Abstract
The presence of DNA in the cytoplasm is a danger signal that triggers immune and inflammatory responses. Cytosolic DNA binds to and activates cyclic GMP-AMP (cGAMP) synthase (cGAS), which produces the second messenger cGAMP. cGAMP binds to the adaptor protein STING and activates a signaling cascade that leads to the production of type-I interferons and other cytokines. Here we report the crystal structures of human cGAS in its apo form, representing its auto-inhibited conformation, as well as cGAMP-bound and sulfate-bound forms. These structures reveal switch-like conformational changes of an activation loop that result in the rearrangement of the catalytic site. The structure of DNA-bound cGAS reveals a complex composed of dimeric cGAS bound to two molecules of DNA. Functional analyses of cGAS mutants demonstrate that both the protein-protein interface and the two DNA binding surfaces are critical for cGAS activation. These results provide new insights into the mechanism of DNA sensing by cGAS.
INTRODUCTION
Infectious microorganisms contain and need nucleic acids in their life cycles. The innate immune system has evolved to recognize microbial DNA and RNA as an essential strategy of host defense (Takeuchi and Akira, 2010). Following endocytosis, microbial DNA and RNA can be detected in the endosome by the transmembrane Toll-like receptors, which then initiate signal transduction cascades in the cytoplasm that lead to the activation of the transcription factors NF-κB and IRFs (e.g, IRF3 and IRF7). These transcription factors then enter the nucleus to induce type-I interferons and other antimicrobial molecules. For those microbes that have succeeded in invading and replicating inside the host cells, the microbial DNA and RNA are detected in the cytoplasm by the innate immune systems. Viral RNA, which usually contains 5’-triphosphate and/or the double-stranded RNA structure, is detected by the RIG-I family of receptors (Rehwinkel and Reis e Sousa, 2010; Yoneyama and Fujita, 2009). RIG-I then activates NF-κB and IRFs through the mitochondrial adaptor protein MAVS (also known as IPS-1, VISA or CARDIF).
We have recently identified cyclic GMP-AMP synthase (cGAS) as the cytosolic DNA sensor that triggers type-I interferon production (Sun et al., 2013; Wu et al., 2013). cGAS binds to microbial DNA as well as self DNA in a sequence-independent manner, which may allow this DNA sensor to detect any DNA that invades the cytoplasm. Upon DNA binding, cGAS is activated to catalyze the synthesis of a unique isomer of cyclic GMP-AMP (cGAMP) from ATP and GTP. This cGAMP isomer contains two phosphodiester bonds, one between 2’-OH of GMP and 5’-phosphate of AMP and the other between 3’-OH of AMP and 5’-phosphate of GMP(Ablasser et al., 2013; Diner et al., 2013; Gao et al., 2013b; Zhang et al., 2013). This cGAMP, termed 2’3’-cGAMP, functions as a second messenger that binds to the endoplasmic reticulum membrane protein STING (also known as MITA, MPYS or ERIS)(Barber, 2011; Wu et al., 2013; Zhang et al., 2013). This binding induces a conformational change of STING, which then recruits the kinases IKK and TBK1 to activate NF-κB and IRF3, respectively (Ishikawa and Barber, 2008; Tanaka and Chen, 2012). Recent genetic studies validate the essential role of cGAS in sensing cytosolic DNA in multiple cell types and in immune defense against DNA viruses in vivo (Li et al., 2013b). In addition, cGAS has been shown to be an innate immune sensor of retroviruses including HIV (Gao et al., 2013a).
In this study, we investigated the mechanism by which cGAS is activated by DNA through crystallographic and biochemical approaches. We determined the human cGAS structures in its apo form, which represents the auto-inhibited conformation, as well as 2’3’-cGAMP bound form and sulfate bound form, which are captured in locally activated conformation as compared to the mouse cGAS-DNA complex. Based on these structures, we identified a conserved activation loop in cGAS, located near the primary DNA binding surface, which exhibits switch-like conformational changes after DNA binding. Surprisingly, unlike the recent structural analyses, which focused on a cGAS-DNA complex that contains one molecule of cGAS and one molecule of DNA (Civril et al., 2013; Gao et al., 2013b; Kranzusch et al., 2013), we found that cGAS forms a 2:2 complex with DNA. Mutagenesis experiments demonstrated that the two DNA binding surfaces and the protein-protein interface of cGAS are important for IRF3 activation and IFNβ induction.
