Summary
Mice carrying mutations in multiple genes are traditionally generated by sequential recombination in embryonic stem cells and/or time-consuming intercrossing of mice with a single mutation. The CRISPR/Cas system has been adapted as an efficient gene-targeting technology with the potential for multiplexed genome editing. We demonstrate that CRISPR/Cas-mediated gene editing allows the simultaneous disruption of five genes (Tet1, 2, 3, Sry, Uty - 8 alleles) in mouse embryonic stem (ES) cells with high efficiency. Coinjection of Cas9 mRNA and single-guide RNAs (sgRNAs) targeting Tet1 and Tet2 into zygotes generated mice with biallelic mutations in both genes with an efficiency of 80%. Finally, we show that coinjection of Cas9 mRNA/sgRNAs with mutant oligos generated precise point mutations simultaneously in two target genes. Thus, the CRISPR/Cas system allows the one-step generation of animals carrying mutations in multiple genes, an approach that will greatly accelerate the in vivo study of functionally redundant genes and of epistatic gene interactions.
Introduction
Genetically modified mice represent a crucial tool for understanding gene function in development and disease. Mutant mice are conventionally generated by insertional mutagenesis (Copeland and Jenkins, 2010; Kool and Berns, 2009) or by gene-targeting methods (Capecchi, 2005). In conventional gene-targeting methods, mutations are introduced through homologous recombination in mouse embryonic stem (ES) cells. Targeted ES cells injected into wild-type (WT) blastocysts can contribute to the germline of chimeric animals, generating mice containing the targeted gene modification (Capecchi, 2005). It is costly and time consuming to produce single-gene knockout mice and even more so to make double-mutant mice. Moreover, in most other mammalian species, no established ES cell lines are available that contribute efficiently to chimeric animals, which greatly limits the genetic studies in many species.
Alternative methods have been developed to accelerate the process of genome modification by directly injecting DNA or mRNA of site-specific nucleases into the one-cell embryo to generate DNA double-strand break (DSB) at a specified locus in various species (Bogdanove and Voytas, 2011; Carroll et al., 2008; Urnov et al., 2010). DSBs induced by these site-specific nucleases can then be repaired by error-prone nonhomologous end joining (NHEJ) resulting in mutant mice and rats carrying deletions or insertions at the cut site (Carbery et al., 2010; Geurts et al., 2009; Sung et al., 2013; Tesson et al., 2011). If a donor plasmid with homology to the ends flanking the DSB is coinjected, high-fidelity homologous recombination can produce animals with targeted integrations (Cui et al., 2011; Meyer et al., 2010). Because these methods require the complex designs of zinc finger nucleases (ZNFs) or Transcription activator-like effector nucleases (TALENs) for each target gene and because the efficiency of targeting may vary substantially, no multiplexed gene targeting in animals has been reported to date. To dissect the functions of gene family members with redundant functions or to analyze epistatic relationships in genetic pathways, mice with two or more mutated genes are required, prompting the development of efficient technology for the generation of animals carrying multiple mutated genes.
Recently, the type II bacterial CRISPR/Cas system has been demonstrated as an efficient gene-targeting technology with the potential for multiplexed genome editing. Bacteria and archaea have evolved an RNA-based adaptive immune system that uses CRISPR (clustered regularly interspaced short palindromic repeat) and Cas (CRISPR-associated) proteins to detect and destroy invading viruses and plasmids (Horvath and Barrangou, 2010; Wiedenheft et al., 2012). Cas proteins, CRISPR RNAs (crRNAs), and trans-activating crRNA (tracrRNA) form ribonucleoprotein complexes, which target and degrade foreign nucleic acids, guided by crRNAs (Gasiunas et al., 2012; Jinek et al., 2012). It was shown that the Cas9 endonuclease from Streptococcus pyogenes type II CRISPR/Cas system can be programmed to produce sequence-specific DSB in vitro by providing a synthetic single-guide RNA (sgRNA) consisting of a fusion of crRNA and tracrRNA (Jinek et al., 2012). More intriguingly, Cas9 and sgRNA are the only components necessary and sufficient for induction of targeted DNA cleavage in cultured human cells (Cho et al., 2013; Cong et al., 2013; Mali et al., 2013) as well as in zebrafish (Chang et al., 2013; Hwang et al., 2013). A recent report also demonstrated disruption of a GFP transgene in mice using the CRISPR/Cas system (Shen et al., 2013). The ease of design, construction, and delivery of multiple sgRNAs suggest the possibility of multiplexed genome editing in mammals. Indeed, one study demonstrated that two loci separated by 119 bp could be cleaved simultaneously in cultured human cells at a low efficiency (Cong et al., 2013). The extent of achievable multiplexed genome editing has yet to be demonstrated in stem cells as well as in animals. Here, we use the CRISPR/Cas system to drive both NHEJ-based gene disruption and homology directed repair (HDR)-based precise gene editing to achieve highly efficient and simultaneous targeting of multiple genes in stem cells and mice.
