Abstract
Targeted nucleases are powerful tools for mediating genome alteration with high precision. The RNA-guided Cas9 nuclease from the microbial clustered regularly interspaced short palindromic repeats (CRISPR) adaptive immune system can be used to facilitate efficient genome engineering in eukaryotic cells by simply specifying a 20-nt targeting sequence within its guide RNA. Here we describe a set of tools for Cas9-mediated genome editing via nonhomologous end joining (NHEJ) or homology-directed repair (HDR) in mammalian cells, as well as generation of modified cell lines for downstream functional studies. to minimize off-target cleavage, we further describe a double-nicking strategy using the Cas9 nickase mutant with paired guide RNAs. This protocol provides experimentally derived guidelines for the selection of target sites, evaluation of cleavage efficiency and analysis of off-target activity. Beginning with target design, gene modifications can be achieved within as little as 1–2 weeks, and modified clonal cell lines can be derived within 2–3 weeks.
INTRODUCTION
The ability to engineer biological systems and organisms holds enormous potential for applications across basic science, medicine and biotechnology. Programmable sequence-specific endonucleases that facilitate precise editing of endogenous genomic loci are now enabling systematic interrogation of genetic elements and causal genetic variations1,2 in a broad range of species, including those that have not previously been genetically tractable3–6. A number of genome editing technologies have emerged in recent years, including zinc-finger nucleases (ZFNs)7–10, transcription activator–like effector nucleases (TALENs)10–17 and the RNA-guided CRISPR-Cas nuclease system18–25. The first two technologies use a strategy of tethering endonuclease catalytic domains to modular DNA-binding proteins for inducing targeted DNA double-stranded breaks (DSBs) at specific genomic loci. By contrast, Cas9 is a nuclease guided by small RNAs through Watson-Crick base pairing with target DNA26–28 (Fig. 1), representing a system that is markedly easier to design, highly specific, efficient and well-suited for high-throughput and multiplexed gene editing for a variety of cell types and organisms.
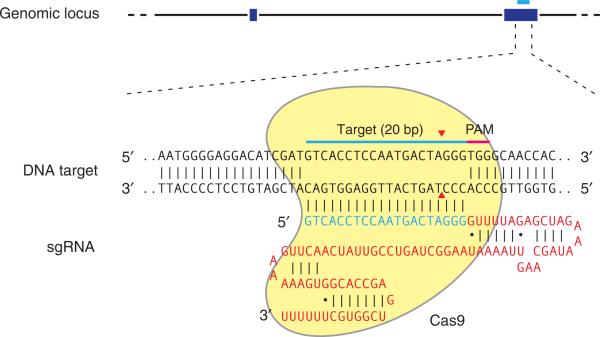
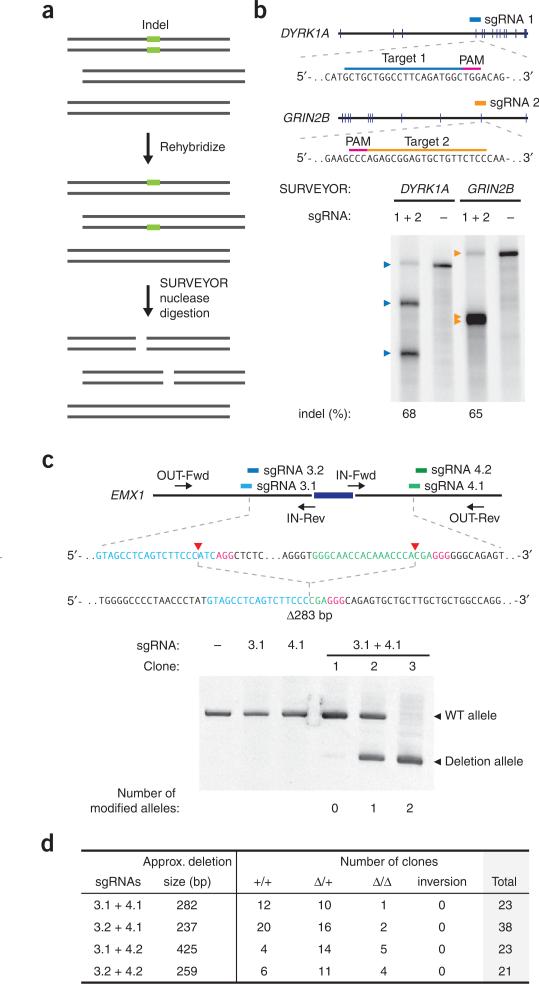
Figure 1.
