Abstract
Genomics is a relatively new scientific discipline, having DNA sequencing as its core technology. As technology has improved the cost and scale of genome characterization over sequencing’s 40-year history, the scope of inquiry has commensurately broadened. Massively parallel sequencing has proven revolutionary, shifting the paradigm of genomics to address biological questions at a genome-wide scale. Sequencing now empowers clinical diagnostics and other aspects of medical care, including disease risk, therapeutic identification, and prenatal testing. This Review explores the current state of genomics in the massively parallel sequencing era.
Prior to the advent of next-generation sequencing (NGS) technology, genomics initially was concerned with studying genomes that were tractable from the standpoint of size and repetitive content (e.g., viruses and bacteria) and with characterization of single genes associated with disease (e.g., Cystic Fibrosis, Huntington disease, and cancer). As the ability to construct large clone-based physical maps improved, the subcloned fragments of the genome contributing to physical map construction could be sequenced as individual projects, and their finished sequences melded together to represent entire chromosomes. Hence, important large genomes, including model organisms and the human genome, were decoded. Indeed, in the era of NGS, the short reads obtained from most platforms absolutely require these reference genomes as a substrate for read alignment prior to variation discovery. The impact of these technologies on genomic variant discovery has been profound, as we will describe. Although we limit the scope of this Review to genomics, an accompanying Review explores the disruptive impact of NGS on studies of the epigenome to further highlight the profound transformation brought on by NGS technology (Rivera and Ren, 2013 [this issue of Cell]).
Genomic Techniques
Although NGS technology initially was used to study whole genomes, a variety of approaches that address defined regions of the genome have emerged. There are essentially two technical preparatory approaches to explore selected regions of the genome with NGS. The first is by PCR, typically involving multiple primer pairs in a mixture that are combined with genomic DNA of interest in a multiplex approach to preserve precious DNA. The use of multiplex primer pairs couples the high throughput of NGS platforms and the fact that each sequence read represents a single DNA product in the mixture due to the nature of the sequencing platforms (Mardis, 2013). Following the PCR, the resulting fragments have platform-specific adapters ligated to their ends to form a library that is suitable for sequencing. The second approach involves hybrid capture, which has been developed by several groups and commercialized (Albert et al., 2007; Gnirke et al., 2009; Hodges et al., 2007). Essentially, hybrid capture takes advantage of the hybridization of DNA fragments from a whole-genome library to complementary sequences that were synthesized and combined into a mixture of probes designed with high specificity for the matching regions in the genome. These probes typically have covalently linked biotin moieties, enabling a secondary “capture” by mixing the probe:library complexes with streptavidin-coated magnetic beads. Hence, the targeted regions of the genome can be selectively captured from solution by applying a magnetic field, whereas most of the remainder of the genome is washed away in the supernatant. Subsequent denaturation releases the captured library fragments from the beads into solution, ready for postcapture amplification, quantitation, and sequencing. When the probes are designed to capture essentially all of the known coding exons in a genome, the capture approach is referred to as “exome sequencing.” Additional probes may be designed, synthesized, and added to an exome reagent, typically referred to as “exome plus.” When only a subset of the exome or of the genome outside of the exome is targeted, this is called a “targeted panel.”
Genomic Analysis
As important as techniques to produce the NGS data that address biological questions are, analytical approaches are equally critical for successful interpretation of those data. Many analytical approaches depend on the digital nature of NGS data, a consequence of the fact that individual DNA fragments of the library are amplified either on beads or on flat surfaces (platform specific) prior to the sequencing reaction. Hence, each sequence read is equivalent to a single DNA fragment. What follows are selected data analysis techniques from a dizzying number of advances published in just the last 18 months. The pace of innovation in analytical approaches to genome-wide data analysis continues to engage and excite the computational biology community as the number of technical applications continues.
Technological advances have often driven the methods for discovering new disease genes. Early studies leveraged families in which a disease was segregating to identify the genetic causes of the phenotype. These linkage analysis studies were successful for highly penetrant, monogenic diseases such as cystic fibrosis. Standard parametric linkage studies of some complex traits were successful, particularly when sampling from extreme ends of the phenotypic distribution. For example, analyzing families segregating early onset Alzheimer’s disease let to the discovery of multiple genes that contribute significantly to the phenotype and shed light on the biological mechanisms (e.g., plaque formation) of disease progression (Goate et al., 1991; Harrington et al., 1995; Pericak-Vance et al., 1991).
Yet, for many complex diseases and traits, this model was not as successful because the genetic predispositions to complex traits are, as their name implies, more difficult to elucidate and require larger numbers of samples to discern signal from noise. Theoretically, it was determined that comparing allele frequencies across the genome between large numbers of cases and controls would be able to capture common disease susceptibility alleles (Risch and Merikangas, 1996), and this ushered in the era of genome-wide association studies (GWAS). It was economically practical to screen thousands of individuals by genotyping hundreds of thousands of common single-nucleotide polymorphisms (SNPs) on microarrays. GWAS are well suited too and have been successful in studying population structure (Price et al., 2010b), anthropomorphic traits (Berndt et al., 2013), targets of natural selection such as variants associated with high-altitude adaptation (Bigham et al., 2009, 2010; Scheinfeldt et al., 2012), and some complex diseases such as Crohn’s disease (Yamazaki et al., 2005) and age-related macular degeneration (Klein et al., 2005). These studies led to hundreds of replicable associated loci that cannot be fully enumerated in this Review. GWAS has perhaps had the most impact in the area of pharmacogenomics, where robust, highly replicable associations have impacted clinical actions. For example, warfarin dose is routinely adjusted based upon VKORC1, CYP2C9, and CYP4F2 genotypes confirmed by GWAS (Takeuchi et al., 2009), which has significantly improved patient outcomes. Yet, most early GWAS yielded few variants with large effect sizes; this was perhaps to be expected, given the heterogeneity of the phenotypes and sample sizes needed to statistically detect signals of association.
The exponentially decreasing cost of next-generation sequencing data generation has put large-scale investigation of rare variation within reach, and there has been a resultant shift in the field of complex disease genetics over the past 5 years. GWAS data strongly suggest that the vast majority of the heritability of complex traits will not be due to a few common variants with low to moderate effects (Schork et al., 2009). Rare variation with large effect sizes is likely contributing a significant proportion to the “missing heritability” of complex traits and disease (Cohen et al., 2006; Manolio, 2009; Zhu et al., 2010). The common disease-common variant versus common disease-rare variant debate remains unresolved. There are still questions that remain as to whether the genetic contribution to common traits can be attributed to an infinite number of common alleles with small effect, a large number of rare alleles with large effects, or some combination of genes and environment (Gibson, 2011). But the evaluation of rare variants in common disease is ongoing.
Variant Detection
The advent of NGS has enabled the inquiry of nearly every base in the genome, and thus techniques to reliably interpret and identify millions of variants are being developed. As will be described below, the advantage of sequencing in this regard is that most variants, common and rare, can be discovered with the appropriate sequencing read coverage, algorithmic methods to identify the variants, and a sufficient careful orthogonal validation to confirm true from false positives. The exception to this discovery potential is due to the reliance on alignment to the Human Genome Reference sequence, which is the first step to analysis of NGS data, as this reference does not contain the entirety of novel genome content across all humans. Numerous variant-calling algorithms have been developed for the detection and genotyping of germline SNPs (DePristo et al., 2011; Koboldt et al., 2009; Li et al., 2008; McKenna et al., 2010; Shen et al., 2010) and small indels (Emde et al., 2012; Leone et al., 2013; Ye et al., 2009) in high-throughput sequencing data. Once detected, these variants can be analyzed in case-control studies using the same methods that have been developed for GWAS.