RESULTS
Overall Structure of cGAS in the Apo Form
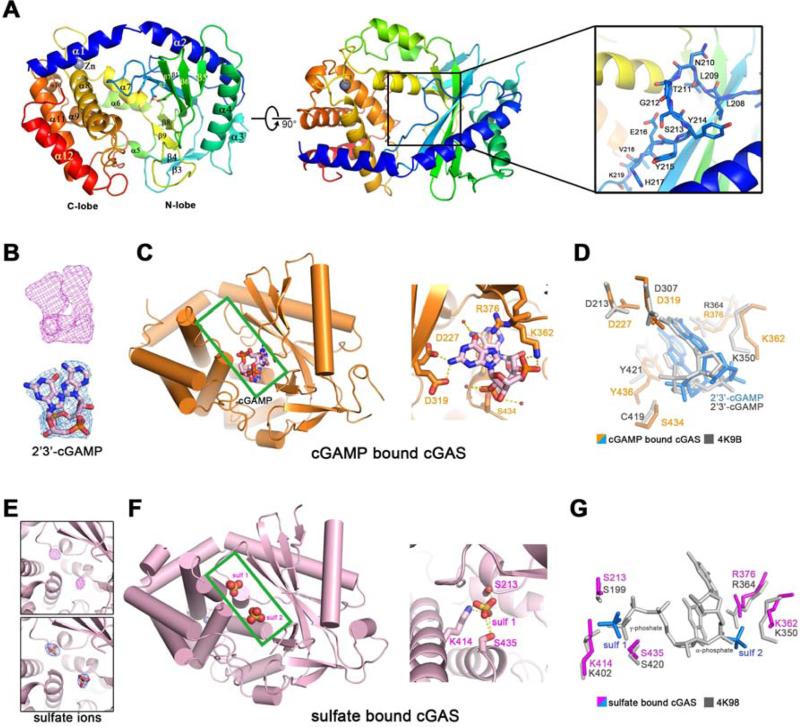
Human cGAS contains 522 amino acid residues, in which the N-terminus containing approximately 160 residues is predicted to be unstructured and was previously shown to be dispensable for DNA-dependent cGAMP synthesis (Sun et al., 2013). We expressed a truncated human cGAS (residues 147-522) in E. coli as a SUMO fusion protein and purified it after removal of the SUMO tag. We determined the crystal structure of this human cGAS protein in its apo form at 2.44 Å using the selenomethionine derivative (Table 1, Figure S1 and S2A). Similar to the recently reported structures of cGAS from different species (Civril et al., 2013; Gao et al., 2013b; Kranzusch et al., 2013), human cGAS adopts an overall fold of mixed α/β topology, which is composed of two lobes (Figure 1A, left and middle panels). The N-terminal lobe comprises a series of central- twisted β-sheets (β1-β9), flanking with α1-α7 helices. The C-terminal lobe is a compact α-helix bundle (α8- α12), including a conserved zinc binding motif. The catalytic residues (Glu225, Asp227 and Asp319) are located on the central β-sheet, the side chains of which point to the cleft between N-lobe and C-lobe. Two residues Gly212 and Ser213 are in a highly conserved loop region, connecting the first and second β strand (β1 and β2) (Figure 1A, right panel). Mutations of Gly212 and Ser213 to alanine abolish the cGAS activity (Sun et al., 2013), underscoring the functional significance of this loop region. Herein, we refer to this loop (residues 210-220 in human cGAS; Figure S1) as the “activation loop” (see details below).
Figure 1. Overall structures of human cGAS in apo-form or in complex with 2’3’-cGAMP or sulfate ions.
(A) Overall structure of human cGAS in the apo-form. The catalytic residues are shown in sticks. Two perpendicular views and a close-up view of the conserved loop connecting the first and second β strand are shown. The residues in the loop are shown in sticks. (B) The simulated annealing omit map (upper panel) and the 2Fo-Fc electron density map (lower pannel) for 2’3’-cGAMP are contoured at 3.0 σ and 1.0 σ, respectively. (C) Overall structure of cGAS in complex with 2’3’-cGAMP (left panel). The cleft between N-lobe and C-lobe is highlighted by green rectangle. 2’3’-cGAMP binds to cGAS by multiple polar contacts (right panel). (D) Residues involved in 2’3’-cGAMP binding are identical to the ones in mouse-cGAS-DNA-2’3’-cGAMP ternary complex (PDB: 4K9B). (E) The simulated annealing omit map (upper panel) and the 2Fo-Fc electron density map (lower panel) for the sulfate ion are contoured at 4.0 σ and 1.5 σ, respectively. (F) Overall structure of cGAS in complex with sulfate ions (left panel). The cleft between N-lobe and C-lobe is highlighted by green rectangle. Sulfate ion 1 is coordinated by several polar residues (right panel). (G) Residues involved in sulfate ion binding are conserved in mouse-cGAS-DNA-linear-2’-GTP-GMP ternary complex (PDB: 4K98). The position of sulfate ion 1 is similar to the γ-phosphate group of GTP moiety, whereas the position of sulfate ion 2 is similar to the α-phosphate group of GMP moiety. All the structure figures are prepared in PyMol (DeLano, 2002).
The Structure of cGAS in Complex with 2’3’-cGAMP
We attempted to co-crystallize human cGAS in complex with dsDNA but failed to obtain well-diffracting crystals. In the meantime, we were able to obtain well-diffracting crystals of cGAS (residues 161-522) bound to its product 2’3’-cGAMP. The structure of this complex was determined by molecular replacement, using apo cGAS as the search model, and refined to 2.44 Å with good Rfree value and stereochemistry (Table 1 and Figure S2B). Simulated annealing omit map and 2Fo-Fc electron density map revealed decent density for 2’3’-cGAMP (Figure 1B). 2’3’-cGAMP sits at the lower part of the cleft between N-lobe and C-lobe (Figure 1C, left panel). The catalytic residues Asp227 and Asp319, in addition to Lys362, Arg376 and Ser434, interact with 2’3’-cGAMP through multiple polar contacts (Figure 1C, right panel). The base group of adenine moiety stacks against the aromatic residue Tyr436 (Figure 1D). The residues involved in 2’3’-cGAMP binding are identical to the corresponding residues identified in the recently determined structure of mouse cGAS that contains a 16-bp dsDNA in addition to 2’3’-cGAMP (PDB: 4K9B, Figure 1D) (Gao et al., 2013b).
