Abstract
Purpose
To investigate the role of endorectal MR imaging and MR spectroscopic imaging in defining the contour of treatable intraprostatic tumor foci in prostate cancer, since targeted therapy requires accurate target volume definition.
Materials and Methods
We retrospectively identified 20 patients with prostate cancer who underwent endorectal MR imaging and MR spectroscopic imaging prior to radical prostatectomy and subsequent creation of detailed histopathological tumor maps from whole-mount step sections. Two experienced radiologists independently reviewed all MR images and electronically contoured all suspected treatable (≥ 0.5 cm3) tumor foci. Deformable co-registration in MATLAB was used to calculate the margin of error between imaging and histopathological contours at both capsular and non-capsular surfaces and the treatment margin required to ensure at least 95% tumor coverage.
Results
Histopathology showed 17 treatable tumor foci in 16 patients, of which 8 were correctly identified by both readers and an additional 2 were correctly identified by reader 2. For all correctly identified lesions, both readers accurately identified that tumor contacted the prostatic capsule, with no error in contour identification. On the non-capsular border, the median distance between the imaging and histopathological contour was 1.4 mm (range, 0 to 12). Expanding the contour by 5 mm at the non-capsular margin included 95% of tumor volume not initially covered within the MR contour.
Conclusions
Endorectal MR imaging and MR spectroscopic imaging can be used to accurately contour treatable intraprostatic tumor foci; adequate tumor coverage is achieved by expanding the treatment contour at the non-capsular margin by 5 mm.
Keywords: MRI, MRSI, prostate cancer, dominant intraprostatic lesion, focal therapy, dose escalation
Introduction
Prostate cancer is the second leading cause of cancer death in men, with an estimated 241,740 new cases and 28,170 deaths in 2012 [15]. The majority of men with prostate cancer opt for active surveillance, radical prostatectomy, or radiation therapy. Of those that undergo radiation therapy, in both the primary and salvage setting [7] there is interest in boosting, or selectively treating, the dominant intraprostatic lesion (DIL). For primary definitive treatment, particularly in the IMRT era, the hotspots can be targeted to this lesion or a simultaneous integrated boost can be given, as a method to increase biochemical control in more aggressive disease [21]. Similarly, prescription of a higher dose in low or high dose rate brachytherapy [4,12] can be targeted to the DIL, potentially increasing local control. At the other end of the disease spectrum, there is interest in focal therapy as a supplementary or salvage therapy, with localized re-irradiation of the DIL with brachytherapy implant for intraprostatic recurrence after external beam radiation [8]. More recently, MR-guided high intensity focused ultrasound has emerged as an exciting method of tumor ablation which offers precisely targeted and monitored treatment delivery [10]. Despite these advances in focal treatment and while endorectal MR imaging and MR spectroscopic imaging have been shown to improve tumor localization, staging, and volume estimation while allowing non-invasive identification of prostate cancer [3,14,23], the role of endorectal MR imaging and MR spectroscopic imaging in defining the margin of dominant intraprostatic tumor has not been reported. Therefore, we undertook this study to investigate the role of endorectal MR imaging and MR spectroscopic imaging in defining the contour of treatable intraprostatic tumor foci in men with prostate cancer. In particular, we sought to determine the amount of imaging “undercall” of treatable (≥ 0.5 cm3) tumor foci, based upon contours drawn by two independent MR readers, and thereby determine the size of radial expansion of the MR lesion contours to encompass 95% of the histopathologically identified lesion.
Materials and Methods
Patient selection
This single institution retrospective study was approved by our Committee on Human Research with a waiver of informed consent and was compliant with the Health Insurance Portability and Accountability Act.
Over a four year period, we identified all patients at our institution who were part of a separate study that provided an MR exam preprostatectomy and whole mount histology of the excised gland. These patients had untreated biopsy-proven prostate cancer and underwent endorectal MR imaging and MR spectroscopic imaging prior to radical prostatectomy with subsequent creation of detailed tumor maps from step-section whole-mount histopathological evaluation (n = 20). MRI was performed more than 6 weeks after biopsy and no evidence of hemorrhage was detected in the T1-weighted MR images. No interval prostate cancer treatment was administered between MR imaging and prostatectomy. The median patient age was 62 years, (range, 54–71). The median serum prostate specific antigen level at diagnosis was 5.8 (range, 1.0–13.4). The median Gleason score was 7 (range, 6–9), and the stage ranged from pT2a to pT3a. (Table 1)
Table 1.
