Abstract
A new Matrix Assisted Laser Desorption Ionization Imaging Mass Spectrometry (MALDI-IMS) method to spatially profile the location and distribution of multiple N-linked glycan species in tissues is described. Application of an endoglycosidase, peptide N-glycosidase F (PNGaseF), directly on tissues followed by incubation releases N-linked glycan species amenable to detection by MALDI-IMS. The method has been designed to simultaneously profile the multiple glycan species released from intracellular organelle and cell surface glycoproteins, while maintaining histopathology compatible preparation workflows. A recombinant PNGaseF enzyme was sprayed uniformly across mouse brain tissue slides, incubated for two hours, then sprayed with 2,5-dihydroxybenzoic acid matrix for MALDI-IMS analysis. Using this basic approach, global snapshots of major cellular N-linked glycoforms were detected, including their tissue localization and distribution, structure and relative abundance. Off-tissue extraction and modification of glycans from similarly processed tissues and further mass spectrometry or HPLC analysis was done to assign structural designations. MALDI-IMS has primarily been utilized to spatially profile proteins, lipids, drug and small molecule metabolites in tissues, but it has not been previously applied to N-linked glycan analysis. The translatable MALDI-IMS glycan profiling workflow described herein can readily be applied to any tissue type of interest. From a clinical diagnostics perspective, the ability to differentially profile N glycans and correlate their molecular expression to histopathological changes can offer new approaches to identifying novel disease related targets for biomarker and therapeutic applications.
Introduction
The majority of proteomic and metabolomic analytical techniques require the macro- or microdissection and subsequent extraction of analytes from the target tissue. This process leads to loss of the spatial distribution and associated histopathology of the tissue. A novel but maturing technology, MALDI imaging mass spectrometry (MALDI- IMS), has been used to generate two- and three-dimensional molecular maps of hundreds of analytes directly from the surface of a tissue section, allowing the display of the relative abundance and spatial distribution of individual analytes1–5. The distribution of the analytes are also readily linkable to molecular histology and pathology data from the same tissues6,7. To date, most applications of MALDI-IMS have focused on profiling of proteins8,9, lipids10,11 and drug metabolites12–14 in tissues, but the technique has not yet been defined for analysis of complex carbohydrates, typified by N-linked glycans. Glycosylation is a major post-translational modification to proteins critical in regulating protein folding and vesicular transport, cell-cell communication and adhesion, immune recognition and other extracellular functions15–17. N-linked glycans are specifically attached to asparagine residues in proteins via a conserved amino acid motif of N-X-S/T, where X represents any amino acid except proline, and represent a diverse but biosynthetically definable group of carbohydrate structures ranging generally from m/z = 1000–5000 in size15. Because MALDI-TOF analysis is one of the most robust and well established methods for profiling multiple species of N-linked glycans18,19, imaging of glycans directly on tissue by MALDI-IMS should be feasible.
Using a recombinant source of peptide N-glycosidase F, which allowed an abundant supply of enzyme for optimization, and adaptation of a molecular spraying method developed for on-tissue protease digestions8, 20, a method workflow for MALDI-IMS of released N-glycans has been developed that maintains their spatial distribution in frozen tissue specimens. Combinations of permethylation derivatization21, 2-aminobenzoate-modified normal phase HPLC separations22,23, glycan standards, and existing structural database resources24 were used to confirm glycan release and initial structural determinations. Examples of the method development and verification workflows for robust on-tissue N-linked glycan profiling by MALDI IMS are presented for mouse brain and human kidney tissues.
Materials and Methods
Materials
The glycan standard A2 and sialidase S were obtained from ProZyme (Hayward, CA). Asialofetuin glycoprotein, 2,5-Dihydroxybenzioc Acid (DHB), trifluoroacetic acid, sodium hydroxide, dimethyl sulfoxide (DMSO), and iodomethane were obtained from Sigma-Aldrich (St. Louis, MO). HPLC grade methanol, ethanol and water were obtained from Fisher Scientific. ITO slides were purchased from Bruker Daltonics (Billerica, MA) for MALDI-IMS experiments.
Tissues
Mouse brains were excised from four euthanized C57BL/6 mice and snap frozen. Mice were housed in an Institutional Animal Care and Use Committee-approved small animal facility at MUSC, and brains were harvested as part of approved projects. Whole excised brains were placed in plastic weigh boats and rapidly frozen at the vapor phase interface of a liquid nitrogen containing dewar. A human kidney tissue was obtained from an Institutional Review Board approved study from the Mayo Clinic (Jacksonville, FL) evaluating the molecular changes associated with matched patient non-tumor and clear cell renal cell carcinoma tissues. A slice of kidney from a non-tumor region was selected for use in this current study. No personal identifier information was provided to the laboratory investigators. This particular tissue section represents a distal section of kidney where there was no evidence of tumor. Coronal sections of mouse brains (10 microns) and the human kidney (10 microns) were prepared using a Thermo Microm HM550 cryostat and stored at −80°C. For each section analyzed, a serial section was collected for histological analysis and staining with hematoxylin and eosin (H & E).
Recombinant Peptide N-Glycosidase F Expression
The entire Peptide N-Glycosidase F (PNGaseF) gene (P21163.2) from the genome of Flavobacterium meningosepticum was cloned, expressed and purified as described in detail in the Supporting Information.