Rare Variation and Burden Testing
However, unlike GWAS (which examines common mutations), sequencing facilitates the discovery of rare mutations that, combined with the continuing unexplained genetic contributions to complex phenotypes from GWAS (Manolio et al., 2009), has sparked intense interest in measuring the association of rare variation with complex phenotypes. This interest has given rise to a variety of statistical tests with varying strategies for detecting association of rare variation with phenotype (Chen et al., 2013; Han and Pan, 2010; Ionita-Laza et al., 2013; Lee et al., 2012a, 2012b; Li and Leal, 2008; Liu and Leal, 2010; Madsen and Browning, 2009; Neale et al., 2011; Oualkacha et al., 2013; Price et al., 2010a; Wu et al., 2011; Zhang et al., 2011). In any single gene, there are a large number of rare variants due to recent human population growth (Coventry et al., 2010; Nelson et al., 2012; Tennessen et al., 2012), and there may be many nonassociated variants in a gene. Furthermore, even in large cohorts, there may not be enough individuals with a given variant to achieve statistical significance.
To deal with the aforementioned challenge, all of these types of tests share the common feature that they group or collapse rare variation, usually by gene, in order to increase statistical power (see Wu et al., 2013 for a recent review). Early tests (such as the cohort allelic sums test [Morgenthaler and Thilly, 2007] and the combined multivariate collapsing method [Li and Leal, 2008]) assumed that each variant had the same direction of effect and, in addition, required a fixed minor allele frequency cutoff to define which variants to include; but these assumptions are not always valid or optimal. Further innovations have allowed for weighting of individual variants (for example, by variant frequency in the weighted sum statistic [Madsen and Browning, 2009] or the data [Han and Pan, 2010; Lin and Tang, 2011; Wu et al., 2011; Zhang et al., 2011]), variants with heterogeneous direction of effect (Han and Pan, 2010; Lin and Tang, 2011; Neale et al., 2011; Wu et al., 2011; Zhang et al., 2011), and selection of the ideal frequency cutoff for rare variants (Price et al., 2010a). Though this remains an active area of research, the SKAT family of tests (Chen et al., 2013; Ionita-Laza et al., 2013; Lee et al., 2012a, 2012b; Oualkacha et al., 2013; Wu et al., 2011) has emerged as one of the most popular. SKAT and its variants allow for inclusion of covariates for managing both case-control and quantitative data and family or unrelated data, and they are computationally undemanding. Although the initial version of SKAT lost power in cases in which all variants in a gene have the same direction of effect, the newer SKAT-O (Lee et al., 2012a) test combines a test handling bidirectional effects and a test handling unidirectional effects to achieve excellent power in either case.
Identifying De Novo Mutations
The rarest of variants are de novo mutations: those variants that arise first in an individual. They have tremendous relevance for disease biology, as they are more likely to have functional consequences in rare diseases. Characterizing these mutations also allows for the estimation of the baseline human mutation rate as well as its correlation to parental age (Abecasis et al., 2010; Kong et al., 2012). An entire class of computational tools has arisen that utilize both sequencing data and pedigree information to identify de novo mutations genome wide. Most of these tools currently deal with trios (mother, father, and child) only and can identify de novo variants arising in the children (Cartwright et al., 2012; http://sourceforge.net/p/denovogear/wiki/Home/; Li et al., 2012; Li, 2011). Because sequencing reads have a higher error rate than traditional genotyping, these tools incorporate information about coverage, the sequencing error rate, the expected de novo mutation rate, and family relationships.
Although all of these tools identify potential de novo mutations, there remain significant feature differences between them, and no single tool has yet emerged as the frontrunner. In addition, only Samtools, DeNovoGear (DNG), and GATK can also predict de novo indels. Both DNG and Polymutt can handle larger pedigrees, with DNG able to handle multiple siblings and Polymutt able to handle arbitrarily large pedigrees.
Studying Rare Mendelian Disorders
Rare monogenic disorders have provided unique opportunities to identify disease genes in humans. Traditionally, such disorders were studied by positional cloning or candidate gene approaches. Determining their molecular basis, however, was often hindered by small kindred sizes, genetic heterogeneity, and diagnostic classifications that may not reflect molecular pathogenesis. However, high-throughput sequencing of the full set of protein-coding genes—the exome—helps to overcome these obstacles by screening thousands of genes in a single experiment. Although this limits the types of mutations that can be discovered, rare coding variants that are predicted to have significant functional consequences can be discovered (Bamshad et al., 2011). In fact, it is estimated that, in ~60% of projects, exome sequencing will identify new Mendelian disease genes (Gilissen et al., 2012), and it is likely this approach also will contribute to complex disease genetics. Hence, the exome represents an enriched target space to identify rare variants with large effect sizes, as opposed to GWAS, wherein variants have low effect sizes.
The analytical approach applied to most exome sequencing studies of rare disorders is relatively straightforward. First, genetic variants shared by affected individuals (or segregating with a phenotype, in family studies) are collected. Hundreds or thousands of variants might be in this initial set. These are filtered using information from public databases (e.g., dbSNP [Sherry et al., 2001]) to remove common polymorphisms, based on the expectation that causal mutations will be extremely rare in human populations. Next, annotation with gene structure information and bioinformatics programs (e.g., SIFT, Polyphen, CONDEL) further restricts the list of candidates to those most likely to affect an encoded protein. Ideally, these sequential filtering steps reduce the list to a handful of candidate causal variants, which can be further evaluated with mutation screening (in other family members or unrelated, affected individuals), pathway analysis, and functional validation.
Somatic Variant Detection
The comparison of an individual’s cancer genome to the normal genome (derived from an unaffected tissue DNA) provides a comprehensive description of the somatic changes that have occurred in the transition from normal to cancerous cells. WGS approaches to somatic variant detection are more challenging due to the size of the data and the numerous types of variants that can be discovered by different algorithmic predictors, relative to WES. However, structural variants, which are most difficult to predict accurately and with a reasonable false positive rate, occur frequently in cancer genomes and only can be discovered from WGS data. With an increasing focus on characterizing cancer heterogeneity, discussed below, the ability of somatic variant detection algorithms to predict low-frequency single-nucleotide variants (SNVs) in cancer cell populations is becoming critically important. There are several new algorithms with this capability, including Strelka (Saunders et al., 2012), Var-Scan 2 (Koboldt et al., 2012), and MuTect (Cibulskis et al., 2013). Strelka implements a Bayesian approach that treats the tumor and normal allele frequencies as continuous variables. In particular, the normal sample is represented as a mixture of diploid germline variation with noise, and the tumor samples are represented as a mixture of the normal sample with somatic variation. This approach is meant to provide robust call sensitivity on low-purity samples and, as such, provides the same robust sensitivity for low-level variants. Accuracy around indel detection is achieved by Strelka by jointly performing indel search and read realignment in the context of both samples. VarScan 2 is a somatic variation version of the original VarScan algorithm that applies heuristic methods and a statistical test to detect SNVs and indels and their somatic status by simultaneously analyzing the tumor and normal data. In addition, VarScan 2 can identify both LOH and somatic copy number alterations as deviations from the log ratio of sequence coverage depth within the pair that are quantified statistically. MuTect takes input data from matched tumor and normal DNA alignments and removes low-quality sequence data. Variant detection is performed in the tumor data by a Bayesian classifier, filters to remove false positives due to sequencing artifacts that are not captured by the prior error-model-based filters, and designates variants as somatic or germline using a second Bayesian classifier. The step to remove rare sources of false positives uses a panel of normal samples filter that represents rare error modes only detectable from the comparison to additional samples.
Exciting Biological Insights from Recent Studies
Rare Inherited Disorders
Although next-generation sequencing has impacted the human genetics field as a whole, few areas have benefited more than the study of rare genetic diseases. Some of the earliest applications of NGS to Mendelian disorders (Table 1) demonstrated that it was possible to identify disease-causing genes by sequencing the exomes of a few unrelated individuals (Gilissen et al., 2010; Hoischen et al., 2010; Lalonde et al., 2010; Ng et al., 2010a, 2010b) or affected family members (Bilgüvar et al., 2010; Bolze et al., 2010; Johnson et al., 2010; Krawitz et al., 2010; Musunuru et al., 2010; Walsh et al., 2010; Wang et al., 2010). Even the exome sequence of a single index case proved sufficient for genetic diagnosis for some disorders when information about the molecular underpinnings of the disease was known. For example, prioritization of mitochondrial proteins helped to identify ACAD9 in a case with complex I deficiency (Haack et al., 2010), whereas prior evidence linking STIM1 to recessive immunodeficiency helped to implicate this gene in a pediatric case with classic Kaposi sarcoma associated with human herpesvirus 8 infection (Byun et al., 2010).