Table of patient and tumor foci characteristics. (Pt 1 had two correctly identified tumors). Tumors correctly identified by reviewers 1 and 2 (R1, R2) are noted. Foci 1a and 1b are two correctly identified tumor foci within the same patient.
| Pt# | Age | Gleason | Stage | PSA | Path Vol (cc) | False Positive | R1 | R2 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| 1a | 54 | 3 | 3 | 6 | pT2c | 8 | 2.3 | X | X | |
| 1b | 54 | 3 | 3 | 6 | pT2c | 8 | 0.4 | X | X | X |
| 2 | 62 | 4 | 3 | 7 | pT3a | 6.3 | 3.9 | X | X | |
| 3 | 67 | 4 | 5 | 9 | pT3a | 1 | 2.3 | X | X | |
| 4 | 66 | 3 | 3 | 6 | pT2a | 4.07 | 1.6 | X | X | |
| 5 | 59 | 3 | 4 | 7 | pT2c | 6.5 | 0.8 | X | X | |
| 6 | 60 | 3 | 3 | 6 | pT3a | 3.5 | 4.5 | X | X | |
| 7 | 63 | 4 | 4 | 8 | pT2a | 3.7 | 0.5 | X | X | |
| 8 | 64 | 4 | 4 | 8 | pT2a | 8.4 | 0.8 | X | X | |
| 9 | 71 | 3 | 4 | 7 | pT2c | 6.2 | 5.1 | X | X | |
| 10 | 65 | 3 | 4 | 7 | pT2c | 4.2 | 6.4 | X | X | |
| 11 | 56 | 3 | 3 | 6 | pT2c | 1.8 | 3.2 | X | ||
| 12 | 55 | 3 | 3 | 6 | pTX | 6.5 | 0.1 | X | X | |
| 13 | 61 | 4 | 5 | 9 | pT3a | 7.85 | 7.0 | X | X | |
| 14 | 69 | 4 | 3 | 7 | pT2a | 4 | 0.9 | X | X | |
| 15 | 68 | 4 | 3 | 7 | pT3a | 13.4 | 0.7 | X | ||
| 16 | 71 | 3 | 3 | 6 | pT2a | 8.5 | 0.0 | |||
| 17 | 60 | 4 | 3 | 7 | pT3a | 6.2 | 0.8 | |||
| 18 | 54 | N/A | N/A | N/A | N/A | 5.5 | 0.0 | |||
| 19 | 49 | 3 | 3 | 6 | pT2c | 3.5 | 1.3 | |||
| 20 | 60 | 3 | 3 | 6 | pT2c | 4.3 | 1.0 | |||
| Median | 61.5 | 7 | 5.85 |
MR technique
MR imaging was performed on a 3T whole-body MR scanner (Signa; GE Healthcare, Waukesha, WI). Patients were imaged in the supine position using a body coil for signal excitation and a pelvic phased-array coil (GE Healthcare, Waukesha, WI) combined with a balloon-covered expandable endorectal coil (Medrad, Pittsburgh, PA) for signal reception. Detailed descriptions of MRI and MRSI sequences can be found in Supplementary Methods.
Briefly, T1-weighted images were acquired with either a spin-echo sequence or a fast spoiled gradient recalled echo sequence. The axial spin-echo T1-weighted images used the following parameters: repetition time/echo time of 950/9 msec, 5-mm section thickness, 1-mm intersection gap, 24 cm field of view, 256 × 160 matrix, transverse frequency direction, and one signal excitation acquired. The fast spoiled gradient recalled sequence had the following parameters: repetition time/echo time of 3.84/2.1 msec, 3D acquisition, 4.2-mm section thickness, 24 cm field of view, 192 × 128 acquisition matrix, transverse frequency direction, and one signal excitation acquired. Thin-section high-spatial-resolution axial and coronal T2-weighted fast spin-echo MR images of the prostate and seminal vesicles were then obtained using the following parameters: repetition time/effective echo time of 6000/95 msec, echo train length of 16, 3-mm section thickness, no intersection gap, 18 cm field of view, 384 × 384 acquisition matrix, and one signal excitation acquired. Three-dimensional 1H MR spectroscopic imaging data were acquired using a water and lipid- suppressed flyback MLEV-PRESS sequence that allows the acquisition of completely upright citrate resonance at TE of 85ms with a 0.16 cc nominal spatial resolution in 8 minutes. The MR spectroscopic imaging parameters were as follows: TR = 2.0s; TE = 85ms; NEX = 1; phase encoding steps = 16 × 10 × 8; FOV = 86 × 54 × 43 mm3; scan time = 11 min. An interleaved echo-planar spectroscopic readout was used in the left-right dimension.