MALDI Imaging Mass Spectrometry
Sectioned mouse brain tissue samples were mounted on ITO coated slides, and desiccated at room temperature for 20 minutes. A sequential ethanol wash was done in 70% ethanol for two minutes, two times, and one wash in 100% ethanol for two minutes, then a final desiccation at room temperature for 10 minutes. An ImagePrep spray station (Bruker Daltonics) was used to coat the slide with a 0.2 ml solution containing 20 ul of 1 mg/ml PNGaseF stock in water. This solution was sprayed by the ImagePrep using settings originally optimized for spraying of trypsin that result in minimal volumes and retention of spatial distribution8,20. If sialidase was used, 0.1 units of enzyme were diluted in 200uL water and sprayed following PNGaseF application using the same ImagePrep settings. Control tissue slices were blocked with a glass slide during the spraying process. Following application of glycosidase, slides were then incubated at 37°C for 2 hrs in a humidified chamber, then dried in a desiccator prior to matrix application. 2,5-Dihydroxybenzoic acid (DHB) matrix at a concentration of 0.2M in 50%MeOH 1%TFA was sprayed on to the slide using the ImagePrep for positive ion mode analysis. Spectra were acquired across the entire tissue section on a Solarix 70 dual source 7T FTICR mass spectrometer (Bruker Daltonics) to detect the N-glycans (m/z = 690–5000) with a SmartBeam II laser operating at 1000 Hz, a laser spot size of 25 µm and a raster width of 125µm unless otherwise indicated. For each laser spot, 1000 spectra were averaged. Images of differentially expressed glycans were generated to view the expression pattern of each analyte of interest using FlexImaging 3.0 software (Bruker Daltonics). Following MS analysis, data was loaded into FlexImaging Software focusing on the range m/z = 1200–4500 and reduced to 0.98 ICR Reduction Noise Threshold. Glycans were identified by selecting peaks that appeared in an average mass spectrum from the tissue that received PNGaseF application but did not appear in the control tissue. Observed glycans were searched against the glycan database provided by the Consortium for Functional Glycomics24 and known glycans that exist in mouse brain.
Permethylation and normal phase HPLC of tissue extracted N-glycans
PNGaseF sprayed mouse brain tissue slides were incubated for 2 hr at 37°C; 50uL water was applied on top of the tissue and incubated for 20 minutes to extract the released native N-glycans. The water was removed from the tissue, then concentrated under vacuum by centrifugation. Permethylation was performed as described21, and glycans analyzed by MALDI. Masses detected in the permethylation experiments were searched against the permethylated glycan database provided by the Consortium for Functional Glycomics24. Alternatively, concentrated glycans were labeled with 2-aminobenzamide (2-AB) for subsequent normal phase HPLC analysis22,23. Glycan identification was made through sequential exoglycosidase digestions as previously described23. Further details for both analysis procedures are provided in Supporting Information.
RESULTS
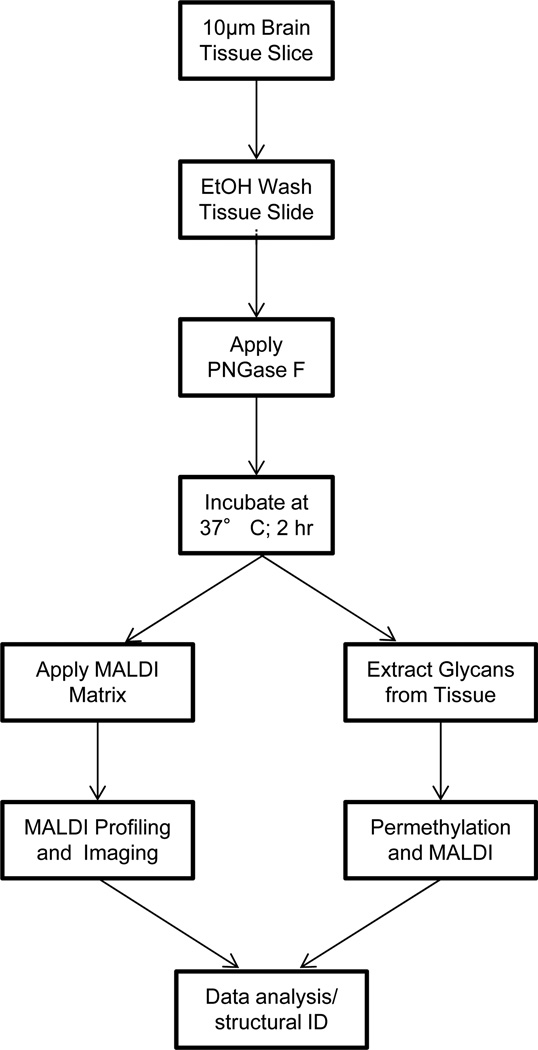
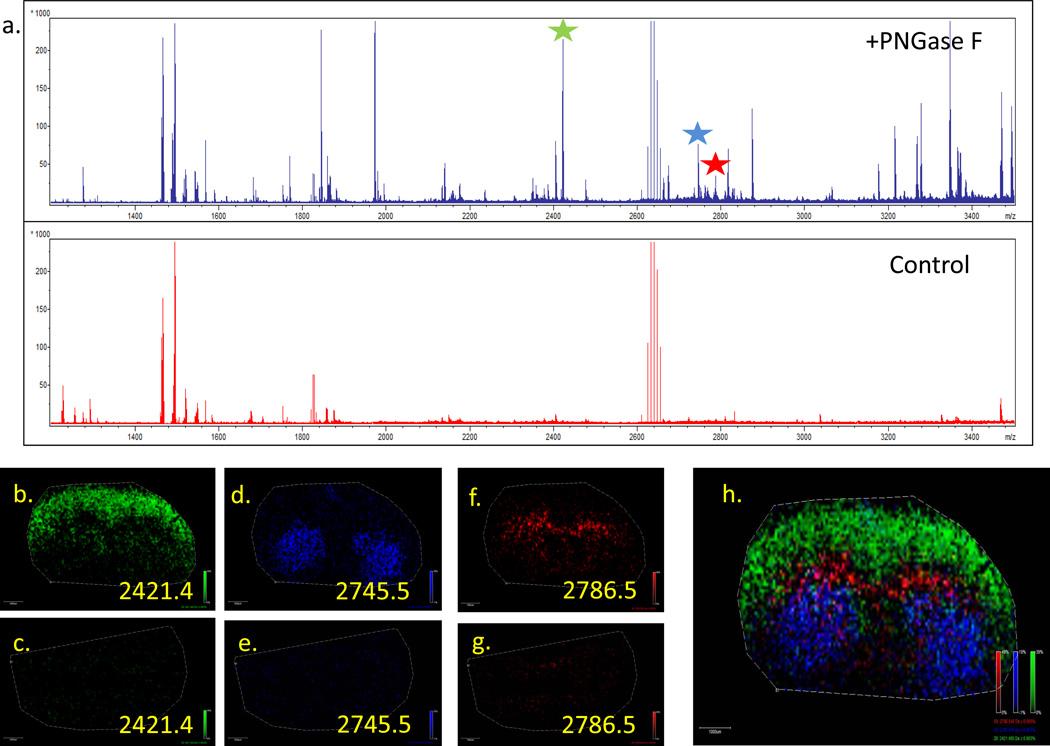
A workflow for on-tissue analysis of PNGaseF released N-glycans was developed, as summarized in Figure 1. Initial experiments were done to verify that PNGaseF is active on mouse brain tissues following application by an ImagePrep sprayer, and that the products of the reaction are N-glycans. Two frozen mouse brain tissue slices (10 micron) were mounted on ITO-coated glass slides and dehydrated in sequential ethanol washes. One brain slice was covered by a glass slide to serve as an experimental control, and the other section was sprayed with 20 ug PNGaseF. The tissues were incubated for 2hr at 37°C in a humidified chamber. After drying, DHB matrix was applied to the entire slide and MALDI-MSI imaging done for each brain slice. The spectra from the PNGaseF sprayed brain slice revealed multiple novel ions peaks that are consistent with release of N-glycans (Figure 2a). Additionally, these ions displayed regional localization following PNGaseF spray (Figure 2b, 2d, 2f, 2h) but were not detected in the absence of PNGaseF spray (Figure 2c, 2e, 2g). These obvious patterns of differential glycan expression distributions associated with brain substructure is a novel finding, and a representative overlay image of PNGaseF released glycans is shown in Figure 2h.
