Abstract
Competition for molecular oxygen (O2) among respiratory microorganisms is intense because O2 is a potent electron acceptor. This competition leads to the formation of microoxic environments wherever microorganisms congregate in aquatic, terrestrial and host-associated communities. Bacteria can harvest O2 present at low, even nanomolar, concentrations using high-affinity terminal oxidases. Here, we report the results of surveys searching for high-affinity terminal oxidase genes in sequenced bacterial genomes and shotgun metagenomes. The results indicate that bacteria with the potential to respire under microoxic conditions are phylogenetically diverse and intriguingly widespread in nature. We explore the implications of these findings by highlighting the importance of microaerobic metabolism in host-associated bacteria related to health and disease.
Microbiologists have long recognized the existence of microaerophiles — that is, bacteria that grow optimally at low levels of molecular oxygen (O2). However, many bacteria that grow optimally at saturating concentrations of O2 also have the potential to respire under microoxic conditions1. For instance, Canfield and colleagues recently demonstrated that Escherichia coli can respire aerobically at nanomolar O2 concentrations, which is more than two orders of magnitude lower than previously observed for aerobes2. Recent studies also suggest that microaerobic lifestyles are important in host-associated microbial communities3,4, but the conceptual framework and terminology for describing life in microoxic environments has not yet been fully developed.
Even some of the traditional definitions of bacterial responses to O2 need to be refined in the light of recently obtained genetic and physiological information5. Historically, bacteria have been assigned to one of five categories on the basis of their requirements for O2 and their ability to metabolize it in different environments6 (TABLE 1). Obligate aerobes require atmospheric O2 concentrations (~20%) for optimal growth. Microaerophiles grow optimally at concentrations well below normal atmospheric concentrations. Facultative anaerobes can respire aerobically, use alternative terminal electron acceptors for anaerobic respiration or grow via fermentation. Aerotolerant anaerobes can tolerate the presence of some O2, but do not gain energy from aerobic respiration and grow optimally without O2. Finally, obligate anaerobes cannot tolerate O2 and grow only under anoxic conditions. A sixth category, nanaerobes, was proposed in 2004 to describe bacteria that can respire O2 at nanomolar concentrations, but grow best under anoxic conditions7. Although this addition to the traditional definitions is helpful, none of these categories encompasses all the microorganisms capable of respiring O2 in microoxic environments. Microaerophile is too constraining a term, as it describes only those organisms that grow best under microoxic conditions and ignores facultative anaerobes and nanaerobes, which can also utilize low concentrations of O2. Consequently, we propose the term microaerobe to define any organism that uses a high-affinity terminal oxidase to respire O2 in a microoxic environment. Microaerobes might grow optimally under anoxic or atmospheric levels of O2, but are distinguished from other bacteria by their capacity to obtain energy from aerobic respiration as O2 concentrations approach zero.
Table 1.
Characterization of microorganisms in relation to O2 metabolism
| Organisms | Aerobic growth |
Low-affinity oxidase |
Microaerobic growth |
High-affinity oxidase |
ROS defence |
Anaerobic growth |
Representative species |
|---|---|---|---|---|---|---|---|
| Obligate aerobes | + | + | − | − | + | − | Mycobacterium leprae |
| Microaerophiles | + | − | + | + | + | − | Helicobacter pylori |
| Facultative anaerobes* | + | + | + | + | + | + | Escherichia coli |
| Nanaerobes | + | − | + | + | + | + | Bacteroides fragilis |
| Aerotolerant anaerobes | − | − | − | − | + | + | Streptococcus pneumoniae |
| Obligate anaerobes‡ | − | − | − | − | − | + | Clostridium tetani |
ROS, reactive oxygen species.
Not all facultative anaerobes have high-affinity oxidases or grow microaerobically.
Many obligate anaerobes tolerate transient or low levels of O2.
In this Analysis article, we document the occurrence of annotated, high-affinity oxidases in the genomes of 1,001 bacterial species, expanding on a previous analysis of terminal oxidase genes in bacterial and archaeal genomes1. We also assess the environmental distribution of high-affinity cytochrome oxidases by analysing metagenomic data sets, and discuss the significance of microaerobes in host-associated microbiomes. Finally, we discuss the role of microaerobic respiration in bacterial pathogenesis, including host invasion and virulence.
The prevalence of microoxic environments
Bacteria are traditionally cultured and studied under strictly anoxic or ambient (ca. 21%) O2 conditions, both of which have revealed remarkable biochemical and physiological diversity among bacteria. However, it is important to note that although bacteria are often described as growing at atmospheric O2 concentrations, the O2 levels actually experienced by microorganisms are not equal to those measured in the atmosphere (BOX 1). Moreover, the natural habitats of bacteria include transition zones between oxic and anoxic environments. Such transition zones are common in microbial communities whenever consumption outstrips diffusion. The resulting O2 gradient includes a microoxic zone of reduced O2 concentration between the oxic and anoxic environments.
Box 1.
The solubility of O2 in water
Microorganisms live in a world driven by the diffusion of molecules in aqueous environments, whether those microorganisms are associated with host tissues, attached to inanimate objects in hydrated biofilms, or free-living in aquatic environments. As a result, these organisms do not experience the concentrations of O2 that are measured in the atmosphere, but rather experience the amount of O2 that diffuses into their immediate environment. To translate atmospheric O2 measurements into concentrations of O2 that are relevant for microorganisms, Henry’s law (p = kHc) can be used. This formula facilitates the calculation of the concentration of a gas in the liquid phase (c) given the partial pressure of the gas in the gas phase (p). The constant kH is known as Henry’s law constant and depends on characteristics of the solute, the solvent and temperature. The table below illustrates the effect of temperature and salinity on the concentration of O2 in fresh water and water with 5% salinity56. As with any gas, the saturating concentration of dissolved O2 decreases with temperature and salinity, and is thus another factor that can influence the requirement for high-affinity terminal oxidases.
| Temperature (°C) |
O2 concentration in fresh water |
O2 concentration in water with 5% salinity |
||||
|---|---|---|---|---|---|---|
| ml per l | mg per l | µM | ml per l | mg per l | µM | |
| 15 | 7.0 | 10.1 | 316 | 5.2 | 7.4 | 231 |
| 25 | 5.8 | 8.3 | 259 | 4.4 | 6.3 | 197 |
| 35 | 4.9 | 7.0 | 219 | 3.7 | 5.3 | 166 |
Microoxic zones have been documented in a wide variety of environments. Soil aggregates8 and marine snow9–11 develop O2 gradients across their radii as microorganisms on and near the surface consume O2 before it can diffuse all the way into the interior of the community. Similarly, biofilms and sediments have microoxic zones. In these environments, O2 levels decrease away from the aqueous interface because diffusion limits the progression of O2 into the film or sediment, and because microorganisms near the surface respire12–14. In the oceans, mixing of the upper water column delivers O2 produced in the photic zone to deeper waters, where biotic consumption results in the formation of a microoxic region called the oxygen minimum zone. O2 concentrations then increase again with depth owing to the circulation of more-oxygenated water from the depths, forming a sandwich-like O2 gradient15.