Schematic of the RNA-guided Cas9 nuclease. The Cas9 nuclease from S. pyogenes (in yellow) is targeted to genomic DNA (shown for example is the human EMX1 locus) by an sgRNA consisting of a 20-nt guide sequence (blue) and a scaffold (red). The guide sequence pairs with the DNA target (blue bar on top strand), directly upstream of a requisite 5′-NGG adjacent motif (PAM; pink). Cas9 mediates a DSB ~3 bp upstream of the PAM (red triangle).
Precise genome editing using engineered nucleases
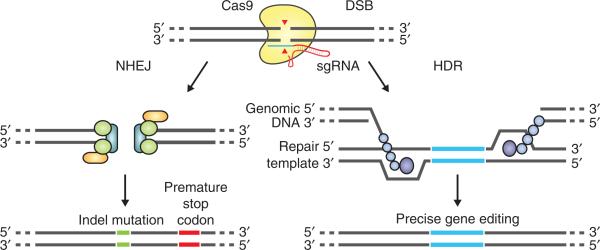
Similarly to ZFNs and TALENs, Cas9 promotes genome editing by stimulating a DSB at a target genomic locus29,30. Upon cleavage by Cas9, the target locus typically undergoes one of two major pathways for DNA damage repair (Fig. 2): the error-prone NHEJ or the high-fidelity HDR pathway, both of which can be used to achieve a desired editing outcome. In the absence of a repair template, DSBs are re-ligated through the NHEJ process, which leaves scars in the form of insertion/deletion (indel) mutations. NHEJ can be harnessed to mediate gene knockouts, as indels occurring within a coding exon can lead to frameshift mutations and premature stop codons31. Multiple DSBs can additionally be exploited to mediate larger deletions in the genome22,32.
Figure 2.
DSB repair promotes gene editing. DSBs induced by Cas9 (yellow) can be repaired in one of two ways. In the error-prone NHEJ pathway, the ends of a DSB are processed by endogenous DNA repair machinery and rejoined, which can result in random indel mutations at the site of junction. Indel mutations occurring within the coding region of a gene can result in frameshifts and the creation of a premature stop codon, resulting in gene knockout. Alternatively, a repair template in the form of a plasmid or ssODN can be supplied to leverage the HDR pathway, which allows high fidelity and precise editing. Single-stranded nicks to the DNA can also induce HDR.
HDR is an alternative major DNA repair pathway. Although HDR typically occurs at lower and substantially more variable frequencies than NHEJ, it can be leveraged to generate precise, defined modifications at a target locus in the presence of an exogenously introduced repair template. The repair template can either be in the form of conventional double-stranded DNA targeting constructs with homology arms flanking the insertion sequence, or single-stranded DNA oligonucleotides (ssODNs). The latter provides an effective and simple method for making small edits in the genome, such as the introduction of single-nucleotide mutations for probing causal genetic variations32. Unlike NHEJ, HDR is generally active only in dividing cells, and its efficiency can vary widely depending on the cell type and state, as well as the genomic locus and repair template33.
Cas9: an RNA-guided nuclease for genome editing
CRISPR-Cas is a microbial adaptive immune system that uses RNA-guided nucleases to cleave foreign genetic elements18–21,26. Three types (I–III) of CRISPR systems have been identified across a wide range of bacterial and archaeal hosts, wherein each system comprises a cluster of CRISPR-associated (Cas) genes, noncoding RNAs and a distinctive array of repetitive elements (direct repeats). These repeats are interspaced by short variable sequences20 derived from exogenous DNA targets known as protospacers, and together they constitute the CRISPR RNA (crRNA) array. Within the DNA target, each protospacer is always associated with a protospacer adjacent motif (PAM), which can vary depending on the specific CRISPR system34–36.