Table 1.
OMIM Phenotypes for which the Molecular Basis Is Known, 2007 and 2013
| Inheritance Pattern | January 2007 | July 2013 |
|---|---|---|
| Autosomal | 1,851 | 3,525 |
| X Linked | 169 | 277 |
| Y Linked | 2 | 4 |
| Mitochondrial | 26 | 28 |
| Total | 2,048 | 3,834 |
The impact of NGS technologies on rare genetic diseases is further evidenced by the growth of the Online Mendelian Inheritance in Man (OMIM) database (McKusick, 2007), in which the number of inherited phenotypes for which the molecular basis is known has nearly doubled since 2007 (Table 2). The number of genes associated with rare diseases, too, has grown at an impressive rate. Yet for many disorders, elucidation of the genetic basis has outstripped an understanding of the molecular and pathological mechanisms of disease. More work will be required to determine the precise relationship between genotype and phenotype.
Table 2.
Disease-Causing Genes Identified by Exome Sequencing Studies, 2009–2010
| Gene | Disorder | Individuals | Citation |
|---|---|---|---|
| DHODH | Miller syndrome | four affected from three kindreds | (Ng et al., 2010b) |
| FLVCR2 | Fowler syndrome | two unrelated | (Lalonde et al., 2010) |
| GPSM2 | Nonsyndromic hearing loss | one proband | (Walsh et al., 2010) |
| MLL2 | Kabuki syndrome | ten unrelated | (Ng et al., 2010a) |
| WDR62 | Severe brain malformations | one proband | (Bilgüvar et al., 2010) |
| PIGV | Hyperphosphatasia mental retardation | three siblings | (Krawitz et al., 2010) |
| WDR35 | Sensenbrenner syndrome | two unrelated | (Gilissen et al., 2010) |
| STIM1 | Kaposi sarcoma | one patient | (Byun et al., 2010) |
| ANGPTL3 | Familial combined hypolipidemia | two family members | (Musunuru et al., 2010) |
| ACAD9 | Complex I deficiency | one patient | (Haack et al., 2010) |
| SETBP1 | Schinzel-Giedion syndrome | four unrelated | (Hoischen et al., 2010) |
| TGM6 | Spinocerebellar ataxia | four family members | (Wang et al., 2010) |
| FADD | Autoimmune lymphoproliferative syndrome | one proband | (Bolze et al., 2010) |
| VCP | Familial ALS | two family members | (Johnson et al., 2010) |
Lessons from Mendelian Disease Studies
Although NGS offers a powerful strategy to search for Mendelian disease genes, it is important to realize that many such studies fail despite sufficient numbers of samples. One failure occurs when the causal variant is found but is deemed nonpathogenic. While the majority of known disease-causing mutations affect highly conserved protein residues, other pathogenic mechanisms—such as synonymous changes of rare codons that affect the rate of cotranslational folding (Kimchi-Sarfaty et al., 2007)— may be responsible but not ascribed importance. This emphasizes the need for better functional assays of discovered variants.
It is also possible to miss a causal variant. Even with NGS and hybrid capture, ~5% of target coding bases do not achieve sufficient coverage for reliable variant detection. Furthermore, with adequate coverage, certain types of mutations (e.g., inversions, duplications, and other structural variants) remain challenging to detect. Causal mutations also may reside outside of the regions targeted for exome sequencing. Nearly half of familial ALS in Finnish populations, for example, is caused by a hexanucleotide repeat expansion in the intron of the C9orf72 gene (Renton et al., 2011).
Failure also can result when one of the underlying assumptions was incorrect. Genetic and phenotypic heterogeneity can hinder correct diagnosis of cases, or the assumed mode of inheritance (and therefore expected genotype pattern) could be incorrect. In retinitis pigmentosa (RP), for example, around 8.5% of families provisionally diagnosed with autosomal dominant disease truly have X-linked RP (Churchill et al., 2013).
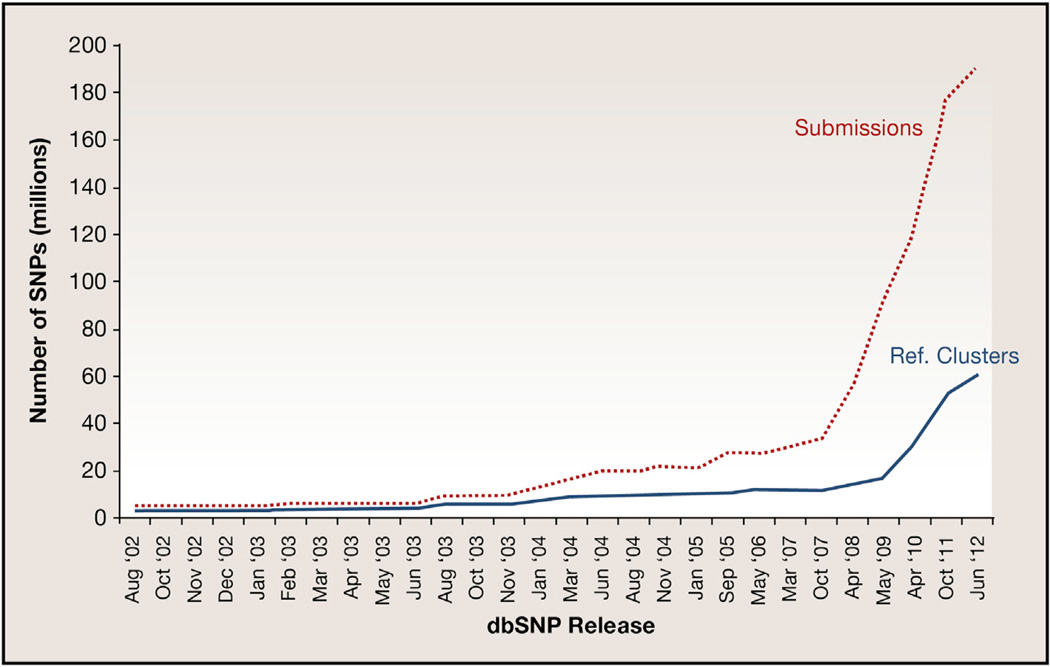
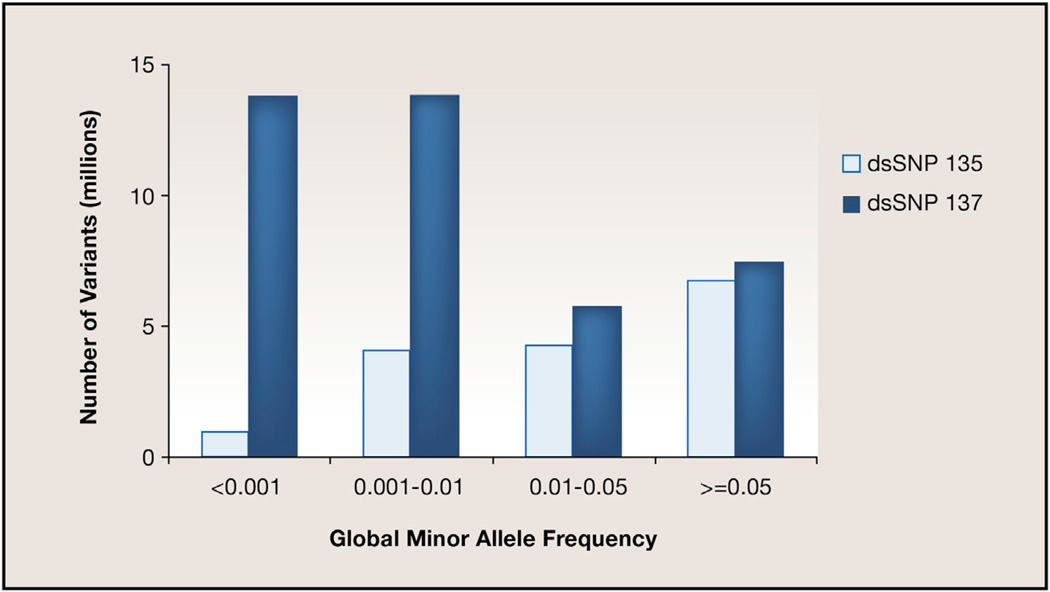
In addition, the reliance on public databases such as dbSNP may confound some analyses of NGS data. The number of known variants in the human genome has risen dramatically over the past decade (Figure 1), fueled in large part by the advent of NGS technologies. Intriguingly, although the submissions from 2007 to 2012 grew almost exponentially, the number of unique reference variants (RefSNPs) followed a more linear growth. Further, a comparison of the global minor allele frequency (GMAF) distribution between dbSNP builds 135 (October 2011) and 137 (June 2012) demonstrates that most of the recent growth came from variants that were rare (GMAF < 0.01) or extremely rare (GMAF < 0.001) in human populations (Figure 2).