Microoxic environments are also present in plants and animals. In plants, an O2 gradient has been measured within N2-fixing alfalfa root nodules. The O2 concentration decreases from ~250 µM at the nodule apex to <1 µM in the regions where bacterial nitrogen fixation takes place16. In soybean nodules, O2 concentrations are in the nanomolar range17. In the gastrointestinal tracts of animals, O2 gradients develop as O2 diffuses from the tissues towards the lumen. This phenomenon has been measured with microelectrodes in termites and terrestrial snails18,19. More recently, an oxygenated zone in the rabbit ileum was visualized using bacteria expressing GFP3. As GFP requires O2 to fold properly, it can be visualized only in the presence of O2. This characteristic was exploited to generate an image that clearly demonstrates the presence of O2 across a 70 µm region reaching from the epithelium into the lumen. Although this method is not sensitive to gradients of O2, the obvious oxic and anoxic layers in the image provide solid evidence of a microoxic zone within the mammalian gut. This finding is particularly noteworthy because the microbial community within the microoxic zone is proximal to the host and therefore physically poised to interact with the host20. Examples of how microoxic environments can affect host–microorganism interactions are discussed below.
Taken together, the above examples illustrate the considerable diversity of environments in which microaerobes can reside. In spite of the fact that microoxic zones are so widespread, the potential for microoxic conditions to influence bacterial physiology and ecology is underappreciated. Recent studies using microoxic conditions to culture organisms have revealed a great diversity in the free-living diazotrophs found in the soil21 and also recovered organisms that did not grow under anoxic conditions from human-associated samples22. Clearly, a better understanding of the selective pressures that low O2 concentrations exert on bacteria is required if we are to develop a comprehensive understanding of microbial communities in nature.
High-affinity terminal oxidases
The potential interactions between microorganisms and the O2 in their environment provide a useful basis for the physiological and ecological characterization of bacteria. Obviously, whether or not an organism can use O2 as a terminal electron acceptor is important, but the answer to this question alone fails to capture vital information about the ability of microorganisms to access O2 in microoxic environments. Terminal oxidases in bacteria have a range of affinities for O2, and as discussed below, only some of these enzymes provide the capacity to grow in low-O2 environments. In addition to characterizing the affinity for O2 as a terminal electron acceptor, it is valuable to consider the ability of a microorganism to mount a defence against the toxic effects of reactive oxygen species (ROS). This characteristic has been used to identify obligate anaerobes, but in fact many bacteria that are traditionally defined as such can escape the toxic effects of O2 exposure, at least for short periods of time23. Together, the capacity for aerobic and anaerobic growth, the characteristics of the terminal oxidases and the response to ROS provide a framework for classifying organisms with respect to O2 metabolism (TABLE 1).
Terminal oxidase families
Terminal oxidases are the final links in the membrane-associated electron transport chains of respiratory bacteria. Like many redoxactive enzymes, they have metal reaction centres that are reduced and then oxidized as electrons are shuttled to a terminal electron acceptor. This study focuses on the subset of terminal oxidases that transfer electrons to O2.
As is common in the scientific literature, we use the term cytochrome to describe the multihaem protein complexes found in bacterial electron transport chains. Specific cytochromes are named according to the combination of protein-bound haem subunits that constitute the cytochrome (a, b, c, d or o). Terminal cytochrome oxidases that transfer electrons to O2 are grouped into two major families: the haem–copper oxidases (HCOs) and the cytochrome bd-type oxidases (TABLE 2). The redox centres of both families include haem, but the catalytic subunits of HCOs contain copper ions as part of a bimetallic centre24. As per convention, the HCO catalytic subunit that transfers electrons to O2 is denoted with a subscript 3 (REF. 25). Functionally, the HCOs are distinguished from the cytochrome bd-oxidases by the capacity of HCOs to translocate protons across the cytoplasmic membrane26,27. Both families contribute to membrane potential through the consumption of protons during the reduction of O2 to water in the cytoplasm28.
Table 2.
Bacterial oxidases that use O2 as a terminal electron acceptor
| Low-affinity terminal oxidases | High-affinity terminal oxidases | |||
|---|---|---|---|---|
| Family | HCOs | HCOs | HCOs | Cytochrome bd-type oxidases |
| Class | A | C | B | No class divisions |
| Representatives | Cytochrome aa3 oxidase and cytochrome bo3 oxidase | Cytochrome cbb3 oxidase | Cytochrome ba3 oxidase | Cytochrome bd oxidase |
| Catalytic subunits* | CtaD or CyoB | FixN (also known as CcoN in some species) | CbaA | CydA |
| Km | 200 nM (REF. 33) | 7 nM (REF. 47) | NA | 3–8 nM (REF. 34) |
| H+ pump | Yes | Yes | Yes | No |
HCOs, haem–copper oxidases; Km, Michaelis constant. NA, not available.
The catalytic subunits listed are those responsible for the transfer of electrons to O2.
Although terminal oxidases are named for the haem groups they contain (for example, cytochrome bd oxidase), they receive electrons from either quinols or other cytochromes. The HCOs receive electrons from both quinols and other cytochromes, depending on the particular enzyme. The cytochrome bd-type oxidases are quinol oxidases, receiving electrons from either ubiquinones or menaquinones, both of which contain a quinone that is reduced to a quinol during electron transport.
On the basis of biochemical differences in their catalytic subunits, HCOs are further divided into three classes: A, B and C29. A-class HCOs, which include the mitochondrial terminal oxidase, have a low affinity for O2 (REFS 29,30). Therefore, for optimal growth, organisms that have only A-class oxidases are predicted to require dissolved O2 concentrations that are typically found under normal atmospheric concentrations of O2.
C-class HCOs have been detected only in bacteria1. The oxidases from this class that have been investigated experimentally have high affinities for O2, and organisms expressing these enzymes are therefore considered to be capable of growth in microoxic environments30. The most common example of a C-class HCO is cytochrome cbb3 oxidase, which is found in many proteobacteria27,31.
B-class HCOs are found in both bacterial and archaeal lineages1. The B-class oxidases include Thermus thermophilus cytochrome ba3 oxidase, which is expressed under microoxic conditions32. On the basis of this expression pattern and structural similarity to the C-class HCOs, B-class oxidases are thought to be high-affinity oxidases used for microaerobic metabolism30.
