correlate with eosinophil numbers (9). The molecular regulation of CCL11 expression in MΦs is not yet fully understood, however, in vivo evidence from parasite infestation and rhinovirus (RSV) models suggests that MΦ-driven eosinophilic inflammation is associated with an alternative MΦ activation phenotype (M2) and requires signal transducer and activator of transcription (STAT)-6 activation (14, 15). Consistent with this, CCL11 expression can be induced by IL-4 and IL-13; however in vitro evidence in various cell lines indicates that cytokines including TNF-α, IL-9, and IL-17 can also stimulate CCL11 expression through activation of STAT-6-independent pathways including nuclear factor (NF)-κB, STAT-3, or MAPK-mediated signaling (16–19).
In the present study, we demonstrate STAT-6 and NF-κB activation in colonic F4/80+CD11b+Ly6Chi monocyte/MΦs during DSS-induced colitis. We show that DSS-induced F4/80+CD11b+Ly6Chi monocyte/MΦ recruitment, CCL11 expression and eosinophilic inflammation can occur in the absence of STAT-6. In contrast, loss of RelA/p65 in the myeloid lineage leads to decreased DSS-induced CCL11 secretion, eosinophil recruitment, IL-6 secretion and histopathology. Purification of F4/80+CD11b+Ly6Chi RelA/p65-deficient monocyte/MΦs from the colon of mice exposed to DSS revealed significantly reduced Ccl11 expression. In vitro studies identify S100a8/S100a9-induced BMMΦ-derived CCL11 production via a p65-dependent mechanism. These studies demonstrate that MΦ-driven eosinophilic inflammation in experimental colitis is regulated by CCL11 and NF-κB-dependent pathways.
Methods and Materials
Mice
Male and female, 6- to 8-week-old strain-, age- and weight-matched Lysozyme M (LysM)cre/creRelA/p65fl/fl (RelA/p65Δmye, C57BL/6/129/SvEv) and LysMcre/creRelA/p65+/+ (wild-type [WT] line for RelA/p65Δmye mice) and STAT-6−/− (C57Bl/6) (20) and WT C57BL/6 mice were used. All mice were housed under specific pathogen-free conditions and treated according to institutional guidelines.
DSS-induced colonic injury and histopathologic examination
DSS (ICN chemicals 40–45 kDa) was administered in the drinking water as a 2.5% (weight/volume) solution for up to 7 days. Disease monitoring and histopathologic changes in the colon were scored as previously described (9).
Western blot
Bone marrow-derived MΦs, spleen, or colonic epithelial lysates were ran on 4%-12% Bis-Tris gels and transferred to a nitrocellulose membrane (Invitrogen; Carlsbad, CA). The following antibodies were used: rabbit-anti-IKKα, IκBβ, c-Rel, p105/p50 (Santa Cruz; Santa Cruz, CA), phosphoserine 536 RelA/p65, IκBα (Cell Signaling; Danvers MA), and total RelA/p65 (Rockland Immunochemicals, Gilbertsville, Pa) followed by goat anti-rabbit peroxidase-conjugated antibody (Calbiochem; Darmstadt, Germany) and ECL-plus detection reagents (GE Healthcare, Buckinghamshire, United Kingdom). Rabbit anti-actin (Sigma, St Louis, MO) was used as loading control.
MΦ Activation
Bone marrow–derived MΦs (BMMΦs) were obtained as described (21). The cells (1×106) were seeded onto 24-well plates and cultured overnight (37°C, 5% CO2). The next day, cells were treated with LPS (1 ug/mL P. gingivalis, Invivogen; San Diego, CA), IL-4 (20 ng/mL), IFN-γ (1000U/mL, PeproTech Inc; Rocky Hill, NJ) for 24 hours and supernatants were assessed for active cleaved caspase-3 or cytokine production by enzyme-linked immunosorbent assay (ELISA). In some experiments WT and p65Δmye BMMΦs were stimulated with calprotectin (S100a8/S100a9 complex; Abcam, Cambridge, MA; 0.5µg/ml) for 24 hours and CCL11 levels were measured in the supernatants as described.
ELISA
CCL11, IL-6, IL-1β and TNF-α levels were measured in the supernatants or lysates using the ELISA Duo-Set kit according to the manufacturer's instructions (R&D Systems, Minneapolis, Minn). IL-12p40 was measured in the supernatants using the ELISA BD OptEIA kit according to the manufacturer’s instructions (BD Biosciences, San Diego, CA).
MBP staining
Eosinophil levels were quantified by anti-MBP immunohistochemistry as previously described (22) and numbers given as eosinophils per high power field (HPF).
Punch biopsies
Colons were excised, flushed with PBS with gentamycin (20 ug/mL) and opened along a longitudinal axis. Thereafter, 3-mm2 punch biopsies were excised and incubated for 24 hours in 24-well plates with RPMI 1640 supplemented with 10% FCS and antibiotics. Supernatants were collected and kept at −80°C until assessed for cytokines/chemokines by ELISA.
Real-time reverse transcription PCR (qRT-PCR) analysis
Mouse Hprt, RelA/p65, Ccl11, Retnla, Arg1, Cxcl2, Cxcl3, Cxcl10, Cxcl9, Il1b,Ccl22, Tnf, Il6, Nfkbia, Gapdh andIl10 mRNA were quantified by qRT-PCR as previously described (23). In brief, the RNA samples were subjected to reverse transcription analysis using SuperScript II reverse transcriptase (Invitrogen; Carlsbad, CA) according to the manufacturer’s instructions and quantified using the iQ5 multicolor real-time PCR detection system (Bio-Rad Laboratories; Hercules, CA) with iQ5 software V2.0 and LightCycler FastStart DNA Master SYBR Green I. Gene expression was determined as relative expression on a linear curve based on a gel-extracted standard and was normalized to Hprt amplified from the same cDNA mix. Results were expressed as the gene of interest/Hprt ratio. Nfkbia expression was normalized to Gapdh and expressed as ΔΔCt.
Intestinal MΦ purification
MΦ populations from the colons were isolated as previously described (9). In brief, the colon segment of the GI tract was removed and flushed with 20 ml of Ca2+- and Mg2+-free HBSS (CMF-HBSS). The colon was cut longitudinally, placed in CMF-HBSS containing 10% FBS/5 mM EDTA/25 mM HEPES and shaken vigorously at 37°C for 30 minutes. The tissue was cut into 1-cm segments and incubated in digestion buffer containing 2.4 mg/ml collagenase A (Roche Diagnostics; Indianapolis, IN) and 0.2 mg/mL DNase I (Roche; Indianapolis, IN) in RPMI 1640 for 45 minutes on a shaker at 37°C. Following incubation, the cell aggregates were dissociated by filtering thorough a 19-gauge needle and 70-µm filter and centrifuged at 1200 rpm for 20 minutes at 4°C. The supernatant was decanted and the cell pellet resuspended in 1% FBS/5 mM EDTA/PBS, and cells were incubated for 30 minutes with biotinylated rat anti-mouse CD11b (1 ug/1×106 cells; BD Pharmingen; San Jose, CA) at 4°C. Cells were subsequently incubated with anti-biotin microbeads (Miltenyi; Auburn, CA) for 15 minutes at 10°C and purified by LS MACS column by positive selection as described by the manufacturer. In brief, 1 mL of cell suspension was added to the LS column, and the column was washed 3 times with 3 mL of 1% FBS/5 mM EDTA/PBS. CD11b+ cells were removed from the column using a plunger. After washing, CD11b+-selected cells were labeled with rat anti-mouse Ly6C-Alexa-647 (AbD Serotec; Raleigh, NC) and Streptavidin-PerCp (BD Pharmingen; San Jose, CA) and immediately sorted for CD11b and Ly6C using a FACSAria cell sorter (BD Biosciences; San Jose, CA). Purity of CD11b+Ly6Chi cells was >95% as assessed by flow cytometry. RNA was isolated using the Qiagen RNeasy micro kit for cDNA synthesis and qRT PCR analysis as described above.
FACS analysis
Single-cell suspensions were washed with FACS buffer (PBS/1% FBS) and incubated with combinations of the following antibodies: PE anti-mouse F4/80 (clone CI:A3-1; Serotec; Raleigh, NC), PE-Cy7 anti-mouse CD11b (clone M1/70; BD Pharmingen; San Jose, CA), and AlexaFluor-647 anti-mouse Ly6C (clone ER-MP20; AbD Serotec; Raleigh, NC). For phospho-flow staining, cells were fixed with 2% formaldehyde/PBS for 10 minutes, permeabilized for 15 minutes with ice cold methanol and stained for 30 minutes at room temperature with AlexaFluor-647 anti-mouse phospho-RelA/p65 (polyclonal; Cell Signaling; Danvers, MA) or AlexaFluor-488 anti-mouse phospho-STAT-6 (polyclonal; Cell Signaling; Danvers, MA). For apoptosis analysis, cells were fixed and permeabilized using the BD cytofix/cytoperm kit followed by staining with the AlexaFluor-647 rabbit anti-active caspase-3 antibody (clone C92–605; BD Pharmingen; San Jose, CA). The following antibodies were used as appropriate isotype controls: FITC rat IgG2a (clone B39-4; BD Pharmingen; San Jose, CA), PE rat IgG2a (clone 53-6.7; BD Pharmingen; San Jose, CA), PE-Cy7 rat IgG2b (clone DTA-1; BD Pharmingen; San Jose, CA) and AlexaFluor 647 rat IgG2a (clone R35–95 BD Pharmingen; San Jose, CA). Cells were analyzed on FACSCalibur (BD Immunocytometry Systems; San Jose, CA), and analysis was performed using Flow Jo software (Tree Star; Ashland, OR).
Microarray analysis
Gene expression profiles from DSS-treated mice [day 6 (n = 6) and day 0 (n = 5)] as reported by Fang, et al (24) were downloaded from the NBCI Gene Expression Omnibus (GEO) database (accession number GSE22307; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE22307) and analyzed using GeneSpring GX ® software (version 11.5.1) (Agilent Technologies, Inc.). Data were baseline transformed to the median of all samples and compared against the DSS day 0 treatment group. Statistical analyses were performed using an unpaired t test and Benjamini Hochberg false discovery rate. Genes showing significant differential expression (p < .05) with greater than 2-fold change were assessed for correlation with Ccl11 expression (Pearson r > .075) and hierarchical clustering was performed. Normalized log2-transformed expression levels of Ear5, S100a8, and S100a9 from individual mice were plotted against Ccl11 levels.
Statistical analysis
Data were analyzed by means of ANOVA, followed by the Tukey or Dunnett’s post-hoc test or a two-tailed unpaired T test with GraphPad Prism 5 (San Diego, CA). Data are presented as the mean ± SE. P values of less than 0.05 were considered statistically significant. ND represents below limit of detection.
Results
STAT-6 and NF-κB-induced genes are elevated in UC patients compared to normal patients
Recently we demonstrated a relationship between intestinal CD68+ MΦ-derived CCL11 and eosinophilic inflammation in pediatric UC patients (9). CCL11 expression is directly regulated by STAT-6 and NF-κB-mediated signaling through interactions with overlapping consensus DNA response elements within the CCL11 promoter (25, 26). To begin to distinguish the involvement of STAT-6 and NF-κB-mediated signaling in intestinal MΦ-derived CCL11 expression and eosinophilic inflammation, we examined for evidence of NF-κB and STAT-6-dependent gene expression in a pediatric UC transcriptome generated from gene array analyses of colonic biopsy samples from pediatric UC and healthy patients (9). Analysis of the pediatric UC transcriptome revealed increased expression of STAT-6-regulated genes, including IL13RA2 (16-fold), SELP, SELE, CD40, COLA1A and COL1A2 (Table I). The promoter regions of all these genes possess STAT-6 binding sites (27). STAT-6-dependent pathways are also involved in the induction of the alternatively activated MΦ phenotype (28) and we also observed increased expression of genes associated with alternative MΦ activation including SERPINE1, CCL18 and SOCS1 (Table I). NF-κB-regulated genes were also significantly elevated in pediatric UC, including CXCL5, IL8, CXCL2 and CXCL3, which are all increased over 10-fold compared to healthy patients (Table II). These data demonstrate elevated levels of both STAT-6- and NF-κB-regulated genes in pediatric UC.