The Type II CRISPR system is one of the best characterized26–28,37,38, consisting of the nuclease Cas9, the crRNA array that encodes the guide RNAs and a required auxiliary trans-activating crRNA (tracrRNA) that facilitates the processing of the crRNA array into discrete units26,28. Each crRNA unit then contains a 20-nt guide sequence and a partial direct repeat, where the former directs Cas9 to a 20-bp DNA target via Watson-Crick base pairing (Fig. 1). In the CRISPR-Cas system derived from Streptococcus pyogenes (which is the system used in this protocol), the target DNA must immediately precede a 5′-NGG PAM27, whereas other Cas9 orthologs may have different PAM requirements, such as those of S. thermophilus (5′-NNAGAA22,26 for CRISPR1 and 5′-NGGNG28,37 for CRISPR3) and Neisseria meningiditis (5′-NNNNGATT)39.
The RNA-guided nuclease function of CRISPR-Cas is reconstituted in mammalian cells through the heterologous expression of human codon–optimized Cas9 and the requisite RNA components22–25. Furthermore, the crRNA and tracrRNA can be fused together to create a chimeric, single-guide RNA (sgRNA)27 (Fig. 1). Cas9 can thus be re-directed toward almost any target of interest in immediate vicinity of the PAM sequence by altering the 20-nt guide sequence within the sgRNA.
Given its ease of implementation and multiplexing capacity, Cas9 has been used to generate engineered eukaryotic cells carrying specific mutations via both NHEJ and HDR22–25,40. Direct injection of sgRNA and mRNA encoding Cas9 into embryos has enabled the rapid generation of transgenic mice with multiple modified alleles41,42. These results hold enormous promise for editing organisms that are otherwise genetically intractable.
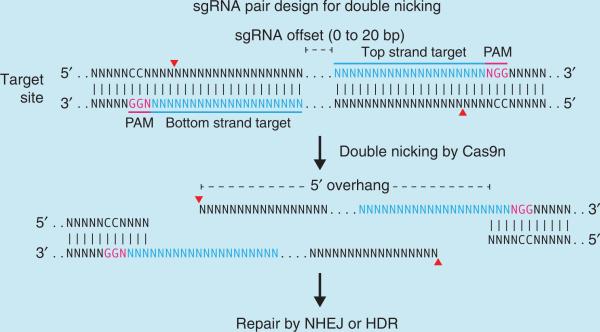
Cas9 nucleases carry out strand-specific cleavage by using the conserved HNH and RuvC nuclease domains, which can be mutated and exploited for additional function37. An aspartate-to-alanine (D10A) mutation in the RuvC catalytic domain27,28 allows the Cas9 nickase mutant (Cas9n) to nick rather than cleave DNA to yield single-stranded breaks, and the subsequent preferential repair through HDR22 can potentially decrease the frequency of unwanted indel mutations from off-target DSBs. Appropriately offset sgRNA pairs can guide Cas9n to simultaneously nick both strands of the target locus to mediate a DSB, thus effectively increasing the specificity of target recognition43. In addition, a Cas9 mutant with both DNA-cleaving catalytic residues mutated has been adapted to enable transcriptional regulation in Escherichia coli44, demonstrating the potential of functionalizing Cas9 for diverse applications, such as recruitment of fluorescent protein labels or chromatin- modifying enzymes to specific genomic loci for reporting or modulating gene function.
Here we explain in detail how to use a human codon–optimized, nuclear localization sequence-flanked wild-type (WT) Cas9 nuclease or mutant Cas9 nickase to facilitate eukaryotic gene editing. We describe considerations for designing the 20-nt guide sequence, protocols for rapid construction and functional validation of sgRNAs and finally the use of the Cas9 nuclease to mediate both NHEJ- and HDR-based genome modifications in human embryonic kidney (HEK 293FT) and human stem cell (HUES9) lines (Fig. 3). The Cas9 system can similarly be applied to other cell types and organisms, including humans22,23,25, mice22,41,45, zebrafish45, Drosophila46 and Caenorhabditis elegans47.
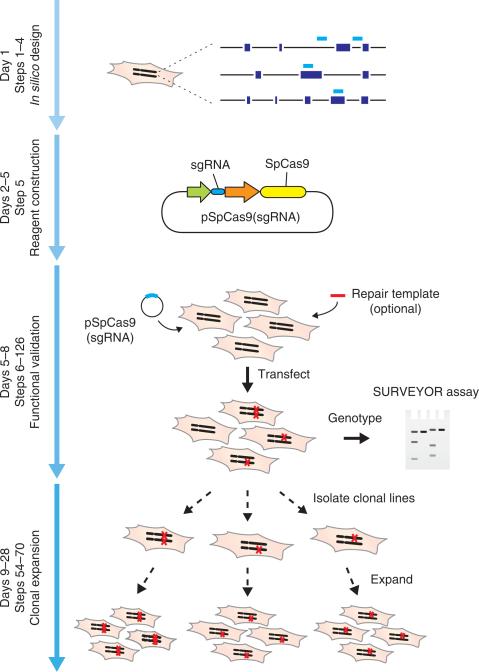
Figure 3.