Figure 1. Growth in the Numbers of dbSNP Variants in the Human Genome.
Increases in the numbers of SNPs submitted (dotted line) and cataloged as unique reference variants (solid line) in dbSNP are charted over the periodic database releases from August 2002 until the most recent release in June 2012. As indicated by the two lines, while overall submissions have increased exponentially since 2008 (when large projects such as the 1,000 Genomes Project began), the number of unique variants has not increased at a comparable rate.
Figure 2. dbSNP Growth due to Rare Variant Discovery.
This graphic illustrates the amount of rare and extremely rare variant discovery in two recent releases of the NCBI dbSNP database, where a global minor allele frequency of >0.05 is considered a common variant. As indicated, rare variant discovery has increased dramatically in the most recent build of dbSNP (137).
These trends suggest that the majority of common sequence variants in humans have already been reported, and those that remain undiscovered tend to be rare, perhaps specific to an individual or population. This has important implications for studies of rare genetic diseases. The ponderous size of dbSNP certainly makes it a powerful discriminatory tool for common variation. However, it also suggests that dbSNP filtering approaches must be applied with caution because dbSNP entries are associated with disease—variants from Online OMIM (McKusick, 2007) or mutations from the Catalogue of Somatic Mutations in Cancer (COSMIC) (Forbes et al., 2010, 2011) —and a growing number are too rare to exclude from consideration.
Sequencing under GWAS Peaks
One way to leverage the results from GWAS and linkage studies to identify rare variation is to perform targeted sequencing of the regions identified under significant peaks. This strategy has been used to identify a rare variant in a gene under a linkage peak where common SNPs could not explain the variance in the phenotype (Bowden et al., 2010). In this study, common polymorphisms in the ADIPOQ gene that are highly associated with circulating plasma adiponectin levels in European populations were minimally associated with plasma adiponectin levels in Hispanic families; however, a rare coding mutation was identified that contributes up to 17% of the observed variance in Hispanic plasma adiponectin levels. Additionally, Wang et al. (2013) sequenced exons of >1,000 genes identified from GWAS linkage peaks that impact human stature. Using a pooled sample strategy of individuals who were significantly shorter than the average population but were not diagnosed with any known syndrome affecting height or with any endocrinological deficiency, the researchers identified unique rare nonsynonymous and splicing mutations. In a similar study design, researchers were able to narrow a large 288 Kbp region identified from GWAS of multiple sclerosis to an 86.5 Kbp haplotype block containing 42 SNPs, using targeted capture and NGS (Cortes et al., 2013).
Family Studies of Complex Disease
There has been a return to family-based experimental designs for complex disease genetics recently, as it is expected that many members of the same family will carry a particular rare variant; hence, the number of individuals needed for rare variant discovery is much smaller than in cohorts of unrelated individuals (Bailey-Wilson and Wilson, 2011). Using a combination of exome and whole-genome sequencing of affected individuals in consanguineous families, researchers can use homozygosity mapping to identify and characterize the variants contributing to genetically heterogeneous disorders. Nonconsanguineous, large multigenerational, and multiplex pedigrees can also be used to identify rare inherited variants. For example, Weedon et al. (2011) identified a novel heterozygous mutation in DYNC1H1, segregating in a four-generation family affected with Charcot-Marie-Tooth disease by using whole-exome sequencing (WES). Similarly, WES was performed on a three-generation family with multiple individuals affected with pulmonary arterial hypertension who did not carry the canonical TGF-b mutation (Austin et al., 2012). WES revealed a frameshift mutation in caveolin-1 (CAV1) that reduced the normal caveo-lin-1 in the endothelial cell layer of the small arteries. In many cases, the variants identified in these studies can also be independently validated in other cohorts. For example, WES of a large pedigree identified a missense mutation in the affected individuals that also segregated with other families suffering from late-onset Parkinson disease (Vilariño-Güell et al., 2011; Zimprich et al., 2011), thus bolstering the significance of the association. In some studies, WES results provide insights into the biological pathways involved in disease susceptibility and/or pathogenicity. For example, Timms et al. (2013) analyzed the exomes of multiplex families with schizophrenia and identified rare coding variants in N-methyl-D-aspartate (NMDA) receptor genes in all of the families. Although the variants were dispersed over many genes, this pathway was significantly enriched for rare, deleterious mutations and suggested a possible role for glutamate signaling in the pathogenesis of schizophrenia.
De Novo Mutation Studies
Although genomic research in the past decade has largely emphasized inherited variation, NGS technologies also allow us to study, at base-pair resolution, the mutational processes that occur in humans from one generation to the next. Family-based WGS studies have shown that each individual’s genome harbors ~74 germline de novo mutations (DNMs) (Conrad et al., 2011). These mutations are potentially more deleterious because they have not been subject to natural selection and therefore are of considerable interest for sporadic diseases.
Neurological and developmental disorders in particular highlight the impact of DNMs on disease risk. Exome sequencing revealed rare de novo protein-altering mutations in seven of ten individuals with idiopathic intellectual disability (ID) affecting nine different genes (Vissers et al., 2010). Four large-scale studies (Iossifov et al., 2012; Neale et al., 2012; O’Roak et al., 2012; Sanders et al., 2012) evaluated the impact of DNMs in autism spectrum disorder (ASD) via exome sequencing of family quartets (patient, parents, and an unaffected sibling). Each study included >100 families and found that DNM rates were consistently higher in patients than in their unaffected siblings. Similar WES approaches have implicated genes expressed in the developing heart for sporadic congenital heart disease (Zaidi et al., 2013) and genes encoding chromatin regulators for sporadic ALS (Chesi et al., 2013). De novo mutational paradigms have also been suggested by exome sequencing in sporadic psychiatric disorders, such as schizophrenia (Girard et al., 2011; Xu et al., 2012). These findings collectively support a role for de novo mutational processes in sporadic disorders and highlight the extraordinary locus heterogeneity underlying susceptibility to complex diseases.
The application of NGS to both rare and common genetic diseases has offered many insights into disease etiology that undoubtedly merit deeper investigation. Taken together, these studies have also served to highlight our incomplete understanding of the molecular mechanisms by which mutations cause disease. Nevertheless, it seems likely that applying NGS to uncover the genetic underpinnings of disease will help us to better understand the complex relationship between genotype and phenotype.