The Structure of cGAS in Complex with Sulfate Ions
We crystallized human cGAS (161-522) in the presence of 2 M (NH4)2SO4, and refined the structure to 2.25 Å (Table 1 and Figure S2C). During the structure refinement, two pieces of spherical electron density were observed in the cleft between N-lobe and C-lobe (Figure 1E). They likely represent sulfate ions because of the crystallization condition. Sulfate ions are often utilized to mimic phosphate groups and interact with phosphate-binding site in order to trap specific conformations of kinase or nucleotide cyclase (Jeong et al., 2003; Tanaka et al., 2010). After fitting two sulfate ions in the cleft, we found that the first sulfate ion, in the upper part of the cleft, is well coordinated by surrounding residues Ser213, Lys414, and Ser435 (Figure 1F). If the sulfate bound cGAS structure is superimposed with mouse-cGAS-DNA-linear-2’-GTP-GMP ternary complex (PDB:4K98), the first sulfate ion locates at the similar position as that of γ-phosphate group in the GTP moiety, whereas the second ion sits near the α-phosphate group in the GMP moiety (Figure 1G). The structural similarity among cGAMP-bound human cGAS, sulfate-bound human cGAS and DNA-bound mouse cGAS suggests that cGAS is trapped in a locally activated conformation in the presence of 2’3’-cGAMP or sulfate ions.
Conformational Changes in the Activation Loop of cGAS
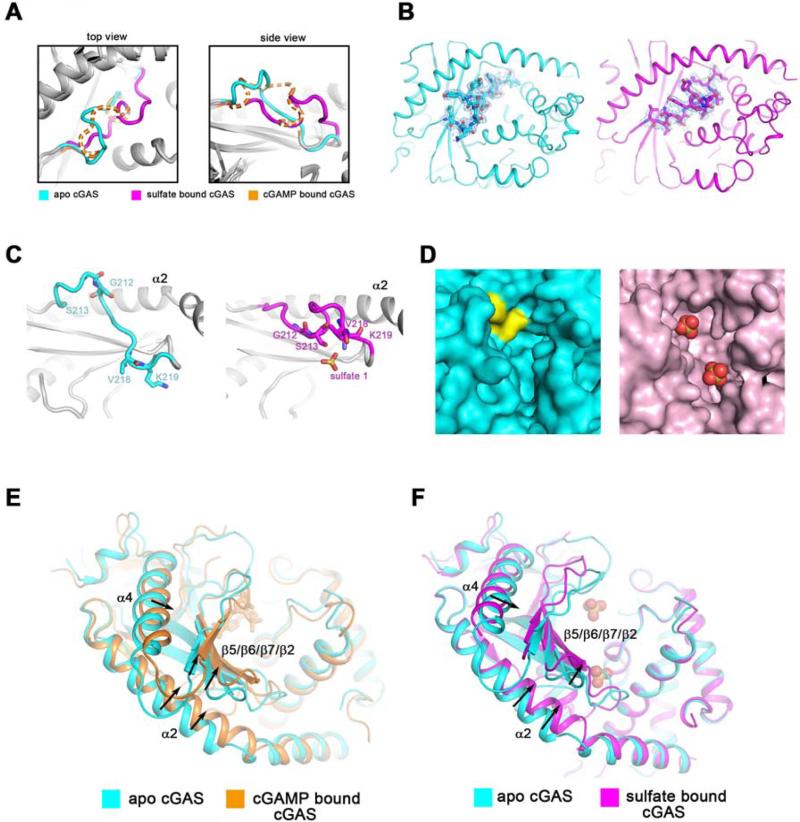
Comparison of the activation loops in apo cGAS and in the other two ligand-bound cGAS structures reveals that the loop undergoes significant conformational changes (Figure 2A). The activation loop is not involved in the crystal packing interface (Figure S3 A-C). 2Fo-Fc electron density of the activation loops of apo cGAS and sulfate-bound cGAS are of decent quality (Figure 2B). The sulfate ion in the upper part of the cleft directly interacts with Ser213 in the activation loop (Figure 1F), resulting in a fixed loop orientation. However, the activation loop in cGAMP-bound cGAS structure appears relatively more flexible (Figure 2A), probably because 2’3’-cGAMP binds to the lower part of the cleft and releases the activation loop to some extent (Figure 1C).
Figure 2. Ligand-induced conformational changes of the activation loop and active site rearrangement of cGAS.
(A) Comparison of the activation loops in apo cGAS (cyan), sulfate bound cGAS (magenta), and cGAMP bound cGAS (orange). The loop of cGAMP bound cGAS is shown in dash line because of its high flexibility. Two perpendicular views are shown. The same color scheme is applied unless indicated otherwise. (B) The 2Fo-Fc electron density maps of the activation loop in apo cGAS (left panel) or sulfate bound cGAS (right panel), shown in blue mesh, are contoured at 0.8 σ. (C) The activation loop undergoes significant conformational changes in the presence of sulfate ions (right panel) when compared to apo-cGAS (left panel). α2 helix is used as the reference. (D) Val218 and Lys219 (colored in yellow) occupy a large part of the cleft between N-lobe and C-lobe in apo cGAS. The sulfate ions are shown in red spheres in the right panel. (E and F) The N lobe of cGAS exhibits distinguishable conformational shifts in the presence of 2’3’-cGAMP (E) or sulfate ions (F), as compared to the apo form.