Table I.
STAT-6-dependent gene expression in pediatric UC.
| Gene Name | Gene Symbol | UC vs. NL (Fold change) |
|---|---|---|
| Interleukin 13 receptor, alpha 2 | IL13RA2 | 16.47 |
| Selectin E | SELE | 7.28 |
| Collagen, type I alpha 1 | COL1A1 | 5.09 |
| Chemokine (C-C motif) ligand 18 | CCL18 | 4.77 |
| Collagen, type I alpha 2 | COL1A2 | 4.36 |
| Selectin P (granule membrane protein 140kDa, antigen CD62) | SELP | 3.37 |
| Suppressor of cytokine signaling 1 | SOCS1 | 2.49 |
| Serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), membrane 1 | SERPINE1 | 2.40 |
| CD40 molecule, TNF receptor superfamily member 5 | CD40 | 2.26 |
Table II.
NFκB-dependent gene expression in pediatric UC.
| Gene Name | Gene Symbol | UC vs. NL (Fold change) |
|---|---|---|
| Chemokine (C-X-C motif) ligand 5 | CXCL5 | 39.49 |
| Interleukin 8 | IL8 | 21.24 |
| Chemokine (C-X-C motif) ligand 2 | CXCL2 | 11.68 |
| Chemokine (C-X-C motif) ligand 3 | CXCL3 | 10.35 |
| Lipocalin 2 (oncogene 24p3) | LCN2 | 7.32 |
| Selectin E | SELE | 7.28 |
| Immunoglobulin heavy constant gamma 4 | IGHG4 | 6.89 |
| Interleukin 1, beta | IL1B | 5.94 |
| Chemokine (C-C motif) ligand 11 | CCL11 | 5.87 |
| Prostaglandin-endoperoxidase synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | PTGS2 | 5.59 |
| Chemokine (C-X-C motif) ligand 10 | CXCL10 | 4.35 |
| Interleukin 6 | IL6 | 4.30 |
| Secretory leukocyte peptidase inhibitor | SLPI | 4.14 |
| Matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) | MMP9 | 3.78 |
| Intercellular adhesion molecule 1 (CD54) | ICAM1 | 3.64 |
| Guanylate binding protein 1, interferon-inducible, 67 kDa | GP1 | 3.24 |
| Chemokine (C-C motif) ligand 2 | CCL2 | 3.01 |
| Nucleotide-binding oligerization domain containing 2 | NOD2 | 2.96 |
CCL11 expression in IL-4-stimulated bone marrow derived-MΦs (BMMΦs) is STAT-6-dependent
Previous in vitro and in vivo studies indicate a role for IL-4 in MΦ-derived STAT-6 activation and in the regulation of pulmonary eosinophilic inflammation (15). To begin to determine the requirement for IL-4-STAT-6 axis in CCL11 expression in MΦs, we examined CCL11 expression in IL-4 stimulated BMMΦs generated in vitro. IL-4-stimulation of BMMΦs induced STAT-6 activation (Supplemental figure S1A–B). STAT-6 activation was associated with increased Ccl11 mRNA expression and significant CCL11 protein release (Supplemental figure S1D–E). Notably, IL-4 stimulation of MΦ induced alternative MΦ activation as demonstrated by increased expression of Chi3l3, Arg1, and Retnla (Supplemental figure S1C). The alternatively activated MΦ phenotype and CCL11 expression were dependent on STAT-6, as indicated by the loss ofCcl11 and Arg1 expression in STAT-6−/− BMDMs (Supplemental figure S1D–E). LPS stimulation of BMMΦs did not induce Ccl11 mRNA expression or significant protein release (results not shown). These data indicate that IL-4 induced MΦ-derived CCL11 expression is STAT-6-dependent.
STAT-6 is not required for DSS-induced CCL11 expression or eosinophil recruitment
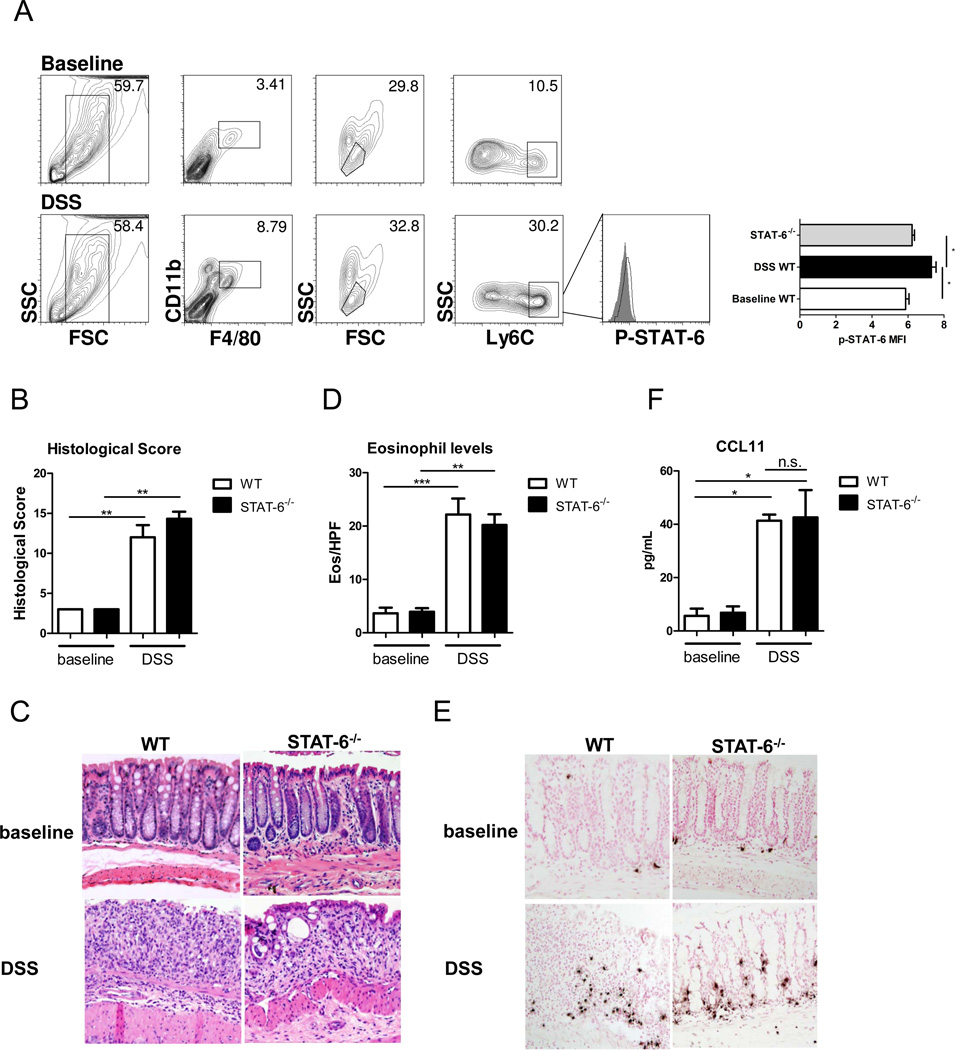
To assess the contribution of STAT-6 signaling to DSS-induced eosinophil recruitment and CCL11 expression, we performed phospho-flow analysis on isolated lamina propria cells from DSS-treated and vehicle (baseline)-treated mice. Levels of phosphorylated STAT-6 in the F4/80+CD11b+Ly6Chi colonic MΦs from DSS-treated WT mice were elevated compared with F4/80+CD11b+Ly6Chi MΦs from vehicle (baseline)-treated WT and DSS-treated STAT-6−/− mice (Figure 1A) [p-STAT6 mean fluorescence intensity (MFI) WT control 6.03 ± 0.21; WT DSS 7.6 ± 0.4*; STAT6−/− 6.5 ± 0.2; mean ± SD; n = 3–7 samples per group; * p < 0.05]. DSS treatment (6 days) of WT mice induced significant colitic disease characterized by cryptitis, epithelial ulceration and pronounced inflammatory infiltrate, including significant eosinophilic inflammation (Figure 1B–E). The absence of STAT-6 did not influence DSS-induced histopathology or eosinophil influx (Figure 1B–E). Importantly, CCL11 levels were not different between WT and STAT-6−/− mice following DSS treatment (Figure 1F). These data indicate that STAT-6 is not required for DSS-induced CCL11 expression, colonic eosinophil influx and histopathology.
Figure 1. Eosinophil recruitment and CCL11 expression are STAT-6-independent in DSS-induced colitis.
A, Representative flow cytometry plots of F4/80+CD11b+Ly6Chi colonic MΦs and representative histograms of phospho-STAT-6 expression in DSS-treated WT mice (open histogram) compared to STAT-6−/− mice (filled histogram) and mean fluorescence intensity of phospho-STAT-6. B, Histological score. C, Representative photomicrographs of H&E-stained colonic sections from baseline and DSS-treated mice (day 7, 2.5% DSS). D, Eosinophil levels in the colon and E, representative photomicrographs of anti-MBP-stained colonic sections from baseline and DSS-treated mice (day 7, 2.5% DSS). F, CCL11 levels in punch biopsy supernatants from WT and STAT-6−/− mice at baseline and following 5 days of DSS. (A). Data represents the mean ± SEM of n = 3–7 mice per group from duplicate experiments. (B-F) Data represents the mean ± SEM of n = 5–6 mice per group. Data is representative of triplicate experiments. Significant differences (*p<0.05; **p<0.01; ***p<0.001) between groups.