Cancer Genomics Discovery
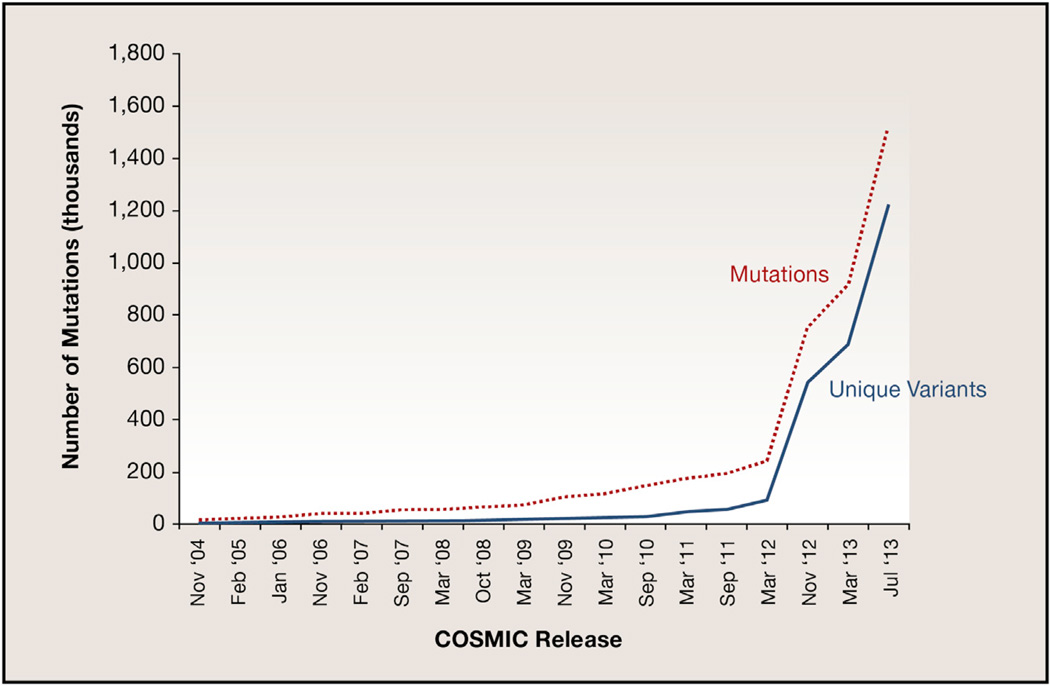
Over the past two years, the growth in cancer genomics discovery due to NGS is unprecedented, with multiple examples of large-scale WGS- or WES-based studies published in the literature for both adult and pediatric cancer types. The growth in our knowledge of the genes frequently mutated in cancer genomes is illustrated in Figure 3, based on the number of new mutations deposited in COSMIC (Forbes et al., 2010, 2011). Here, the number of unique variants identified in tumor genomes stands in stark contrast to those in germline DNA shown in Figure 1. Namely, in dbSNP, there is a clear saturation effect because the majority of variants in any individual genome are shared with other members of the population (and thus already in dbSNP). In COSMIC, however, the number of unique variants closely mirrors the number of mutations submitted, reflecting the fact that most mutations in a tumor genome are private to that tumor.
Figure 3. Growth in COSMIC Database Reports of Identified and Unique Mutations.
Increases in the numbers of mutations and unique variants identified from DNA sequencing of cancer samples as cataloged in the COSMIC database, from November 2004 until the most recent release in July 2013. Note that the numbers of unique variants identified are increasing at a rate equal to the numbers of mutations discovered.
Cancer Genome Heterogeneity
For >100 years, the view of cancer cells through the pathologist’s microscope has indicated that not all cancer cells in a tissue block are entirely similar. Several groups, using the digital nature of NGS data, now have proven this “heterogeneity” of cancer cells at the genomic level. Initially, genomic heterogeneity was demonstrated by copy number comparisons between primary and metastatic disease (Campbell et al., 2010) and by whole-genome amplification and low-coverage sequencing of amplified genomic DNA from single breast cancer cells (Navin et al., 2011). Within the past year, published studies using either WES or WGS have demonstrated the changes in genomic heterogeneity in cancers over the primary-to-relapse/metastatic transition or have characterized heterogeneity with primary tumor specimens. Specifically, these changes are determined by comparing the associated changes in the percentage of tumor cells carrying specific mutations detected by deep coverage NGS data during disease progression. These studies evoke an evolutionary aspect to cancer’s response to survival pressures, including therapy, and have fueled interest in better under- standing the genomes of patients who are likely to recur in their disease.
Early in 2012, Ding et al. described changes in heterogeneity and subclonal architecture of primary acute myeloid leukemia (AML) samples compared to their matched first relapse samples for eight patients (Ding et al., 2012). Using WGS coupled with secondary deep hybrid capture-based NGS data on variant sites, clusters of mutations defining the genotypes of a founding clone and derived subclones were identified. In each case studied, the primary AML sample was either mono- or multiclonal, whereas the relapse sample was monoclonal and carried the somatic profile of one of the primary subclones, as well as new mutations that were acquired during chemotherapy. An analysis of transversion and transition mutations indicated that all types of transversions were elevated in the relapse samples, a DNA damage phenomenon that is attributable to the use of DNA-damaging chemotherapy agents.
In genomic analyses of renal cell cancers, Gerlinger and colleagues (Gerlinger et al., 2012) studied regional heterogeneity in four advanced tumors and metastases from a clinical trial of everolimus (an inhibitor of mTOR) to evaluate the similarities and differences in the genomic landscapes. Their approach included WES, SNP arrays, and gene expression arrays. Their results indicated a branching evolution of the primary and metastatic tumors studied, with a combination of universally shared and primary region-specific or metastasis-specific private mutations. Unlike the previous study, everolimus was shown to not impact the number and types of new mutations in posttreatment samples studied. A case was made for phenotypic convergent evolution due to spatially separated, distinct mutations in SETD2, KDM5C, and PTEN.
A study in breast cancer heterogeneity utilized data from 20 breast cancers selected across the different molecular subtypes, one of which was sequenced to 188-fold depth to provide sufficient sensitivity for heterogeneity analysis (Nik-Zainal et al., 2012). Much like the AML study mentioned above, clustering of mutations sharing similar variant fractions from high-coverage data was performed to identify the subclones. The clusters were further refined by application of a Bayesian Dirichlet process, and further associations were made to identify a hierarchy of mutational events in the natural history of the cancer’s development.
Prediction of Targeted Therapy/Actionable Mutations
Since the earliest descriptions of specific mutations in EGFR predicting response to small-molecule inhibitors such as tyro-sine kinase inhibitors (Lynch et al., 2004; Paez et al., 2004; Pao et al., 2004), the association of somatic mutations to drug response has been of increasing interest. The use of NGS technologies in this regard has several advantages over the original methods (PCR and Sanger fluorescent sequencing) used to acquire these data. Namely, the NGS-based inquiries required for discovering the gene-therapy association can be less hypothesis driven and examine all genes, the associated cost to generate the data for each patient sample is both less expensive and more rapidly obtained, and the ability to detect specific types of mutations such as insertions or deletions of one or several nucleotides is facilitated by NGS. The first aspect is important because most small-molecule therapies target a range of mutated proteins, so multiple genes must be tested in each patient. The second aspect is important because these queries are now approaching clinical usage wherein identification of appropriate therapy(ies) must happen in a 2–3 week period to be applicable to patient care. Lastly, although small insertion/ deletion mutations are rarer than single-nucleotide substitutions, their impact on the resulting protein may be more profound. Because Sanger sequencing typically fails to detect these variants, it is both likely that the frequency of these mutations is underestimated and certain that their response to therapy is less well understood as a result.
One downside of the use of targeted small-molecule inhibitors is that many patients experience an initial complete pathologic response or at least stable disease but then acquire resistance to the therapy and progress (Engelman et al., 2007). This phenomenon has mainly been studied at the protein level (Girotti et al., 2013; Prahallad et al., 2012) or by focused sequencing (Sequist et al., 2011). Here, results often demonstrate that the cellular pathway blockade affected by targeted therapy is circumvented by new mutations and/or overexpression either of the targeted gene or of another gene in the same pathway. Given these discoveries, it remains to be demonstrated by deep NGS or single-cell sequencing of progression disease biopsies whether the mutations that enable circumvention of the blockade are pre-existing in a minor proportion of tumor cells or are new mutations that arise in response to the pathway blockade.
Circulating Tumor DNA Analysis
Many solid tumors shed cells and/or DNA into the blood stream at very low levels that are thought to fluctuate with increases or decreases in the disease burden of the patient. Hence, the ability to detect these changes with high sensitivity poses an interesting and potentially powerful disease-monitoring capability that likely would complement imaging modalities such as CT or MRI but at much lower cost and with lower associated morbidities (Diehl et al., 2005; Diehl et al., 2008; Swisher et al., 2005). In this regard, several groups have recently published manuscripts describing the selective capture of circulating tumor cells (CTCs) or the amplification and sequencing of circulating tumor DNA or RNA. This so-called “liquid biopsy” approach using plasma can detect the predominant somatic mutations for that tumor type (Forshew et al., 2012), or if chromosomal translocations or structural variants already are known from prior characterization of the cancer genome, PCR primers can be designed to amplify the tumor-specific products for NGS and analysis (Dawson et al., 2013; Leary et al., 2012). Another recently published example of this type of detection by NGS involved the detection of ovarian or endometrial cancer by gene-specific assays of PAP test samples (Kinde et al., 2013).