Results
Simultaneous Targeting up to Five Genes in ES Cells
To test the possibility of targeting functionally redundant genes from the same gene family, we designed sgRNAs targeting the Ten-eleven translocation (Tet) family members, Tet1, Tet2, and Tet3 (Figure 1A). Tet proteins (Tet1/2/3) convert 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) in various embryonic and adult tissues and mutant mice for each of these three genes have been produced by homologous recombination in ES cells (Dawlaty et al., 2011; Gu et al., 2011; Li et al., 2011; Moran-Crusio et al., 2011). To test whether the CRISPR/Cas system could produce targeted cleavage in the mouse genome, we transfected plasmids expressing both the mammalian-codon-optimized Cas9 and a sgRNA targeting each gene (Cong et al., 2013; Mali et al., 2013) into mouse ES cells and determined the targeted cleavage efficiency by the Surveyor assay (Guschin et al., 2010). All three Cas9-sgRNA transfections produced cleavage at target loci with high efficiency of 36% at Tet1, 48% at Tet2, and 36% at Tet3 (Figure 1B). Because each target locus contains a restriction enzyme recognition site (Figure 1A), we PCR amplified an ∼500 bp fragment around each target site and digested the PCR products with the respective enzyme. A correctly targeted allele will lose the restriction site, which can be detected by failure to cleave upon enzyme treatment. Using this restriction fragment length polymorphism (RFLP) assay, we screened 48 ES cell clones from each single-targeting experiment. Consistent with the Surveyor analysis, a high percentage of ES cell clones were targeted, with a high probability of having both alleles mutated (Figure S1A available online). The results summarized in Table 1 demonstrate that between 65% and 81% of the tested ES cell clones carried mutations in the Tet genes with up to 77% having mutations in both alleles.
Figure 1. Multiplexed Gene Targeting in mouse ES cells.
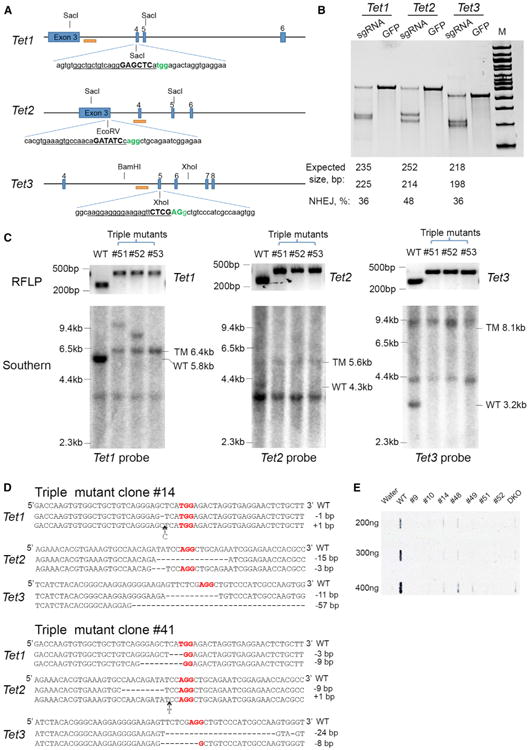
(A) Schematic of the Cas9/sgRNA-targeting sites in Tet1, 2, and 3. The sgRNA-targeting sequence is underlined, and the protospacer-adjacent motif (PAM) sequence is labeled in green. The restriction sites at the target regions are bold and capitalized. Restriction enzymes used for RFLP and Southern blot analysis are shown, and the Southern blot probes are shown as orange boxes.
(B) Surveyor assay for Cas9-mediated cleavage at Tet1, 2, and 3 loci in ES cells.
(C) Genotyping of triple-targeted ES cells, clones 51, 52, and 53 are shown. Upper: RFLP analysis. Tet1 PCR products were digested with SacI, Tet2 PCR products were digested with EcoRV, and Tet3 PCR products were digested with XhoI. Lower: Southern blot analysis. For the Tet1 locus, SacI digested genomic DNA was hybridized with a 5′ probe. Expected fragment size: WT = 5.8 kb, TM (targeted mutation) = 6.4 kb. For the Tet2 locus, SacI, and EcoRV double-digested genomic DNA was hybridized with a 3′ probe. Expected fragment size: WT = 4.3 kb, TM = 5.6 kb. For the Tet3 locus, BamHI and XhoI double-digested genomic DNA was hybridized with a 5′ probe. Expected fragment size: WT = 3.2 kb, TM = 8.1 kb.
(D) The sequence of six mutant alleles in triple-targeted ES cell clone 14 and 41. PAM sequence is labeled in red.
(E) Analysis of 5hmC levels in DNA isolated from triple-targeted ES cell clones by dot blot assay using anti-5hmC antibody. A previously characterized DKO clone derived using traditional method is used as a control. See also Figure S1.
Table 1. CRISPR/Cas-Mediated Gene Targeting in V6.5 ES Cells.
| Gene | Mutant Alleles per Clone/Total Clones Tested | ||||||
|---|---|---|---|---|---|---|---|
| 6 | 5 | 4 | 3 | 2 | 1 | 0 | |
| Tet1 | N/A | 27/48 | 4/48 | 17/48 | |||
| Tet2 | 37/48 | 2/48 | 9/48 | ||||
| Tet3 | 32/48 | 3/48 | 13/48 | ||||
| Tet1+ Tet2 + Tet3 | 20/96 | 16/96 | 2/96 | 2/96 | 1/96 | 0/96 | 55/96 |
Plasmids encoding Cas9 and sgRNAs targeting Tet1, Tet2, and Tet3 were transfected separately (single targeting) or in a pool (triple targeting) into ES cells. The number of total alleles mutated in each ES cell clone is listed from 0 to 2 for single-targeting experiment, and 0 to 6 for triple-targeting experiment. The number of clones containing each specific number of mutated alleles is shown in relation to the total number of clones screened in each experiment. See also Table S1.