Myeloid cell-specific deletion of the RelA/p65 gene in mice (RelA/p65Δmye)
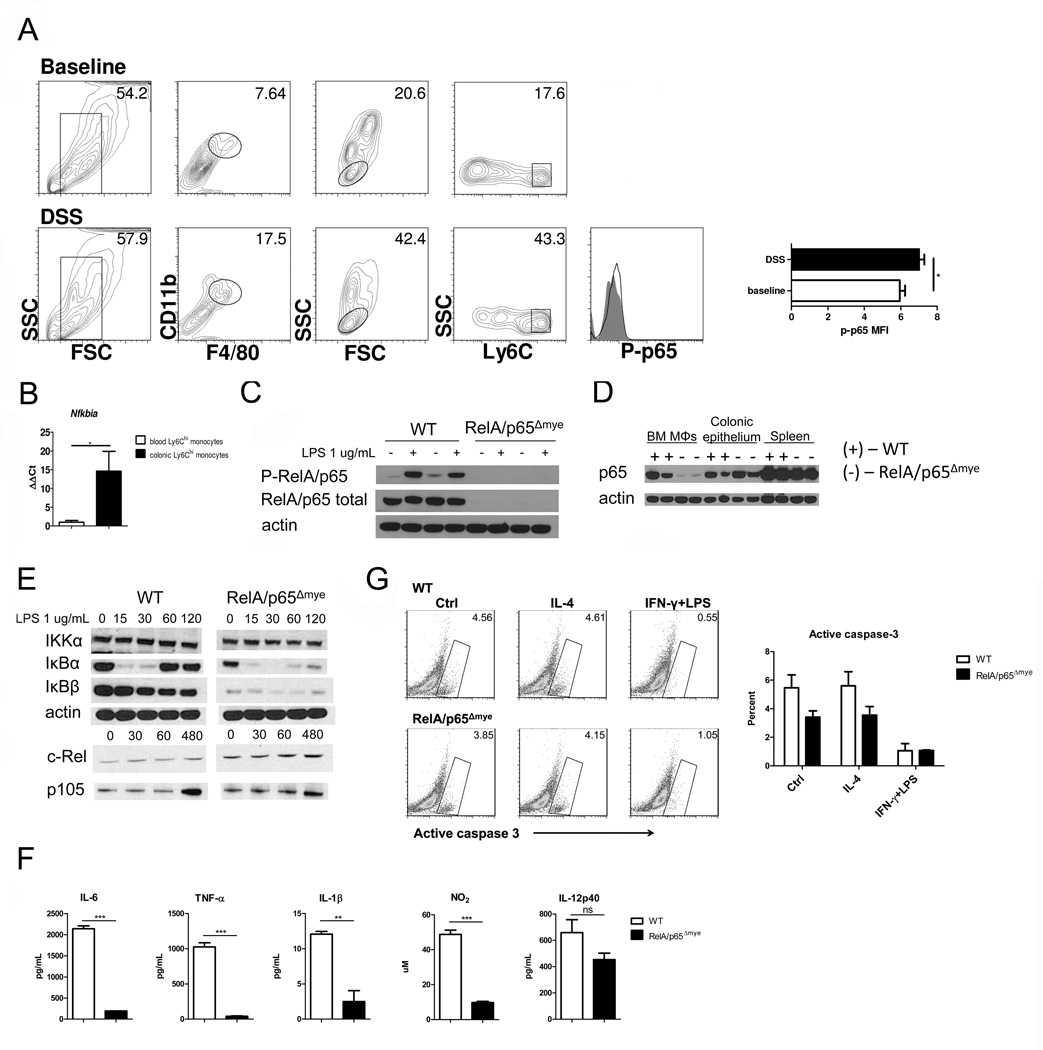
We have previously reported increased classical- and alternatively activated-MΦ gene expression in purified CCL11+F4/80+CD11b+Ly6Chi colonic MΦs from DSS-treated mice (13), suggesting activation of both STAT-6- and NF-κB-regulated pathways. Given our demonstration that CCL11 expression and colonic eosinophilic inflammation could occur in the absence of STAT-6 we next assessed the role of NF-kB.Phospho-flow analysis revealed increased levels of phosphorylated RelA/p65 in colonic F4/80+CD11b+Ly6Chi MΦs from DSS-treated mice compared to RelA/p65-deficient mice (Figure 2A). Consistent with this, qRT-PCR analysis revealed a 10-fold increase in the mRNA levels of the NF-κB-dependent gene, Nfkbia, in this population compared with blood monocytes from DSS-treated mice (Figure 2B). In order to delineate the requirement of NF-κB to the intestinal MΦ:CCL11:eosinophil pathway in colitis, we backcrossed RelA/p65fl/fl mice (29) onto the LysM-Cre mice (30) to specifically delete RelA/p65 in myeloid cells. To demonstrate the efficiency of the LysM-Cre-mediated deletion of RelA/p65, we assessed presence and activation (phosphorylation) of RelA/p65 in BMMΦs (Figure 2C). Western blot analyses revealed the loss of total RelA/p65 protein in RelA/p65Δmye BMDMs (Figure 2C). Further, RelA/p65 activation was ablated in RelA/p65Δmye BMMΦs following 1 hour of LPS stimulation (Figure 2C). RelA/p65 deletion was specific for the myeloid lineage as we observed normal levels of total RelA/p65 in the spleen and colonic epithelium of RelA/p65Δmye mice (Figure 2D).
Figure 2. Specificity of RelA/p65 deletion in RelA/p65Δmye mice.
A, Representative flow cytometry plots and histograms of phospho-RelA/p65 expression in F4/80+CD11b+Ly6Chi colonic MΦs from DSS-treated WT (open histogram) and RelA/p65Δmye mice (filled histogram). B. Quantification of Nfkbia expression from Ly6Chi MΦs/monocytes sorted from the peripheral blood or colon of DSS-treated mice. Data represent the mean ± SEM of n = 5 mice per group. Representative western blot analysis of WT (+) and RelA/p65Δmye (−) (C) BMDMs stimulated with 1 ug/mL LPS for 1 hour, (D), colonic epithelium and spleen of total RelA/p65 and/or phospho-RelA/p65. E, representative western blot analysis of IKKα, IκBα, IκBβ, c-Rel, p105 and actin expression in BMDMs stimulated with LPS for indicated times (minutes). F, WT and RelA/p65Δmye BMDMs were stimulated for 24 hours with vehicle or 1 ug/mL LPS and levels of IL-6, TNF-α, IL-1β, NO2 and IL-12p40 were measured in the supernatants. G, Representative flow cytometry plots and graph of BMDMs stimulated for 24 hours with control, IL-4 or IFN-γ + LPS and assessed for active caspase-3. (F and G) Data represent the mean ± SEM of n = 3 individual samples per group. (A, C, E–G) Data is representative of duplicate experiments. Significant differences (*p<0.05**, p < 0.01; ***p < 0.001) between groups.
To determine the effect of RelA/p65 deletion in myeloid cells on other components of the NF-κB signaling cascade, we assessed activation of IκB kinase (IKK)α, c-Rel and p105 expression in BMDMs (Figure 2E). We observed comparable levels of IKKα, c-Rel and p105 expression between WT and RelA/p65Δmye BMMΦs following LPS-stimulation (Figure 2E). As expected, total levels of inhibitor of κB (IκB)α and IκBβ were decreased in RelA/p65Δmye compared to WT BMMΦs at time 0 (29, 31–33); however, these inhibitory proteins underwent comparable degradation following LPS stimulation, indicating that signaling components upstream of RelA/p65 remained intact (Figure 2E). These data indicate selective RelA/p65 deletion in myeloid cells and that this is independent of effects of other components of the NF-κB signaling pathway.
LPS-induced pro-inflammatory cytokine production in RelA/p65-deficient MΦs
To delineate the effect of RelA/p65 deficiency on MΦ pro-inflammatory cytokine production, we generated BMMΦs from WT and RelA/p65Δmye mice and examined responsiveness to LPS stimulation. BMMΦs from RelA/p65Δmye mice develop normally, with comparable FSC, SSC, F4/80 and CD11b expression compared to WT (data not shown). LPS-induced secretion of TNF-α, IL-6, IL-1β and NO was significantly attenuated in RelA/p65Δmye BMMΦs compared to WT (Figure 2F). However, IL-12p40 secretion was similar between RelA/p65Δmye and WT BMMΦs (Figure 2F), which is consistent with functional c-Rel-dependent transcription (34). To assess NF-κB-independent pathway function in RelA/p65Δmye BMMΦs, we measured Type I interferon (IFN)-induced chemokine (CXCL10) production in RelA/p65Δmye and WT BMMΦs. Levels of Type I IFN-induced STAT-1 phosphorylation and CXCL10 secretion were similar between RelA/p65Δmye and WT BMMΦs (Supplemental figure S2A–B). Furthermore, assessment of active caspase-3 in IL-4 or IFN-γ+ LPS-stimulated BMMΦs revealed that deletion of RelA/p65 in MΦ does not alter MΦ apoptosis (Figure 2G). These data indicate that RelA/p65Δmye MΦs have attenuated NF-κB-dependent pro-inflammatory cytokine responses; however NF-κB-independent signaling such as type I IFN-induced STAT-1-dependent chemokine production remains intact.
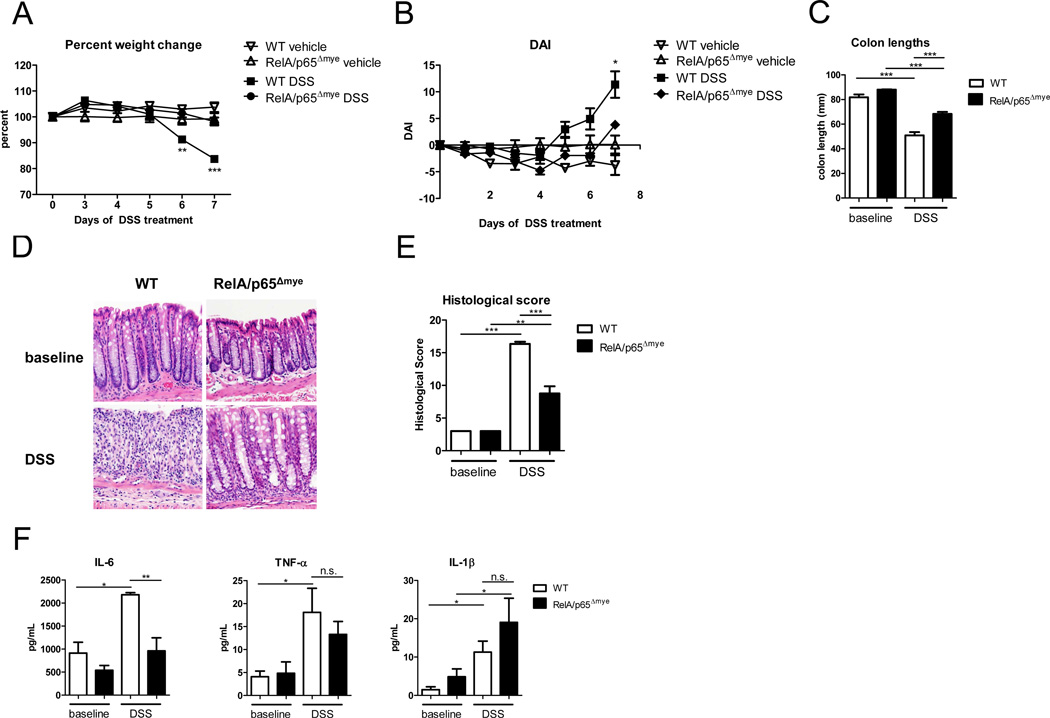
DSS-induced colitis is attenuated in RelA/p65Δmye mice
To assess the requirement for myeloid expression of RelA/p65 in DSS-induced colonic injury, we exposed RelA/p65Δmye and aged- and strain-matched WT mice to 2.5% DSS for 6 days and examined colitic disease. RelA/p65 deletion in myeloid cells protected the mice from DSS-induced colonic injury (Figure 3). DSS-induced weight loss, development of diarrhea, rectal bleeding and colon shortening were significantly attenuated in RelA/p65Δmye mice compared to WT mice (Figure 3A–C). Histological assessment of the colon revealed that the reduction in clinical symptoms of disease in RelA/p65Δmye mice was accompanied by a significant reduction in intestinal epithelial crypt loss, erosion, inflammatory infiltrate and the pro-inflammatory cytokine IL-6 (Figure 3D–F). Surprisingly, we observed no significant reduction in TNF-α or IL-1β in colonic punch biopsy samples between DSS-treated WT and RelA/p65Δmye mice (Figure 3F). We observed no differences in intestinal immune and epithelial architecture between naïve WT and RelA/p65Δmye mice, indicating no homeostatic effects of myeloid p65 deletion on intestinal function. Collectively, these studies identify that myeloid expression of RelA/p65 is required for DSS-induced increases in IL-6, intestinal inflammation and colonic injury.
Figure 3. DSS-induced colitis is attenuated in RelA/p65Δmye mice.
A, Percent weight change and (B) Disease activity index (DAI) of WT and p65Δmye mice following DSS exposure. C, colon lengths D, representative photomicrographs and (E) quantitative histological score of H&E-stained colonic sections from of WT and p65Δmye mice following 7 days of DSS exposure. F, Cytokine Profile of colonic punch biopsy samples from WT and p65Δmye mice following DSS-exposure. Data represent the mean ± SEM of n = 3–5 mice per group. Data is representative of triplicate (A–E) and duplicate (F) experiments. Significant differences (*p < 0.05; **p < 0.01; ***p < 0.001) compared with WT vehicle or as indicated. Magnification of photomicrographs is ×100. n.s. – not significant.