The conformational changes of the activation loop include not only an inward movement of the protruded loop (Gly212 and Ser213), but also an outward shift of Val218 and Lys219 in order to empty the pocket for the substrates, in this case the sulfate ions that mimic the phosphate moieties of nucleotides (Figure 2C, 2D and Movie S1). Besides the rearrangement in the cleft of the active site, the N-lobe also exhibits conformational shifts, especially in α2, α4 and the β sheet formed by β5, β6, β7 and β2 (Figure 2E, 2F and Movie S2 and S3). This region exhibits high B factors, suggesting relatively high mobility (Figure S3D).
The large conformational change of the activation loop from apo cGAS to sulfur-bound cGAS raises the question of how such conformational change is triggered. On the backside of the cleft, there is one positively charged patch which has recently been shown to bind DNA. Interestingly, the activation loop, pointing outward in apo cGAS, hinders the interaction between human cGAS and DNA (Figure 3A). The sulfate ions trap a conformation that displays several positively charged patches, which is able to accommodate the modeled double stranded DNA helix (Figure 3B).
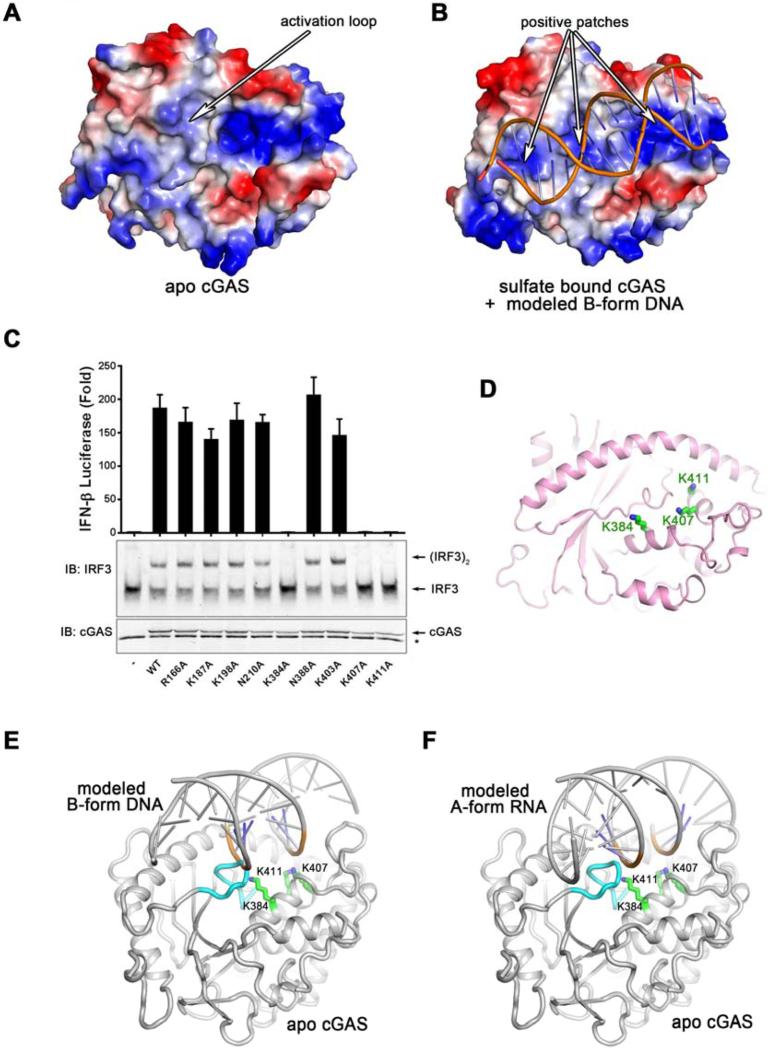
Figure 3. The primary DNA-binding surface of cGAS is essential for IFNβ induction.
(A and B) The electrostatic representations of apo cGAS (A) and sulfate bound cGAS (B). A series of positively charged patches in sulfate bound cGAS indicate the potential primary DNA binding surface. (C) Expression plasmids encoding WT and various mutants of human cGAS fragments (161-522) containing alanine substitutions of positively charged residues shown in (B) were transfected into HEK293T-STING-IFNβ luciferase reporter cells followed by luciferase assays to measure IFNβ induction. Aliquots of the cell extracts were immunoblotted with an IRF3 antibody following native gel electrophoresis (middle panel) or with a cGAS antibody flowing SDS-PAGE (bottom panel). The error bars represent variation ranges of duplicate experiments. (D) Functionally important positively charged residues, shown in green sticks, are located in the center of the primary binding surface. (E and F) Docking B-form DNA to apo cGAS (E) results in a steric clash between the activation loop and the DNA, which likely triggers the inward movement of the activation loop (E). However, docking A-form RNA to apo cGAS does not reveal the steric clash or the movement of the activation loop (F). The activation loop is colored in cyan. Loss-of-function mutations on the primary DNA binding surface are shown in sticks and colored in green.
We examined a series of conserved positively charged residues located on the potential DNA binding surface by mutagenesis. Expression plasmids encoding wild-type or mutant human cGAS were transfected into HEK293T cells stably expressing STING and an IFNβ-luciferase reporter. Luciferase activity and IRF3 dimerization assays show that Lys384, Lys407 and Lys411 are critical for cGAS function (Figure 3C). Lys407 and Lys411 are located on the pre-existing positively charged patch, whereas Lys384 is near the activation loop and is exposed only in sulfate-bound cGAS (Figure 3D). Notably, mutations of several positively charged residues in the first and second α helices (R166A, K187A, K198A) of cGAS did not abrogate its ability to induce IFNβ or activate IRF3 (Figure 3C).