The high efficiency of single-gene modification prompted us to test the possibility of targeting all three genes simultaneously. For this we cotransfected ES cells with the constructs expressing Cas9 and three sgRNAs targeting Tet1, 2, and 3. Of 96 clones screened using the RFLP assay, 20 clones were identified as having mutations in all six alleles of the three genes (Figures 1C and S1B and Table 1). To exclude that a PCR bias could give false positive results, we performed Southern blot analysis and confirmed complete agreement with the RFLP results (Figure 1C). We subcloned and sequenced the PCR products of Tet1-, Tet2-, and Tet3-targeted regions to verify that all of eight tested clones carried biallelic mutations in all three genes with most clones displaying two mutant alleles for each gene with small insertions or deletions (indels) at the target site (Figure 1D). To test whether these mutant alleles would abolish the function of Tet proteins, we compared the 5hmC level of targeted clones to WT ES cells. Previously, we reported a depletion of 5hmC in Tet1/Tet2 double-knockout ES cells derived using traditional gene-targeting methods (Dawlaty et al., 2013). As expected from loss of function alleles, we found a significant reduction of 5hmC levels in all clones carrying biallelic mutations in the three genes (Figure 1E).
To further test the potential of multiplexed gene targeting by CRISPR/Cas system, we designed sgRNAs targeting two Y-linked genes, Sry and Uty (Figure S1C). Short PCR products encoding sgRNAs targeting all five genes (Tet1, Tet2, Tet3, Sry, and Uty) were pooled and cotransfected with a Cas9 expressing plasmid and the PGK puroR cassette into ES cells. Of 96 clones that were screened using the RFLP assay, 10% carried mutations in all eight alleles of the five genes (Figure S1D and Table S1), demonstrating the capacity of the CRISP/Cas9 system for highly efficient multiplexed gene targeting.
One-Step Generation of Single-Gene Mutant Mice by Zygote Injection
We tested whether mutant mice could be generated in vivo by direct embryo manipulation. Capped polyadenylated Cas9 mRNA was produced by in vitro transcription and coinjected with sgRNAs. Initially, to determine the optimal concentration of Cas9 mRNA for targeting in vivo, we microinjected varying amounts of Cas9-encoding mRNA with Tet1 targeting sgRNA at constant concentration (20 ng/μl) into pronuclear (PN) stage one-cell mouse embryos and assessed the frequency of altered alleles at the blastocyst stage using the RFLP assay. As expected, higher concentration of Cas9 mRNA led to more efficient gene disruption (Figure S2A). Nevertheless, even embryos injected with the highest amount of Cas9 mRNA (200 ng/μl) showed normal blastocyst development, suggesting low toxicity.
To investigate whether postnatal mice carrying targeted mutations could be generated, we coinjected sgRNAs targeting Tet1or Tet2 with different concentrations of Cas9 mRNA. Blastocysts derived from the injected embryos were transplanted into foster mothers and newborn pups were obtained. As summarized in Table 2, about 10% of the transferred blastocysts developed to birth independent of the RNA concentrations used for injection suggesting low fetal toxicity of the Cas9 mRNA and sgRNA. RFLP, Southern blot, and sequencing analysis demonstrated that between 50 and 90% of the postnatal mice carried biallelic mutations in either target gene (Figures 2A, 2B, and 2C and Table 2).
Table 2. CRISPR/Cas-Mediated Single-Gene Targeting in BDF2 Mice.
| Gene | Cas9/sg RNA (ng/μl) | Blastocysts/Injected Zygotes | Transferred Embryos (Recipients) | Newborns (Dead) | Mutant Alleles per Mouse/Total Mice Testeda | ||
|---|---|---|---|---|---|---|---|
| 2 | 1 | 0 | |||||
| Tet1 | 200/20 | 38/50 | 19 (1) | 2 (0) | 2/2 | 0/2 | 0/2 |
| 100/20 | 50/60 | 25 (1) | 3 (0) | 2/3 | 0/3 | 1/3 | |
| 50/20 | 40/50 | 40 (2) | 8 (3) | 4/7 | 2/7 | 1/7 | |
| 100/50 | 167/198 | 60 (3) | 12 (2) | 9/11 | 1/11 | 1/11 | |
| Tet2 | 100/50 | 176/203 | 108 (5) | 22 (3) | 19/20 | 0/20 | 1/20 |
| Tet3 | 100/50 | 85/112 | 64 (4) | 15 (13) | 9/13 | 2/13 | 2/13 |
Cas9 mRNA and sgRNAs targeting Tet1, Tet2, or Tet3 were injected into fertilized eggs. The blastocysts derived from injected embryos were transplanted into foster mothers and newborn pups were obtained and genotyped. The number of total alleles mutated in each mouse is listed from 0 to 2. The number of mice containing each specific number of mutated alleles is shown in relation to the total number of mice screened in each experiment. See also Table S2.
Some of the pups were cannibalized.
Figure 2. Single- and Double-Gene Targeting In Vivo by Injection into Fertilized Eggs.