Cytochrome bd-type oxidases have also been found in both bacterial and archaeal genomes1. Like B- and C-class HCOs, cytochrome bd-type oxidases generally have a strong affinity for O2. For example, the high-affinity oxidase found in E. coli is a cytochrome bd-type oxidase with a Michaelis constant (Km) of 3–8 nM (REFS 33,34). The nanaerobe Bacteroides fragilis also uses a cytochrome bd-type oxidase for microaerobic metabolism7. However, the oxidase previously classified as cytochrome bd-type in Campylobacter jejuni is actually a low-affinity oxidase, and it has been suggested that this oxidase instead be referred to as a cyanide-insensitive oxidase and the genes be annotated cio, as in Pseudomonas spp.35,36. Comparison between the genes encoding the E. coli high-affinity cytochrome bd-type oxidase catalytic subunit (CydA) and the Pseudomonas aeruginosa low-affinity oxidase catalytic subunit (CioA) indicates that the two catalytic subunits can be distinguished by the length of the sequence for the conserved periplasmic Q-loop36,37.
Distribution of high-affinity oxidases
Although direct measurements of the oxygen affinity of intact cells or purified terminal oxidases are limited to a few microorganisms, as described above, the conservation of gene sequences in each family of high-affinity oxidase genes offers the prospect of identifying homologues in bacterial genomes and shotgun metagenomes. Although homologous genes do not necessarily encode proteins with similar activity profiles, the presence of homologues of high-affinity terminal oxidases in a genome indicates a potential for the encoded enzyme to be active at low O2 concentrations. The phylogenetic and environmental distribution of these homologues, as reported below, highlights the potential importance of microaerobes and helps identify specific organisms and environments for experimental investigation of microaerobic metabolism.
Microaerobic potential in sequenced genomes
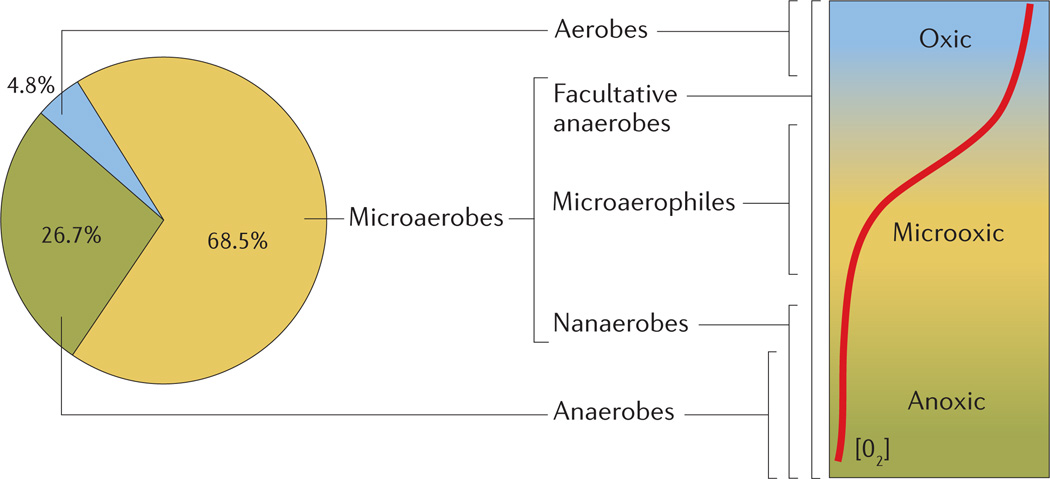
Building on the results of a previous study that investigated the evolutionary histories of terminal oxidase genes in 673 sequenced bacterial and archaeal genomes1, we explored the microaerobic potential of 1,001 bacterial species. The genomes were queried, using tBLASTx, with four sets of reference genes, each set representing the catalytic subunit of a terminal oxidase class38. We excluded archaeal genomes because there is little experimental data available regarding the oxygen affinity of archaeal versions of cytochrome bd-type oxidases and B-class HCOs. Species were classified as being aerobes, microaerobes or anaerobes on the basis of the terminal oxidase genes detected: organisms encoding homologues of high-affinity oxidases, either alone or along with low-affinity oxidase matches, were categorized as microaerobes, those with only A-class oxidases as aerobes and those with no annotated oxidases as anaerobes. When multiple genomes were available for a single species, the presence of terminal oxidase genes in any of the genomes contributing to the pangenome was ascribed to the species. According to our definitions and analysis, 4.8% of species analysed were aerobes, 26.7% were anaerobes, and the remaining 68.5% were microaerobes (FIG. 1). These percentages are similar to those calculated from the previous study, even though the number of bacterial genomes included in our analysis was nearly double that of the previous study. Therefore, on the basis of our analysis of available genomes, bacterial species with the capacity for microaerobic growth are more common than obligately aerobic or anaerobic species.
Figure 1. The distribution of terminal oxidases in bacterial genomes linked to physiological groups of microorganisms and their distribution in an O2 gradient.
Relationships between the occurrence of high- and low-affinity terminal oxidases in bacterial genomes, bacterial groups named according to their response to O2 (TABLE 1), and the environments in which each group of bacteria can be found. The pie chart shows the percentage of bacterial species in which our analysis detected genes encoding the various oxidases. Microaerobes are defined genomically by the presence of a high-affinity cytochrome oxidase, either alone or in combination with a low-affinity oxidase, and encompass bacterial groups that can be found in the entire range of O2 concentrations which occur in nature. Organisms with only low-affinity oxidases are classed as aerobes and are found only in oxic environments. Anaerobes encode no oxidases and are found in anoxic environments. The red line depicts an O2 gradient, decreasing from top to bottom.
Although this analysis was constrained by the bacterial genomes that have been sequenced, the predicted genes for high-affinity terminal oxidases were widely distributed across bacterial phyla (Supplementary information S1 (table)). So, although there is debate regarding the specific evolutionary origins of the different terminal oxidase families1,29,39, microaerobic metabolism does not appear to be limited to any single phylogenetic lineage within bacteria.
Distribution of high-affinity oxidases in nature
Wherever O2 gradients exist, the zone between the oxic and anoxic layers provides an environment that selects for organisms adapted to low O2 tension13. Although high-affinity oxidase genes are found in a majority of sequenced genomes from cultivated bacteria, cultivation captures only a fraction of the true bacterial diversity40. As a result, the abundance of bacteria with the capacity for microaerobic growth in most environments is unknown. To assess the distribution of bacterial microaerobes in complex microbial communities, we searched a collection of shotgun metagenomes for homologues of genes encoding the catalytic subunits of high-affinity terminal oxidases from both families.