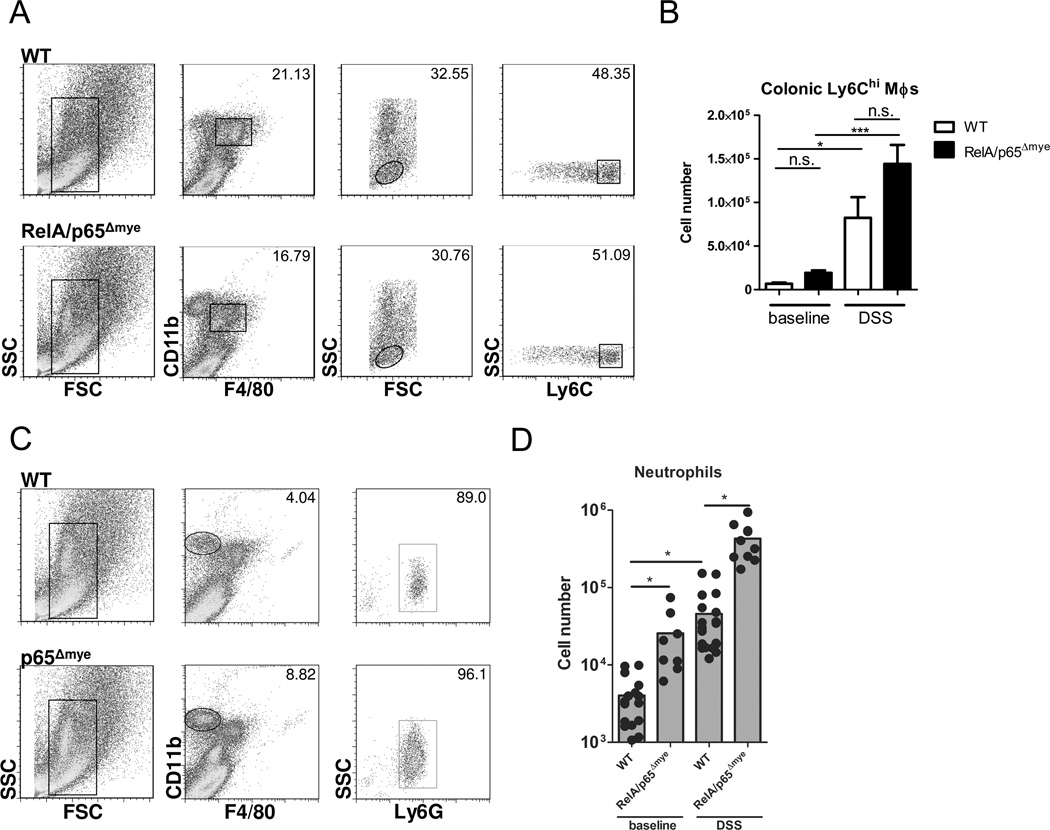
Attenuation of DSS-induced colitis is not due to decreased Ly6Chi monocyte or neutrophil recruitment
Previous studies demonstrated that blockade of the recruitment of inflammatory MΦs to the colon via a CCR2-dependent pathway can attenuate DSS-induced colitis disease (13). We show that DSS exposure (5 days) induced a significant influx of F4/80+CD11b+Ly6Chi monocytes in WT mice (Figure 4 A and B) (13). Importantly, loss of myeloid RelA/p65 did not dysregulate recruitment of Ly6Chi monocytes to the colon at baseline or following DSS treatment (Figure 4 A and B). Similarly, we observed no reduction in neutrophil recruitment into the colon in RelA/p65Δmye mice (Figure 4 C and D). Peripheral blood levels of neutrophils and Ly6Chi monocytes were also equivalent between WT and RelA/p65Δmye mice (Supplemental figure S3). These data indicate that myeloid trafficking to the colon during chronic inflammatory conditions does not require RelA/p65 signaling in myeloid cells.
Figure 4. Colonic recruitment of myeloid cells is not impaired in p65Δmye mice.
A, Representative flow cytometry plots and B, quantification of the F4/80+CD11b+Ly6Chi MΦ populations from the colon of WT and RelA/p65Δmye mice following 5 days of vehicle (baseline) or DSS exposure. MΦs were initially gated by SSC vs. FSC followed by F4/80+, CD11b+ and Ly6Chi. C, Representative flow cytometry plots and D, quantification of the F4/80−CD11bhi neutrophil population from the colon of WT and RelA/p65Δmye mice following 5 days of vehicle (baseline) or DSS exposure. MΦs were initially gated by SSC vs. FSC followed by F4/80− and CD11bhi. B and D, data represents the mean ± SEM of n = 5–6 mice per group and is representative of duplicate experiments. Significant differences (*p < 0.05; ***p < 0.001) between groups. n.s. – not significant.
Colonic eosinophilic inflammation is decreased in RelA/p65Δmye mice
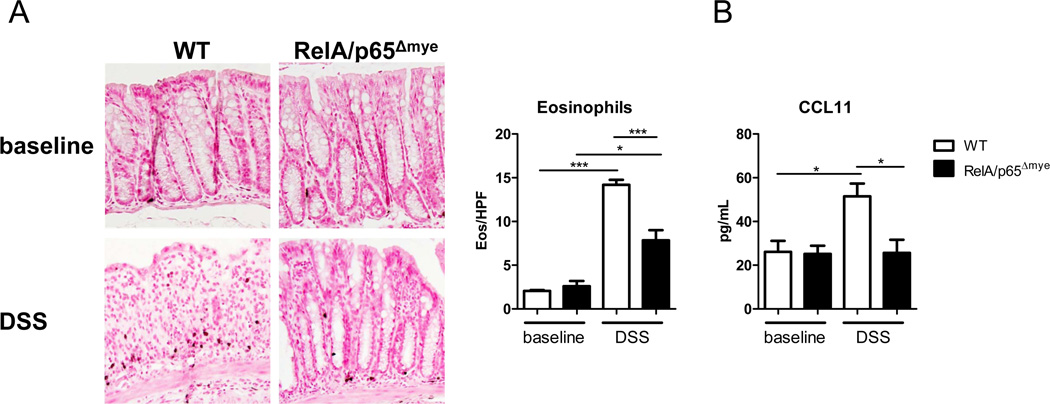
Assessment of colonic eosinophil levels in DSS-treated RelA/p65Δmye mice revealed a significant decrease in induction of eosinophil levels compared to DSS-treated WT mice (Figure 5A). The blunting of the DSS-induced increase in eosinophil number in RelA/p65Δmye mice was associated with a similar and significant lack of DSS-induced increase in CCL11 levels in colonic punch biopsies from DSS-treated RelA/p65Δmye mice (Figure 5B). We have previously demonstrated that eosinophils are localized to the gastrointestinal tract under homeostatic conditions and that the recruitment of this cell population is regulated by CCL11 (35). Deletion of RelA/p65 in myeloid cells did not alter steady-state CCL11 and eosinophil levels indicating that myeloid RelA/p65 does not regulate homeostatic CCL11 production or eosinophil recruitment. These data implicate the specificity of myeloid RelA/p65 in regulating the increase in CCL11 expression and colonic eosinophilic inflammation during colonic injury.
Figure 5. Lack of induction of CCL11 and eosinophils in RelA/p65Δmye mice during DSS-induced colitis.
A, Representative photomicrographs of anti-MBP-stained colonic sections from baseline and DSS-treated mice (day 6, 2.5%) and colonic eosinophil levels in the colon. B, CCL11 levels in punch biopsy supernatants from WT and RelA/p65Δmye mice in vehicle- (baseline) and DSS-treated mice (day 5, 2.5%) Data indicates the mean ± SEM of n = 6–8 mice per group from triplicate experiments. Significant differences (*p < 0.05; ***p < 0.001) between groups. Magnification of photomicrographs is ×100.
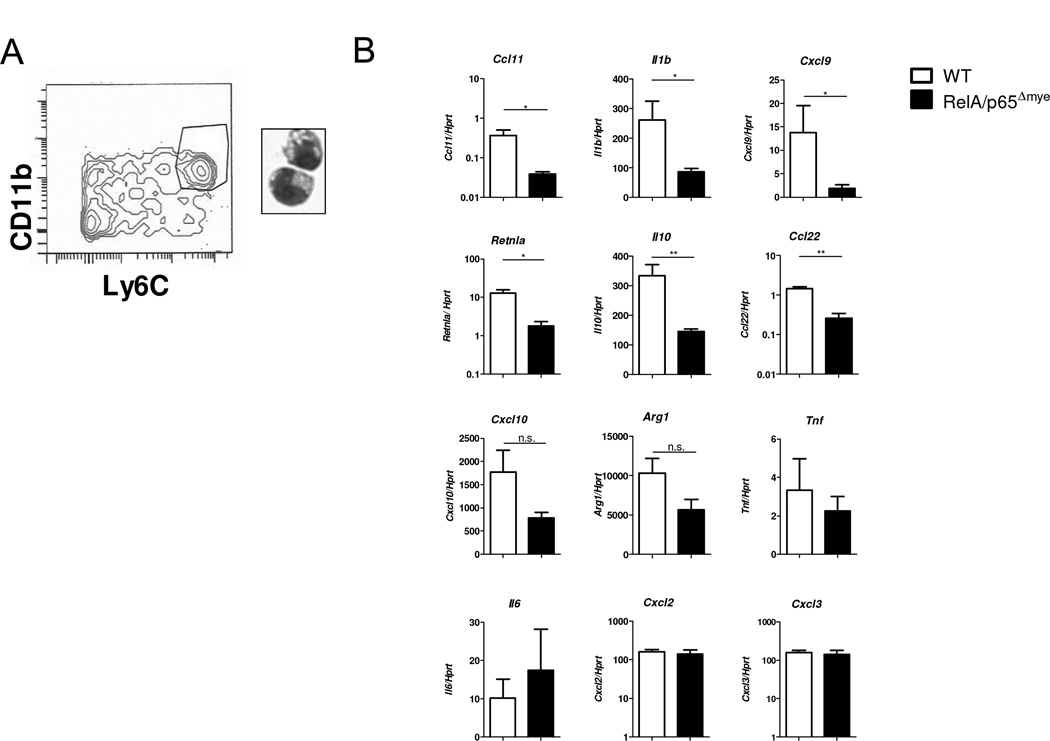
To directly determine whether the reduction in DSS-induced CCL11 in the colon of RelA/p65Δmye mice was a consequence of reduced CCL11 production from Ly6Chi colonic MΦs, we used flow sorting to purify Ly6Chi MΦs from the colon of DSS-treated mice using CD11b and Ly6C (Figure 6A). PCR analyses revealed decreased Ccl11 expression in DSS-treated RelA/p65Δmye Ly6Chi colonic MΦs compared to WT (Figure 6B). Importantly, the reduction in Ccl11 expression in RelA/p65Δmye MΦs was not due to an inability to express CCL11, as IL-4 treatment induced equivalent levels of CCL11 expression in both WT and RelA/p65Δmye BMDMs (Supplemental figure S4).
Figure 6. Phenotype Ly6Chi colonic MΦs in WT vs. RelA/p65Δmye mice during DSS-induced colonic injury.
A, Representative flow cytometry plot of CD11b+Ly6Chi colonic MΦs flow sorted on day 5 of DSS treatment. B, gene expression was analyzed by qRT-PCR. 3–4 mice were pooled per sample from DSS-treated WT and RelA/p65Δmye mice (3–4 samples per group). Data represents the mean ± SEM. Data is representative of duplicate experiments. Significant differences (*p < 0.05; **p < 0.01) between groups. n.s. – not significant.