Figure 1. A summary schematic of the on-tissue PNGaseF digestion for MALDI-IMS.
Figure 2. Mouse brain tissue slices treated with or without on-tissue PNGaseF digestion and MALDI-IMS.
The PNGase F workflow was applied to one mouse brain while the other mouse brain was prepared with an identical workflow without PNGase F. (a). The upper spectrum is from the brain slice incubated with PNGase F and the lower spectrum is from the non-PNGase F control brain slice. Example masses that were detected following PNGase F treatment are shown for three ion peaks that also displayed regional expression distributions in the mouse brain. The m/z = 2421.5 (b and c), m/z = 2745.5 (d and e) and m/z = 2786.5 (f and g) were only detected after PNGase F digestion (panels b,d,f,h). An overlay of these three peaks in the PNGase Fdigested brain slice is shown in panel h. A raster distance of 75 microns was used per MALDI laser acquisition across the tissues.
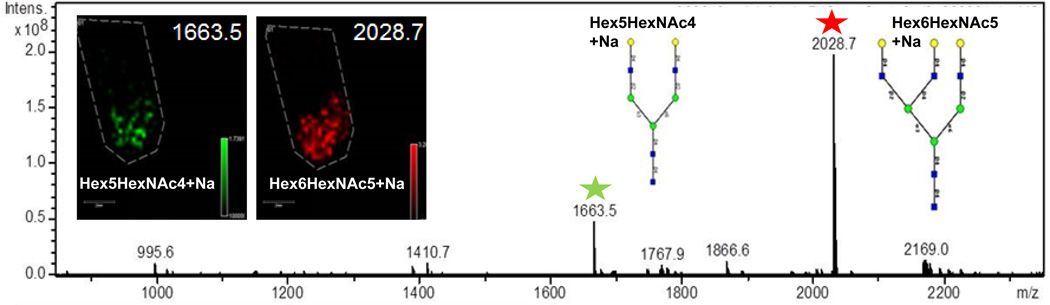
Purified glycoprotein and glycan standards were added to mouse brain tissues prior to PNGaseF digestion and matrix application, to further verify the on-tissue PNGaseF activity and the ability to detect native N-glycans on tissue. A common purified glycoprotein standard, asialofetuin, was mixed with brain tissue homogenate and added back to the brain for refreezing and MALDI-MS imaging analysis of the tissue. Focusing on two known N-linked glycans of asialofetuin, Hex5HexNAc4+Na (m/z = 1663.5) and Hex6HexNAc5+Na (m/z = 2028.7), these glycans were detected following in-solution digest of asialofetuin alone (Figure 3). Following on-tissue PNGaseF digestion, these same glycans were readily detected in the supplemented brain tissue following PNGaseF digestion (inset of Figure 3), but not in the tissue without the enzyme sprayed (data not shown). Furthermore, addition of a glycan A2 standard (m/z = 2223.8, Hex5HexNAc4NeuAc2) directly on mouse brain tissues revealed multiple glycan ion peaks that differed in loss of sialic acids (−1 NeuAc, m/z = 1954.7; −2 NeuAc, m/z = 1663.5) (Supplemental Figure 1), a common occurrence in MALDI analysis of sialylated glycan structures25, and multiple sodium ion adducts. The corresponding MALDI imaging distribution of the standard glycan peaks are shown for each ion in Supplemental Figure 1. Analysis of these standards further verified detection of N-linked glycans by MALDI-IMS.
Figure 3. Addition of a glycoprotein standard following PNGaseF digestion and MALDI-IMS.
Asialofetuin was incubated with PNGase F and the native glycans were analyzed by FT-MS. Hex5HexNAc4 + Na (m/z = 1663.5) and Hex6HexNAc5 + Na (m/z = 2028.7) were detected. Asialofetuin was incorporated into a mouse brain and the PNGase F imaging method was performed. The two inset images show the same N-glycans at m/z = 1663.5 and m/z = 2028.7 m/z detected on-tissue following PNGase F digestion.
Two off-tissue approaches were used to further verify the release of N-glycans and begin to identify specific glycan structures associated with on-tissue native glycan peaks. Brain tissues were digested with PNGaseF on-slide as already described. Released glycans were extracted from the on-slide digestion in water. On-slide extracted glycans were treated with 2-aminobenzoate (2AB) for subsequent normal-phase HPLC analysis22,23. or permethylated for further MALDI analysis and comparison to structure and reference spectra databases24. Reference spectra for these extracted glycans following permethylation and MALDI profiling and normal phase HPLC separation of 2AB glycans are presented in Supplemental Figures 2A and 2B. Structural identification of the permethylated N-glycans were assigned by searching for the same ion in the MALDI structural databases and reference spectra of mouse brain glycans provided by the Consortium for Functional Glycomics24. A majority of the permethylated glycans extracted from the tissues were the same masses as the glycans listed in the Consortium for Functional Glycomics mouse brain tissue spectral libraries.