We analysed a subset of the shotgun metagenomes that are publicly available through MG-RAST41. The metagenomes were chosen to encompass a range of terrestrial, aquatic and host-associated environments. We also included a set of shotgun metagenomes from the Kellogg Biological Station Long-Term Ecological Research (KBS LTER) field sites. These metagenomes are uncommon in that they are derived from experimentally replicated field sites and so can be used for testing hypotheses.
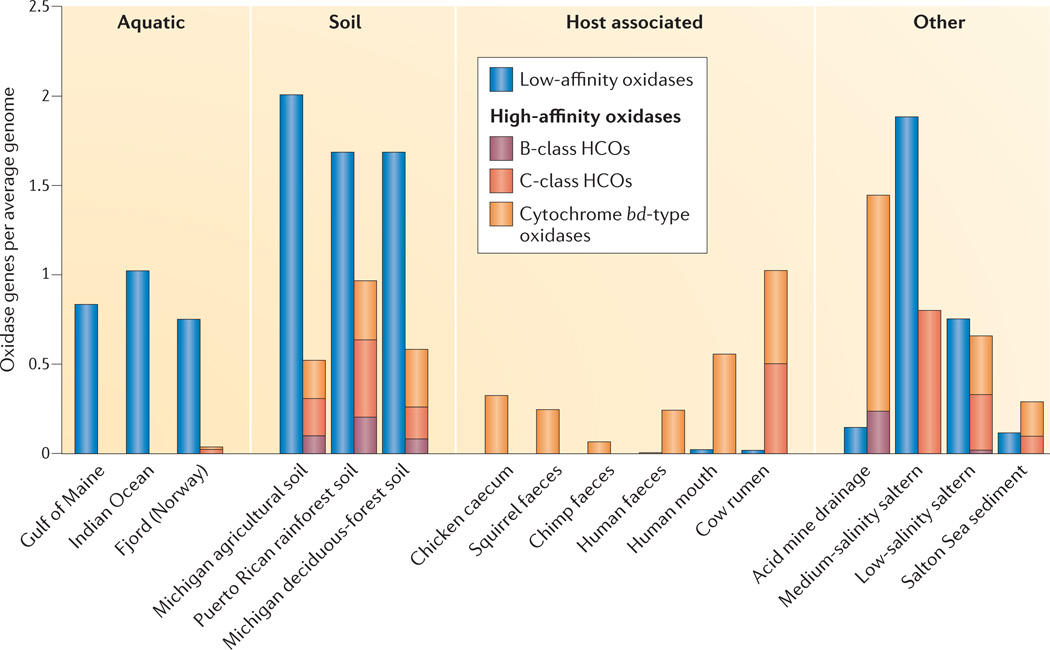
All the selected metagenomes were first assessed for the presence of genes encoding the catalytic subunits of bacterial terminal oxidases (TABLE 2). Genes from at least one class of high-affinity oxidases were detected in most metagenomes, clearly demonstrating that bacteria with the potential for microaerobic metabolism are widespread in nature. Then, to estimate the abundance of genomes with the potential for microaerobic respiration, we compared the abundance of high-affinity terminal oxidase genes with that of low-affinity terminal oxidase genes for each metagenome. Genes encoding terminal oxidases predicted to have either high or low affinity for O2, as well as a set of four housekeeping genes, were counted and normalized to gene length. The arithmetic mean of the housekeeping gene counts was then used as an estimate of the average number of genomes per metagenome. In order to determine that this estimate was as robust as possible, the coefficient of variation (CV) for the count of each set of housekeeping genes was calculated, and metagenomes with CV < 15% are shown in FIG. 2 (see Supplementary information S2,S3 (box; table) for complete details). Many bacteria express either high- or low-affinity oxidases as environmental conditions warrant, and some even encode multiple oxidases from the same class1, so this normalization was performed to facilitate comparison among the metagenomes. When this method was used to estimate the proportion of bacteria from various environments with high- or low-affinity terminal oxidase genes, distinct patterns emerged in the ratios of the two groups (FIG. 2).
Figure 2. Distribution of terminal oxidase genes in shotgun metagenomes.
An average genome is an estimate based on the mean of the normalized gene lengths of four bacterial housekeeping genes (the RNA polymerase genes rpoA, rpoB and rpoC, and the recombinase gene recA) from each metagenome. Metagenomes from various environments were included, for comparison. See Supplementary information S2,S3 (box; table) for a detailed description of this analysis. HCOs, haem–copper oxidases.
The only metagenomes in which high-affinity oxidase genes were not detected were from the open ocean. Genomic analysis suggests that certain abundant unattached marine bacteria are under selective pressure to streamline their genomes42. Therefore, the absence of high-affinity oxidase genes was expected (FIG. 2). Filtration of the samples before sequencing might also have contributed to this result by removing marine snow particles in which microoxic zones are likely to be found9–11,15. In terrestrial soils, the ratios of low- to high-affinity oxidases were much lower, ranging between 2:1 and 1:1. Soil aggregates are prime locations for the formation of O2 gradients, creating habitats for microaerobes. Host-associated metagenomes were dominated by high-affinity oxidases, especially the gut-associated metagenomes, where few low-affinity oxidases were detected. Aerobic respiration is not advantageous in the largely anoxic lumen, but in the microoxic zone along the mucosa, microaerobic respiration might give microorganisms a competitive fitness advantage.
When sorted by class, the normalized high-affinity oxidase gene counts revealed another set of interesting patterns, this time in the abundance of the different classes of terminal oxidases present in certain environments (FIG. 2). For example, in gut-associated metagenomes, the only high-affinity oxidase genes detected were those predicted to encode cytochrome bd-type oxidases. It has been suggested that cytochrome bd-type oxidases are selected in bacterial pathogens because, unlike HCOs, they are relatively insensitive to nitric oxide, an important component of the immune response by macrophages43. Nitric oxide is also present in small amounts in the healthy large intestine, which may provide the selective pressure for microaerobes in the gastrointestinal tract to encode cytochrome bd-type oxidases instead of HCOs44.