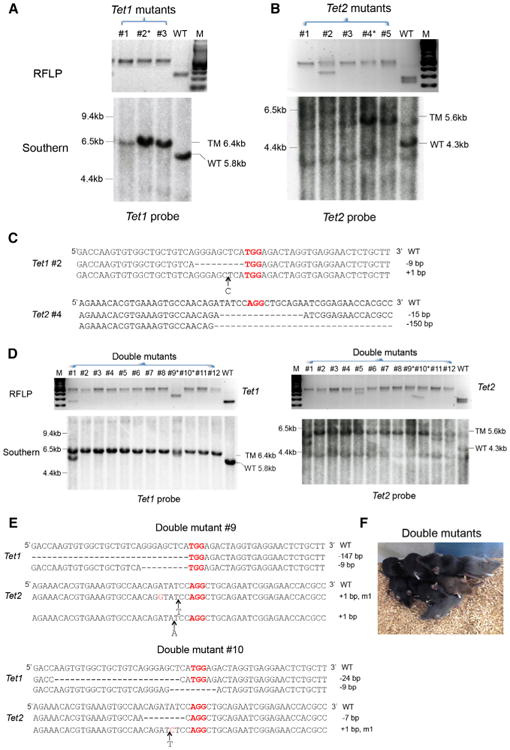
(A) Genotyping of Tet1 single-targeted mice.
(B) Upper: genotyping of Tet2 single-targeted mice. RFLP analysis; lower: Southern blot analysis.
(C) The sequence of both alleles of targeted gene in Tet1 biallelic mutant mouse 2 and Tet2 biallelic mutant mouse 4.
(D) Genotyping of Tet1/Tet2 double-mutant mice. Analysis of mice 1 to 12 is shown. Upper: RFLP analysis; lower: southern blot analysis. The Tet1 locus is displayed on the left and the Tet2 locus on the right.
(E) The sequence of four mutant alleles from double-mutant mouse 9 and 10. PAM sequences are labeled in red.
(F) Three-week-old double-mutant mice. All RFLP and Southern digestions and probes are the same as those used in Figure 1. See also Figures S2 and S3.
Surprisingly, specific Δ9 Tet1 and specific Δ8 and Δ15 Tet2 mutant alleles were repeatedly recovered in independently derived mice. Preferential generation of these alleles is likely caused by a short sequence repeat flanking the DSB (see Figure S2B) consistent with previous reports demonstrating that perfect microhomology sequences flanking the cleavage sites can generate microhomology-mediated precise deletions by end repair mechanism (MMEJ) (McVey and Lee, 2008; Symington and Gautier, 2011) (Figure S2B). A similar observation was also made when TALEN mRNA was injected into one-cell rat embryos (Tesson et al., 2011).
We also derived blastocysts from zygotes injected with Cas9 mRNA and Tet3 sgRNA. Genotyping of the blastocysts demonstrated that of eight embryos three were homozygous and three were heterozygous Tet3 mutants (two failed to amplify) (Figure S2C). Some blastocysts were implanted into foster mothers and, upon C section, we readily identified multiple mice of smaller size (Figure S2D), many of which died soon after delivery. Genotyping shown in Figure S2E indicated that all pups with mutations in both Tet3 alleles died neonatally. Only 2 out of 15 mice survived that were either Tet3 heterozygous mutants or WT (Figure S2F). These results are consistent with the lethal neonatal phenotype of Tet3 knockout mice generated using traditional methods (Gu et al., 2011), although we have not yet established which of the Tet3 mutations produced loss of function rather than hypomorphic alleles.
One-Step Generation of Double-Gene Mutant Mice by Zygote Injection
To test whether Tet1/Tet2 double-mutant mice could be produced from single embryos, we coinjected Tet1 and Tet2 sgRNAs with 20 or 100 ng/μl Cas9 mRNA into zygotes. A total of 28 pups were born from 144 embryos transferred into foster mothers (21% live-birth rate) that had been injected at the zygote stage with high concentrations of RNA (Cas9 mRNA at 100 ng/μl, sgRNAs at 50 ng/μl), consistent with low or no toxicity of the Cas9 mRNA and sgRNAs (Table 3). RFLP, Southern blot analysis, and sequencing identified 22 mice carrying targeted mutations at all four alleles of the Tet1 and Tet2 genes (Figures 2D and 2E) with the remaining mice carrying mutations in a subset of alleles (Table 3). Injection of zygotes with low concentration of RNA (Cas9 mRNA at 20 ng/μl, sgRNAs at 20 ng/μl) yielded 19 pups from 75 transferred embryos (25% live-birth rate), which is a higher survival rate than from embryos injected with 100 ng/μl of Cas9 RNA. Nevertheless, more than 50% of the pups were biallelic Tet1/Tet2 double mutants (Table 3). These results demonstrate that postnatal mice carrying biallelic mutations in two different genes can be generated within one month with high efficiency (Figure 2F).
Table 3. CRISPR/Cas-Mediated Double-Gene Targeting in BDF2 Mice.
| Gene | Cas9/sgRNA (ng/μl) | Blastocyst/Injected Zygotes | Transferred Embryos (Recipients) | Newborns (Dead) | Mutant Alleles per Mouse/Total Mice Testeda | ||||
|---|---|---|---|---|---|---|---|---|---|
| 4 | 3 | 2 | 1 | 0 | |||||
| Tet1 + Tet2 | 100/50 | 194/229 | 144(7) | 31(8) | 22/28 | 4/28 | 1/28 | 1/28 | 0/28 |
| 20/20 | 92/109 | 75(5) | 19(3) | 11/19 | 1/19 | 2/19 | 3/19 | 2/19 | |
Cas9 mRNA and sgRNAs targeting Tet1 and Tet2 were coinjected into fertilized eggs. The blastocysts derived from the injected embryos were transplanted into foster mothers and newborn pups were obtained and genotyped. The number of total alleles mutated in each mouse is listed from 0 to 4 for Tet1 and Tet2. The number of mice containing each specific number of mutated alleles is shown in relation to the number of total mice screened in each experiment.