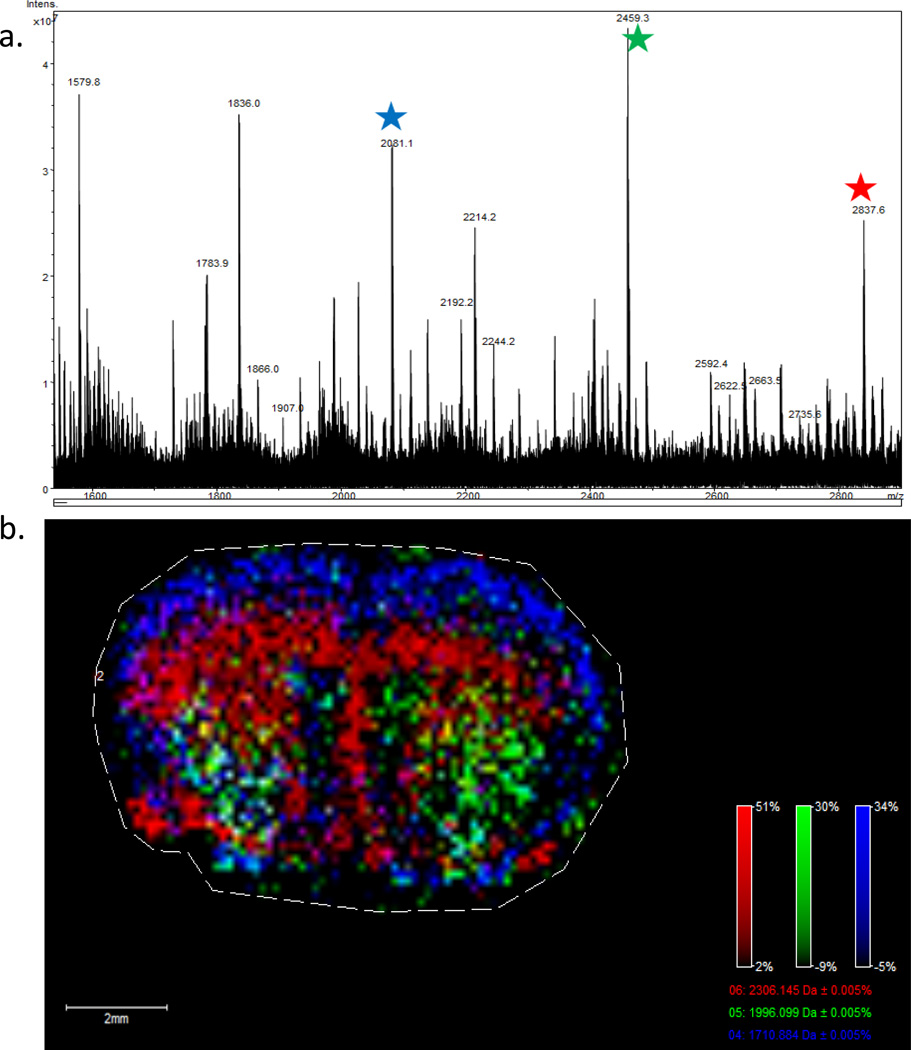
These permethylated masses and structures were used to correlate back to the expected native mass/structure of the glycan. As an example, an overlay of Hex5dHex3HexNAc5 + Na, Hex4dHex2HexNAc5 + Na, and Hex3dHex1HexNAc5 + Na demonstrates the spatial pattern of expression for these specific glycans by MALDI-IMS linked to the corresponding extracted permethylated peaks (Figure 4). A reference panel of 28 glycan structures was generated that lists structural composition, expected and observed masses of the permethylated glycans, and the masses of their corresponding native glycan masses for cross-referencing permethylated glycan spectra and native tissue glycan masses. (Supplemental Figure 3) Additionally, the majority of the identities of the off-tissue native glycan species reacted with 2AB and separated by normalphase HPLC are also common to those listed in Supplemental Figure 3 (Supplemental Figure 2a). The majority of the detected masses were within 0.5 mass units of the predicted masses (Supplemental Figure 3). The few discrepancies of more than 0.5 mass units generally occurred with native glycans of larger mass (>2500) and were associated with complex overlapping ion signals in those specific tissue regions.
Figure 4. Linkage of permethylated glycan masses with native tissue glycan ion distribution.
The N-glycan imaging method was applied to a mouse brain for tissue imaging and an off tissue permethylation was performed for glycan verification. (a.) Permethylated glycans Hex5dHex3HexNAc5 + Na (m/z = 2837.6), Hex4dHex2HexNAc5 + Na (m/z = 2459.3), and Hex3dHex1HexNAc5 +2Na (m/z = 2081.1) were identified and the corresponding glycans displayed specific localization in the tissue image (b). M/z = 2306.1 corresponds to Hex5dHex3HexNAc5 + 2Na (red), m/z = 1996.1 corresponds to Hex4dHex2HexNAc5 + Na (green), and m/z = 1710.9 corresponds to Hex3dHex1HexNAc5 + 2Na (blue). A raster distance of 125 microns was used per MALDI laser acquisition across the tissues.