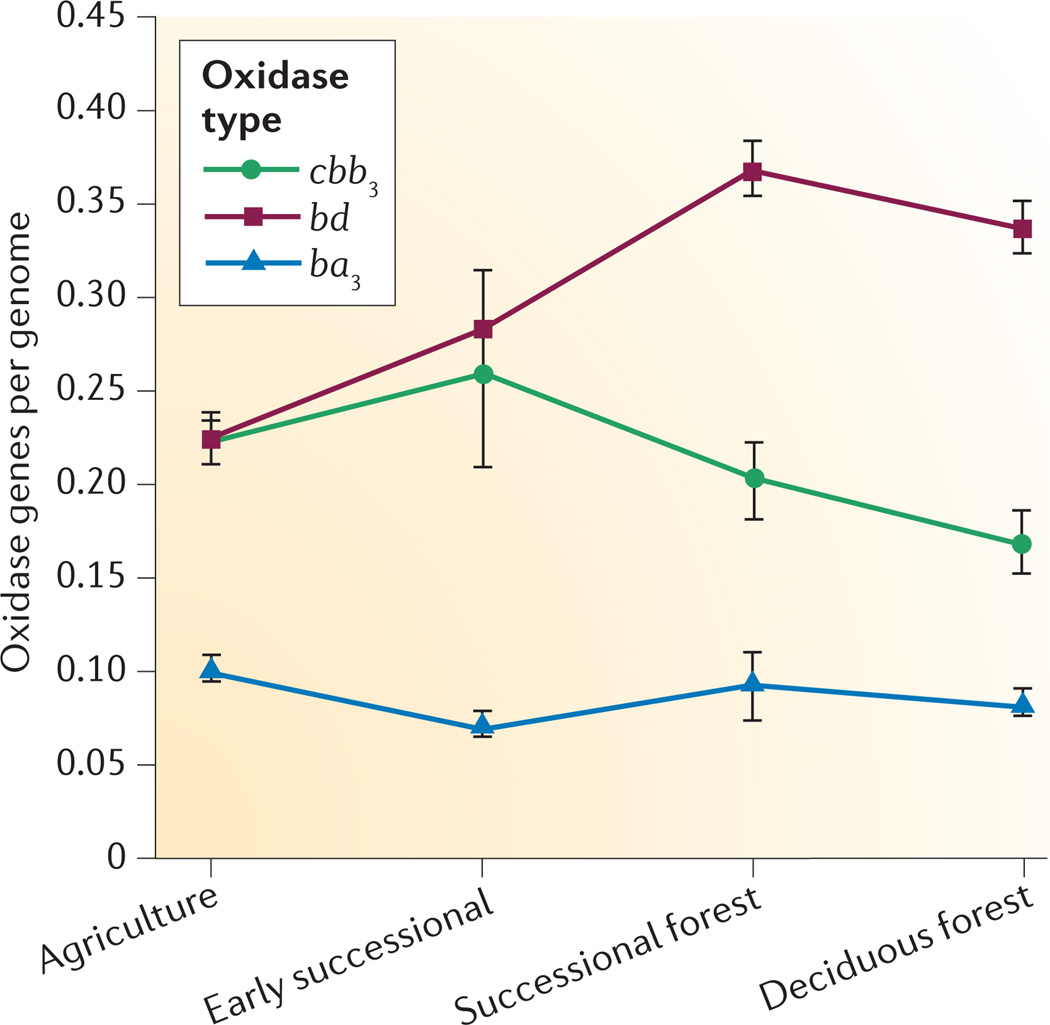
Soil metagenomes, however, contained genes for all three classes of high-affinity oxidase, and the replicate metagenomes from the KBS LTER field sites revealed differences in the abundances of these three classes across a land use gradient that comprises four different treatments: agricultural plots were used for traditional row crop agriculture, early successional plots had been released from agricultural use 20 years before being sampled, successional forest plots were released from agriculture 40–60 years before being sampled, and deciduous forest plots were never disrupted for agriculture (FIG. 3). The proportion of cytochrome cbb3 oxidase- and cytochrome ba3 oxidase-type genes was similar in metagenomes from all four treatments, but the proportion of cytochrome bd-type oxidase genes varied significantly (P < 0.001 using analysis of variance), with highest values in the forested (successional and deciduous) plots. This pattern might reflect a major difference between the two types of enzyme: cytochrome cbb3-type and cytochrome ba3-type oxidases pump protons, whereas cytochrome bd-type oxidases do not28,45. In carbon-poor soils such as the agricultural plots, proton-pumping oxidases might be selected because they generate more ATP per electron. In environments where carbon is more plentiful, as in the forested plots or the gastrointestinal tract of animals, cytochrome bd-type high-affinity oxidases would provide a fitness advantage if the flux of electrons to O2 is more rapid.
Figure 3. Distribution of high-affinity terminal oxidase genes across terrestrial landscapes following release of the land from agriculture.
Agriculture (AG) plots were used for traditional row crop agriculture at the time of sampling, early successional (ES) plots were released from agricultural use 20 years before our sampling, successional forest (SF) plots were released from agriculture 40–60 years prior to sampling, and deciduous forest (DF) plots were never disrupted for agriculture. Error bars show standard error of the mean.
The patterns in the relative abundance of high-affinity oxidase genes revealed by our analysis of metagenomes provide intriguing evidence supporting the importance of microaerobic lifestyles in a wide variety of environments. Further evidence of the impact of microaerobes can be seen in their physiological roles in host-associated microbiomes. Below, we review some salient studies that demonstrate the role of microaerobic metabolism in both healthy microbiomes and pathogenesis.
Host-associated microaerobes
Microaerobes live in a variety of host-associated environments. For example, Stenoxybacter acetivorans is abundant in termite hindguts. This bacterium utilizes a cytochrome cbb3-type oxidase for respiration in the microoxic region along the hindgut wall46. By consuming O2 that diffuses into the gut from the host tissue, populations of S. acetivorans and other microaerobes create an anoxic environment in the lumen of the termite hindgut. Maintenance of this anoxic environment is essential for the community of microorganisms that resides there and ferments complex plant matter to acetate, the primary resource for the termite host.
Low-O2 environments are also necessary for the mutualistic relationships between plants and their nitrogen-fixing symbionts. Symbiotic nitrogen fixation is both energetically costly and oxygen sensitive. Leghaemoglobin produced by the plant host keeps O2 concentrations in the root nodule within the nanomolar range17, protecting the key enzyme in nitrogen fixation, nitrogenase, from inhibition by O2. To generate sufficient ATP to support nitrogen fixation and other anabolic reactions in this microoxic environment, Bradyrhizobium japonicum uses a high-affinity oxidase for microaerobic respiration during symbiosis47.
Healthy microbiomes
Evidence is accumulating to suggest that high-affinity oxidase genes are also important in the microbiota present in the mammalian gastrointestinal tract. Although the mammalian large intestine is often described as an anoxic environment, it does contain O2 near its periphery3,48. O2 diffuses from the epithelium, providing a constant, but low flux that establishes an O2 gradient across the gut mucosa as O2 is consumed. The availability of a low concentration of O2 is likely to provide a selective growth advantage to bacteria that can utilize it20. Expression of high-affinity terminal oxidases would provide access to a high-potential electron acceptor for respiration and provide some protection against ROS through O2 scavenging23,49.