Noninvasive Prenatal Testing
As mentioned, clinical use of NGS in cancer diagnosis, therapeutic decision making, and progression monitoring is poised for introduction. Several large academic centers and a handful of commercial entities are offering NGS-based assays in the CLIA-regulated environment. An NGS-based clinical assay that already has received widespread adoption is noninvasive prenatal testing for chromosomal abnormality diagnosis using samples such as maternal blood. In 1997, Lo et al. demonstrated that male sex could be determined from circulating fetal DNA in maternal plasma and serum samples and that the level of circulating fetal DNA increases with gestational age (Lo et al., 1997). However, achieving high sensitivity and specificity of fetal genotype was difficult, given the low levels of fetal DNA and the cost of high-depth sequencing.
With the advent of NGS, resolving the whole genome of a fetus from maternal blood sources became possible. In 2010, Lo et al. sequenced maternal plasma genomic DNA to 65× coverage and then used the parental SNP genotypes (from SNP array data) to distinguish fetal versus maternal sequencing reads (Lo et al., 2010). This elegant proof-of-concept study demonstrated that the entire fetal genome is represented in the maternal plasma. Yet, this approach was limited by the use of a chorionic villus sample and the somewhat circular logic by which parental haplotypes were inferred from common heterozygous SNP genotypes and then used to predict the fetal haplotype, thereby missing a large proportion of the rare variation. In addition, the authors were unable to detect de novo mutations. To overcome these obstacles, Kitzman et al. used WGS with maternal plasma as well as fosmid clone pooling to resolve long haplotype blocks in the mother (i.e., “phasing”; Kitzman et al., 2012). The paternal genome was sequenced but not phased. This approach achieved >99% genotype accuracy at maternal heterozygous sites when predicting the fetal genotype. In addition, de novo mutations and recombination switch breakpoints were detected using a Hidden Markov model. The results were confirmed by WGS from cord blood after birth. Similarly, Fan et al. (2012) performed WGS and WES with maternal plasma and maternal haplotype resolution via direct deterministic phasing using single cells. The paternal genome was inferred using detection of paternal-specific alleles and imputation, and the fetal genome was resolved to >99% accuracy using molecular counting of parental genotypes in the maternal plasma.
These studies demonstrate the feasibility of prenatal testing at single-nucleotide resolution, but major limitations likely will hinder clinical translation. For example, sequencing to sufficient depth to detect fetal DNA genotypes is still quite expensive. In addition, it is prohibitively expensive and time consuming to routinely create and sequence maternal fosmid pools. As single-molecule sequencing technologies improve, it may be realistic to routinely resolve extended parental haplotypes to assist in fetal genotyping. For the time being, commercial noninvasive NGS prenatal tests are offered, but these only detect common chromosomal aneuploidies such as Trisomy 21.
Concluding Remarks
In summary, next-generation sequencing technologies have had an incredible impact on our knowledge of human genetic diseases over a very short time frame. Whether this trend will continue rests on a variety of issues, some quite complex. For example, the size of whole-human-genome data sets remains large, and this poses significant challenges for data download and storage and for computational infrastructure. Data privacy of human subjects is paramount but is increasingly difficult to control, raising concerns in the public that may inhibit consent by individuals to participate in genetic studies. Ethical aspects overshadow the return of information to study participants and individuals seeking genetic diagnosis due to our remaining ignorance about the pathologic and functional consequences of variation in the human genome. The next few years will determine which applications of NGS are incorporated into the clinical diagnostic setting, many of which may benefit patients but yet may not be covered by insurers. Even as this scenario plays out, it is undoubtedly the case that NGS will continue to be a revolutionary force in basic biomedical and biological genomics inquiry for some time to come.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, et al. Direct selection of human genomic loci by microarray hybridization. Nat. Methods. 2007;4:903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- Austin ED, Ma L, LeDuc C, Berman Rosenzweig E, Borczuk A, Phillips JA, Palomero T, 3rd, Sumazin P, Kim HR, Talati MH, et al. Whole exome sequencing to identify a novel gene (caveolin-1) associated with human pulmonary arterial hypertension. Circ. Cardiovasc. Genet. 2012;5:336–343. doi: 10.1161/CIRCGENETICS.111.961888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Wilson JE, Wilson AF. Linkage analysis in the next-generation sequencing era. Hum. Hered. 2011;72:228–236. doi: 10.1159/000334381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- Berndt SI, Gustafsson S, Mägi R, Ganna A, Wheeler E, Feitosa MF, Justice AE, Monda KL, Croteau-Chonka DC, Day FR, et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat. Genet. 2013;45:501–512. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham AW, Mao X, Mei R, Brutsaert T, Wilson MJ, Julian CG, Parra EJ, Akey JM, Moore LG, Shriver MD. Identifying positive selection candidate loci for high-altitude adaptation in Andean populations. Hum. Genomics. 2009;4:79–90. doi: 10.1186/1479-7364-4-2-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham A, Bauchet M, Pinto D, Mao X, Akey JM, Mei R, Scherer SW, Julian CG, Wilson MJ, López Herráez D, et al. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 2010;6:e1001116. doi: 10.1371/journal.pgen.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgüvar K, Oztürk AK, Louvi A, Kwan KY, Choi M, Tatli B, Yalnizoğlu D, Tüysüz B, Cağlayan AO, Gökben S, et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolze A, Byun M, McDonald D, Morgan NV, Abhyankar A, Premkumar L, Puel A, Bacon CM, Rieux-Laucat F, Pang K, et al. Whole-exome-sequencing-based discovery of human FADD deficiency. Am. J. Hum. Genet. 2010;87:873–881. doi: 10.1016/j.ajhg.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden DW, An SS, Palmer ND, Brown WM, Norris JM, Haffner SM, Hawkins GA, Guo X, Rotter JI, Chen YD, et al. Molecular basis of a linkage peak: exome sequencing and family-based analysis identify a rare genetic variant in the ADIPOQ gene in the IRAS Family Study. Hum. Mol. Genet. 2010;19:4112–4120. doi: 10.1093/hmg/ddq327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun M, Abhyankar A, Lelarge V, Plancoulaine S, Palanduz A, Telhan L, Boisson B, Picard C, Dewell S, Zhao C, et al. Whole-exome sequencing-based discovery of STIM1 deficiency in a child with fatal classic Kaposi sarcoma. J. Exp. Med. 2010;207:2307–2312. doi: 10.1084/jem.20101597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin ML, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright RA, Hussin J, Keebler JE, Stone EA, Awadalla P. A family-based probabilistic method for capturing de novo mutations from high-throughput short-read sequencing data. Stat. Appl. Genet. Mol. Biol. 2012;11:11. doi: 10.2202/1544-6115.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Meigs JB, Dupuis J. Sequence kernel association test for quantitative traits in family samples. Genet. Epidemiol. 2013;37:196–204. doi: 10.1002/gepi.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesi A, Staahl BT, Jovičić A, Couthouis J, Fasolino M, Raphael AR, Yamazaki T, Elias L, Polak M, Kelly C, et al. Exome sequencing to identify de novo mutations in sporadic ALS trios. Nat. Neurosci. 2013;16:851–855. doi: 10.1038/nn.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill JD, Bowne SJ, Sullivan LS, Lewis RA, Wheaton DK, Birch DG, Branham KE, Heckenlively JR, Daiger SP. Mutations in the X-linked retinitis pigmentosa genes RPGR and RP2 found in 8.5% of families with a provisional diagnosis of autosomal dominant retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 2013;54:1411–1416. doi: 10.1167/iovs.12-11541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JC, Pertsemlidis A, Fahmi S, Esmail S, Vega GL, Grundy SM, Hobbs HH. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc. Natl. Acad. Sci. USA. 2006;103:1810–1815. doi: 10.1073/pnas.0508483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Keebler JE, DePristo MA, Lindsay SJ, Zhang Y, Casals F, Idaghdour Y, Hartl CL, Torroja C, Garimella KV, et al. 1000 Genomes Project. Variation in genome-wide mutation rates within and between human families. Nat. Genet. 2011;43:712–714. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes A, Field J, Glazov EA, Hadler J, Stankovich J, Brown MA, Brown MA ANZgene Consortium. Resequencing and fine-mapping of the chromosome 12q13-14 locus associated with multiple sclerosis refines the number of implicated genes. Hum. Mol. Genet. 2013;22:2283–2292. doi: 10.1093/hmg/ddt062. [DOI] [PubMed] [Google Scholar]