Image interpretation
Two attending radiologists (---, --), each with over 5 years of experience in the interpretation of endorectal MR imaging and MR spectroscopic imaging of the prostate, independently reviewed all MR and MR spectroscopic images on our picture archiving and communication system workstation. Readers knew that patients had biopsy-proven prostate cancer, but were unaware of any other clinical data or histopathological results. Readers identified all suspected treatable (≥ 0.5 cm3) tumor foci [16] meeting previously described imaging criteria [1] (all visible T2-weighted lesions with at least 0.54 cm3 of concordant spectroscopic abnormality, Figure 1a). It should be noted that the spectroscopic findings were used to identify the tumor focus, but not the tumor boundary. Each reader selected the T2 slice that contained the largest cross-section of the treatable tumor, and contoured the tumor margin.
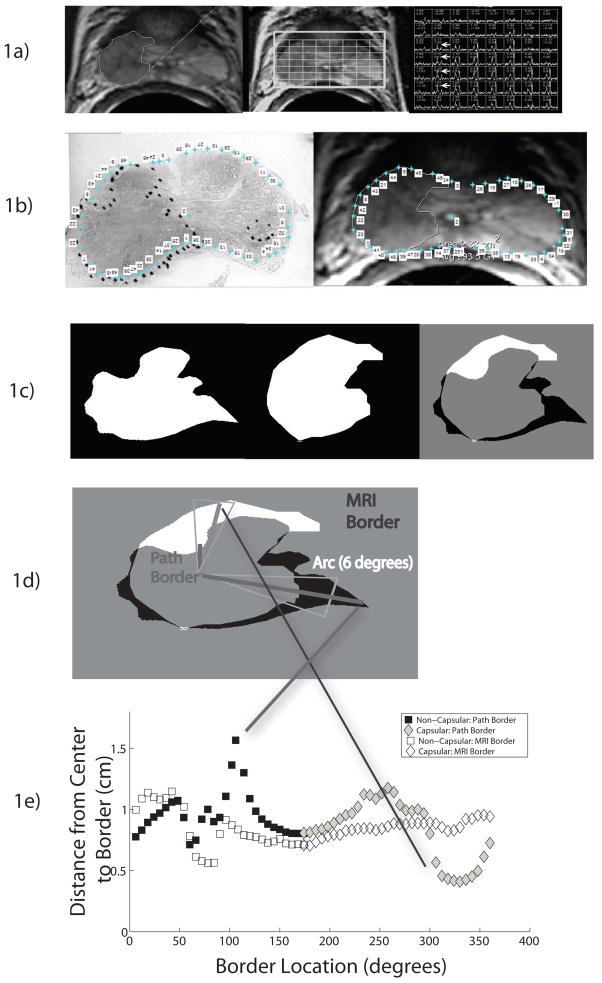
Figure 1.
Figure 1A: Photomontage of axial T2-weighted MR image, overlaid MR spectral grid, and corresponding MR spectra array in a 62 year old with recently diagnosed Gleason 4+3 prostate cancer and a baseline serum prostate specific antigen level of 6.3 ng/mL. A large mass-like focus of reduced T2 signal intensity in the peripheral zone of the right mid-gland is associated with multiple voxels demonstrating high choline peaks (white arrows). This was considered a treatable tumor focus by both readers. The electronic contour generated by reader 1 is shown on the axial T2-weighted image.
Figure 1B: Photomontage of whole mount step section slide, with tumor focus outlined, and the corresponding axial image from Figure 1A. Selected pairs of matching points have been selected along the outline of the prostate on both histopathological and MR images in MATLAB.