We previously reported that Ly6Chi colonic MΦs from DSS-treated mice had a mixed M1 and M2 pro-inflammatory phenotype (13). To assess the effect of myeloid cell-specific RelA/p65 deletion to this phenotype, we examined expression of M1 and M2 genes in purified Ly6Chi colonic MΦs from WT and RelA/p65Δmye DSS-treated mice. qPCR analyses of p65 expression confirmed RelA/p65 deletion in purified Ly6Chi colonic MΦs from RelA/p65Δmye DSS-treated mice (Ly6Chi colonic MΦs p65/hprt ratio WT 1.41 ± 0.19 vs p65Δmye 0.37 ± 0.04 ; n = 3 purified F4/80+ CD11b+ Ly6Chi colonic MΦ’s preparations per group; p < 0.05). Il1b, Cxcl9 (M1),Retnla, Il10 andCcl22 (M2) gene expression was decreased in colonic Ly6Chi MΦs from DSS-treated RelA/p65Δmye mice compared to DSS-treated WT mice, suggesting that these genes are positively regulated by RelA/p65 (Figure 6B). Cxcl10 and Arg1 expression was also decreased, although not significantly. Suprisingly, Tnf, Il6, Cxcl2 and Cxcl3 expression was not decreased in colonic Ly6Chi MΦs from RelA/p65Δmye mice compared to WT mice (Figure 6B), indicating loss of RelA/p65 may not regulate their expression at the time examined.
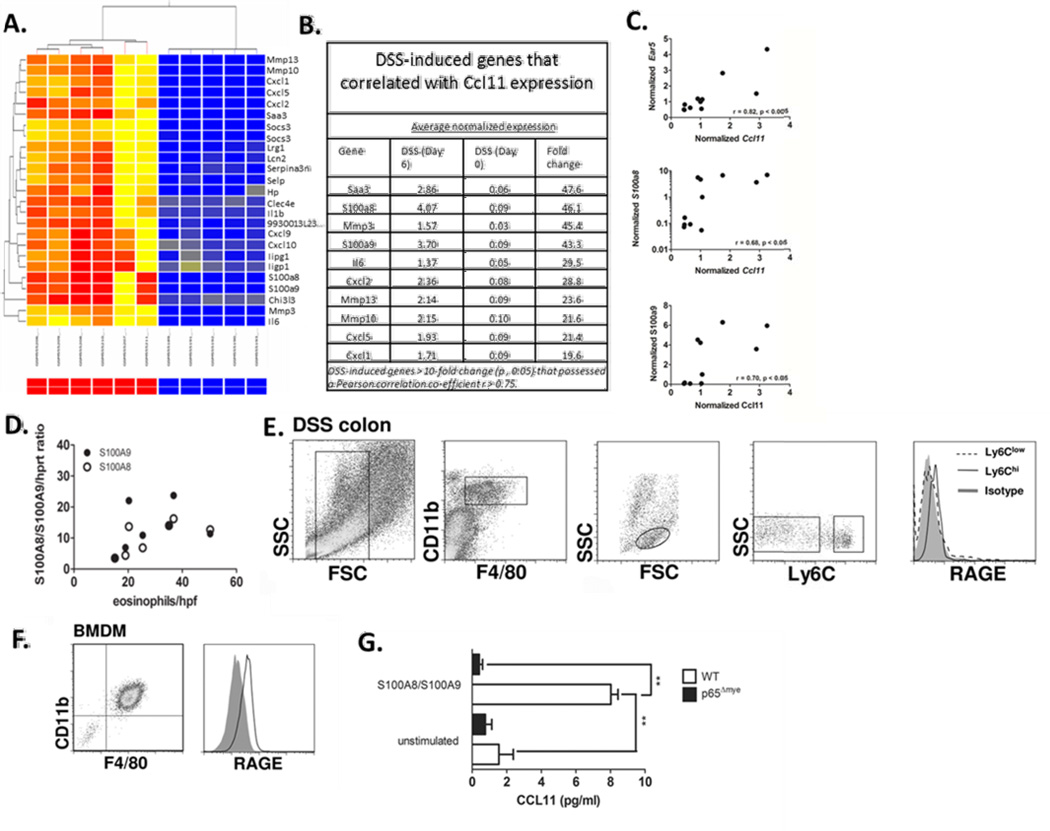
Calprotectin-RAGE involvement in macrophage-derived CCL11 production
To identify potential candidates involved in the stimulation of CCL11 expression and secretion in intestinal inflammatory MΦs in DSS colitis we re-analyzed microarray profiling analyses performed on the colon of control and DSS-treated C57BL/6 mice (data accessible at NCBI GEO database ((24)), accession GSE22307). These array data are of the colon of C57BL/6 mice following 0 and 6 days of DSS (3%), and changes in gene expression have been validated by qRT-PCR with r2=0.925122 (24). We compared gene expression at day 0 and 6 as we have previously demonstrated maximal inflammatory MΦ recruitment into the colon on day 6 of DSS exposure (9). 3% DSS exposure induced the upregulation of 1008 genes and down regulation 173 genes (results not shown). Consistent with the previous gene array analysis comparing gene expression between Day 0 vs Day 6 of DSS exposure revealed increased expression of inflammatory genes, including Il6, Cxcl2, Cxcl1, Il33 and Il1β (results not shown (24)). Assessment of CCL11 and eosinophil-specific genes (e.g. Ear5- eosinophil-associated, ribonuclease A family, member 5) revealed significant elevated levels of Ccl11 and Ear5 (3.2 and 2.7-fold increase respectively). Correlative analyses demonstrated a positive correlation between Ccl11 and Ear5 expression (r = 0.82; p < 0.001 Figure 7 C). These data are consistent with our previous studies in C57BL/6 demonstrating a direct relationship between CCL11 and eosinophils (9, 13). The top DSS-induced genes (between Day 0 vs Day 6 of DSS exposure) that correlated with Ccl11 expression were Saa3 (47.6 fold increase), Mmp3 (45.4 fold increase) and the S100 proteins, S100a8 (46.1 fold increase) and S100a9 (43.3 fold-increase) (Figure 7 B). Notably, S100a8 and S100a9 were the highest upregulated genes that possessed the strongest positive correlation with Ccl11 (S100a8 r = 0.68; p < 0.03 and S100a9 r = 0.70; p < 0.02 Fig 7 B and C). These studies revealed a relationship between S100a8 and S100a9, colonic CCL11 expression and eosinophilic inflammation in DSS-induced colitis. To confirm these observations, we assessed S100a8 and S100A9 mRNA expression and eosinophil numbers in the colon following 0, 3 and 7 days of DSS exposure (Fig. 7D). We demonstrate a positive correlation between S100a8 and 9 mRNA expression and eosinophil numbers/hpf in the colon during DSS exposure (Fig. 7D; p < 0.01). The S100a8/S100a9 (calprotectin) receptor is not fully delineated, however there is evidence that the 35kDa multiligand receptor Receptor for Advanced Glycation Endproducts (RAGE) acts as the primary receptor for calprotectin (36, 37). Assessment of the calprotectin receptor RAGE on Ly6Chi colonic MΦs by flow cytometry revealed on the colon of mice following 6 days of DSS exposure. We show that Ly6Chi colonic MΦs and not the resident Ly6Clow colonic MΦs express RAGE (Fig. 7E). To assess if calprotectin induced CCL11 expression in macrophages we assessed S100a8/S100a9 stimulation of BMMΦs. Firstly, we show that BMMΦs express the RAGE (Fig. 7 E). Stimulation of BMMΦs with heterodimeric calprotectin complex induced CCL11 secretion. Notably, the calprotectin response was dependent on NfkB signaling as the calprotectin-induced CCL11 secretion was ablated p65-deficient BMMΦs. These studies indicate that calprotectin (S100a8/S100a9) induce CCL11 secretion in macrophages via NfkB-dependent pathway (Fig. 7F).
Figure 7. Relationship between eosinophil-specific gene (Ear5), Ccl11 and Calprotectin (S100a8 and S100a9) expression in DSS-induced colitis.
A. Heat map showing the clustering of the top 25 differentially-expressed genes following DSS-induced colitis (day 6 vs. day 0, p < 0.05). B. Top 10 differentially-expressed genes following DSS-induced colitis (day 6 vs. day 0, p < 0.05, fold change > 10) that correlated with Ccl11 expression (r > 0.75). C. Pearson product-moment correlation between normalized microarray expression values of Ear5, S100a8 and S100a9 are plotted against Ccl11 from individual animals. (D) Pearson product-moment correlation between S100a8 and S100a9/Hprt ratio and eosinophils/hpf in the colon of WT C57BL6 mice on day 3, 5 and 7 after DSS exposure. E. Representative flow plots of RAGE expression on (E.) colonic F4/80+ CD11b+ Ly6Chi monocytes/MΦ from DSS-treated mice and (F.) BMMΦ. G. CCL11 levels in supernatants from WT and RelA/p65Δmye BMMΦ following 24 hour stimulation with S100a8/S100a9 complex. G. Data represents the mean ± SEM. Data is representative of duplicate experiments. Significant differences (**p < 0.01) between groups.
Discussion
In the present study, we investigated the contribution of STAT-6 and NF-κB RelA/p65 in myeloid cells in the regulation of colonic eosinophilic inflammation and histopathology of DSS-induced colitis. Expression of both NF-κB and STAT-6-dependent genes are increased in pediatric UC colonic biopsies. We report that NF-κB and STAT-6 are activated in colonic Ly6Chi MΦs during DSS-induced colitis in mice. We show that loss of STAT-6 does not alter susceptibility to colitic disease, whereas loss of RelA/p65 in myeloid cells leads to decreased susceptibility to DSS-induced colitis. Notably, attenuated DSS-induced clinical symptoms and histopathology in RelA/p65Δmye mice was associated with decreased induction of the pro-inflammatory cytokine, IL-6, CCL11 expression and eosinophil recruitment and not due to reduced recruitment of myeloid cells. We show that CCL11 levels in DSS-treated RelA/p65Δmye mice was linked with attenuated Ccl11 expression in Ly6Chi colonic MΦs. We identify a positive correlation between calprotectin (S100A8 and S100A9) mRNA expression and colonic eosinophil numbers and that calprotectin stimulation of BMMΦs lead to p65-dependent-CCL11 secretion. Collectively, these data indicate that myeloid RelA/p65 plays a central role in the pro-inflammatory cytokine response and CCL11:eosinophil axis in experimental colitis.
Previous in vivo studies in models of heart transplant rejection (38), pulmonary hypertension (39), rhinovirus (15) and helminth infection (14) have identified a role for MΦs in eosinophil recruitment. The majority of these studies show that MΦ-mediated eosinophil recruitment is associated with M2 MΦ alternate activation and CCL11 expression. Consistent with this finding, we show that IL-4 stimulation of BMDMs induces the M2 MΦ phenotype and CCL11 expression and that this pathway is dependent on STAT-6. However, our in vivo analyses revealed that CCL11-dependent eosinophil recruitment can occur in the absence of STAT-6 signaling. Previous studies have reported STAT-6-independent CCL11 expression in airway epithelial cells and that this was mediated by TNF-α-induced NF-κB binding to the CCL11 promoter (25). Consistent with this, in a mouse model of allergic lung inflammation, NF-κB inhibition by NF-κB decoy oligodeoxynucleotides or by genetic approaches (transgenic CC10-IκBα-super-repressor mice) attenuated lung inflammation and reduced CCL11 expression (40, 41). While these studies identified an interaction between NF-κB-regulated pathways and CCL11 expression, they did not elucidate the role of NF-κB in specific cell types. Our study indicates that at least in DSS-induced colitis, Ly6Chi colonic MΦ-derived RelA/p65 regulates CCL11 levels and eosinophilic inflammation; however, it still remains to be determined whether RelA/p65 directly binds to the CCL11 promoter in colonic MΦs.