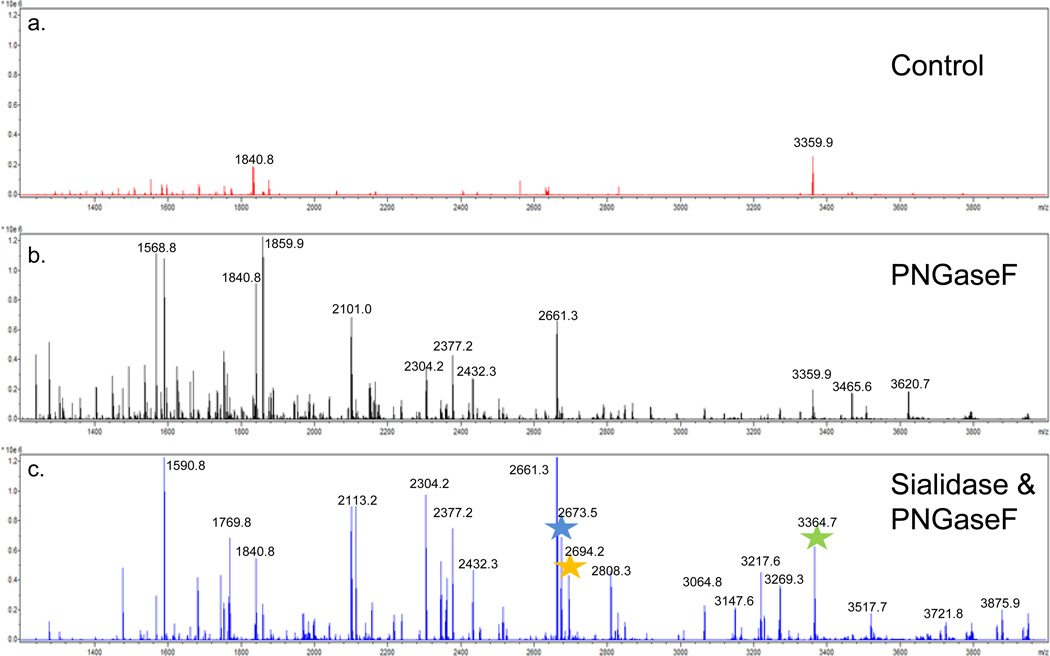
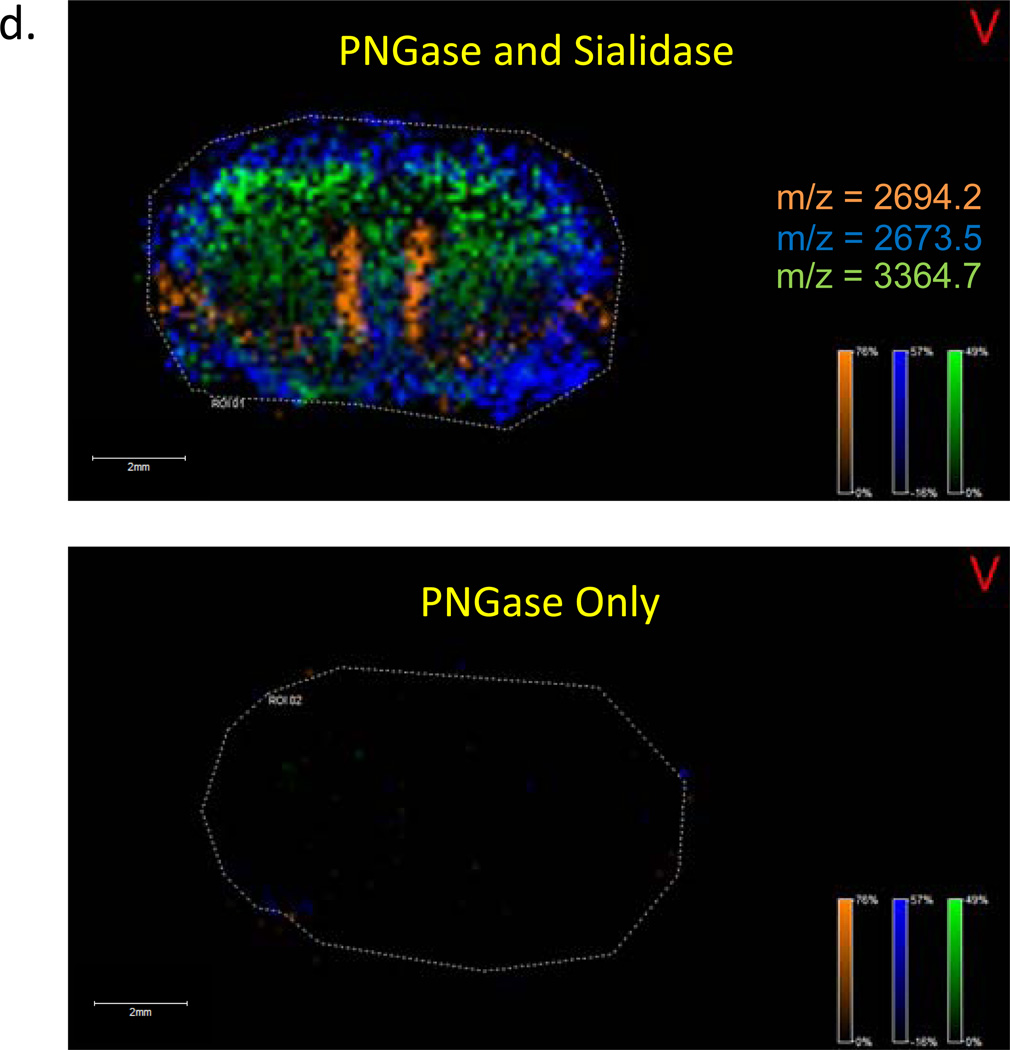
Many larger glycans are branch-chained with terminal sialic acid residues, thus a comparison was done for detection of on-tissue glycans released by PNGaseF digestion alone versus a combination of PNGaseF and sialidase. If larger sialylated structures were present, then sialidase digestion would result in new glycan peaks detected in the spectra relative to PNGaseF alone. As shown in Figure 5, the on-tissue analysis indicated an increase in the levels of larger mass native glycan ions (m/z > 2500) detected in the double digest of PNGaseF and sialidase (Figure 5A–C). These structures are consistent with the sialidase products of glycans that contained tri- and tetra branched chains with multiple N-acetylglucosamine-galactose (LacNAc) dimers26. A representative overlay image of three of these structures detected only in the combined sialidase/PNGaseF digest is shown in Figure 5D. Sialidase digestion of 2AB modified glycans also demonstrated shifts in the detection of smaller glycan species following normal phase HPLC separation (Supplemental Figure S2a).
Figure 5. Comparative profiles of released N-glycans following on-tissue glycosidase digests of mouse brain tissues.