Figure 1C: Photomontage of digitally extracted and matched tumor outlines on after deformable co-registration of histopathological and axial T2 images. The first image shows the histopathological tumor outline, the second image shows the MR tumor outline, and the final image shows the differences betlackween the two outlines (white indicates areas where the MR outline overestimated the tumor and white indicates areas where the MR outline underestimated the tumor).
Figure 1D: Photomontage illustrating quantification of contour differences between histopathological and MR contours.
Figure 1E: Quantification of differences in MRI and Path contours as a function of capsular and non-capsular borders.
Histopathological review
Specimens removed at radical prostatectomy were marked with ink and fixed overnight in 10% buffered formalin. Six of the cases were molded during fixing to replicate the mechanical compression effect of the endorectal probe [17]. Transverse step sections were obtained at 3 mm intervals in a plane perpendicular to the prostatic urethra. Paraffin sections were cut at 3 microns and stained with hematoxylin and eosin. An attending pathologist (---) outlined all tumor foci on the whole mount slides. Histopathological tumor volumes were estimated using the formula for tumor volume of (4/3)π (D/2)3, where D is the average of the maximum and minimum axial diameters of the histopathological tumor obtained from the slide demonstrating the maximum tumor area [18]. The recorded pathological tumor stages were pT2a (n = 5), pT2c (n = 7), pT3a (n = 6). In one patient, tumor stage was indeterminate because the specimen was positive at the inked margin. For another patient, no tumor was found in the prostatectomy specimen. The whole mount pathology slides of each radical prostatectomy specimen were optically scanned at high resolution and imported into MATLAB.
Image pair selection, co-registration, and measurement of registration error
The first author (--, not one of the MR readers) compared all the MR contoured lesions with the corresponding whole mount step sections to determine if the MR-identified lesion was a true or false positive. Using previously established criteria [6], MR-identified tumors were considered true positives if they were in the same anatomic location as histopathologically identified tumor and if the MR transverse diameter was not more than twice the histopathological transverse diameter. In addition, contoured lesions were considered false positives if the corresponding histopathological tumor volume was less than 0.5 cm3. Spectroscopy was used to confirm the presence of tumor, but not for contouring due to its low resolution. Next, for true positive MR-identified tumor foci, the first author selected the pathology slide that contained the largest cross-sectional area of the treatable tumor focus and matched it to the corresponding MR image selected by the readers. Using image processing software (MATLAB), at least 30 corresponding points including the capsular border and easily identifiable intraprostatic features such as the urethra (but not any points related to the tumor) were selected on the pathology slide and the MR image (Figure 1b). A two dimensional, best-fit, spatial transformation (e.g. deformable co-registration) was then applied to the pathology image to align it to the MR image (Figure 1c). The error in the deformable co-registration was quantified by measuring the distance between the user defined registration points and the best-fit transformed registration points (median error 2 mm).
Quantification of tumor contour matching
To quantify the difference in contour between MR and histopathology on the matched and co-registered images, the contours were transformed and superimposed using the registration process described above (Figure 1c–d). In order to determine the radial margin expansion needed to ensure that the MR contour adequately encompasses the histopathological contour, a quantification of the radial difference between the MR and histopathological contours was needed. Since voxels are discrete points in space, a line connecting the border points to the center of the tumor may not encompass exactly one point on each border. Therefore, the contour was subdivided into discrete six degree arcs, within which the average radial distance was calculated. The first author classified each arc as capsular or non-capsular, depending on whether the arc involved a tumor segment where the margin abutted the capsule or not. Within each arc, the mean difference in distance between the MR and histopathological contours was calculated, resulting in 60 margin difference measurements for each tumor focus (Figure 1e). We assigned a negative value to differences where the MR contour extended beyond the histopathological contour (“MR overcall”) and a positive value to differences where the MR contour did not reach the histopathological contour (“MR undercall”). We separately calculated the margin of error between imaging and histopathological contours at both capsular and non-capsular surfaces and the treatment margin required to ensure at least 95% tumor coverage in tumors where MR had underestimated tumor extent (Figure 2).