Clinical and experimental evidence identifies an important contribution by MΦs through the production of pro-inflammatory cytokines, including TNF-α, IL-6, IL-1β and IL-23 in the exacerbation of the chronic inflammatory response in IBD and experimental colitis (3, 13, 42, 43). We show that myeloid deletion of RelA/p65 signaling attenuated DSS-induced colitic disease and IL-6 levels in punch biopsy samples. However, we did not observe reduced secreted TNF-α levels between WT and RelA/p65Δmye mice. This was somewhat surprising as we identified an important role for RelA/p65 in LPS-induced MΦ-derived TNF-α expression in vitro. Assessment of total TNF-α expression (secreted and non-secreted) by analyzing whole colonic lysates did reveal a decrease in TNF-α expression in RelA/p65Δmye mice compared to WT (results not shown). These potential conflicting observations maybe attributed to the fact that MΦs may not be the primary source of TNF-α in the colon during DSS-induced colitis, as human intestinal epithelial cells, mast cells and NK cells are also able to produce TNF-α (44, 45). Gene expression analysis of purified Ly6Chi colonic MΦs from RelA/p65Δmye mice revealed that decreased susceptibility to colitic disease was associated with lower levels of a number of pro-inflammatory genes Il1b, Retnla, Cxcl9, Cxcl10, Arg1 and Ccl22. CXCL9 and CXCL10 are both increased in IBD patients (46, 47), and inhibition of CXCL10 protected mice from DSS-induced colitis (48). Furthermore, we have previously demonstrated that Retnla−/− mice had decreased susceptibility to DSS-induced colitis (21). Ccl22 expression is increased in DSS-induced colitis and in the sera of UC and CD patients, but its role in IBD is not known (49, 50). These data indicate that MΦs express many pro-inflammatory genes which may contribute chronic intestinal inflammatory phenotypes and disease pathology.
Previous studies have demonstrated that DSS exposure stimulates neutrophil recruitment into the colon (51). Consistent with this, we observed a significant increase in the level of neutrophil infiltration into the colon following DSS exposure in both the WT (~ 12-fold) and RelA/p65Δmye (16-fold) mice. However, neutrophil levels in the colon of RelA/p65Δmye mice were greater than that observed in WT mice under both steady state and following DSS exposure. There are a number of potential explanations for this observation. Firstly, that LysMcre-mediated deletion of p65 is occurring in neutrophils and effecting neutrophil migration or apoptosis. RelA/p65 has been implicated in neutrophil apoptosis and initial description of the LysMcre transgene revealed efficient LysMcre expression and deletion of floxed targeted genes in neutrophils (30). Alternatively, elevated levels of neutrophils in the RelA/p65Δmye mice maybe attributed to decreased macrophage function and activity. Interestingly, deletion of macrophages in the LysMcre c-FLIPfl/fl mice was associated with heightened neutrophilia (52). The authors demonstrated that the elevated neutrophil levels was a consequence of heightened G-CSF and IL-1β and not necessarily due to c-FLIP deletion in neutrophils as neutralization of G-CSF and IL-1β (anti-G-CSF and IL-1R antagonist) reduced neutrophil levels back to baseline levels (52). Notably, we observed increased levels of IL-1β in the colon of RelA/p65Δmye mice following DSS-treatment. The molecular basis for the heightened neutrophil response in the RelA/p65Δmye mice is under current investigation.
Experimental analyses suggest that NF-κB activity in different cell populations has differential roles in the maintenance of tissue homeostasis vs. pro-inflammatory function (29, 53–58). For example, intestinal epithelial cell specific deletion of IKK complex proteins or RelA/p65 increased susceptibility to infection, spontaneous inflammation, and DSS-induced colitis (29, 53, 59). Conversely, transgenic overexpression of constitutively active IKKβ in intestinal epithelial cells drives intestinal inflammation and cancer (54). In contrast, whole animal or hematopoietic cell inhibition of NF-κB using genetic, anti-sense oligonucleotides, or small molecule inhibitors ameliorated mucosal inflammation (55–58). We demonstrate that selective deletion of RelA/p65 in myeloid cells attenuates intestinal inflammation and colitic disease, further emphasizing tissue-specific functions for the NF-κB-pathway in homeostasis vs inflammation.
Previous studies employing inducible IKKβ knockout mice (Mxcre/IKKβfl/fl) have demonstrated that TNF-α-dependent apoptosis in the absence of IKKβ results in increased IL-1β protein secretion and granulocytosis (60), and LPS-induced apoptosis in myeloid cells was increased in IKKβΔmye BMDMs (61). Notably, a number of IKKβ substrates and effector pathways are NF-κB-independent and control proliferation, apoptosis, and cytokine production. IKKβ regulation of MAPK activation, mTOR signaling, expression of autophagy genes, and proteins which regulate cell division and death such as Aurora A, p53, and FOXO3a can occur independently of NF-κB activity (62). Thus, one may predict a partial phenotypic overlap between mice lacking myeloid RelA/p65 and those deficient in myeloid IKKβ. Consistent with this, we show that genetic deletion of RelA/p65 in myeloid cells did not affect peripheral MΦ or neutrophil numbers, indicating that RelA/p65 is not required for MΦ and neutrophil anti-apoptosis. Furthermore, apoptosis was not increased following LPS-stimulation of RelA/p65Δmye BMDMs compared to WT. Collectively, these data indicate the presence of RelA/p65-independent IKKβ substrates and activity in the regulation of myeloid cell function.
NF-κB activity is tightly controlled by interactions with the NF-κB inhibitory proteins, IκBα and IκBβ. We show that the absence of RelA/p65 in BMDMs reduced baseline levels of the IκBα and IκBβ. Further, the levels of IκBβ were significantly more attenuated than that of IκBα suggesting preferential loss of IκBβ. Consistent with this deletion of RelA/p65 in mouse embryonic fibroblasts (MEF’s) and primary liver fetals cells is associated with reduced levels of IκBα and IκBβ and, similar to our data, the reduction in IκBβ levels was greater than that of IκBα ((32). The preferential reduction in IκBβ is attributed to the greater importance of RelA/p65 in stabilizing IκBβ protein via RelA/p65 carboxyl terminus from 26S proteasome degradation (32).
We have previously demonstrated in pediatric UC that CCL11 is derived from both intestinal epithelial cells and CD68+ myeloid cells (9). In contrast, in the murine system employing in situ-hybridization and immunofluorescence technologies we show that CCL11 is derived from mononuclear cells, primarily Ly6Chi colonic MΦ’s (13). One possible explanation for this discrepancy is that the peak DSS-induced colonic eosinophilic inflammation (day 6) is observed in the presence of a pronounced colonic epithelial ulceration and shedding and that this may eliminate any intestinal epithelial CCL11 signal. Assessment of murine colonic intestinal epithelial cells from mice treated with DSS for four days revealed a positive CCL11 signal suggesting that murine intestinal epithelial cells may express CCL11 (results not shown). However, BM chimera experiments demonstrate that Ly6Chi colonic MΦ’s are sufficient drive CCL11-dependent colonic eosinophilic inflammation (13).
One limitation of these analyses is that deletion of RelA/p65 in myeloid cells using the LysMCre system leads to deletion of RelA/p65 in all monocyte/macrophage subpopulations including the Ly6Clow colonic tissue resident MΦ’s and the Ly6Chi inflammatory MΦ’s. Thus we cannot exclude the contribution of RelA/p65-signaling in Ly6Clow colonic tissue resident MΦ’s to DSS-induced CCL11 expression and eosinophilic inflammation. However, we have previously demonstrated that DSS exposure does not alter the levels of resident Ly6Clow colonic tissue MΦ’s (13) and that loss of the Ly6Chi inflammatory MΦ’s and not resident Ly6Clow colonic tissue MΦ’s in the colon was associated with reduced CCL11 levels and eosinophilic inflammation (13) indicating that Ly6Chi inflammatory MΦ’s are likely the MΦ subpopulation responsible for CCL11 and eosinophilic inflammation in the colon during epithelial injury.
Bioinformatic analyses and in vitro studies on BMMΦs reveal that calprotectin, the S100a8/S100a9 heterodimeric complex may be the stimulus for MΦ-derived CCL11 secretion. Indeed, we show that BMMΦ and inflammatory recruited Ly6Chi colonic MΦs express the calprotectin receptor RAGE and that the expression of S100a9 and S100a8 in the colon of DSS-treated mice positively correlated with eosinophil numbers. S100A8 and S100A9 are members of the S100 protein family and the EF-hand protein superfamily (63) and are intracellular proteins that exist mainly as a heterodimer (termed calprotectin). Recent work indicates that S100a8 and S100a9 also possess extracellular functions and regulate leukocyte migration, cytokine expression and innate immune activity (64–66). The S100A8/S100A9 (calprotectin) receptor is not fully delineated, however there is evidence that the 35kDa multiligand receptor Receptor for Advanced Glycation Endproducts (RAGE) acts as the primary receptor for calprotectin (36, 37). Previous experimental evidence indicates that calprotectin induces NFkB activation in RAGE+ cells (37, 67) and that calprotectin-induced myeloid-derived cell migration and accumulation was RAGE and NFkB-dependent (68). We show that calprotectin-induced BMMΦ-derived CCL11 secretion and that this was dependent on NFkB signaling. There is significant clinical evidence suggesting calprotectin and RAGE involvement in IBD. Fecal calprotectin levels are elevated in pediatric UC and CD and levels positively correlate with disease severity and mucosal inflammation (69, 70). Notably, fecal calprotectin levels are one of the most accurate measurements for the presence of active mucosal inflammation and likelihood of IBD relapse (71). Furthermore, Calprotectin is a stronger predictive marker of relapse in UC than CD as patients under clinical remission who recorded a higher initial concentration of fecal calprotectin (> 150 ug/g) are 2 times and 14 times more likely to relapse in CD and UC, respectively (72). RAGE mRNA and protein levels are also increased in colonic samples of CD patients, and the functional RAGE -374T/A polymorphism has been linked with CD (73). RAGE is expressed on many hemapoietic (MΦs, neutrophils, DC and B and T lymphocytes) and non-hemapoietic cell populations (74) and small interfering (si) RNA (si-S100A9) knockdown of S100A9 reduced DSS-induced granulocyte infiltration and colitis disease activity (75).
Recent clinical evidence suggesting a central function for MΦs in the exacerbation of the chronic inflammatory response and manifestations of IBD has led to intense focus on the identification of MΦ-mediated intestinal inflammatory cascades. We have highlighted the importance of recently recruited Ly6Chi colonic MΦs in eosinophil recruitment and CCL11 expression. We now demonstrate that this is regulated by myeloid expression of RelA/p65 and is surprisingly STAT-6-independent. These studies provide significant rationale for the assessment of RelA/p65 activation in the expression of monocyte/MΦ-derived CCL11 in human IBD and further highlights the importance of targeting of the monocyte/MΦ:RelA/p65 pathway as a therapeutic modality for the treatment and prevention of IBD.
Supplementary Material
Acknowledgments
We thank Drs Patricia Fulkerson and DeBroski Herbert and members of the Division of Allergy and Immunology and Gastroenterology, Hepatology, and Nutrition, Cincinnati Children’s Hospital Medical Center for critical review of the manuscript and insightful conversations. We thank Jamie and Nancy Lee for the generous provision of anti-MBP antibody. We would also like to thank Shawna Hottinger for editorial assistance and manuscript preparation.
Grant support: This work was supported by The Crohn’s and Colitis Foundation of America Career Development Award (S.P.H.), NIH R01 AI073553 and DK090119 (S.P.H.), and American Gastroenterological Association Foundation Graduate Student Research Fellowship Award (A.W.).
Abbreviations
- BMDM
bone marrow-derived macrophage
- DSS
dextran sodium sulphate
- GI
gastrointestinal
- IFN
interferon
- IKK
activation of IκB kinase
- IL
interleukin
- IκB
inhibitor of κB
- LPS
lipopolysaccharide
- LysM
Lysozyme M
- MΦ
macrophage
- NF-κB
nuclear factor κB
- STAT
signal transducer and activator of transcription
- TNF
tumor necrosis factor
- WT
wild-type
Footnotes
Disclosures: S.P.H. is a consultant for Immune Pharmaceuticals. The other authors have declared that they have no conflict of interest.