Modeling of the cGAS structures bound to the B-form dsDNA shows that the DNA binding clashes with the protruded activation loop, causing this loop to move inward and rearrange the active site (Figure 3E and Movie S1). In contrast, modeling using the A-from dsRNA shows that the activation loop inserts into the major groove of dsRNA without causing detectable conformational changes (Figure 3F), which may explain why cGAS cannot be activated by dsRNA.
cGAS forms a 2:2 functional complex with DNA
To further understand how DNA binding activates cGAS, we solved the crystal structure of mouse cGAS in the presence of DNA. The same mouse cGAS protein and DNA were previously used to obtain the crystal structure of a cGAS-DNA complex, which was interpreted to contain one cGAS and one DNA (Gao et al., 2013b). However, we found that each asymmetric unit contains two cGAS molecules and two DNA molecules (Figure 4A). Besides the primary DNA binding surface (surface 1) which is identical to the ones reported previously (see also Figure 3B and 3D) (Civril et al., 2013; Gao et al., 2013b), cGAS has another surface area, composed of two positive patches, that interacts with DNA (Figure 4B).
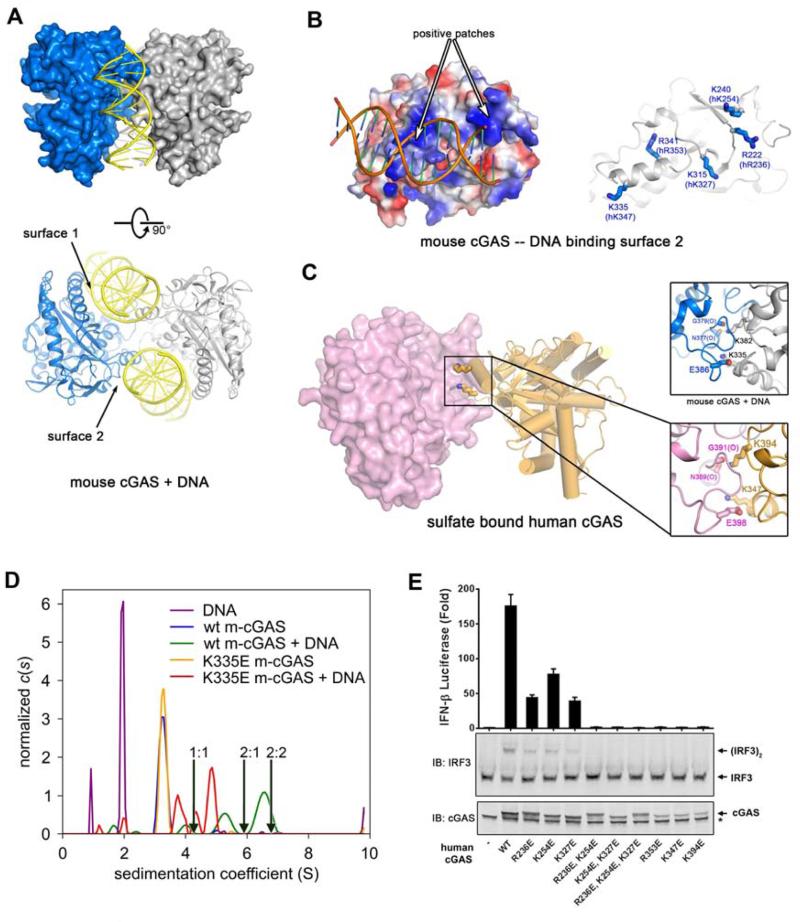
Figure 4. cGAS forms a functional 2:2 complex with DNA.
(A) Crystal structure of mouse cGAS in complex with a 16 bp dsDNA. Each asymmetric unit contains one 2:2 complex, composed of two protein molecules and two DNA molecules. Two perpendicular views are shown. (B) The electrostatic representations of DNA binding surface 2 of mouse cGAS (left). Conserved positively charged residues are shown on the right. with the corresponding residues in human cGAS shown in parentheses. (C) Sulfate bound human cGAS forms a dimer in the crystal (left); residues involved in the dimer interface of this structure (lower right panel) are identical to the ones in mouse cGAS-DNA complex (upper right panel). (D) Analytical ultracentrifugation of m-cGAS and DNA. Shown are c(s) distributions for various combinations of wild-type (“wt”) or K335E m-cGAS in the presence or absence of DNA. A distribution for DNA alone is also shown. The concentrations used were: DNA alone, 120 μM; wt m-cGAS, 80 μM; wt m-cGAS/DNA, 80 and 120 μM, respectively; K335E m-cGAS, 94 μM; K335E m-cGAS/DNA, 94 μM and 120 μM, respectively. All of the distributions have been normalized by the total interference signal (in fringes) present in the respective experiments. The arrows show the theoretical sedimentation coefficients of 1:1, 2:1, and 2:2 protein/DNA complexes. (E) The second DNA binding surface and the protein-protein interface are important for cGAS activity. Expression vectors encoding WT or mutant human cGAS proteins as indicated were transfected into HEK293T-STING-IFNβ luciferase reporter cells, followed by measurement of luciferase activities (upper panel). Aliquots of the cell extracts were immunoblotted with an IRF3 antibody following native gel electrophoresis (middle panel) or with a cGAS antibody flowing SDS-PAGE (lower panel).