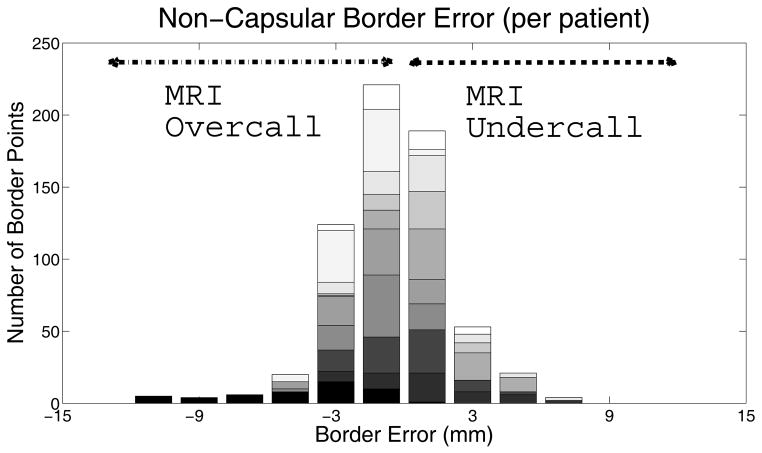
Figure 2.
This figure shows the distribution of the absolute error between the non-capsular border as drawn on MRI and delineated on the corresponding path slice, color-coded by patient. 647 arcs along the non-capsular edge are plotted, with 267 corresponding to points of MRI undercall. To encompass 95% of these 267 points, a margin of 5 mm on the non-capsular border is needed.
Results
Histopathology and identification of treatable tumor foci
Histopathology showed 17 treatable tumor foci in 16 patients, with a mean tumor volume of 2.5cm3 (range, 0.5 to 7.0). Of these 17 treatable foci, 8 were correctly identified by both readers and an additional two foci were correctly identified by reader 2. Both readers identified another treatable tumor focus that corresponded to a histopathological tumor focus of 0.4cm3, and was therefore considered a false positive lesion. Reader 1 identified an additional 2 lesions with no corresponding tumor at histopathology, while reader 2 identified an additional 3 lesions with no corresponding tumor at histopathology. There were five additional foci that did not meet size inclusion criteria.
Quantification of tumor contour matching
A representative example illustrating the process for image registration and quantification of tumor contour matching is shown in Figure 1. Tumor contours were analyzed for the correctly identified lesions. Both readers accurately identified that tumor contacted the prostatic capsule, with no error in contour identification. On the non-capsular border, the median distance between the imaging and histopathological contour (MR overcall and undercall) was 1.4 mm (range, 0 to 12, N=647). The distribution of margin difference measurements at the non-capsular border is shown in Figure 2. Expanding the contour by 5 mm at the non-capsular margin included 95% of tumor volume not initially covered within the MR contour (N = 267). To encompass all tumor identified on pathology, a margin of 8 mm is needed (Figure 2).
Discussion
Several aspects of this study deserve discussion. Our results suggest that MR imaging and MR spectroscopic imaging can identify the contour of correctly identified foci of treatable prostate cancer with high accuracy. In particular, the margin of tumor seen abutting the capsule can be assumed to lie on the capsule. The margin of tumor not abutting the capsule should be extended by 5 mm to ensure adequate tumor coverage. It should be noted that this suggested non-capsular treatment margin is purely geometric, and may not be sufficient for biological tumor control, where an additional margin may be appropriate to provide greater confidence in eradication of microscopic spread or to allow somewhat more latitude for additional error during treatment setup, planning, and delivery. One previous report [5] did attempt to correlate tumor volumes on prostate MR imaging to histopathology, with registration errors of 2 to 3 mm and a suggested margin of 5 mm, but this was a small study using only 5 lesions. We are unaware of any other prior study that quantitatively analyzed the error in contours drawn by MR imaging, stratified by capsular and non-capsular borders, as compared to the histopathological contour, a metric essential to determine what margins are necessary to cover the tumor.
The purpose of this study was to evaluate the accuracy of contours drawn for correctly identified tumor foci, but not the radiographic selection criteria themselves, and therefore analysis was restricted to the correctly selected tumor foci. While MRSI was used in this study, other groups have studied the use of diffusion weighted imaging (DWI) and dynamic contrast enhancement (DCE) in identifying tumor foci. These studies demonstrated increased positive predictive value when using multiparametric MRI [19,20] but they did not specifically address contour accuracy, an essential aspect in radiotherapy planning. Additionally, since the analysis performed here was done on treatment naïve patients, the results cannot necessarily be extended to the salvage setting.