Timeline and overview of experiments. Steps for reagent design, construction, validation and cell line expansion are depicted. Custom sgRNAs (light blue bars) for each target, as well as genotyping primers, are designed in silico via the CRISPR Design Tool (http://tools.genome-engineering.org). sgRNA guide sequences can be cloned into an expression plasmid bearing both sgRNA scaffold backbone (BB) and Cas9, pSpCas9(BB). The resulting plasmid is annotated as pSpCas9(sgRNA). Completed and sequence-verified pSpCas9(sgRNA) plasmids and optional repair templates for facilitating HDR are then transfected into cells and assayed for their ability to mediate targeted cleavage. Finally, transfected cells can be clonally expanded to derive isogenic cell lines with defined mutations.
Comparison with other genome editing technologies
As with other designer nuclease technologies such as ZFNs and TALENs, Cas9 can facilitate targeted DNA DSBs at specific loci of interest in the mammalian genome and stimulate genome editing via NHEJ or HDR. Cas9 offers several potential advantages over ZFNs and TALENs, including the ease of customization, higher targeting efficiency and the ability to facilitate multiplex genome editing. As custom ZFNs are often difficult to engineer, we will primarily compare Cas9 with TALEN.
• Ease of customization
Cas9 can be easily retargeted to new DNA sequences by simply purchasing a pair of oligos encoding the 20-nt guide sequence. In contrast, retargeting of TALEN for a new DNA sequence requires the construction of two new TALEN genes. Although a variety of protocols exist for TALEN construction14,17,48,49, it takes substantially more hands-on time to construct a new pair of TALENs.
• Cleavage pattern
WT S. pyogenes Cas9 (SpCas9) is known to make a blunt cut between the 17th and 18th bases in the target sequence (3 bp 5′ of the PAM)27. Mutating catalytic residues in either the RuvC or the HNH nuclease domain of SpCas9 converts the enzyme into a DNA nicking enzyme22,27. In contrast, TALENs cleave nonspecifically in the 12–24-bp linker between the pair of TALEN monomer-binding sites50.
• Editing efficiency
SpCas9 and TALENs have both been shown to facilitate efficient genome editing in a variety of cell types and organisms. However, owing to the ease of targeting, Cas9 can be used to target multiple genomic loci simultaneously, by co-delivering a combination of sgRNAs to the cells of interest.
Limitations of the Cas9 system
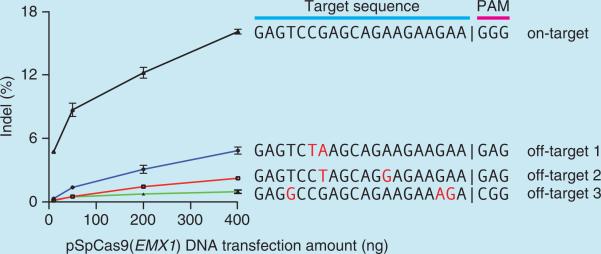
Cas9 can be targeted to specific genomic loci via a 20-nt guide sequence on the sgRNA. The only requirement for the selection of Cas9 target sites is the presence of a PAM sequence directly 3′ of the 20-bp target sequence. Each Cas9 ortholog has a unique PAM sequence; for example, SpCas9 requires a 5′-NGG PAM sequence. This PAM requirement does not severely limit the targeting range of SpCas9—in the human genome, such target sites can be found on average every 8–12 bp (refs. 22,51). In addition to the targeting range, another possible limitation is the potential for off-target mutagenesis; please see Boxes 1 and 2 for details and strategies on minimizing off- target modifications.