Three serial sections from a mouse brain were prepared and either (a) untreated, (b) digestion with PNGaseF, or (c) a combination of PNGaseF and Sialidase S. Recombinant PNGaseF (20 mU) alone or in combination with 100 mU Sialidase S (Prozyme) were applied using a Bruker ImagePrep prior to 2 hr incubation at 37° C. In (d), a representative overlay image of the unique glycans detected with the sialidase and PNGaseF combination is shown for m/z values of 2673.5 (blue), 2694.2 (orange) and 3364.7 (green). A raster distance of 125 microns was used per MALDI laser acquisition across the tissues.
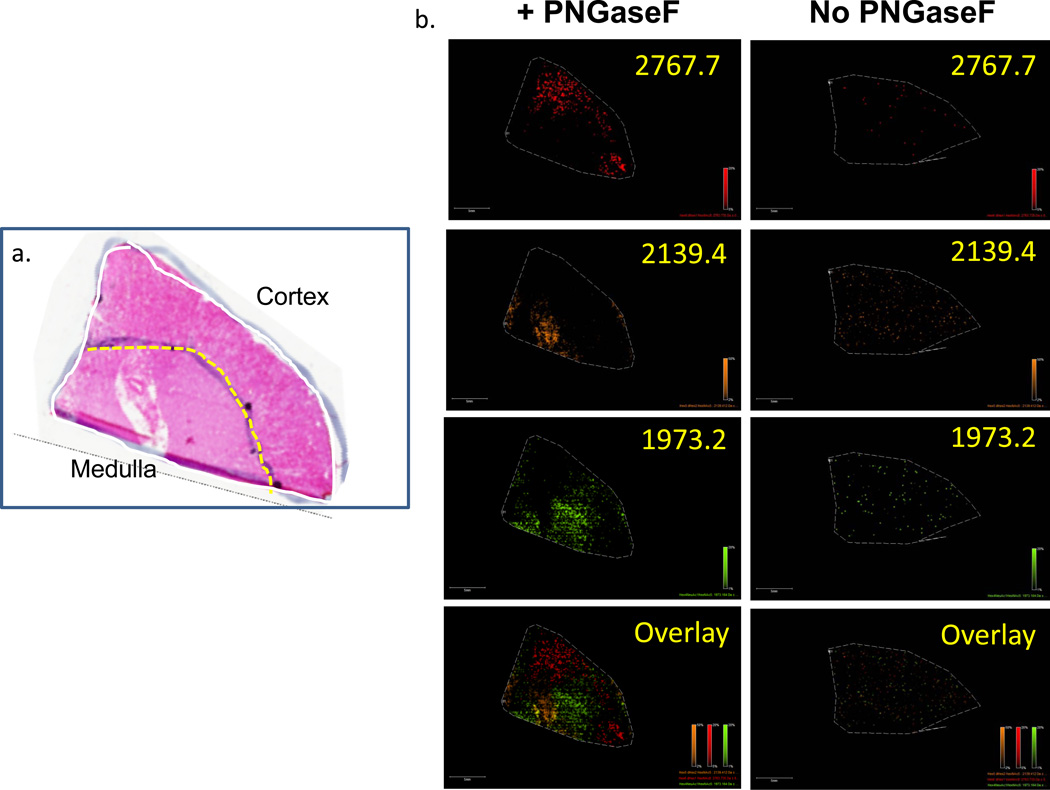
The methods developed for mouse brain tissues were applied to a frozen slice of human kidney containing both medulla and cortex regions. As shown in Figure 6, representative N-glycans released by on-tissue PNGaseF digestion can be detected that are specific to either cortex or medulla regions. Compared to the mouse brains analyzed, the total number of glycan species was slightly reduced in the human kidney sections. Beyond these kidney tissues and mouse brain tissues, we have found it is necessary to optimize the length of incubation and amount of PNGaseF digestion conditions for each tissue type being tested. Overall, these baseline conditions described for mouse brains have worked as useful starting points for any new tissue being analyzed.
Figure 6. N-glycan MALDI-IMS of normal human kidney.
Three serial sections from a human kidney were prepared and either untreated (data not shown), digested with PNGaseF, or analyzed by H&E staining. Cortex and medulla regions were confirmed by a pathologist. Recombinant PNGaseF (20 mU) was applied using a Bruker ImagePrep followed by a 2 hr incubation at 37° C. Following MALDI-MSI, m/z values that are specific to PNGaseF treated tissue are observed. Shown are the results for an m/z value of 2767.7 (a) localized in the cortex, while the m/z values of 2139.4 (b) and 1973.2 (c) are localized primarily in the medulla when compared to the H&E stain (d). An overlay of the three m/z values is provided (e). A raster distance of 125 microns was used per MALDI laser acquisition across the tissues.
Discussion
We describe a new mass spectrometry-based glycan imaging approach in combination with on-tissue PNGaseF digestion to spatially profile released N-linked glycans in their local microenvironment. The method has been designed to facilitate the tissue analysis of the cell surface glycans in frozen tissues while maintaining histopathology compatible preparation workflows. While MALDI-IMS has primarily been utilized to spatially profile proteins, lipids, drug and small molecule metabolites in tissues, it has not been previously applied to N-linked glycan analysis. Coronal slices of mouse brain tissues were ideal for method development because of the robust glycan signal and the existence of an N-glycan catalogue, provided by the Consortium for Functional Glycomics, that documents known N-glycans present in mouse brain tissue24. Therefore the off-tissue extraction of glycans from PNGaseF digested brain slices for permethylation and mass spectrometry profiling or normal phase HPLC analyses could be readily compared to on-tissue glycan species, in particular for assigning glycan structure and composition. The Consortium of Functional Glycomics reference spectra library of permethylated N-glycan species from mouse brains as detected by MALDI lists over 40 identified structures. We estimate that 30 or more N-glycan species can be detected on-tissue in mouse brains by our current MALDI-IMS workflows, including the 28 structures listed in the panels of Supplemental Figure 3. This number varies depending on the location of the tissue cut within the brain and the number of sub-regions present. MALDI-IMS detection of brain substructure regions of protein, small molecule metabolites and lipid distributions in brain substructure regions is well documented10,27–29, so detection of specific N-linked glycan isoform distribution is not surprising, and represents to our knowledge the first demonstration of this. The specific distribution of N-glycans in the medulla and cortex regions of the human kidney tissue (Figure 6) also demonstrates sub-organ structure specificity of glycan species expression.