Closer inspection of the apo- and sulfate-bound human cGAS also reveals a dimer in crystalline (Figure S4A and S4B). Interestingly, Lys394 within the zinc finger of human cGAS, corresponding to Lys382 in mouse cGAS, interacts with the carbonyl oxygen atoms of Asn389 and Gly391 (Figure 4C). Lys347 in human cGAS (Lys335 in mouse cGAS) not only mediates DNA binding (Figure 4B), but also interacts with Glu398 from the adjacent protomer (Figure 4C). The interaction area is about 678 Å2 in total, suggesting a weak interaction, which may be reinforced by the binding of cGAS to DNA. Although not reported previously, the same dimer interface exists in the previously published crystal structures of apo- and DNA-bound cGAS proteins from different species and crystallized under different conditions (Figure S4C-F) (Civril et al., 2013; Gao et al., 2013a; Kranzusch et al., 2013), strongly suggesting that the formation of cGAS dimer is not a crystallographic artifact. Indeed, we found that Flag-cGAS and HA-cGAS co-immunoprecipitated from HEK293T cells that expressed both of these proteins, suggesting that cGAS may form a dimer or oligomer in mammalian cells, perhaps in the presence of the transfected plasmid DNA (Figure S4G).
Analytical ultracentrifugation sedimentation velocity (SV) experiments were performed to examine the hydrodynamic behavior of mouse cGAS (m-cGAS) in the absence and presence of DNA. Without DNA in solution, m-cGAS is a monomer that sediments at 3.2 S at all concentrations (0.8-80 μM) studied (Figure S4H). Thus, the m-cGAS dimer observed in the crystal structures in the absence of DNA may be due to the very high concentrations of cGAS under the crystallographic conditions. However, in the presence of a 16-mer dsDNA, faster-sedimenting complexes are evident at 4.0 S, 5.3 S, and 6.5 S (Figure 4D). The latter peak is close to that expected for a 2:2 complex. A multisignal SV (Balbo et al., 2005; Padrick et al., 2010) analysis demonstrates that the molar ratio of protein:DNA in this peak is 0.9:1 (Figure S4I). Thus, the hydrodynamics and composition of this peak are consistent with the formation of the postulated 2:2 complex. The high concentrations needed to achieve this complex (80 μM protein, 120 μM DNA) reflect the apparently poor association constant of complex formation. The other fast-sedimenting peaks mentioned here are likely DNA and protein whose time-average sedimentation coefficients have been increased (“advanced species”) by brief residencies in complexes between the two.
The conclusion that the 6.5-S peak represents 2:2 complexes is further buttressed by parallel experiments carried out with the m-cGAS point mutant K335E (corresponding to K347 in human cGAS). As mentioned above, this residue mediates both cGAS:cGAS and cGAS:DNA interactions (Figure 4C). Under similar conditions (94 μM protein, 120 μM DNA), this mutant protein does not form fast-sedimenting complexes as efficiently as the wild-type m-cGAS (Figure 4D). Although this mutant efficiently binds DNA, its ability to form large complexes is greatly diminished, and thus the population of 2:2 complexes must be significantly smaller than that observed in the experiment with wild-type cGAS. It appears, therefore, that Lys335 is very important for the formation of the protein:protein interface, but its mutation does not disrupt DNA binding, likely because the intact DNA binding surface 1 and the other residues in the DNA binding surface 2 can still mediate DNA binding. The resolved peaks in this analysis likely consist of a combination of effective particles (Schuck, 2010) containing various populations of possible complexes and advanced species (Figure 4D, green curve).
In cell-based functional assays, point mutations of Lys347 (Lys335 in m-cGAS), Arg353 or Lys394 to Glu in the human cGAS completely abolished its ability to activate IRF3 and induce IFNβ (Figure 4E). Although single mutations of Arg232, Lys254 and Lys327 to Glu only partially impaired the activity of cGAS, double (R236E and K254E; K254E and K327E) and triple mutations (R236E, K354E and K327E) abolished the activity. These results indicate that the second DNA binding site and the protein-protein interface in cGAS are important for activation of the type-I interferon pathway (Figure S4J, Movie S4).
DISCUSSION
In this study, we determined the crystal structures of human cGAS in its apo form, 2’3’-cGAMP-bound form and sulfate-bound form. Structural alignment with the DNA-bound mouse cGAS shows that the ligand-bound human cGAS resembles the activated conformation induced by DNA binding. In apo cGAS, the activation loop points to the outside. Once cGAS encounters cytosolic B-form DNA, the pre-existing positively charged patch (Lys407, Lys411 and other DNA-binding residues on α1 and α2 helices) ‘grabs’ the DNA. The steric clash between the DNA and the activation loop ‘switches on’ dramatic conformational changes in the loop region such that new positively charged patches are formed, including the exposure of Lys384, allowing further DNA binding (Figure 3E and Movie S1). The DNA binding causes the activation loop to move inward, thus rearranging the active site to catalyze cGAMP formation (Movie S4).