Experimental design
Target selection for sgRNA
The specificity of the Cas9 nuclease is determined by the 20-nt guide sequence within the sgRNA. For the S. pyogenes system, the target sequence (e.g., 5′-GTCACCTCCAATGACTAGGG-3′) must immediately precede (i.e., be 5′ to) a 5′-NGG PAM, and the 20-nt guide sequence base pairs with the opposite strand to mediate Cas9 cleavage at ~3 bp upstream of the PAM (Figs. 1 and 4a top strand example). Note that the PAM sequence is required to immediately follow the target DNA locus, but that it is not a part of the 20-nt guide sequence within the sgRNA.
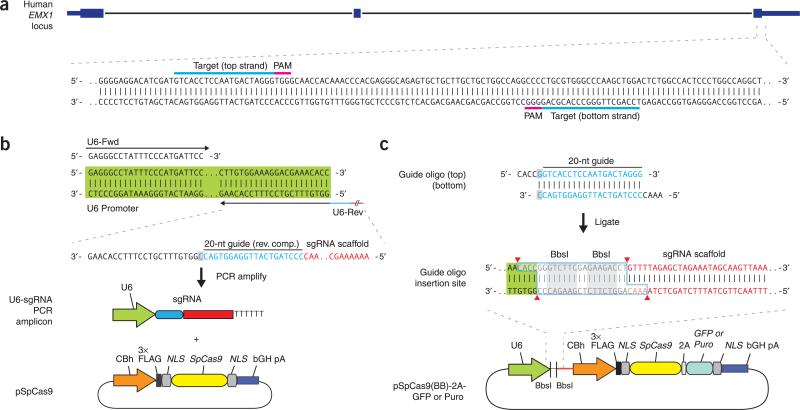
Figure 4.
Target selection and reagent preparation. (a) For S. pyogenes Cas9, 20-bp targets (highlighted in blue) must be followed at their 3′ends by 5′-NGG, which can occur in either the top or the bottom strand of genomic DNA, as in the example from the human EMX1 gene. We recommend using the CRISPR Design Tool (http://tools.genome-engineering.org) to facilitate target selection. (b) Schematic for co-transfection of the Cas9 expression plasmid (pSpCas9) and a PCR-amplified U6-driven sgRNA expression cassette. By using a U6 promoter-containing PCR template and a fixed forward primer (U6-Fwd), sgRNA-encoding DNA can be appended onto the U6 reverse primer (U6-Rev) and synthesized as an extended DNA oligo (Ultramer oligos from IDT). Note that the guide sequence in the U6-Rev primer, designed against an example target from the top strand (blue), is the reverse complement of the 20-bp target sequence preceding the 5′-NGG PAM. An additional cytosine (‘C’ in gray rectangle) is appended in the reverse primer directly 3′ to the target sequence to allow guanine as the first base of the U6 transcript. (c) Schematic for scarless cloning of the guide sequence oligos into a plasmid containing Cas9 and the sgRNA scaffold (pSpCas9(BB)). The guide oligos for the top strand example (blue) contain overhangs for ligation into the pair of BbsI sites in pSpCas9(BB), with the top and bottom strand orientations matching those of the genomic target (i.e., the top oligo is the 20-bp sequence preceding 5′-NGG in genomic DNA). Digestion of pSpCas9(BB) with BbsI allows the replacement of the Type II restriction sites (blue outline) with direct insertion of annealed oligos. Likewise, a G-C base pair (gray rectangle) is added at the 5′ end of the guide sequence for U6 transcription, which does not adversely affect targeting efficiency. Alternate versions of pSpCas9(BB) also contain markers such as GFP or a puromycin resistance gene to aid the selection of transfected cells.
Thus, there are two main considerations in the selection of the 20-nt guide sequence for gene targeting: (i) the 5′-NGG PAM for S. pyogenes Cas9 and (ii) the minimization of off-target activity51,52. We provide an online CRISPR Design Tool (http://tools.genome-engineering.org) that takes a genomic sequence of interest and identifies suitable target sites. To experimentally assess off-target genomic modifications for each sgRNA, we also provide computationally predicted off-target sites (for a detailed discussion, see Box 1) for each intended target, ranked according to our quantitative specificity analysis on the effects of base-pairing mismatch identity, position and distribution. For increased targeting specificity, an alternative strategy using the D10A nickase mutant of Cas9 (Cas9n) along with a pair of sgRNAs may be used. The design criteria for orientation and spacing of such sgRNA pairs are described in Box 2.