MALDI mass spectrometry is a common detection method for analysis of complex carbohydrates, including routine analysis of N-linked glycans released from glycoproteins by PNGaseF. From a practical perspective, our approach used a recombinant PNGaseF enzyme engineered for increased stability and activity. In addition, the amount of commercially available preparations of PNGaseF that would be required to perform the described experiments would not be economically feasible for most laboratories. The recombinant enzyme is also not formulated with detergents and additives that are common components of commercial enzyme, many of which can interfere with MALDI ionization. However, glycan analysis by MALDI is notoriously complicated by problems with glycan dissociation and sodium adduct additions. It is common to observe the loss of sialic acid when native glycans are analyzed by MALDI MS, hence the need for modifications like permethylation or 2-aminobenzoate that stabilize and help ionize sialic acid containing glycans25. Most common chemical modification methods require organic solvents and the need to separate reaction products, thus these are not feasible for on-tissue derivitization and retention of spatial distributions. The MALDI-IMS profiling of the A2 glycan standard on-tissue was similarly affected, with the number of Na ions and loss of sialic acid being a factor for an effective prediction of structure. In addition to those glycans listed in Supplemental Figure 3 that we have cross-referenced for structure identification, there are multiple PNGaseF-released ions detected that have not been correlated to reported structures. Many of these are larger species m/z > 3500, and like those glycan ions assigned structures, all will ultimately require confirmatory structural characterization involving tandem MS identifications off tissue30,31. Use of MALDI alone may be difficult to structurally identify very high mass poly-lactosamino-glycan species26 on-tissue, but some of these higher mass glycans are potentially profilable by MALDI-IMS (Figure 5). Differential exoglycosidase digestion strategies more amenable to native glycan identification on-tissue and preservation of spatial distribution, analogous to that described for sialidase (Figure 5), are ongoing. Preliminary experiments in negative ion mode using other matrices have yielded promising results, and we expect this could be a partial solution for more effective detection of native sialylated glycan species.
Overall, we present a functional MALDI-IMS workflow for on-tissue analysis and distribution of N-glycan species, and there remains room for continued improvement of the assay and analysis workflow. We only report the use of one type of MALDI matrix, data was only collected in positive ion mode, and only fresh frozen tissues were used. Except for those glycans directly compared with reference standards, the structural assignments are correlative to existing databases and subject to known limitations of MALDI analysis. Further verification of these assigned structures by tandem mass spectrometry approaches is ongoing. Evaluation of different chemical matrices for either positive or negative ion mode analysis are ongoing, as well as refinement of methods to profile N-glycans in formalin-fixed paraffin-embedded (FFPE) tissues. As MALDI-IMS has been used to effectively profile proteins in FFPE tissues32,33, we expect that these methods should be adaptable for glycan analysis in FFPE tissues. We believe this capability of profiling N-glycans spatially on tissue will have significant application to tissues representing many types of human diseases, especially the changes in glycosylation associated with oncogenesis. It is well documented that malignant transformation and cancer progression result in fundamental changes in the glycosylation patterns of cell surface and secreted glycoproteins34,35, and the majority of current FDA-approved tumor markers are glycoproteins or glycan antigens30,36. Therefore, development of a molecular assay like the MALDI-IMS approach we describe could directly profile these pathologically relevant carbohydrates in their local microenvironment and accelerate approaches to aid in the diagnosis, prognosis, staging, and treatment of many types of cancers. Coupled with many new improved high resolution mass spectrometry options for intact glycopeptides37,38 and small molecule carbohydrate binding probes39,40, there is potential to link the tissue distribution of glycans from MALDI-IMS to the glycoproteins carrying them.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health/National Cancer Institute grants R21CA137704 and R01CA135087, and the state of South Carolina SmartState Endowed Research program to R.R.D; This work was supported by grants R01 CA120206 and U01 CA168856 from the National Cancer Institute (NCI), the Hepatitis B Foundation, and an appropriation from The Commonwealth of Pennsylvania to A.M. This work was supported by NIH/NCI grants R01CA104505 and R01CA104505-05S1, a generous gift from the David & Lois Stulberg Endowed Fund for Kidney Cancer Research, Kidney Cancer Research at Mayo Clinic in Florida and The Fraternal Order of Eagles, State of Florida to J.A.C.
Footnotes
Conflict of Interest Disclosure: The authors declare no competing financial interest.