Colonization experiments with knock-out mutations of high-affinity cytochrome oxidases in E. coli (a cytochrome bd oxidase)50,51 and C. jejuni (a cytochrome cbb3 oxidase)52 indicate that these organisms must access O2 at low concentrations in order to colonize the mouse and chicken intestine, respectively. Knocking out the low-affinity oxidases did not affect colonization by either organism. Commensal organisms that occupy the microoxic environment of the mucosa would be uniquely positioned to communicate with the host. Therefore, the relationship between colonization and high-affinity oxidases could provide important insights into the establishment and stability of microbial communities adjacent to host cells.
Pathogens and microaerobic metabolism
High-affinity oxidases also have a role in disease and in disruption of the normal microbiota. For example, the enteric pathogen Shigella flexneri responds to low O2 concentrations in the gastrointestinal tract by upregulating virulence genes3, presumably because low O2 levels indicate to the pathogen that it is in close proximity to host epithelial cells. The cytochrome bd-type high-affinity oxidase of S. flexneri is important for virulence, allowing the bacterium to respire in the host cell cytoplasm and the microoxic zone of the mucosa53. In fact, many human intestinal pathogens have high-affinity oxidases1. In the case of pathogens such as S. flexneri and Salmonella enterica, microaerobic metabolism can facilitate host invasion and competition with the normal microbiota.
High-affinity cytochrome oxidases also seem to be important to the lifestyle of pathogens that cause infections outside the gastrointestinal tract. It has been suggested that microaerobic respiration facilitates P. aeruginosa infections in patients with cystic fibrosis, helping the organism to survive the microoxic conditions in the thick mucus of the cystic fibrosis lung54. Mycobacterium tuberculosis has the genes for a cytochrome bd-type terminal oxidase, and the expression of this oxidase has been linked to the ability of the organism to reside in the host55. Recently, 17 pathogenicity-related genes were identified by tracking mutations in Burkholderia dolosa during an outbreak in patients with cystic fibrosis. Three of the candidate genes (fnr, fixL, and fixJ) are homologues of genes that regulate the expression of high-affinity cytochrome oxidases (both cytochrome cbb3 oxidases and cytochrome bd-type oxidases)4.
Taken together, the above studies provide intriguing evidence that microaerobic metabolism contributes to the development and maintenance of host microbiomes. Understanding the role of high-affinity oxidases in shaping the structure and function of microbial communities will advance our ability to build predictive models of the interactions between the host and the microbiota. Such an understanding will provide information that is important not only in the gastrointestinal tract, but at all mucosal surfaces where O2 gradients exist. Accurate models of the interactions between the host and the microbiota could be used to guide the manipulation of microbial communities, with the goal of establishing or restoring healthy microbiomes.
Conclusions and future directions
Our analyses of sequenced genomes and metagenomes suggest that microaerobes are phylogenetically diverse and found in most environments. However, although we know that O2 is an important ecological force, little is understood about the ‘shallow breathing’ lifestyle of the bacteria that live in these microoxic environments. To fill this gap in our knowledge, culturing techniques and physiological studies of host-associated and free-living bacteria must be expanded to include microoxic conditions. By studying microaerobes under the conditions that they experience in nature, we reduce the risk of missing metabolic strategies that are crucial components of fitness in natural environments.
Biochemical and genetic characterizations of high-affinity oxidase classes have provided much useful information, but the ecological implications of the ability of microaerobes to grow and even thrive at low O2 concentrations has not been examined in depth. The advanced O2-sensing technology now available should be used in environmental studies to help define low-O2 environments and provide accurate and sensitive measures of O2 gradients (BOX 2). Considering the abundance and environmental distribution of putative microaerobes, we need to learn how life works in microoxic zones, as this knowledge may well provide key insights into how microbial communities form and function both outside and within us.
Box 2.
Advances in oxygen-sensing technology
Our understanding of the microoxic world continues to expand as technological developments improve our ability to measure O2. Previously, Clark-type electrochemical electrodes, the gold standard of O2 measurement, could at best measure O2 concentrations of around 1–2 µmol per litre, but improved microelectrodes that can measure nanomolar concentrations have been developed in the past few years57. The impact of these improved sensors was recently demonstrated during a study in which the O2 concentration at which microorganisms can respire aerobically was shown to be orders of magnitude lower than previously reported2.
Optodes, which are an optics-based technology, pair O2 measurement with quenching of fluorescence from indicator molecules and are also being used to expand our view of the microoxic world. Less fragile than microelectrodes, optodes provide stable and specific measurements when imbedded in optic fibres to form probes or when attached to planar surfaces58,59. They can provide sensitive and accurate measurements of O2 gradients, which can be particularly useful for determining the rates of oxygen consumption in microoxic environments. This strategy was used recently in the discovery of the remarkably slow rates of respiration occurring for tens of metres into deep-sea sediments60.
In addition to measuring O2 concentrations, these sensors provide a full two-dimensional view of O2 gradients in situ, as demonstrated when optodes were spin-coated onto coverslips supporting the growth of biofilms14 and when the sensors were used at water–sediment interfaces58.
Supplementary Material
Acknowledgements
The authors thank J. Breznak for seminal conversations about the ecological importance of microaerobes; K. D. Noel and C. Waldron for critical reading of this manuscript; T. Teal and B. Klahn for their expert assistance with the genomic and metagenomic analyses; and members of the Schmidt laboratory for many helpful discussions concerning the potential consequences of microaerobes and microoxic environments. This work was supported by grants from the US National Institutes of Health (R01 HG004906 and UH3 DK083993) and the US National Science Foundation (MCB 0731913 and DEB 1027253).
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Thomas M. Schmidt’s homepage: http://microbiomes.msu.edu
KBS LTER: http://lter.kbs.msu.edu
MG-RAST: http://metagenomics.anl.gov
SUPPLEMENTARY INFORMATION
See online article: S1 (table) | S2 (box) | S3 (table)
References
- 1. Brochier-Armanet C, Talla E, Gribaldo S. The multiple evolutionary histories of dioxygen reductases: implications for the origin and evolution of aerobic respiration. Mol. Biol. Evol. 2009;26:285–297. doi: 10.1093/molbev/msn246. This article includes genomic surveys of terminal oxidase genes and presents a model for the evolutionary origin of aerobic respiration in bacteria and archaea.
- 2. Stolper DA, Revsbech NP, Canfield DE. Aerobic growth at nanomolar oxygen concentrations. Proc. Natl Acad. Sci. USA. 2010;107:18755–18760. doi: 10.1073/pnas.1013435107. This study demonstrates aerobic growth of an E. coli strain at nanomolar concentrations of O2 — concentrations that are orders of magnitude lower than those previously reported.