While we were refining the cGAS structures, three groups published the crystal structures of cGAS (Civril et al., 2013; Gao et al., 2013b; Kranzusch et al., 2013). Gao et al and Civril et al determined the crystal structures of mouse cGAS and porcine cGAS, respectively, in both the apo form and DNA-bound form. Both groups observed that the first and second helices bind to DNA and undergoes conformational shifts. Although Gao et al showed that none of the single point mutations of the positive residues on α1 and α2 helices abrogated the DNA-dependent activity of cGAS (Gao et al., 2013b), Civril et al found that the double mutant K173A/R176A on α1 helix of human cGAS had greatly reduced activity (Civril et al., 2013). More recently, Li et al showed that R158E of mouse cGAS (equivalent to K173 of human cGAS) was largely defective in cGAMP synthesis and IFNβ induction (Li et al., 2013a). Thus, DNA binding to the first two α helices of cGAS likely contributes to its activation. However, this is not sufficient. Here we show that the switch-like movement of the activation loop, which is triggered in part by DNA binding to the positive charged residues (K384, K407 and K411) located in α7 and α8 surrounding the zinc finger, mediates cGAS activation. Contrary to our results which showed that K384A or K407A mutation abolished cGAS activity, Gao et al found that mutations of the corresponding residues in mouse cGAS did not impair its activity (Gao et al., 2013b). However, two recent reports presented evidence showing that point mutations of K384, K407 and K411 in human cGAS or the corresponding residues in mouse cGAS abolished its ability to induce IFNβ (Kato et al., 2013; Li et al., 2013a).
Interestingly, mouse cGAS is able to tolerate G199A mutation, but G199P or G199AS200A largely abolishes its activity (Sun et al., 2013; Gao et al, 2013b), suggesting a crucial role of the loop flexibility in the activation process. The S200A mutation has little effect on the cGAS activity, presumably because it does not affect the loop flexibility and the binding of the γ-phosphate group from the substrate (sulfate ion in our structure) involves multiple polar contacts besides the interaction with S200 (S213 in human cGAS). Kranzusch et al determined the crystal structure of human cGAS in its apo form (Kranzusch et al., 2013). The activation loop in this structure is also protruded to the outside, even though the cGAS protein fragment and the crystallization conditions were distinct from what were used in our study, suggesting that the outward protrusion of the activation loop in human cGAS is reproducible in different crystals. Although the protrusion of the activation loop is less obvious in mouse cGAS, the significant ligand-induced conformational switch of the activation loop occurs in both human and mouse cGAS structures.
cGAS is not the only example of proteins possessing the activation loop. The conserved activation segment, starting with the DFG motif and ending with the APE motif, exists in all protein kinase families and is also crucial to the catalytic activity (Endicott et al., 2012). The DFG loop has been shown to be an important drug target. The identification of the activation loop in cGAS reveals a new mechanism of its activation by DNA and may assist in the development of cGAS inhibitors.
Surprisingly, we found that the cGAS structures that we have determined and those that have been published previously all form an identical dimer in crystalline (Figure 5 and Figure S4) (Civril et al., 2013; Gao et al., 2013a; Kranzusch et al., 2013). These findings indicate that cGAS itself may form a dimer. However, our AUC analysis shows that the mouse cGAS protein does not form a dimer in the absence of DNA at all concentrations tested. In the presence of DNA, cGAS forms a 2:2 complex with DNA in solution, consistent with the crystal structure. A significant amount of this complex was detectable only when both cGAS and DNA were present at high concentrations, suggesting that the complex has a poor association constant. This is consistent with the fact that very few residues, notably K347 and K394 in human cGAS, are involved in the protein – protein interface. Despite this weak interaction, point mutation of either K347 or K394 completely abolishes the ability of cGAS to induce IFNβ, underscoring the functional importance of forming the oligomeric complex. While our paper was in revision, Li et al reported that DNA binding induces oligomerization of cGAS that is important for its activation (Li et al., 2013a). Consistent with our results, these authors showed that mutations of cGAS at residues on the second DNA binding surfaces and at the protein dimer interface abolish cGAMP synthesis and IFNβ induction. While both studies independently identified a common set of residues involved in protein:DNA and protein:protein interactions, we have identified and validated additional key residues that are crucial for cGAS’ functions. For example, we show that K411 and R353 are critical residues in DNA binding surface 1 and 2, respectively. In addition, our results reveal the activation loop of cGAS that is important for switching on the active site rearrangement in response to DNA binding. Future studies should determine whether and how the formation of the cGAS oligomeric complex is regulated in cells to provide a sensitive and timely response to DNA that invades the cytoplasm.
EXPERIMENTAL PROCEDURES
Truncated human cGAS (147-522 in apo form, 161-522 in 2’3’-cGAMP bound form and sulfate bound form) and mouse cGAS (147-507) were cloned into pET-SUMO vector (Invitrogen™) and overexpressed in E. coli BL21 (DE3) pLysS. The recombinant proteins were purified by Ni2+-nitrilotriacetate affinity resin (Qiagen) followed by SUMO protease digestion at 4°C overnight. Further purification was applied on Heparin column (GE Healthcare). The peak fractions of the protein were collected and concentrated to 6 mg/ml for crystallization trials. The selenomethionine derivatized protein was purified similarly as described above. All the crystals were grown at 20°C by the hanging-drop vapor diffusion method. The data were collected at 19-ID at the Advanced Photon Source (APS) or using Rigaku FR-E copper rotating-anode generator and an R-AXIS IV++ imaging-plate area detector (Rigaku Americas, Houston, Texas, USA). The data were integrated and scaled with HKL2000 package (Otwinowski and Minor, 1997). Initial phases were generated using anomalous differences of a 3.0 Å Se derivative dataset by SAD. The phase information was improved by the MR-SAD using another 2.45 Å Se derivative dataset. The initial model was built using the Autobuild suite aided by manual correction of the coordinates in the COOT (Emsley et al., 2010). The cGAMP bound cGAS and the sulfate bound cGAS structures were determined by molecular replacement using apo cGAS as the search model. The mouse cGAS DNA complex structure was solved by molecular replacement using 4K98 as the search model. AUC experiments were carried out using Beckman Coulter Optima XL-I ultracentrifuge with samples in a buffer containing 25 mM Tris pH 8.0 and 25 mM NaCl. Data were acquired using both interference optics and absorbance optics. To test the IFNβ inducing ability of different cGAS mutants, expression plasmids were transfected into HEK293T-STING-IFNβ luciferase reporter cells. After 24 hours transfection, the lysate was used to measure firefly luciferase activity (Promega).