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Caprioli RM, Farmer TB, Gile J. Anal. Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 2.Seeley EH, Schwamborn K, Caprioli RM. J. Biol. Chem. 2011;286:25459–25466. doi: 10.1074/jbc.R111.225854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Römpp A, Spengler B. Histochem. Cell Biol. 2013;139:759–783. doi: 10.1007/s00418-013-1097-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seeley EH, Caprioli RM. Anal. Chem. 2012;84:2105–2110. doi: 10.1021/ac2032707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thiele H, Heldmann S, Trede D, Strehlow J, Wirtz S, Dreher W, Berger J, Oetjen J, Kobarg JH, Fischer B, Maass P. Biochim. Biophys. Acta. 2013 doi: 10.1016/j.bbapap.2013.01.040. [DOI] [PubMed] [Google Scholar]
- 6.Schwamborn K, Caprioli RM. Nat. Rev. Cancer. 2010;10:639–646. doi: 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]
- 7.Balluff B, Schöne C, Höfler H, Walch A. Histochem. Cell Biol. 2011;136:227–244. doi: 10.1007/s00418-011-0843-x. [DOI] [PubMed] [Google Scholar]
- 8.Cazares LH, Troyer D, Mendrinos S, Lance RA, Nyalwidhe JO, Beydoun HA, Clements MA, Drake RR, Semmes OJ. Clin. Cancer Res. 2009;15:5541–5551. doi: 10.1158/1078-0432.CCR-08-2892. [DOI] [PubMed] [Google Scholar]
- 9.Chaurand P, Norris JL, Cornett DS, Mobley JA, Caprioli RM. J. Prot. Res. 2006;5:2889–2900. doi: 10.1021/pr060346u. [DOI] [PubMed] [Google Scholar]
- 10.Chaurand P, Cornett DS, Angel PM, Caprioli RM. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.O110.004259. O110.004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berry KA, Hankin JA, Barkley RM, Spraggins JM, Caprioli RM, Murphy RC. Chem. Rev. 2011;111:6491–6512. doi: 10.1021/cr200280p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castellino S, Groseclose MR, Wagner D. Bioanalysis. 2011;3:2427–2441. doi: 10.4155/bio.11.232. [DOI] [PubMed] [Google Scholar]
- 13.Cornett DS, Frappier SL, Caprioli RM. Anal. Chem. 2008;80:5648–5653. doi: 10.1021/ac800617s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nilsson A, Fehniger TE, Gustavsson L, Andersson M, Kenne K, Marko-Varga G, Andrén PE. PLoS One. 2010;5:e11411. doi: 10.1371/journal.pone.0011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varki A, Cummings R, Esko J, Freeze H, Stanley P, Bertozzi CR, Hart GW, Etzler ME. Essentials of Glycobiology. 2nd Ed. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- 16.Dennis JW, Nabi IR, Demetriou M. Cell. 2009;139:1229–1241. doi: 10.1016/j.cell.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leymarie N, Zaia J. Anal. Chem. 2012;84:3040–3048. doi: 10.1021/ac3000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haslam SM, North SJ, Dell A. Curr. Opin. Struct. Biol. 2006;16:584–591. doi: 10.1016/j.sbi.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Wada Y, Azadi P, Costello CE, Dell A, Dwek RA, Geyer H, Geyer R, Kakehi K, Karlsson NG, Kato K, Kawasaki N, Khoo KH, Kim S, Kondo A, Lattova E, Mechref Y, Miyoshi E, Nakamura K, Narimatsu H, Novotny MV, Packer NH, Perreault H, Peter-Katalinic J, Pohlentz G, Reinhold VN, Rudd PM, Suzuki A, Taniguchi N. Glycobiology. 2007;17:411–422. doi: 10.1093/glycob/cwl086. [DOI] [PubMed] [Google Scholar]
- 20.Clemis EJ, Smith DS, Camenzind AG, Danell RM, Parker CE, Borchers CH. Anal. Chem. 2012;84:3514–3522. doi: 10.1021/ac202875d. [DOI] [PubMed] [Google Scholar]
- 21.Mechref Y, Kang P, Novotny MV. Methods Mol. Biol. 2009;534:53–64. doi: 10.1007/978-1-59745-022-5_4. [DOI] [PubMed] [Google Scholar]
- 22.Mehta A, Carrouee S, Conyers B, Jordan R, Butters T, Dwek RA, Block TM. Hepatology. 2001;33:1488–1495. doi: 10.1053/jhep.2001.25103. [DOI] [PubMed] [Google Scholar]
- 23.Guile GR, Rudd PM, Wing DR, Prime SB, Dwek RA. Anal. Biochem. 1996;240:210–226. doi: 10.1006/abio.1996.0351. [DOI] [PubMed] [Google Scholar]
- 24.Consortium for Functional Glycomics. http://www.functionalglycomics.org. [Google Scholar]
- 25.Harvey DJ. Proteomics. 2005;5:1774–1786. doi: 10.1002/pmic.200401248. [DOI] [PubMed] [Google Scholar]
- 26.Bern M, Brito AE, Pang PC, Rekhi A, Dell A, Haslam SM. Mol. Cell. Proteomics. 2013;12:996–1004. doi: 10.1074/mcp.O112.026377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lagarrigue M, Alexandrov T, Dieuset G, Perrin A, Lavigne R, Baulac S, Thiele H, Martin B, Pineau C. J. Proteome Res. 2012;11:5453–5463. doi: 10.1021/pr3006974. [DOI] [PubMed] [Google Scholar]
- 28.Källback P, Shariatgorji M, Nilsson A, Andrén PE. J. Proteomics. 2012;75:4941–4945. doi: 10.1016/j.jprot.2012.07.034. [DOI] [PubMed] [Google Scholar]
- 29.Jones EA, Shyti R, van Zeijl RJ, van Heiningen SH, Ferrari MD, Deelder AM, Tolner EA, van den Maagdenberg AM, McDonnell LA. J. Proteomics. 2012;75:5027–5035. doi: 10.1016/j.jprot.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 30.Mechref Y, Hu Y, Garcia A, Zhou S, Desantos-Garcia JL, Hussein A. Bioanalysis. 2012;4:2457–2469. doi: 10.4155/bio.12.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wuhrer M. Glycoconj. J. 2013;30:11–22. doi: 10.1007/s10719-012-9376-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemaire R, Desmons A, Tabet JC, Day R, Salzet M, Fournier I. J. Proteome Res. 2007;6:1295–1305. doi: 10.1021/pr060549i. [DOI] [PubMed] [Google Scholar]
- 33.Casadonte R, Caprioli RM. Nat. Protoc. 2011;6:1695–1709. doi: 10.1038/nprot.2011.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schultz MJ, Swindall AF, Bellis SL. Cancer Metastasis Rev. 2012;31:501–518. doi: 10.1007/s10555-012-9359-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miwa HE, Song Y, Alvarez R, Cummings RD, Stanley P. Glycoconj. J. 2012;8–9:609–618. doi: 10.1007/s10719-012-9373-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ludwig JA, Weinstein JN. Nat. Rev. Cancer. 2005;5:845–860. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 37.Yin X, Bern M, Xing Q, Ho J, Viner R, Mayr M. Mol. Cell. Proteomics. 2013;12:956–978. doi: 10.1074/mcp.M112.024018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saba J, Dutta S, Hemenway E, Viner R. Int. J. Proteomics. 2012 doi: 10.1155/2012/560391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cazares LH, Troyer DA, Wang B, Drake RR, Semmes OJ. Bioanalyt. Anal. Chem. 2011;401:17–27. doi: 10.1007/s00216-011-5003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai C, Cazares LH, Wang L, Chu Y, Wang SL, Troyer DA, Semmes OJ, Drake RR, Wang B. Chem. Comm. 2011;47:10338–10340. doi: 10.1039/c1cc11814e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.