Some of the pups were cannibalized.
Although the high live-birth rate and normal development of mutant mice suggest low toxicity of CRISPR/Cas9 system, we sought to determine the off-target effects in vivo. Previous work in vitro, in bacteria, and in cultured human cells suggested that the protospacer-adjacent motif (PAM) sequence NGG and the 8 to 12 base “seed sequence” at the 3′ end of the sgRNA are most important for determining the DNA cleavage specificity (Cong et al., 2013; Jiang et al., 2013; Jinek et al., 2012). Based on this rule, only three and four potential off targets exist in mouse genome for Tet1 and Tet2 sgRNA, respectively (Table S2 and Experimental Procedures), with each of them perfectly matching the 12 bp seed sequence at the 3′ end and the NGG PAM sequence of the sgRNA (there is no potential off-target site for Tet3 sgRNA using this prediction rule). From seven double-mutant mice produced from injection with high RNA concentration we PCR amplified 400 to 500 bp fragments from all seven potential off-target loci and found no cleavage in the Surveyor assay (Figure S3), suggesting a high specificity of CRISPR/Cas system.
Multiplexed Precise HDR-Mediated Genome Editing In Vivo
The NHEJ-mediated gene mutations described above produced mutant alleles with different and unpredictable insertions and deletions of variable size. We explored the possibility of precise homology directed repair (HDR)-mediated genome editing by coinjecting Cas9 mRNA, sgRNAs, and single-stranded DNA oligos into one-cell embryos. For this we designed an oligo targeting Tet1 so as to change two base pairs of a SacI restriction site and creating instead an EcoRI site and a second oligo targeting Tet2 with two base pair changes that would convert an EcoRV site into an EcoRI site (Figure 3A). Blastocysts were derived from zygotes injected with Cas9 mRNA and sgRNAs and oligos targeting Tet1 orTet2, respectively. DNA was isolated, amplified, and digested with EcoRI to detect oligo-mediated HDR events. Six out of nine Tet1-targeted embryos and 9 out of 15 Tet2-targeted embryos incorporated an EcoRI site at the respective target locus, with several embryos having both alleles modified (Figure S4A). When Cas9 mRNA, sgRNAs, and single-stranded DNA oligos targeting both Tet1 and Tet2 were coinjected into zygotes, out of 14 embryos, four were identified that were targeted with the oligo at the Tet1 locus, seven that were targeted with the oligo at the Tet2 locus and one embryo (2) that had one allele of each gene correctly modified (Figure S4B). All four alleles of embryo 2 were sequenced, confirming that one allele of each gene contained the 2 bp changes directed by the oligo, whereas the other alleles were disrupted by NHEJ-mediated deletion and insertion (Figure S4C).
Figure 3. Multiplexed HDR-Mediated Genome Editing In Vivo.
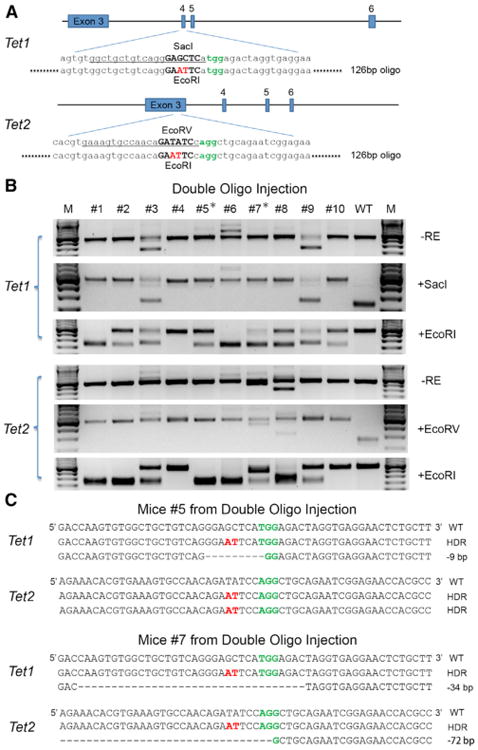
(A) Schematic of the oligo-targeting sites at Tet1 and Tet2 loci. The sgRNA-targeting sequence is underlined, and the PAM sequence is labeled in green. Oligo targeting each gene is shown under the target site, with 2 bp changes labeled in red. Restriction enzyme sites used for RFLP analysis are bold and capitalized.
(B) RFLP analysis of double oligo injection mice with HDR-mediated targeting at the Tet1 and Tet2 loci.
(C) The sequences of both alleles of Tet1 and Tet2 in mouse 5 and 7 show simultaneously HDR-mediated targeting at one allele or two alleles of each gene, and NHEJ-mediated disruption at the other alleles. See also Figure S4.