References
- 1.Heinsbroek SE, Gordon S. The role of macrophages in inflammatory bowel diseases. Expert Rev Mol Med. 2009;11:1–19. doi: 10.1017/S1462399409001069. [DOI] [PubMed] [Google Scholar]
- 2.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 3.Kamada N, Hisamatsu T, Okamoto S, Chinen H, Kobayashi T, Sato T, Sakuraba A, Kitazume MT, Sugita A, Koganei K, Akagawa KS, Hibi T. Unique CD14 intestinal macrophages contribute to the pathogenesis of Crohn disease via IL-23/IFN-gamma axis. J Clin Invest. 2008;118:2269–2280. doi: 10.1172/JCI34610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rugtveit J, Haraldsen G, Hogasen AK, Bakka A, Brandtzaeg P, Scott H. Respiratory burst of intestinal macrophages in inflammatory bowel disease is mainly caused by CD14+L1+ monocyte derived cells. Gut. 1995;37:367–373. doi: 10.1136/gut.37.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reinecker HC, Steffen M, Witthoeft T, Pflueger I, Schreiber S, MacDermott RP, Raedler A. Enhanced secretion of tumour necrosis factor-alpha, IL-6, and IL-1 beta by isolated lamina propria mononuclear cells from patients with ulcerative colitis and Crohn's disease. Clin Exp Immunol. 1993;94:174–181. doi: 10.1111/j.1365-2249.1993.tb05997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghia JE, Galeazzi F, Ford DC, Hogaboam CM, Vallance BA, Collins S. Role of M-CSF-dependent macrophages in colitis is driven by the nature of the inflammatory stimulus. Am J Physiol Gastrointest Liver Physiol. 2008;294:G770–G777. doi: 10.1152/ajpgi.00453.2007. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe N, Ikuta K, Okazaki K, Nakase H, Tabata Y, Matsuura M, Tamaki H, Kawanami C, Honjo T, Chiba T. Elimination of local macrophages in intestine prevents chronic colitis in interleukin-10-deficient mice. Dig Dis Sci. 2003;48:408–414. doi: 10.1023/a:1021960401290. [DOI] [PubMed] [Google Scholar]
- 8.Takeda K, Clausen BE, Kaisho T, Tsujimura T, Terada N, Forster I, Akira S. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of STAT3 in macrophages and neutrophils. Immunity. 1999;10:39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- 9.Ahrens R, Waddell A, Seidu L, Blanchard C, Carey R, Forbes E, Lampinen M, Wilson T, Cohen E, Stringer K, Ballard E, Munitz A, Xu H, Lee N, Lee JJ, Rothenberg ME, Denson L, Hogan SP. Intestinal macrophage/epithelial cell-derived CCL11/eotaxin-1 mediates eosinophil recruitment and function in pediatric ulcerative colitis. J Immunol. 2008;181:7390–7399. doi: 10.4049/jimmunol.181.10.7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodruff SA, Masterson JC, Fillon S, Robinson ZD, Furuta GT. Role of eosinophils in inflammatory bowel and gastrointestinal diseases. J Pediatr Gastroenterol Nutr. 2011;52:650–661. doi: 10.1097/MPG.0b013e3182128512. [DOI] [PubMed] [Google Scholar]
- 11.Takedatsu H, Mitsuyama K, Matsumoto S, Handa K, Suzuki A, Funabashi H, Okabe Y, Hara T, Toyonaga A, Sata M. Interleukin-5 participates in the pathogenesis of ileitis in SAMP1/Yit mice. Eur J Immunol. 2004;34:1561–1569. doi: 10.1002/eji.200324680. [DOI] [PubMed] [Google Scholar]
- 12.Specht S, Arriens S, Hoefrauf A. Induction of chronic colitis in IL-10 deficient mice requires IL-4. Microbes and Infection. 2006;8:694–703. doi: 10.1016/j.micinf.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Waddell A, Ahrens R, Steinbrecher K, Donovan B, Rothenberg ME, Munitz A, Hogan SP. Colonic eosinophilic inflammation in experimental colitis is mediated by Ly6C(high) CCR2(+) inflammatory monocyte/macrophage-derived CCL11. J Immunol. 2011;186:5993–6003. doi: 10.4049/jimmunol.1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voehringer D, Van Rooijen N, Locksley RM. Eosinophils develop in distinct stages and are recruited to peripheral sites by alternatively activated macrophages. J. Leuk. Biol. 2007;81:1434–1444. doi: 10.1189/jlb.1106686. [DOI] [PubMed] [Google Scholar]
- 15.Nagarkar DR, Bowman ER, Schneider D, Wang Q, Shim J, Zhao Y, Linn MJ, McHenry CL, Gosangi B, Bentley JK, Tsai WC, Sajjan US, Lukacs NW, Hershenson MB. Rhinovirus infection of allergen-sensitized and -challenged mice induces eotaxin release from functionally polarized macrophages. J Immunol. 2010;185:2525–2535. doi: 10.4049/jimmunol.1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamasaki A, Saleh A, Koussih L, Muro S, Halayko AJ, Gounni AS. IL-9 induces CCL11 expression via STAT3 signalling in human airway smooth muscle cells. PLoS One. 2010;5:e9178. doi: 10.1371/journal.pone.0009178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saleh A, Shan L, Halayko AJ, Kung S, Gounni AS. Critical role for STAT3 in IL-17A-mediated CCL11 expression in human airway smooth muscle cells. J Immunol. 2009;182:3357–3365. doi: 10.4049/jimmunol.0801882. [DOI] [PubMed] [Google Scholar]
- 18.Rahman MS, Yamasaki A, Yang J, Shan L, Halayko AJ, Gounni AS. IL-17A induces eotaxin-1/CC chemokine ligand 11 expression in human airway smooth muscle cells: role of MAPK (Erk1/2, JNK, and p38) pathways. J Immunol. 2006;177:4064–4071. doi: 10.4049/jimmunol.177.6.4064. [DOI] [PubMed] [Google Scholar]
- 19.Matsukura S, Odaka M, Kurokawa M, Kuga H, Homma T, Takeuchi H, Notomi K, Kokubu F, Kawaguchi M, Schleimer RP, Johnson MW, Adachi M. Transforming growth factor-beta stimulates the expression of eotaxin/CC chemokine ligand 11 and its promoter activity through binding site for nuclear factor-kappaB in airway smooth muscle cells. Clin Exp Allergy. 2010;40:763–771. doi: 10.1111/j.1365-2222.2010.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urban JF, Noben-Trauth N, Donaldson DD, Madden KB, Morris SC, Collins M, Finkelman FD. IL-13, IL-4Ralpha, and Stat6 are required for the expulsion of the gastrointestinal nematode parasite Nippostrongylus brasiliensis. Immunity. 1998;8:255–264. doi: 10.1016/s1074-7613(00)80477-x. [DOI] [PubMed] [Google Scholar]
- 21.Munitz A, Waddell A, Seidu L, Cole ET, Ahrens R, Hogan SP, Rothenberg ME. Resistin-like molecule alpha enhances myeloid cell activation and promotes colitis. J Allergy Clin Immunol. 2008;122:1200–1207. doi: 10.1016/j.jaci.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forbes E, Murase T, Yang M, Matthaei KI, Lee JJ, Lee NA, Foster PS, Hogan SP. Immunopathogenesis of experimental ulcerative colitis is mediated by eosinophil peroxidase. J. Immunol. 2004;172:5664–5675. doi: 10.4049/jimmunol.172.9.5664. [DOI] [PubMed] [Google Scholar]
- 23.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, Jameson SC, Kirby C, Konikoff MR, Collins MH, Cohen MB, Akers R, Hogan SP, Assa'ad AH, Putnam PE, Aronow BJ, Rothenberg ME. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–547. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang K, Bruce M, Pattillo CB, Zhang S, Stone R, 2nd, Clifford J, Kevil CG. Temporal genomewide expression profiling of DSS colitis reveals novel inflammatory and angiogenesis genes similar to ulcerative colitis. Physiol Genomics. 2011;43:43–56. doi: 10.1152/physiolgenomics.00138.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsukura S, Stellato C, Plitt JR, Bickel C, Miura K, Georas SN, Casolaro V, Schleimer RP. Activation of eotaxin gene transcription by NF-kappa B and STAT6 in human airway epithelial cells. J Immunol. 1999;163:6876–6883. [PubMed] [Google Scholar]
- 26.Hoeck J, Woisetschlager M. STAT6 Mediates Eotaxin-1 Expression in IL-4 or TNF-alpha-Induced Fibroblasts. J Immunol. 2001;166:4507–4515. doi: 10.4049/jimmunol.166.7.4507. [DOI] [PubMed] [Google Scholar]
- 27.Hebenstreit D, Wirnsberger G, Horejs-Hoeck J, Duschl A. Signaling mechanisms, interaction partners, and target genes of STAT6. Cytokine Growth Factor Rev. 2006;17:173–188. doi: 10.1016/j.cytogfr.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 28.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 29.Steinbrecher KA, Harmel-Laws E, Sitcheran R, Baldwin AS. Loss of epithelial RelA results in deregulated intestinal proliferative/apoptotic homeostasis and susceptibility to inflammation. J Immunol. 2008;180:2588–2599. doi: 10.4049/jimmunol.180.4.2588. [DOI] [PubMed] [Google Scholar]
- 30.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 31.Scott ML, Fujita T, Liou HC, Nolan GP, Baltimore D. The p65 subunit of NF-kappa B regulates I kappa B by two distinct mechanisms. Genes Dev. 1993;7:1266–1276. doi: 10.1101/gad.7.7a.1266. [DOI] [PubMed] [Google Scholar]
- 32.Hertlein E, Wang J, Ladner KJ, Bakkar N, Guttridge DC. RelA/p65 regulation of IkappaBbeta. Mol Cell Biol. 2005;25:4956–4968. doi: 10.1128/MCB.25.12.4956-4968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun SC, Ganchi PA, Ballard DW, Greene WC. NF-kappa B controls expression of inhibitor I kappa B alpha: evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- 34.Sanjabi S, Hoffmann A, Liou HC, Baltimore D, Smale ST. Selective requirement for c-Rel during IL-12 P40 gene induction in macrophages. Proc Natl Acad Sci U S A. 2000;97:12705–12710. doi: 10.1073/pnas.230436397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mishra A, Hogan SP, Lee JJ, Foster PS, Rothenberg ME. Fundamental signals that regulate eosinophil homing to the gastrointestinal tract. J Clin Invest. 1999;103:1719–1727. doi: 10.1172/JCI6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gebhardt C, Riehl A, Durchdewald M, Nemeth J, Furstenberger G, Muller-Decker K, Enk A, Arnold B, Bierhaus A, Nawroth PP, Hess J, Angel P. RAGE signaling sustains inflammation and promotes tumor development. J Exp Med. 2008;205:275–285. doi: 10.1084/jem.20070679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turovskaya O, Foell D, Sinha P, Vogl T, Newlin R, Nayak J, Nguyen M, Olsson A, Nawroth PP, Bierhaus A, Varki N, Kronenberg M, Freeze HH, Srikrishna G. RAGE, carboxylated glycans and S100A8/A9 play essential roles in colitis-associated carcinogenesis. Carcinogenesis. 2008;29:2035–2043. doi: 10.1093/carcin/bgn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zweifel M, Matozan K, Dahinden C, Schaffner T, Mohacsi P. Eotaxin/CCL11 levels correlate with myocardial fibrosis and mast cell density in native and transplanted rat hearts. Transplant Proc. 2010;42:2763–2766. doi: 10.1016/j.transproceed.2010.05.152. [DOI] [PubMed] [Google Scholar]