- 3. Marteyn B, et al. Modulation of Shigella virulence in response to available oxygen in vivo. Nature. 2010;465:355–358. doi: 10.1038/nature08970. This investigation of the effects of O2 on virulence in Shigella includes the visualization of an oxygenated zone adjacent to the intestinal mucosa.
- 4.Lieberman TD, et al. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nature Genet. 2011;43:1275–1280. doi: 10.1038/ng.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abreu IA, Xavier AV, LeGall J, Cabelli DE, Teixeira M. Superoxide scavenging by neelaredoxin: dismutation and reduction activities in anaerobes. J. Biol. Inorg. Chem. 2002;7:668–674. doi: 10.1007/s00775-002-0363-1. [DOI] [PubMed] [Google Scholar]
- 6.Madigan MT, Martinko JM, Stahl DA, Clark DP. Brock Biology of Microorganisms. 13th edn. Benjamin Cummings; 2010. pp. 117–149. [Google Scholar]
- 7.Baughn AD, Malamy MH. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature. 2004;427:441–444. doi: 10.1038/nature02285. [DOI] [PubMed] [Google Scholar]
- 8.Tiedje J, Sexstone A, Parkin T, Revsbech N, Shelton D. Anaerobic processes in soil. Plant Soils. 1984;76:197–212. [Google Scholar]
- 9.Ploug H, Iversen MH, Fischer G. Ballast, sinking velocity, and apparent diffusivity within marine snow and zooplankton fecal pellets: implications for substrate turnover by attached bacteria. Limnol. Oceanogr. 2008;53:1878–1886. [Google Scholar]
- 10.Alldredge AL, Cohen Y. Can microscale chemical patches persist in the sea? Microelectrode study of marine snow, fecal pellets. Science. 1987;235:689–691. doi: 10.1126/science.235.4789.689. [DOI] [PubMed] [Google Scholar]
- 11.Ploug H. Small-scale oxygen fluxes and remineralization in sinking aggregates. Limnol. Oceanogr. 2001;46:1624–1631. [Google Scholar]
- 12.Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nature Rev. Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 13.Fenchel T, Finlay B. Oxygen and the spatial structure of microbial communities. Biol. Rev. Camb. Philos. Soc. 2008;83:553–569. doi: 10.1111/j.1469-185X.2008.00054.x. [DOI] [PubMed] [Google Scholar]
- 14.Kühl M, Rickelt LF, Thar R. Combined imaging of bacteria and oxygen in biofilms. Appl. Environ. Microbiol. 2007;73:6289–6295. doi: 10.1128/AEM.01574-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright JJ, Konwar KM, Hallam SJ. Microbial ecology of expanding oxygen minimum zones. Nature Rev. Microbiol. 2012;10:381–394. doi: 10.1038/nrmicro2778. [DOI] [PubMed] [Google Scholar]
- 16.Soupène E, Foussard M, Boistard P, Truchet G, Batut J. Oxygen as a key developmental regulator of Rhizobium meliloti N2-fixation gene expression within the alfalfa root nodule. Proc. Natl Acad. Sci. USA. 1995;92:3759–3763. doi: 10.1073/pnas.92.9.3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuzma MM, Hunt S, Layzell DB. Role of oxygen in the limitation and inhibition of nitrogenase activity and respiration rate in individual soybean nodules. Plant Physiol. 1993;101:161–169. doi: 10.1104/pp.101.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charrier M, Brune A. The gut microenvironment of helicid snails (Gastropoda: Pulmonata): in-situ profiles of pH, oxygen, and hydrogen determined by microsensors. Can. J. Zool. 2003;81:928–935. [Google Scholar]
- 19.Brune A, Emerson D, Breznak JA. The termite gut microflora as an oxygen sink: microelectrode determination of oxygen and pH gradients in guts of lower and higher termites. Appl. Environ. Microbiol. 1995;61:2681–2687. doi: 10.1128/aem.61.7.2681-2687.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van den Abbeele P, Van de Wiele T, Verstraete W, Possemiers S. The host selects mucosal and luminal associations of coevolved gut microorganisms: a novel concept. FEMS Microbiol. Rev. 2011;35:681–704. doi: 10.1111/j.1574-6976.2011.00270.x. [DOI] [PubMed] [Google Scholar]
- 21.Mirza BS, Rodrigues JL. Development of a direct isolation procedure for free-living diazotrophs under controlled hypoxic conditions. Appl. Environ. Microbiol. 2012;16:5542–5549. doi: 10.1128/AEM.00714-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young VB, et al. Multiphasic analysis of the temporal development of the distal gut microbiota in patients following ileal pouch anal anastomosis. Microbiome. doi: 10.1186/2049-2618-1-9. (in the press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brioukhanov A, Netrusov A. Aerotolerance of strictly anaerobic microorganisms and factors of defense against oxidative stress: a review. Appl. Biochem. Microbiol. 2007;43:567–582. [PubMed] [Google Scholar]
- 24.Hemp J, et al. Evolutionary migration of a post-translationally modified active-site residue in the proton-pumping heme-copper oxygen reductases. Biochemistry. 2006;45:15405–15410. doi: 10.1021/bi062026u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calhoun MW, Thomas JW, Gennis RB. The cytochrome oxidase superfamily of redox-driven proton pumps. Trends Biochem. Sci. 1994;19:325–330. doi: 10.1016/0968-0004(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 26.Pitcher RS, Watmough NJ. The bacterial cytochrome cbb3 oxidases. Biochim. Biophys. Acta. 2004;1655:388–399. doi: 10.1016/j.bbabio.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 27.Brändén G, Gennis RB, Brzezinski P. Transmembrane proton translocation by cytochrome c oxidase. Biochim. Biophys. Acta. 2006;1757:1052–1063. doi: 10.1016/j.bbabio.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 28.Puustinen A, Finel M, Haltia T, Gennis RB, Wikström M. Properties of the two terminal oxidases of Escherichia coli. Biochemistry. 1991;30:3936–3942. doi: 10.1021/bi00230a019. [DOI] [PubMed] [Google Scholar]
- 29.Pereira MM, Santana M, Teixeira M. A novel scenario for the evolution of haem–copper oxygen reductases. Biochim. Biophys. Acta. 2001;1505:185–208. doi: 10.1016/s0005-2728(01)00169-4. [DOI] [PubMed] [Google Scholar]
- 30. Han H, et al. Adaptation of aerobic respiration to low O2 environments. Proc. Natl Acad. Sci. USA. 2011;108:14109–14114. doi: 10.1073/pnas.1018958108. This report discusses the implications of the adaptation of aerobic respiration to low-O2 environments.