Blastocysts with double oligo injections were implanted into foster mothers and a total of 10 pups were born from 48 embryos transferred (21% live-birth rate). Upon RFLP analysis using EcoRI, we identified seven mice containing EcoRI sites at the Tet1 locus and eight mice containing EcoRI sites at the Tet2 locus, with six mice containing EcoRI sites at both Tet1 and Tet2 loci (Figure 3B). We also applied RFLP analysis using SacI and EcoRV to Tet1 and Tet2 loci, respectively, showing that all alleles not targeted by oligos contained disruptions, which is in consistent with the high biallelic mutation rate by Cas9 mRNA and sgRNAs injection. These results were confirmed by sequencing demonstrating mutations in all four alleles of mouse 5 and 7 (Figure 3C). Our results demonstrate that mice with HDR-mediated precise mutations in multiple genes can be generated in one step by CRISPR/Cas-mediated genome editing.
Discussion
The genetic manipulation of mice is a crucial approach for the study of development and disease. However, the generation of mice with specific mutations is labor intensive and involves gene targeting by homologous recombination in ES cells, the production of chimeric mice, and, after germline transmission of the targeted ES cells, the interbreeding of heterozygous mice to produce the homozygous experimental animals, a process that may take 6 to 12 months or longer (Capecchi, 2005). To produce mice carrying mutations in several genes requires time-consuming intercrossing of single-mutant mice. Similarly, the generation of ES cells carrying homozygous mutations in several genes is usually achieved by sequential targeting, a process that is labor intensive, necessitating multiple consecutive cloning steps to target the genes and to delete the selectable markers.
As summarized in Figure 4, we have established three different approaches for the generation of mice carrying multiple genetic alterations. We demonstrate that CRISPR/Cas-mediated genome editing in ES cells can generate the simultaneous mutations of several genes with high efficiency, a single-step approach allowing the production of cells with mutations in five different genes (Figure 4A). We chose the three Tet genes as targets because the respective mutant phenotypes have been well defined previously (Dawlaty et al., 2011, 2013; Gu et al., 2011). Cells mutant for Tet1, 2 and 3 were depleted of 5hmC as would be expected for loss of function mutations of the genes (Dawlaty et al., 2013). However, we have not as yet established, which of the Cas9-mediated gene mutations produced loss of function rather than hypomorphic alleles.
Figure 4. Mutiplexed Genome Editing in ES Cells and Mouse.
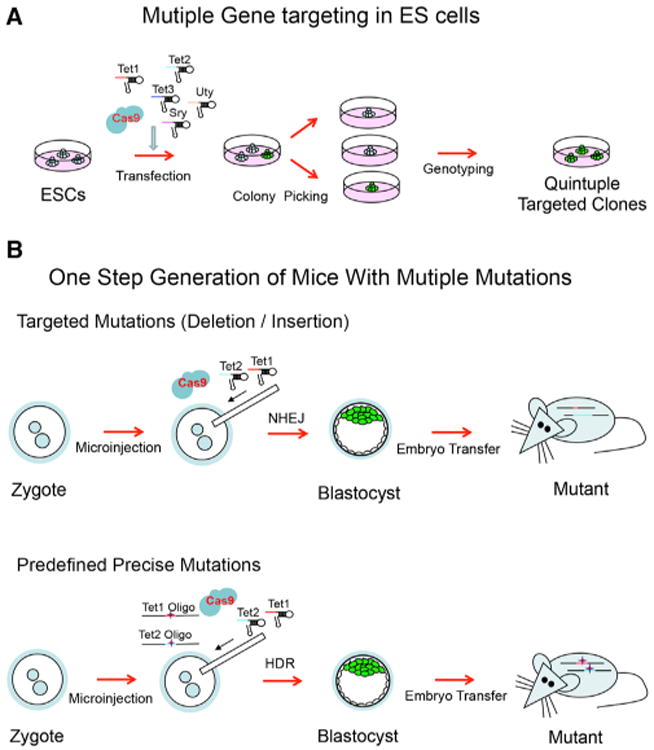
(A) Multiple gene targeting in ES cells.
(B) One-step generation of mice with multiple mutations. Upper: multiple targeted mutations with random indels introduced through NHEJ. Lower: multiple predefined mutations introduced through HDR-mediated repair.
We also show that mouse embryos can be directly modified by injection of Cas9 mRNA and sgRNA into the fertilized egg resulting in the efficient production of mice carrying biallelic mutations in a given gene. More significantly, coinjection of Cas9 with Tet1 and Tet2 sgRNAs into zygotes produced mice that carried mutations in both genes (Figure 4B, upper). We found that up to 95% of newborn mice were biallelic mutant in the targeted gene when single sgRNA was injected and when coinjected with two different sgRNAs, up to 80% carried biallelic mutations in both targeted genes. Thus, mice carrying multiple mutations can be generated within 4 weeks, which is a much shorter time frame than can be achieved by conventional consecutive targeting of genes in ES cells and avoids time-consuming intercrossing of single-mutant mice.
The introduction of DSBs by CRISPR/Cas generates mutant alleles with varying deletions or insertions in contrast to designed precise mutations created by homologous recombination. The introduction of point mutations into human ES cells, cancer cell lines, and mouse by ZNF or TALEN along with DNA oligo has been demonstrated previously (Chen et al., 2011; Soldner et al., 2011; Wefers et al., 2013). We demonstrate that CRISPR/Cas-mediated targeting is useful to generate mutant alleles with predetermined alterations, and coinjection of single-stranded oligos can introduce designed point mutations into two target genes in one step, allowing for multiplexed gene editing in a strictly controlled manner (Figure 4B, lower). It will be of great interest to assess whether this targeting system allows for the production of conditional alleles, or precise insertion of larger DNA fragments such as GFP markers so as to generate conditional knockout and reporter mice for specific genes.