The CRISPR Design Tool provides the sequences for all oligos and primers necessary for (i) preparing the sgRNA constructs, (ii) assaying target modification efficiency and (iii) assessing cleavage at potential off-target sites. It is worth noting that because the U6 RNA polymerase III promoter used to express the sgRNA prefers a guanine (G) nucleotide as the first base of its transcript59, an extra G is appended at the 5′ of the sgRNA where the 20-nt guide sequence does not begin with G (Fig. 4b,c). On rare occasions, certain sgRNAs may not work for reasons yet unknown; therefore, we recommend designing at least two sgRNAs for each locus and testing their efficiencies in the intended cell type.
Approaches for sgRNA construction and delivery
Depending on the desired application, sgRNAs can be delivered as either PCR amplicons containing an expression cassette (Fig. 4b) or sgRNA-expressing plasmids (Fig. 4c). PCR-based sgRNA delivery appends the custom sgRNA sequence onto the reverse PCR primer used to amplify a U6 promoter template (Fig. 4b). The resulting amplicon could be co-transfected with a Cas9 expression plasmid pSpCas9. This method is optimal for rapid screening of multiple candidate sgRNAs, as cell transfections for functional testing can be performed shortly after obtaining the sgRNA-encoding primers. Because this simple method obviates the need for plasmid-based cloning and sequence verification, it is well suited for testing or co-transfecting a large number of sgRNAs for generating large knockout libraries or other scale-sensitive applications. Note that the sgRNA-encoding primers are over 100 bp long, compared with the ~20-bp-long oligos required for plasmid-based sgRNA delivery.
Construction of an expression plasmid for sgRNA is also simple and rapid, involving a single cloning step with a pair of partially complementary oligonucleotides. The oligo pairs encoding the 20-nt guide sequences are annealed and ligated into a plasmid (pSpCas9(BB), Fig. 4c) bearing both Cas9 and the remainder of the sgRNA as an invariant scaffold immediately following the oligo cloning site. The transfection plasmids can also be modified to enable virus production for in vivo delivery. For these approaches, the following plasmids are used within this protocol: Cas9 alone (pSpCas9) or Cas9 with an invariant sgRNA scaffold and cloning sites for inserting a guide sequence (pSpCas9(BB)). For the backbone cloning construct, we have also fused 2A-GFP or 2A-Puro to Cas9 to allow screening or selection of transfected cells (pSpCas9(BB)-2A-GFP or pSpCas9(BB)-2A-Puro, respectively). Finally, we provide pSpCas9n(BB), a D10A nickase mutant of Cas9 for HDR and for double-nicking applications (Box 2), along with the 2A-GFP and 2A-Puro fusion constructs (pSpCas9n(BB)-2A-GFP, pSpCas9n(BB)-2A-Puro).
In addition to PCR and plasmid-based delivery methods, Cas9 and sgRNAs can be introduced into cells as mRNA and RNA, respectively.
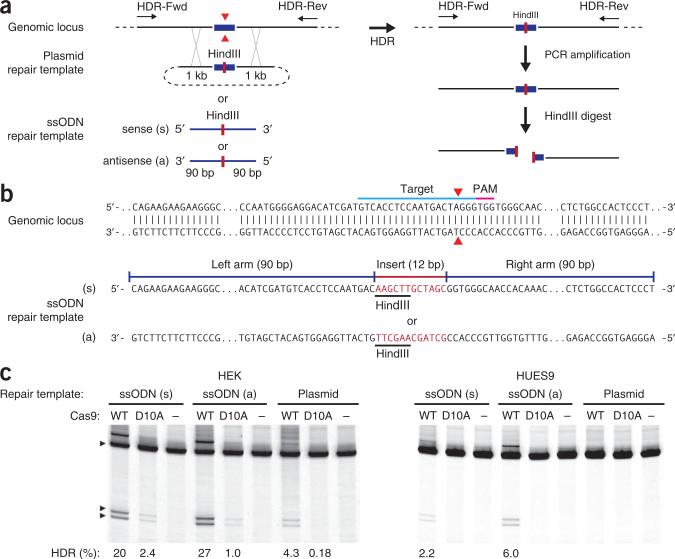
Design of repair template
Traditionally, targeted DNA modifications have required the use of plasmid-based donor repair templates that contain homology arms flanking the site of alteration54,55 (Fig. 2). The homology arms on each side can vary in length, but are typically longer than 500 bp (refs. 55,56). This method can be used to generate large modifications, including insertion of reporter genes such as fluorescent proteins or antibiotic resistance markers. The design and construction of targeting plasmids has been described elsewhere57.