See Supplemental Experimental Procedures for details.
Supplementary Material
Highlights.
DNA binding activates cGAS through switching on an activation loop
DNA binding induces dimerization of cGAS
cGAS dimerization is essential for its activation
Two DNA binding surfaces of cGAS are critical for inducing type-I interferons
ACKNOWLEDGEMENT
Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by University of Chicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. We thank Drs. Diana Tomchick and Thomas Scheuermann at UT Southwestern Medical Center for assistance in crystallography and AUC experiments, respectively. This work was supported by an NIH grant (AI-093967) to Z.J.C, and an international graduate student fellowship from Howard Hughes Medical Institute (HHMI) to J.W. Z.J.C is an Investigator at HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes a Table, four supplemental figures, four supplemental movies and supplemental experimental procedures.
ACCESSION NUMBER
The coordinates of cGAS structures in its apo- and ligand-bound forms have been deposited in the RCSB protein data bank. The PDB codes are: human cGAS alone: 4O68; sulfate-bound human cGAS: 4O69; 2’3’-cGAMP bound human cGAS: 4O67; DNA-bound mouse cGAS: 4O6A.
REFERENCES
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2'-5'-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013 doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbo A, Minor KH, Velikovsky CA, Mariuzza RA, Peterson CB, Schuck P. Studying multiprotein complexes by multisignal sedimentation velocity analytical ultracentrifugation. Proc Natl Acad Sci U S A. 2005;102:81–86. doi: 10.1073/pnas.0408399102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber GN. Cytoplasmic DNA innate immune pathways. Immunol Rev. 2011;243:99–108. doi: 10.1111/j.1600-065X.2011.01051.x. [DOI] [PubMed] [Google Scholar]
- Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, Hopfner KP. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. 2002 on World Wide Web http://wwwpymolorg.
- Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. The Innate Immune DNA Sensor cGAS Produces a Noncanonical Cyclic Dinucleotide that Activates Human STING. Cell Rep. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endicott JA, Noble ME, Johnson LN. The structural basis for control of eukaryotic protein kinases. Annu Rev Biochem. 2012;81:587–613. doi: 10.1146/annurev-biochem-052410-090317. [DOI] [PubMed] [Google Scholar]
- Gao D, Wu J, Wu Y-T, Du F, Aroh C, Yan N, Sun L, Chen ZJ. Cyclic GMP-AMP Synthase Is an Innate Immune Sensor of HIV and Other Retroviruses. Science. 2013a;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, et al. Cyclic [G(2′,5′)pA(3′,5′)p] Is the Metazoan Second Messenger Produced by DNA-Activated Cyclic GMP-AMP Synthase. Cell. 2013b;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SY, Kumagai A, Lee J, Dunphy WG. Phosphorylated claspin interacts with a phosphate-binding site in the kinase domain of Chk1 during ATR-mediated activation. J Biol Chem. 2003;278:46782–46788. doi: 10.1074/jbc.M304551200. [DOI] [PubMed] [Google Scholar]
- Kato K, Ishii R, Goto E, Ishitani R, Tokunaga F, Nureki O. Structural and Functional Analyses of DNA-Sensing and Immune Activation by Human cGAS. PLoS One. 2013;8:e76983. doi: 10.1371/journal.pone.0076983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzusch PJ, Lee AS, Berger JM, Doudna JA. Structure of Human cGAS Reveals a Conserved Family of Second-Messenger Enzymes in Innate Immunity. Cell Rep. 2013;3:1362–1368. doi: 10.1016/j.celrep.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Shu C, Yi G, Chaton CT, Shelton CL, Diao J, Zuo X, Kao CC, Herr AB, Li P. Cyclic GMP-AMP Synthase Is Activated by Double-Stranded DNA-Induced Oligomerization. Immunity. 2013a;39:1019–1031. doi: 10.1016/j.immuni.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-D, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal Roles of cGAS-cGAMP Signaling in Antiviral Defense and Immune Adjuvant Effects. Science. 2013b;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Padrick SB, Deka RK, Chuang JL, Wynn RM, Chuang DT, Norgard MV, Rosen MK, Brautigam CA. Determination of protein complex stoichiometry through multisignal sedimentation velocity experiments. Anal Biochem. 2010;407:89–103. doi: 10.1016/j.ab.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Reis e Sousa C. RIGorous detection: exposing virus through RNA sensing. Science. 2010;327:284–286. doi: 10.1126/science.1185068. [DOI] [PubMed] [Google Scholar]
- Schuck P. Sedimentation patterns of rapidly reversible protein interactions. Biophys J. 2010;98:2005–2013. doi: 10.1016/j.bpj.2009.12.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Smith P, Shuman S. Structure of the RNA 3′-phosphate cyclase-adenylate intermediate illuminates nucleotide specificity and covalent nucleotidyl transfer. Structure. 2010;18:449–457. doi: 10.1016/j.str.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Chen ZJ. STING Specifies IRF3 Phosphorylation by TBK1 in the Cytosolic DNA Signaling Pathway. Sci Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP Containing Mixed Phosphodiester Linkages Is An Endogenous High-Affinity Ligand for STING. Mol Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.