There are several potential limitations of the CRISPR/Cas technology. First, the requirement for a NGG PAM sequence of S. pyogenes Cas9 limits the target space in the mouse genome. It has been shown that the Streptococcus thermophilus LMD-9 Cas9 using different PAM sequence can also induce targeted DNA cleavage in mammalian cells (Cong et al., 2013). Therefore, exploiting different Cas9 proteins may enable to target most of the mouse genome. Second, although the sgRNAs used here showed high targeting efficiency, much work is needed to elucidate the rules for designing sgRNAs with consistent high targeting efficiency, which is essential for multiplexed genome engineering. Third, although our off-target analysis for the seven most likely off targets of Tet1 and Tet2 sgRNAs failed to detect mutations in these loci, it is possible that other mutations were induced following as yet unidentified rules. A more thorough sequencing analysis for a large number of sgRNAs will provide more information about the potential off-target cleavage of the CRISPR/Cas system and lead to a better prediction of potential off-target sites. Last, oligo-mediated repair allows for precise genome editing, but the other allele is often mutated through NHEJ (Figures 3B, 3C, and S4C). We have shown that using lower Cas9 mRNA concentration generates more mice with heterozygous mutations. Therefore, it may be possible to optimize the system for more efficient generation of mice with only one oligo -modified allele. In addition, employment of Cas9 nickase will likely avoid this complication because it mainly induces DNA single-strand break, which is typically repaired through HDR (Cong et al., 2013; Mali et al., 2013).
It is likely that a much larger number of genomic loci than targeted in the present work can be modified simultaneously when pooled sgRNAs are introduced. The methods presented here open up the possibility of systematic genome engineering in mice, facilitating the investigation of entire signaling pathways, of synthetic lethal phenotypes or of genes that have redundant functions. A particularly interesting application is the possibility to produce mice carrying multiple alterations in candidate loci that have been identified in GWAS studies to play a role in the genesis of multigenic diseases. In summary, CRISPR/Cas-mediated genome editing makes possible the generation of ES cells and mice carrying multiple genetic alterations and will facilitate the genetic dissection of development and complex diseases.
Experimental Procedures
Procedures for Generating sgRNAs Expressing Vector
Bicistronic expression vector expressing Cas9 and sgRNA (Cong et al., 2013) were digested with BbsI and treated with Antarctic Phosphatase, and the linearized vector was gel purified. A pair of oligos for each targeting site (Table S3) was annealed, phosphorylated, and ligated to linearized vector.
Cell Culture and Transfection
V6.5 ES cells (on a 129/Sv × C57BL/6 F1 hybrid background) were cultured on gelatin-coated plates with standard ES cell culture conditions. Cells were transfected with a plasmid expressing mammalian-codon-optimized Cas9 and sgRNA (single targeting), three plasmids expressing Cas9 and sgRNAs targeting Tet1, Tet2, and Tet3 (triple targeting), or five PCR products each coding for sgRNA targeting Tet1, Tet2, Tet3, Sry, and Uty, along with a plasmid expressing PGK-puroR using FuGENE HD reagent (Promega) according to the manufacturer's instructions. Twelve hours after transfection, ES cells were replated at a low density on DR4 MEF feeder layers. Puromycin (2 μg/ml) was added 1 day after replating and taken off after 48 hr. After recovering for 4 to 6 days, individual colonies were picked and genotyped by RFLP and Southern blot analysis, and the leftover ES cells on plate were collected for Suveryor assay.
Suveryor Assay and RFLP Analysis for Genome Modification
Suveryor assay was performed as described by (Guschin et al., 2010). Genomic DNA from treated and control ES cells or targeted and control mice was extracted. Mouse genomic DNA samples were prepared from tail biopsies. PCR was performed using Tet1-, 2-, and 3-specific primers (Table S3) under the following conditions: 95°C for 5 min; 35× (95°C for 30 s, 60°C for 30 s, 68°C for 40 s); 68°C for 2 min; hold at 4°C. PCR products were then denatured, annealed, and treated with Suveryor nuclease (Transgenomic). DNA concentration of each band was measured on an ethidium bromide-stained 10% acrylamide Criterion TBE gel (BioRad) and quantified using ImageJ software. The same PCR products for Suveryor assay were used for RFLP analysis. Ten microliters of Tet1, Tet2, or Tet3 PCR product was digested with SacI, EcoRV, or XhoI, respectively. Digested DNA was separated on an ethidium bromide-stained agarose gel (2%). For sequencing, PCR products were cloned using the Original TA Cloning Kit (Invitrogen), and mutations were identified by Sanger sequencing.
Dot Blot
DNA was extracted from ES cells following standard procedures. DNA was transferred to nylon membrane using BioRad slot blot vacuum manifold apparatus. Anti-5hmC (Active Motif 1:10,000) was used to detect 5hmC following manufacturer's protocol.
Production of Cas9 mRNA and sgRNA
T7 promoter was added to Cas9 coding region by PCR amplification using primer Cas9 F and R (Table S3). T7-Cas9 PCR product was gel purified and used as the template for in vitro transcription (IVT) using mMESSAGE mMACHINE T7 ULTRA kit (Life Technologies). T7 promoter was added to sgRNAs template by PCR amplification using primer Tet1 F and R, Tet2 F and R, and Tet3 F and R (Table S3). The T7-sgRNA PCR product was gel purified and used as the template for IVT using MEGAshortscript T7 kit (Life Technologies). Both the Cas9 mRNA and the sgRNAs were purified using MEGAclear kit (Life Technologies) and eluted in RNase-free water.