More recently, ssODNs have been used in place of targeting plasmids for short modifications within a defined locus without cloning32. To achieve high HDR efficiencies, ssODNs contain flanking sequences of at least 40 bp on each side that are homologous to the target region, and they can be oriented in either the sense or antisense direction relative to the target locus. It is worth noting that targeting efficiencies can vary widely depending on cell type, target locus, type of repair donor and location of modification relative to the DSB site. As a rule of thumb, single-base correction rates drop approximately fourfold at 100 bp away from the DSB site, and beyond 200 bp away drug selection markers may be required58.
Clonal isolation of cell lines
Isolation of clonal cell lines with specific modifications is often desired. This can be achieved after transfection by isolating single cells through either FACS (Steps 54–65) or serial dilutions (Steps 66–70), followed by an expansion period to establish a new clonal cell line. It is worth noting that cell types can vary substantially in their responses to single-cell isolation, and literature specific to the cell type of interest should be consulted.
Functional testing
SURVEYOR nuclease assay
In cells co-transfected with a pair of sgRNAs to mediate a genomic (micro)deletion or inversion, indel mutations can be detected either by the SURVEYOR nuclease assay59 or by sequencing (Fig. 5a). Our online CRISPR Design Tool provides recommended primers for both approaches. However, SURVEYOR or sequencing primers can also be designed manually to amplify the region of interest from genomic DNA. Custom primers are chosen using the National Center for Biotechnology Information (NCBI) Primer-BLAST in order to avoid nonspecific amplification. SURVEYOR primers should be designed to amplify 200–400 bp on either side of the Cas9 target (for a total amplicon 400–800 bp long) to allow clear visualization of cleavage bands by gel electrophoresis (Fig. 5b). To prevent excessive primer dimer formation, SURVEYOR primers should be designed to be typically 18 to 25 nt long with melting temperatures of ~60 °C. For SURVEYOR assay or sequencing analysis, we recommend testing that each pair of candidate primers produces a single PCR product, as well as testing for the absence of nonspecific cleavage during the SURVEYOR nuclease digestion process (Fig. 5).
Figure 5.
Anticipated results for multiplex-sgRNA-targeted NHEJ. (a) Schematic of the SURVEYOR assay used to determine the indel percentage. First, genomic DNA from the heterogeneous population of Cas9-targeted cells is amplified by PCR. Amplicons are then reannealed slowly to generate heteroduplexes. The reannealed heteroduplexes are cleaved by SURVEYOR nuclease, whereas homoduplexes are left intact. Cas9-mediated cleavage efficiency (percentage indel) is calculated on the basis of the fraction of cleaved DNA, as determined by integrated intensity of gel bands. (b) Two sgRNAs (orange and dark blue bars) are designed to target the human GRIN2B and DYRK1A loci. SURVEYOR gel shows modification at both loci in transfected cells. Colored arrowheads indicate expected fragment sizes for each locus. (c) Paired sgRNAs (light blue and green bars) are designed to excise an exon (dark blue) in the human EMX1 locus. Target sequences and PAMs (pink) are shown in respective colors, and sites of cleavage by Cas9 are indicated by red triangles. A predicted junction is shown below. Individual clones isolated from cell populations transfected with sgRNA 3, 4 or both are assayed by PCR (using the Out-Fwd and Out-Rev primers), reflecting a deletion of ~270 bp long. Representative clones with no modification (12/23), mono-allelic modification (10/23) and bi-allelic (1/23) modification are shown. (d) Quantification of clonal lines with EMX1 exon deletions. Two pairs of sgRNAs (3.1 and 3.2, left-flanking sgRNAs; 4.1 and 4.2, right flanking sgRNAs) are used to mediate deletions of various sizes around one EMX1 exon. Transfected cells are clonally isolated and expanded for genotyping analysis of deletions and inversion events. Of the 105 clones screened, 51 (49%) and 12 (11%) are carrying heterozygous and homozygous deletions, respectively. Only approximate deletion sizes are given, as deletion junctions may be variable.