- Coventry A, Bull-Otterson LM, Liu X, Clark AG, Maxwell TJ, Crosby J, Hixson JE, Rea TJ, Muzny DM, Lewis LR, et al. Deep rese-quencing reveals excess rare recent variants consistent with explosive population growth. Nat. Commun. 2010;1:131. doi: 10.1038/ncomms1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, Diaz LA, Jr, Goodman SN, David KA, Juhl H, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl. Acad. Sci. USA. 2005;102:16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emde AK, Schulz MH, Weese D, Sun R, Vingron M, Kalscheuer VM, Haas SA, Reinert K. Detecting genomic indel variants with exact breakpoints in single- and paired-end sequencing data using SplazerS. Bioin-formatics. 2012;28:619–627. doi: 10.1093/bioinformatics/bts019. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Fan HC, Gu W, Wang J, Blumenfeld YJ, El-Sayed YY, Quake SR. Non-invasive prenatal measurement of the fetal genome. Nature. 2012;487:320–324. doi: 10.1038/nature11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, Kok CY, Jia M, Ewing R, Menzies A, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38(Database issue):D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(Database issue):D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, Daw-son SJ, Piskorz AM, Jimenez-Linan M, Bentley D, et al. Noninva-sive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 2012;4:136ra168. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. Rare and common variants: twenty arguments. Nat. Rev. Genet. 2011;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilissen C, Arts HH, Hoischen A, Spruijt L, Mans DA, Arts P, van Lier B, Steehouwer M, van Reeuwijk J, Kant SG, et al. Exome sequencing identifies WDR35 variants involved in Sensenbrenner syndrome. Am. J. Hum. Genet. 2010;87:418–423. doi: 10.1016/j.ajhg.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilissen C, Hoischen A, Brunner HG, Veltman JA. Disease gene identification strategies for exome sequencing. Eur. J. Hum. Genet. 2012;20:490–497. doi: 10.1038/ejhg.2011.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard SL, Gauthier J, Noreau A, Xiong L, Zhou S, Jouan L, Dionne-Laporte A, Spiegelman D, Henrion E, Diallo O, et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat. Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- Girotti MR, Pedersen M, Sanchez-Laorden B, Viros A, Turajlic S, Niculescu-Duvaz D, Zambon A, Sinclair J, Hayes A, Gore M, et al. Inhibiting EGF receptor or SRC family kinase signaling overcomes BRAF inhibitor resistance in melanoma. Cancer Discov. 2013;3:158–167. doi: 10.1158/2159-8290.CD-12-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- Haack TB, Danhauser K, Haberberger B, Hoser J, Strecker V, Boehm D, Uziel G, Lamantea E, Invernizzi F, Poulton J, et al. Exome sequencing identifies ACAD9 mutations as a cause of complex I deficiency. Nat. Genet. 2010;42:1131–1134. doi: 10.1038/ng.706. [DOI] [PubMed] [Google Scholar]
- Han F, Pan W. A data-adaptive sum test for disease association with multiple common or rare variants. Hum. Hered. 2010;70:42–54. doi: 10.1159/000288704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington CR, Anderson JR, Chan KK. Apolipoprotein E type epsilon 4 allele frequency is not increased in patients with sporadic inclusion-body myositis. Neurosci. Lett. 1995;183:35–38. doi: 10.1016/0304-3940(94)11108-u. [DOI] [PubMed] [Google Scholar]
- Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, Middle CM, Rodesch MJ, Albert TJ, Hannon GJ, McCombie WR. Genome-wide in situ exon capture for selective resequencing. Nat. Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- Hoischen A, van Bon BW, Gilissen C, Arts P, van Lier B, Steehouwer M, de Vries P, de Reuver R, Wieskamp N, Mortier G, et al. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat. Genet. 2010;42:483–485. doi: 10.1038/ng.581. [DOI] [PubMed] [Google Scholar]
- Ionita-Laza I, Lee S, Makarov V, Buxbaum JD, Lin X. Sequence Kernel Association Tests for the Combined Effect of Rare and Common Variants. Am. J. Hum. Genet. 2013 doi: 10.1016/j.ajhg.2013.04.015. Published online May, 14, 2013. http://dx.doi.org/10.1016/j.ajhg.2013.04.015. [DOI] [PMC free article] [PubMed]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, et al. ITALSGEN Consortium. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Kinde I, Bettegowda C, Wang Y, Wu J, Agrawal N, Shih Ie M, Kurman R, Dao F, Levine DA, Giuntoli R, et al. Evaluation of DNA from the Papanicolaou test to detect ovarian and endometrial cancers. Sci. Transl. Med. 2013;5:167ra164. doi: 10.1126/scitranslmed.3004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzman JO, Snyder MW, Ventura M, Lewis AP, Qiu R, Simmons LE, Gammill HS, Rubens CE, Santillan DA, Murray JC, et al. Nonin-vasive whole-genome sequencing of a human fetus. Sci. Transl. Med. 2012;4:137ra176. doi: 10.1126/scitranslmed.3004323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, Weinstock GM, Wilson RK, Ding L. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinfor-matics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, Gudjonsson SA, Sigurdsson A, Jonasdottir A, Jonasdottir A, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawitz PM, Schweiger MR, Roödelsperger C, Marcelis C, Kölsch U, Meisel C, Stephani F, Kinoshita T, Murakami Y, Bauer S, et al. Identity-by-descent filtering of exome sequence data identifies PIGV mutations in hyperphosphatasia mental retardation syndrome. Nat. Genet. 2010;42:827–829. doi: 10.1038/ng.653. [DOI] [PubMed] [Google Scholar]
- Lalonde E, Albrecht S, Ha KC, Jacob K, Bolduc N, Polychronakos C, Dechelotte P, Majewski J, Jabado N. Unexpectedallelic heterogeneity and spectrum of mutations in Fowler syndrome revealed by next-generation exome sequencing. Hum. Mutat. 2010;31:918–923. doi: 10.1002/humu.21293. [DOI] [PubMed] [Google Scholar]
- Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, O’Shaughnessy J, Kinzler KW, Parmigiani G, Vogelstein B, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci. Transl. Med. 2012;4:162ra154. doi: 10.1126/scitranslmed.3004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, Christiani DC, Wurfel MM, Lin X NHLBI GOExome Sequencing Project—ESP Lung Project Team. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am. J. Hum. Genet. 2012a;91:224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Wu MC, Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics. 2012b;13:762–775. doi: 10.1093/biostatistics/kxs014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone MA, Barizzone N, Esposito F, Lucenti A, Harbo HF, Goris A, Kockum I, Oturai AB, Celius EG, Mero IL, et al. International Multiple Sclerosis Genetics Consortium; Wellcome Trust Case Control Consortium 2; PROGEMUS Group; PROGRESSO Group. Association of genetic markers with CSF oligoclonal bands in multiple sclerosis patients. PLoS ONE. 2013;8:e64408. doi: 10.1371/journal.pone.0064408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Chen W, Zhan X, Busonero F, Sanna S, Sidore C, Cucca F, Kang HM, Abecasis GR. A likelihood-based framework for variant calling and de novo mutation detection in families. PLoS Genet. 2012;8:e1002944. doi: 10.1371/journal.pgen.1002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DY, Tang ZZ. A general framework for detecting disease associations with rare variants in sequencing studies. Am. J. Hum. Genet. 2011;89:354–367. doi: 10.1016/j.ajhg.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DJ, Leal SM. A novel adaptive method for the analysis of next-generation sequencing data to detect complex trait associations with rare variants due to gene main effects and interactions. PLoS Genet. 2010;6:e1001156. doi: 10.1371/journal.pgen.1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, Wainscoat JS. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- Lo YM, Chan KC, Sun H, Chen EZ, Jiang P, Lun FM, Zheng YW, Leung TY, Lau TK, Cantor CR, Chiu RW. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med. 2010;2:61ra91. doi: 10.1126/scitranslmed.3001720. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA. Cohort studies and the genetics of complex disease. Nat. Genet. 2009;41:5–6. doi: 10.1038/ng0109-5. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. Next-generation sequencing platforms. Annu. Rev. Anal. Chem. 2013;6:287–303. doi: 10.1146/annurev-anchem-062012-092628. (Palo Alto Calif.) [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKusick VA. Mendelian Inheritance in Man and its online version, OMIM. Am. J. Hum. Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenthaler S, Thilly WG. A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: a cohort allelic sums test (CAST) Mutat. Res. 2007;615:28–56. doi: 10.1016/j.mrfmmm.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Musunuru K, Pirruccello JP, Do R, Peloso GM, Guiducci C, Sougnez C, Garimella KV, Fisher S, Abreu J, Barry AJ, et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N. Engl. J. Med. 2010;363:2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Rivas MA, Voight BF, Altshuler D, Devlin B, Orho-Melander M, Kathiresan S, Purcell SM, Roeder K, Daly MJ. Testing for an unusual distribution of rare variants. PLoS Genet. 2011;7:e1001322. doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MR, Wegmann D, Ehm MG, Kessner D, St Jean P, Verzilli C, Shen J, Tang Z, Bacanu SA, Fraser D, et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337:100–104. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gil-dersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 2010a;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet. 2010b;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, Raine K, Jones D, Marshall J, Ramakrishna M, et al. Breast Cancer Working Group of the International Cancer Genome Consortium. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oualkacha K, Dastani Z, Li R, Cingolani PE, Spector TD, Hammond CJ, Richards JB, Ciampi A, Greenwood CM. Adjusted sequence kernel association test for rare variants controlling for cryptic and family relatedness. Genet. Epidemiol. 2013;37:366–376. doi: 10.1002/gepi.21725. [DOI] [PubMed] [Google Scholar]
- Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl. Acad. Sci. USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pericak-Vance MA, Bebout JL, Gaskell PC, Jr, Yamaoka LH, Hung WY, Alberts MJ, Walker AP, Bartlett RJ, Haynes CA, Welsh KA, et al. Linkage studies in familial Alzheimer disease: evidence for chromosome 19 linkage. Am. J. Hum. Genet. 1991;48:1034–1050. [PMC free article] [PubMed] [Google Scholar]
- Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, Beijersbergen RL, Bardelli A, Bernards R. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- Price AL, Kryukov GV, de Bakker PI, Purcell SM, Staples J, Wei LJ, Sunyaev SR. Pooled association tests for rare variants in exon-resequencing studies. Am. J. Hum. Genet. 2010a;86:832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet. 2010b;11:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. ITALSGEN Consortium. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Rivera CM, Ren B. Mapping human epigenomes. Cell. 2013;155 doi: 10.1016/j.cell.2013.09.011. this issue, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Will-sey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Chee-tham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- Scheinfeldt LB, Soi S, Thompson S, Ranciaro A, Woldemeskel D, Beggs W, Lambert C, Jarvis JP, Abate D, Belay G, Tishkoff SA. Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biol. 2012;13:R1. doi: 10.1186/gb-2012-13-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schork NJ, Murray SS, Frazer KA, Topol EJ. Common vs. rare allele hypotheses for complex diseases. Curr. Opin. Genet. Dev. 2009;19:212–219. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci. Transl. Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Wan Z, Coarfa C, Drabek R, Chen L, Ostrowski EA, Liu Y, Weinstock GM, Wheeler DA, Gibbs RA, Yu F. A SNP discovery method to assess variant allele probability from next-generation resequencing data. Genome Res. 2010;20:273–280. doi: 10.1101/gr.096388.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisher EM, Wollan M, Mahtani SM, Willner JB, Garcia R, Goff BA, King MC. Tumor-specific p53 sequences in blood and peritoneal fluid of women with epithelial ovarian cancer. Am. J. Obstet. Gynecol. 2005;193:662–667. doi: 10.1016/j.ajog.2005.01.054. [DOI] [PubMed] [Google Scholar]
- Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, Whittaker P, Ranganath V, Kumanduri V, McLaren W, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5:e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, et al. Broad GO; Seattle GO; NHLBI Exome Sequencing Project. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timms AE, Dorschner MO, Wechsler J, Choi KY, Kirkwood R, Girirajan S, Baker C, Eichler EE, Korvatska O, Roche KW, et al. Support for the N -methyl-d-aspartate receptor hypofunction hypothesis of schizophrenia from exome sequencing in multiplex families. JAMA Psychiatry. 2013;70:582–590. doi: 10.1001/jamapsychiatry.2013.1195. [DOI] [PubMed] [Google Scholar]
- Vilariño-Güell C, Wider C, Ross OA, Dachsel JC, Kachergus JM, Lincoln SJ, Soto-Ortolaza AI, Cobb SA, Wilhoite GJ, Bacon JA, et al. VPS35 mutations in Parkinson disease. Am. J. Hum. Genet. 2011;89:162–167. doi: 10.1016/j.ajhg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, van Lier B, Arts P, Wieskamp N, del Rosario M, et al. A de novo paradigm for mental retardation. Nat. Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- Walsh T, Shahin H, Elkan-Miller T, Lee MK, Thornton AM, Roeb W, Abu Rayyan A, Loulus S, Avraham KB, King MC, Kanaan M. Whole exome sequencing and homozygosity mapping identify mutation in the cell polarity protein GPSM2 as the cause of nonsyndromic hearing loss DFNB82. Am. J. Hum. Genet. 2010;87:90–94. doi: 10.1016/j.ajhg.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JL, Yang X, Xia K, Hu ZM, Weng L, Jin X, Jiang H, Zhang P, Shen L, Guo JF, et al. TGM6 identified as a novel causative gene of spinocerebellar ataxias using exome sequencing. Brain. 2010;133:3510–3518. doi: 10.1093/brain/awq323. [DOI] [PubMed] [Google Scholar]
- Wang SR, Carmichael H, Andrew SF, Miller TC, Moon JE, Derr MA, Hwa V, Hirschhorn JN, Dauber A. Large Scale Pooled Next-Generation sequencing of 1077 genes to identify genetic causes of short stature. J. Clin. Endocrinol. Metab. 2013;98:E1428–E1437. doi: 10.1210/jc.2013-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN, Hastings R, Caswell R, Xie W, Paszkiewicz K, Antoniadi T, Williams M, King C, Greenhalgh L, Newbury-Ecob R, Ellard S. Exome sequencing identifies a DYNC1H1 mutation in a large pedigree with dominant axonal Charcot-Marie-Tooth disease. Am. J. Hum. Genet. 2011;89:308–312. doi: 10.1016/j.ajhg.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Wang L, Ye Y, Aakre JA, Pu X, Chang GC, Yang PC, Roth JA, Marks RS, Lippman SM, et al. Genome-wide association study of genetic predictors of overall survival for non-small cell lung cancer in never smokers. Cancer Res. 2013;73:4028–4038. doi: 10.1158/0008-5472.CAN-12-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Ionita-Laza I, Roos JL, Boone B, Woodrick S, Sun Y, Levy S, Gogos JA, Karayiorgou M. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat. Genet. 2012;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki K, McGovern D, Ragoussis J, Paolucci M, Butler H, Jewell D, Cardon L, Takazoe M, Tanaka T, Ichimori T, et al. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn’s disease. Hum. Mol. Genet. 2005;14:3499–3506. doi: 10.1093/hmg/ddi379. [DOI] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Irvin MR, Arnett DK, Province MA, Borecki I. A data-driven method for identifying rare variants with heterogeneous trait effects. Genet. Epidemiol. 2011;35:679–685. doi: 10.1002/gepi.20618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Feng T, Li Y, Lu Q, Elston RC. Detecting rare variants for complex traits using family and unrelated data. Genet. Epidemiol. 2010;34:171–187. doi: 10.1002/gepi.20449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A, Benet-Pagès A, Struhal W, Graf E, Eck SH, Offman MN, Haubenberger D, Spielberger S, Schulte EC, Lichtner P, et al. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am. J. Hum. Genet. 2011;89:168–175. doi: 10.1016/j.ajhg.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]