One-Cell Embryo Injection
All animal procedures were performed according to NIH guidelines and approved by the Committee on Animal Care at MIT. B6D2F1 (C57BL/6 × DBA2) female mice and ICR mouse strains were used as embryo donors and foster mothers, respectively. Superovulated female B6D2F1 mice (7-8 weeks old) were mated to B6D2F1 stud males, and fertilized embryos were collected from oviducts. Cas9 mRNAs (from 20 ng/μl to 200 ng/μl) and sgRNA (from 20 ng/μl to 50 ng/μl) was injected into the cytoplasm of fertilized eggs with well recognized pronuclei in M2 medium (Sigma). For oligos injection, Cas mRNA (100 ng/μl), sgRNA (50 ng/μl), and donor oligos (100 ng/μl) were mixed and injected into zygotes at the pronuclei stage. The injected zygotes were cultured in KSOM with amino acids at 37°C under 5% CO2 in air until blastocyst stage by 3.5 days. Thereafter, 15–25 blastocysts were transferred into uterus of pseudopregnant ICR females at 2.5 dpc.
Southern Blotting
Genomic DNA was separated on a 0.8% agarose gel after restriction digests with the appropriate enzymes, transferred to a nylon membrane (Amersham) and hybridized with 32P random primer (Stratagene)-labeled probes.
Prediction of Potential Off Targets
Potential targets of CRISPR sgRNAs were found using the rules outline in (Mali et al., 2013). For a 20 nt sgRNA targeting sequence of nnnnn nnMMM MMMMM MMMMM, where M are the seed bases preceding the PAM sequence NGG, four search sequences (MMM MMMMM MMMMM AGG; MMM MMMMM MMMMM CGG; MMM MMMMM MMMMM GGG; MMM MMMMM MMMMM TGG) were generated. Exact matches to these search sequences in the mouse genome (mm9) were found using bowtie and reported as potential targets of the CRISPR sgRNA.
Supplementary Material
Acknowledgments
We thank Ruth Flannery and Kibibi Ganz for support with animal care and experiments. We thank Jaenisch lab members for helpful discussions on the manuscript. We are also grateful to G. Grant Welstead and Daniel B. Dadon for the help of editing the manuscript. M.M.D is a Damon Runyon Postdoctoral Fellow. A.W.C. is supported by a Croucher scholarship. R.J. is an adviser to Stemgent and a cofounder of Fate Therapeutics. This work was supported by NIH grants R37-HD045022 and R01-CA084198 to RJ.
Footnotes
Supplemental Information: Supplemental Information includes four figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.04.025.
References
- Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6:507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- Carbery ID, Ji D, Harrington A, Brown V, Weinstein EJ, Liaw L, Cui X. Targeted genome modification in mice using zinc-finger nucleases. Genetics. 2010;186:451–459. doi: 10.1534/genetics.110.117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D, Beumer KJ, Morton JJ, Bozas A, Trautman JK. Gene targeting in Drosophila and Caenorhabditis elegans with zinc-finger nucleases. Methods Mol Biol. 2008;435:63–77. doi: 10.1007/978-1-59745-232-8_5. [DOI] [PubMed] [Google Scholar]
- Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ. Genome editing with RNA-guided Cas9 nuclease in Zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Pruett-Miller SM, Huang Y, Gjoka M, Duda K, Taunton J, Collingwood TN, Frodin M, Davis GD. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA. Harnessing transposons for cancer gene discovery. Nat Rev Cancer. 2010;10:696–706. doi: 10.1038/nrc2916. [DOI] [PubMed] [Google Scholar]
- Cui X, Ji D, Fisher DA, Wu Y, Briner DM, Weinstein EJ. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat Biotechnol. 2011;29:64–67. doi: 10.1038/nbt.1731. [DOI] [PubMed] [Google Scholar]
- Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, Jaenisch R. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell. 2013;24:310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool J, Berns A. High-throughput insertional mutagenesis screens in mice to identify oncogenic networks. Nat Rev Cancer. 2009;9:389–399. doi: 10.1038/nrc2647. [DOI] [PubMed] [Google Scholar]
- Li Z, Cai X, Cai CL, Wang J, Zhang W, Petersen BE, Yang FC, Xu M. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M, Lee SE. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, de Angelis MH, Wurst W, Kühn R. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc Natl Acad Sci USA. 2010;107:15022–15026. doi: 10.1073/pnas.1009424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, Zhang X, Zhang P, Huang X. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013 doi: 10.1038/cr.2013.46. in press. Published online April 2, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Laganière J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, Khurana V, Golbe LI, Myers RH, Lindquist S, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YH, Baek IJ, Kim DH, Jeon J, Lee J, Lee K, Jeong D, Kim JS, Lee HW. Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol. 2013;31:23–24. doi: 10.1038/nbt.2477. [DOI] [PubMed] [Google Scholar]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Tesson L, Usal C, Ménoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L, et al. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29:695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Wefers B, Meyer M, Ortiz O, Hrabé de Angelis M, Hansen J, Wurst W, Kühn R. Direct production of mouse disease models by embryo microinjection of TALENs and oligodeoxynucleotides. Proc Natl Acad Sci USA. 2013;110:3782–3787. doi: 10.1073/pnas.1218721110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