Although tumor selection criteria for focal therapy are discussed elsewhere and were not the focus of our study, our results on this issue are noteworthy; all 9 tumor foci independently identified by both readers as treatable did correspond to histopathological tumor foci (although one focus was under 0.5 cm3 at histopathology and was therefore considered a false positive with respect to identification as a treatable focus). The 5 lesions identified by only a single reader were all false positives. This suggests that identification of treatable tumor foci should also require independent confirmation of tumor presence and location by two readers, although only a single clinician is necessary for accurate contour delineation. Importantly, we utilized the T2-weighted MRI sequence to delineate the tumor border, versus specialized sequences such as dynamic contrast enhancement (DCE) and diffusion weighted (DW) MRI, since T2 is a standard MRI sequence in community practice, making our results widely applicable. Spectroscopy was used to assist in tumor identification and localization, but due to its lower resolution, was not used to contour the tumor borders. Another important aspect of our study is that we have described a method for deformable two dimensional image registration from whole mount step sections to MR images using available scientific software. While previous efforts have focused on creating customized molds for prostatectomy specimens to create good correspondence between MRI and histopathology slices [20], there are several challenges when registering the MR imaging image to the pathology image: the formalin fixed sample can contract, the shape of the prostate itself can be distorted by the insertion of an endorectal coil, and the precise position of the slice and orientation may differ from the MR imaging sequence. We have addressed these issues by using MATLAB to perform a deformable registration of the digitized whole mount image onto the MR image using at least 30 corresponding pairs of hand-picked corresponding points (which averages out user error). Quantification of the registration error showed it to be 2 mm or less in all cases, which generates confidence in our analysis. The border was subdivided into 60 six degree arcs, allowing for precise measurement of contour differences between imaging and histopathology. While we analyzed the margin difference using absolute values, it should be noted that there is a clinical difference between underestimation and overestimation of tumor. That is, the primary effect of an MR overcall (i.e., MR suggests the tumor is bigger than it really is) is excess treatment of tissue that is actually benign. Unless such tissue consists of the neurovascular bundle, urethra or other critical structure, this may be of relatively limited clinical consequence. Conversely, the primary effect of an MR undercall (i.e., MR suggests the tumor is smaller than it really is) is inadequate treatment of tissue that is actually malignant. This has potentially catastrophic consequences, if untreated cancer continues to progress and is unrecognized. So when calculating tumor margin, our analysis only included data on MR undercalls.
Our study has several limitations. First, the study entry criteria combined with the requirement of having whole mount pathology specimens limited the number of possible patients to 20, of which 16 patients with 17 treatable foci met radiographic criteria, and more patients would have increased the robustness of the analysis. Even though tumor selection criteria are not the focus of this study, as noted, more stringent selection criteria could be imposed, including integration of DCE and DWI to the identification of the DIL. Since the goal of this study was to analyze contour accuracy, patient entry criteria was based on only the visible tumor properties, and not Gleason or PSA values. Additionally, the registration was done in two dimensions, relying on the MRI and pathology images being the same orientation. We did not use a mold to orient the majority of the prostatectomy specimens in the exact orientation of the MR imaging, although both were oriented axially, perpendicular to the axis of the urethra. Bias may also be introduced when selecting the slices for comparison; therefore two authors verified the choice of pathology and MRI slices for each patient. Furthermore, the deformable registration algorithm relies primarily on the prostate outline and does not take into account the different areas of stiffness within the sample [22], instead stretching it uniformly, and is subject to both the thickness and the orientation of the slices of the radical prostatectomy specimen, allowing for an alignment error equal to the width of the slice. The pathologic specimen may contract during fixation, and would be uniformly expanded by the image registration algorithm. Additionally, the alignment is reliant on manually picking corresponding points, although the selection of a large number of registration points (> 30) allows for averaging out of errors in placement of registration points. With additional patients and more lesions, these errors would be reduced, but will still contribute to the estimation of the margin error. Future work can take advantage of on-going efforts in improving registration centered on automated registration techniques as well as modeling the mechanical tissue properties of the prostate to improve image registration with whole mount pathology [2,9,11,13]. The cohort of patients in our study may not be representative of the general population, as patient selection methodology may have skewed our population to those with larger tumor volumes. Finally, the contours of tumor on both pathology slice and MR imaging are physician dependent, making the tumor margin potentially user dependent.