- 31.Pitcher RS, Brittain T, Watmough NJ. Cytochrome cbb3 oxidase and bacterial microaerobic metabolism. Biochem. Soc. Trans. 2002;30:653–658. doi: 10.1042/bst0300653. [DOI] [PubMed] [Google Scholar]
- 32.Keightley JA, et al. Molecular genetic and protein chemical characterization of the cytochrome ba3 from Thermus thermophilus HB8. J. Biol. Chem. 1995;270:20345–20358. doi: 10.1074/jbc.270.35.20345. [DOI] [PubMed] [Google Scholar]
- 33.Rice CW, Hempfling WP. Oxygen-limited continuous culture and respiratory energy conservation in Escherichia coli. J. Bacteriol. 1978;134:115–124. doi: 10.1128/jb.134.1.115-124.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D’mello R, Hill S, Poole RK. The cytochrome bd quinol oxidase in Escherichia coli has an extremely high oxygen affinity and two oxygen-binding haems: implications for regulation of activity in vivo by oxygen inhibition. Microbiology. 1996;142:755–763. doi: 10.1099/00221287-142-4-755. [DOI] [PubMed] [Google Scholar]
- 35.Jackson RJ, et al. Oxygen reactivity of both respiratory oxidases in Campylobacter jejuni: the cydAB genes encode a cyanide-resistant, low-affinity oxidase that is not of the cytochrome bd type. J. Bacteriol. 2007;189:1604–1615. doi: 10.1128/JB.00897-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cunningham L, Pitt M, Williams HD. The cioAB genes from Pseudomonas aeruginosa code for a novel cyanide-insensitive terminal oxidase related to the cytochrome bd quinol oxidases. Mol. Microbiol. 1997;24:579–591. doi: 10.1046/j.1365-2958.1997.3561728.x. [DOI] [PubMed] [Google Scholar]
- 37.Arai H. Regulation and function of versatile aerobic and anaerobic respiratory metabolism in Pseudomonas aeruginosa. Front. Microbiol. 2011;2:103. doi: 10.3389/fmicb.2011.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 39.Ducluzeau AL, Ouchane S, Nitschke W. The cbb3 oxidases are an ancient innovation of the domain Bacteria. Mol. Biol. Evol. 2008;25:1158–1166. doi: 10.1093/molbev/msn062. [DOI] [PubMed] [Google Scholar]
- 40.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 41.Meyer F, et al. The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giovannoni SJ, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 43.Borisov VB, et al. Redox control of fast ligand dissociation from Escherichia coli cytochrome bd. Biochem. Biophys. Res. Commun. 2007;355:97–102. doi: 10.1016/j.bbrc.2007.01.118. [DOI] [PubMed] [Google Scholar]
- 44.Reinders CA, et al. Rectal nitric oxide and fecal calprotectin in inflammatory bowel disease. Scand. J. Gastroenterol. 2007;42:1151–1157. doi: 10.1080/00365520701320505. [DOI] [PubMed] [Google Scholar]
- 45.Borisov VB, et al. Aerobic respiratory chain of Escherichia coli is not allowed to work in fully uncoupled mode. Proc. Natl Acad. Sci. USA. 2011;108:17320–17324. doi: 10.1073/pnas.1108217108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wertz JT, Breznak JA. Physiological ecology of Stenoxybacter acetivorans, an obligate microaerophile in termite guts. Appl. Environ. Microbiol. 2007;73:6829–6841. doi: 10.1128/AEM.00787-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Preisig O, Zufferey R, Thöny-Meyer L, Appleby CA, Hennecke H. A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J. Bacteriol. 1996;178:1532–1538. doi: 10.1128/jb.178.6.1532-1538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiles S, Pickard KM, Peng K, MacDonald TT, Frankel G. In vivo bioluminescence imaging of the murine pathogen Citrobacter rodentium. Infect. Immun. 2006;74:5391–5396. doi: 10.1128/IAI.00848-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goldman BS, Gabbert KK, Kranz RG. The temperature-sensitive growth and survival phenotypes of Escherichia coli cydDC and cydAB strains are due to deficiencies in cytochrome bd and are corrected by exogenous catalase and reducing agents. J. Bacteriol. 1996;178:6348–6351. doi: 10.1128/jb.178.21.6348-6351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones SA, et al. Respiration of Escherichia coli in the mouse intestine. Infect. Immun. 2007;75:4891–4899. doi: 10.1128/IAI.00484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones SA, et al. Anaerobic respiration of Escherichia coli in the mouse intestine. Infect. Immun. 2011;79:4218–4226. doi: 10.1128/IAI.05395-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weingarten RA, Grimes JL, Olson JW. Role of Campylobacter jejuni respiratory oxidases and reductases in host colonization. Appl. Environ. Microbiol. 2008;74:1367–1375. doi: 10.1128/AEM.02261-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Way SS, Sallustio S, Magliozzo RS, Goldberg MB. Impact of either elevated or decreased levels of cytochrome bd expression on Shigella flexneri virulence. J. Bacteriol. 1999;181:1229–1237. doi: 10.1128/jb.181.4.1229-1237.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alvarez-Ortega C, Harwood CS. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol. Microbiol. 2007;65:153–165. doi: 10.1111/j.1365-2958.2007.05772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi L, et al. Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc. Natl Acad. Sci. USA. 2005;102:15629–15634. doi: 10.1073/pnas.0507850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Battino R, Rettich TR, Tominaga T. The solubility of oxygen and ozone in liquids. J. Phys. Chem. Ref. Data. 1983;12:163–178. [Google Scholar]
- 57.Revsbech NP, Thamdrup B, Dalsgaard T, Canfield DE. Construction of STOX oxygen sensors and their application for determination of O2 concentrations in oxygen minimum zones. Methods Enzymol. 2011;486:325–341. doi: 10.1016/B978-0-12-381294-0.00014-6. [DOI] [PubMed] [Google Scholar]
- 58.Klimant I, Kuhl M, Glud RN, Holst G. Optical measurement of oxygen and temperature in micro scale: strategies and biological applications. Sens. Actuators B Chem. 1997;38:29–37. [Google Scholar]
- 59.Grate JW, Kelly RT, Suter J, Anheier NC. Silicon-on-glass pore network micromodels with oxygen-sensing fluorophore films for chemical imaging and defined spatial structure. Lab Chip. 2012;12:4796–4801. doi: 10.1039/c2lc40776k. [DOI] [PubMed] [Google Scholar]
- 60.Røy H, et al. Aerobic microbial respiration in 86-million-year-old deep-sea red clay. Science. 2012;336:922–925. doi: 10.1126/science.1219424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.