- 39.Weng M, Baron DM, Bloch KD, Luster AD, Lee JJ, Medoff BD. Eosinophils are Necessary for Pulmonary Arterial Remodeling in a Mouse Model of Eosinophilic-Inflammation Induced Pulmonary Hypertension. Am J Physiol Lung Cell Mol Physiol. 2011;301:L927–L936. doi: 10.1152/ajplung.00049.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poynter ME, Cloots R, van Woerkom T, Butnor KJ, Vacek P, Taatjes DJ, Irvin CG, Janssen-Heininger YM. NF-kappa B activation in airways modulates allergic inflammation but not hyperresponsiveness. J Immunol. 2004;173:7003–7009. doi: 10.4049/jimmunol.173.11.7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desmet C, Gosset P, Pajak B, Cataldo D, Bentires-Alj M, Lekeux P, Bureau F. Selective blockade of NF-kappa B activity in airway immune cells inhibits the effector phase of experimental asthma. J Immunol. 2004;173:5766–5775. doi: 10.4049/jimmunol.173.9.5766. [DOI] [PubMed] [Google Scholar]
- 42.Schenk M, Bouchon A, Seibold F, Mueller C. TREM-1--expressing intestinal macrophages crucially amplify chronic inflammation in experimental colitis and inflammatory bowel diseases. J Clin Invest. 2007;117:3097–3106. doi: 10.1172/JCI30602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Platt AM, Bain CC, Bordon Y, Sester DP, Mowat AM. An independent subset of TLR expressing CCR2-dependent macrophages promotes colonic inflammation. J Immunol. 2010;184:6843–6854. doi: 10.4049/jimmunol.0903987. [DOI] [PubMed] [Google Scholar]
- 44.Bischoff SC, Lorentz A, Schwengberg S, Weier G, Raab R, Manns MP. Mast cells are an important cellular source of tumour necrosis factor alpha in human intestinal tissue. Gut. 1999;44:643–652. doi: 10.1136/gut.44.5.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jung HC, Eckmann L, Yang SK, Panja A, Fierer J, Morzycka-Wroblewska E, Kagnoff MF. A distinct array of proinflammatory cytokines is expressed in human colon epithelial cells in response to bacterial invasion. J Clin Invest. 1995;95:55–65. doi: 10.1172/JCI117676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Egesten A, Eliasson M, Olin AI, Erjefalt JS, Bjartell A, Sangfelt P, Carlson M. The proinflammatory CXC-chemokines GRO-alpha/CXCL1 and MIG/CXCL9 are concomitantly expressed in ulcerative colitis and decrease during treatment with topical corticosteroids. Int J Colorectal Dis. 2007;22:1421–1427. doi: 10.1007/s00384-007-0370-3. [DOI] [PubMed] [Google Scholar]
- 47.Noguchi A, Watanabe K, Narumi S, Yamagami H, Fujiwara Y, Higuchi K, Oshitani N, Arakawa T. The production of interferon-gamma-inducible protein 10 by granulocytes and monocytes is associated with ulcerative colitis disease activity. J Gastroenterol. 2007;42:947–956. doi: 10.1007/s00535-007-2118-9. [DOI] [PubMed] [Google Scholar]
- 48.Sasaki S, Yoneyama H, Suzuki K, Suriki H, Aiba T, Watanabe S, Kawauchi Y, Kawachi H, Shimizu F, Matsushima K, Asakura H, Narumi S. Blockade of CXCL10 protects mice from acute colitis and enhances crypt cell survival. Eur J Immunol. 2002;32:3197–3205. doi: 10.1002/1521-4141(200211)32:11<3197::AID-IMMU3197>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 49.Melgar S, Drmotova M, Rehnstrom E, Jansson L, Michaelsson E. Local production of chemokines and prostaglandin E2 in the acute, chronic and recovery phase of murine experimental colitis. Cytokine. 2006;35:275–283. doi: 10.1016/j.cyto.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 50.Jugde F, Alizadeh M, Boissier C, Chantry D, Siproudhis L, Corbinais S, Quelvennec E, Dyard F, Campion JP, Gosselin M, Bretagne JF, Semana G, Heresbach D. Quantitation of chemokines (MDC, TARC) expression in mucosa from Crohn's disease and ulcerative colitis. Eur Cytokine Netw. 2001;12:468–477. [PubMed] [Google Scholar]
- 51.Qualls JE, Kaplan AM, van Rooijen N, Cohen DA. Suppression of experimental colitis by intestinal mononuclear phagocytes. J Leukoc Biol. 2006;80:802–815. doi: 10.1189/jlb.1205734. [DOI] [PubMed] [Google Scholar]
- 52.Gordy C, Pua H, Sempowski GD, He YW. Regulation of steady-state neutrophil homeostasis by macrophages. Blood. 2011;117:618–629. doi: 10.1182/blood-2010-01-265959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaph C, Troy AE, Taylor BC, Berman-Booty LD, Guild KJ, Du Y, Yost EA, Gruber AD, May MJ, Greten FR, Eckmann L, Karin M, Artis D. Epithelial-cell-intrinsic IKK-beta expression regulates intestinal immune homeostasis. Nature. 2007;446:552–556. doi: 10.1038/nature05590. [DOI] [PubMed] [Google Scholar]
- 54.Vlantis K, Wullaert A, Sasaki Y, Schmidt-Supprian M, Rajewsky K, Roskams T, Pasparakis M. Constitutive IKK2 activation in intestinal epithelial cells induces intestinal tumors in mice. J Clin Invest. 2011;121:2781–2793. doi: 10.1172/JCI45349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, Kagnoff MF, Karin M. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 56.Murano M, Maemura K, Hirata I, Toshina K, Nishikawa T, Hamamoto N, Sasaki S, Saitoh O, Katsu K. Therapeutic effect of intracolonically administered nuclear factor kappa B (p65) antisense oligonucleotide on mouse dextran sulphate sodium (DSS)-induced colitis. Clin Exp Immunol. 2000;120:51–58. doi: 10.1046/j.1365-2249.2000.01183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dave SH, Tilstra JS, Matsuoka K, Li F, Karrasch T, Uno JK, Sepulveda AR, Jobin C, Baldwin AS, Robbins PD, Plevy SE. Amelioration of chronic murine colitis by peptide-mediated transduction of the IkappaB kinase inhibitor NEMO binding domain peptide. J Immunol. 2007;179:7852–7859. doi: 10.4049/jimmunol.179.11.7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eckmann L, Nebelsiek T, Fingerle AA, Dann SM, Mages J, Lang R, Robine S, Kagnoff MF, Schmid RM, Karin M, Arkan MC, Greten FR. Opposing functions of IKKbeta during acute and chronic intestinal inflammation. Proc Natl Acad Sci U S A. 2008;105:15058–15063. doi: 10.1073/pnas.0808216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nenci A, Becker C, Wullaert A, Gareus R, van Loo G, Danese S, Huth M, Nikolaev A, Neufert C, Madison B, Gumucio D, Neurath MF, Pasparakis M. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557–561. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- 60.Mankan AK, Canli O, Schwitalla S, Ziegler P, Tschopp J, Korn T, Greten FR. TNF-alpha-dependent loss of IKKbeta-deficient myeloid progenitors triggers a cytokine loop culminating in granulocytosis. Proc Natl Acad Sci U S A. 2011;108:6567–6572. doi: 10.1073/pnas.1018331108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park JM, Greten FR, Wong A, Westrick RJ, Arthur JS, Otsu K, Hoffmann A, Montminy M, Karin M. Signaling pathways and genes that inhibit pathogen-induced macrophage apoptosis--CREB and NF-kappaB as key regulators. Immunity. 2005;23:319–329. doi: 10.1016/j.immuni.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 62.Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-kappaB signaling pathways. Nat Immunol. 2011;12:695–708. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- 63.Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature) Biochem Biophys Res Commun. 2004;322:1111–1122. doi: 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- 64.Sunahori K, Yamamura M, Yamana J, Takasugi K, Kawashima M, Yamamoto H, Chazin WJ, Nakatani Y, Yui S, Makino H. The S100A8/A9 heterodimer amplifies proinflammatory cytokine production by macrophages via activation of nuclear factor kappa B and p38 mitogen-activated protein kinase in rheumatoid arthritis. Arthritis Res Ther. 2006;8:R69. doi: 10.1186/ar1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kerkhoff C, Eue I, Sorg C. The regulatory role of MRP8 (S100A8) and MRP14 (S100A9) in the transendothelial migration of human leukocytes. Pathobiology. 1999;67:230–232. doi: 10.1159/000028098. [DOI] [PubMed] [Google Scholar]
- 66.Kerkhoff C, Klempt M, Sorg C. Novel insights into structure and function of MRP8 (S100A8) and MRP14 (S100A9) Biochim Biophys Acta. 1998;1448:200–211. doi: 10.1016/s0167-4889(98)00144-x. [DOI] [PubMed] [Google Scholar]
- 67.Boyd JH, Kan B, Roberts H, Wang Y, Walley KR. S100A8 and S100A9 mediate endotoxin-induced cardiomyocyte dysfunction via the receptor for advanced glycation end products. Circ Res. 2008;102:1239–1246. doi: 10.1161/CIRCRESAHA.107.167544. [DOI] [PubMed] [Google Scholar]
- 68.Sinha P, Okoro C, Foell D, Freeze HH, Ostrand-Rosenberg S, Srikrishna G. Proinflammatory S100 proteins regulate the accumulation of myeloid-derived suppressor cells. J Immunol. 2008;181:4666–4675. doi: 10.4049/jimmunol.181.7.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aomatsu T, Yoden A, Matsumoto K, Kimura E, Inoue K, Andoh A, Tamai H. Fecal calprotectin is a useful marker for disease activity in pediatric patients with inflammatory bowel disease. Dig Dis Sci. 2011;56:2372–2377. doi: 10.1007/s10620-011-1633-y. [DOI] [PubMed] [Google Scholar]
- 70.Lewis JD. The utility of biomarkers in the diagnosis and therapy of inflammatory bowel disease. Gastroenterology. 2011;140:1817–1826. e1812. doi: 10.1053/j.gastro.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Canani RB, de Horatio LT, Terrin G, Romano MT, Miele E, Staiano A, Rapacciuolo L, Polito G, Bisesti V, Manguso F, Vallone G, Sodano A, Troncone R. Combined use of noninvasive tests is useful in the initial diagnostic approach to a child with suspected inflammatory bowel disease. J Pediatr Gastroenterol Nutr. 2006;42:9–15. doi: 10.1097/01.mpg.0000187818.76954.9a. [DOI] [PubMed] [Google Scholar]
- 72.Costa F, Mumolo MG, Ceccarelli L, Bellini M, Romano MR, Sterpi C, Ricchiuti A, Marchi S, Bottai M. Calprotectin is a stronger predictive marker of relapse in ulcerative colitis than in Crohn's disease. Gut. 2005;54:364–368. doi: 10.1136/gut.2004.043406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dabritz J, Friedrichs F, Weinhage T, Hampe J, Kucharzik T, Lugering A, Broeckel U, Schreiber S, Spieker T, Stoll M, Foell D. The functional-374T/A polymorphism of the receptor for advanced glycation end products may modulate Crohn's disease. Am J Physiol Gastrointest Liver Physiol. 2011;300:G823–G832. doi: 10.1152/ajpgi.00115.2010. [DOI] [PubMed] [Google Scholar]
- 74.Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2010;28:367–388. doi: 10.1146/annurev.immunol.021908.132603. [DOI] [PubMed] [Google Scholar]
- 75.Lee MJ, Lee JK, Choi JW, Lee CS, Sim JH, Cho CH, Lee KH, Cho IH, Chung MH, Kim HR, Ye SK. Interleukin-6 induces S100A9 expression in colonic epithelial cells through STAT3 activation in experimental ulcerative colitis. PLoS One. 2012;7:e38801. doi: 10.1371/journal.pone.0038801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.