Accurate delineation of the treatable intraprostatic tumor foci may allow radiotherapy dose escalation while sparing neighboring critical structures, potentially increasing biochemical control. Endorectal MR imaging and MR spectroscopic imaging can be used to accurately contour these tumor foci; complete tumor coverage is achieved by expanding the treatment contour at the non-capsular margin by 8 mm.
Supplementary Material
Acknowledgments
Work supported by NIH/NCRR/OD UCSF-CTSI Grant Number KL2 RR024130 (ACW) and NIH Grant Number 1S10RR028911-01 (FVC) and NIH Grant Number R01 CA148708 (SMN) and American Cancer Society Grant Number MRSG-CCE-05-087-01(SMN). The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or of the American Cancer Society. FVC receives grant funding from General Electric and Insightec.
Footnotes
Conflict of Interest: None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chang ST, Westphalen AC, Jha P, et al. Endorectal MRI and MR spectroscopic imaging of prostate cancer: Developing selection criteria for MR-guided focal therapy. Journal of Magnetic Resonance Imaging. 2013:n/a–n/a. doi: 10.1002/jmri.24187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chappelow J, Bloch BN, Rofsky N, et al. Elastic registration of multimodal prostate MRI and histology via multiattribute combined mutual information. Medical Physics. 2011;38:2005–2018. doi: 10.1118/1.3560879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coakley FV, Kurhanewicz J, Lu Y, et al. Prostate Cancer Tumor Volume: Measurement with Endorectal MR and MR Spectroscopic Imaging1. Radiology. 2002;223:91–97. doi: 10.1148/radiol.2231010575. [DOI] [PubMed] [Google Scholar]
- 4.DiBiase SJ, Hosseinzadeh K, Gullapalli RP, et al. Magnetic resonance spectroscopic imaging-guided brachytherapy for localized prostate cancer. International Journal of Radiation Oncology*Biology*Physics. 2002;52:429–438. doi: 10.1016/s0360-3016(01)02609-8. [DOI] [PubMed] [Google Scholar]
- 5.Groenendaal G, Moman MR, Korporaal JG, et al. Validation of functional imaging with pathology for tumor delineation in the prostate. Radiotherapy and Oncology. 2010;94:145–150. doi: 10.1016/j.radonc.2009.12.034. [DOI] [PubMed] [Google Scholar]
- 6.Hom JJ, Coakley FV, Simko JP, et al. Prostate Cancer: Endorectal MR Imaging and MR Spectroscopic Imaging—Distinction of True-Positive Results from Chance-detected Lesions1. Radiology. 2006;238:192–199. doi: 10.1148/radiol.2381041675. [DOI] [PubMed] [Google Scholar]
- 7.Hsu CC, Hsu H, Pickett B, et al. Feasibility of MR Imaging/MR Spectroscopy-Planned Focal Partial Salvage Permanent Prostate Implant (PPI) for Localized Recurrence After Initial PPI for Prostate Cancer. International Journal of Radiation Oncology*Biology*Physics. 2012 doi: 10.1016/j.ijrobp.2012.04.028. [DOI] [PubMed] [Google Scholar]
- 8.Lee B, Shinohara K, Weinberg V, et al. Feasibility of high-dose-rate brachytherapy salvage for local prostate cancer recurrence after radiotherapy: The University of California–San Francisco experience. International Journal of Radiation Oncology*Biology*Physics. 2007;67:1106–1112. doi: 10.1016/j.ijrobp.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Mazaheri Y, Bokacheva L, Kroon D-J, et al. Semi-automatic deformable registration of prostate MR images to pathological slices. Journal of Magnetic Resonance Imaging. 2010;32:1149–1157. doi: 10.1002/jmri.22347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muto S, Yoshii T, Saito K, Kamiyama Y, Ide H, Horie S. Focal Therapy with High-intensity-focused Ultrasound in the Treatment of Localized Prostate Cancer. Japanese Journal of Clinical Oncology. 2008;38:192–199. doi: 10.1093/jjco/hym173. [DOI] [PubMed] [Google Scholar]
- 11.Park H, Piert MR, Khan A, et al. Registration methodology for histological sections and in vivo imaging of human prostate. Academic radiology. 2008;15:1027–1039. doi: 10.1016/j.acra.2008.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pouliot J, Kim Y, Lessard E, Hsu IC, Vigneron DB, Kurhanewicz J. Inverse planning for HDR prostate brachytherapy used to boost dominant intraprostatic lesions defined by magnetic resonance spectroscopy imaging. International Journal of Radiation Oncology*Biology*Physics. 2004;59:1196–1207. doi: 10.1016/j.ijrobp.2004.02.055. [DOI] [PubMed] [Google Scholar]
- 13.Samavati N, McGrath D, Lee J, et al. Biomechanical model-based deformable registration of MRI and histopathology for clinical prostatectomy. Journal of Pathology Informatics. 2011:2. doi: 10.4103/2153-3539.92035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheidler J, Hricak H, Vigneron DB, et al. Prostate Cancer: Localization with Three-dimensional Proton MR Spectroscopic Imaging—Clinicopathologic Study1. Radiology. 1999;213:473–480. doi: 10.1148/radiology.213.2.r99nv23473. [DOI] [PubMed] [Google Scholar]
- 15.Society AC; Society AC, editor. Facts and Figures for 2012. 2012. [Google Scholar]
- 16.Stamey T, Freiha F, McNeal J, Redwine E, Whittemore A, Schmid H. Localized prostate cancer: relationship of tumor volume to clinical significance for treatment of prostate cancer. Cancer. 1993;71:933–938. doi: 10.1002/1097-0142(19930201)71:3+<933::aid-cncr2820711408>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 17.Starobinets O, Guo R, Simko J, et al. Semi-automatic registration of digital histopathology to in-vivo MR images in molded and unmolded prostates. Journal of Magnetic Resonance Imaging. 2013 doi: 10.1002/jmri.24287. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taouli B, Goh JSK, Lu Y, et al. Growth Rate of Hepatocellular Carcinoma: Evaluation With Serial Computed Tomography or Magnetic Resonance Imaging. Journal of Computer Assisted Tomography. 2005;29:425–429. doi: 10.1097/01.rct.0000164036.85327.05. [DOI] [PubMed] [Google Scholar]
- 19.Turkbey B, Mani H, Aras O, et al. Correlation of Magnetic Resonance Imaging Tumor Volume with Histopathology. The Journal of Urology. 2012;188:1157–1163. doi: 10.1016/j.juro.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turkbey B, Mani H, Shah V, et al. Multiparametric 3T Prostate Magnetic Resonance Imaging to Detect Cancer: Histopathological Correlation Using Prostatectomy Specimens Processed in Customized Magnetic Resonance Imaging Based Molds. The Journal of Urology. 2011;186:1818–1824. doi: 10.1016/j.juro.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Lin ENJT, Fütterer JJ, Heijmink SWTPJ, et al. IMRT boost dose planning on dominant intraprostatic lesions: Gold marker-based three-dimensional fusion of CT with dynamic contrast-enhanced and 1H-spectroscopic MRI. International Journal of Radiation Oncology*Biology*Physics. 2006;65:291–303. doi: 10.1016/j.ijrobp.2005.12.046. [DOI] [PubMed] [Google Scholar]
- 22.Ward A, Crukley C, McKenzie C, et al. Registration of In Vivo Prostate Magnetic Resonance Images to Digital Histopathology Images Prostate Cancer Imaging. In: Madabhushi A, Dowling J, Yan P, Fenster A, Abolmaesumi P, Hata N, editors. Computer-Aided Diagnosis, Prognosis, and Intervention. Vol. 6367. Springer; Berlin/Heidelberg: 2010. pp. 66–76. [Google Scholar]
- 23.Yu KK, Scheidler J, Hricak H, et al. Prostate Cancer: Prediction of Extracapsular Extension with Endorectal MR Imaging and Three-dimensional Proton MR Spectroscopic Imaging1. Radiology. 1999;213:481–488. doi: 10.1148/radiology.213.2.r99nv26481. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.