Plasmid- or ssODN-mediated HDR
HDR can be detected via PCR amplification, followed by either sequencing of the modified region or restriction-fragment length polymorphism (RFLP) analysis. PCR primers for these purposes should anneal outside the region spanned by the homology arms to avoid false detection of residual repair template (primers HDR-Fwd and HDR-Rev; Table 1 and Fig. 6a). For ssODN-mediated HDR, SURVEYOR PCR primers may be used. Either the WT Cas9 nuclease or mutant Cas9 nickase can be used to mediate HDR, although the efficiency of the latter can vary widely by cell type.
TABLE 1.
Primer sequences for sgRNA cloning and validation.
| Step | Primer | Sequence (5′-3′) | Purpose |
|---|---|---|---|
| 5A(iii) | U6-Fwd | GAGGGCCTATTTCCCATGATTCC | Amplify any U6-sgRNA |
| 5A(iii) | U6-Rev | AAAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAcNNNNNNNNNNNNNNNNNNNCCGGTGTTTCGTCCTTTCCACAAG | Amplify specifically designed U6-sgRNA; N is the reverse complement of target; appended cytosine (complementary to appended guanine) in lowercase |
| 5B(i) | sgRNA-top | CACCgNNNNNNNNNNNNNNNNNNN | Clone sgRNA into pSpCas9(BB); appended guanine in lowercase |
| 5B(i) | sgRNA-bottom | AAACNNNNNNNNNNNNNNNNNNNc | Clone sgRNA into pSpCas9(BB); appended cytosine (complementary to appended guanine) in lowercase |
| 117 | pUC-Fwd (M13 -20 primer) | GTAAAACGACGGCCAGT | Sanger sequencing of modified genomic regions cloned into pUC19 |
| 117 | pUC-Rev (M13 -26 primer) | CAGGAAACAGCTGTAAC | Sanger sequencing of modified genomic regions cloned into pUC19 |
Figure 6.
Anticipated results for HDR in HEK and HUES9 cells. (a) Either a targeting plasmid or an ssODN (sense or antisense) with homology arms can be used to edit the sequence at a target genomic locus cleaved by Cas9 (red triangle). To assay the efficiency of HDR, we introduced a HindIII site (red bar) into the target locus, which was PCR-amplified with primers that anneal outside of the region of homology. Digestion of the PCR product with HindIII reveals the occurrence of HDR events. (b) ssODNs, oriented in either the sense or the antisense (s or a) direction relative to the locus of interest, can be used in combination with Cas9 to achieve efficient HDR-mediated editing at the target locus. A minimal homology region of 40 bp, and preferably 90 bp, is recommended on either side of the modification (red bar). (c) Example of the effect of ssODNs on HDR in the EMX1 locus is shown using both wild-type Cas9 and Cas9 nickase (D10A). Each ssODN contains homology arms of 90 bp flanking a 12-bp insertion of two restriction sites.
Detection of indels or HDR by sequencing
Targeted genome modifications can also be detected by either Sanger or deep sequencing. For the former, genomic DNA from the modified region can be amplified with either SURVEYOR or HDR primers. Amplicons should be subcloned into a plasmid such as pUC19 for transformation, and individual colonies should be sequenced to reveal the clonal genotype.
Alternatively, deep sequencing is suitable for sampling a large number of samples or target sites. NGS primers are designed for shorter amplicons, typically in the 100–200-bp size range. For the detection of NHEJ mutations, it is important to design primers situated at least 50 bp from the Cas9 target site to allow for the detection of longer indels. For larger deletions mediated by multiple sgRNAs, priming sites should be designed outside the deleted region. We provide guidelines for a two-step PCR fusion method to attach bar-coded sequencing adaptors for multiplex deep sequencing. We recommend the Illumina platform for its generally low levels of false positive indel detection. By comparison, Ion Torrent is less suitable for indel analysis owing to high sequencing error rate with homo-polymers60. Detailed descriptions of NGS optimization and troubleshooting can be found in the Illumina user manual. Off-target indel analysis (Box 1) can then be performed through read-alignment programs such as ClustalW, Geneious or simple custom sequence analysis scripts.
MATERIALS
REAGENTS
sgRNA preparation
Plasmids: pSpCas9 (Addgene plasmid ID: 48137), pSpCas9(BB) (formerly pX330; Addgene plasmid ID: 42230), pSpCas9(BB)-2A-GFP (Addgene plasmid ID: 48138), pSpCas9(BB)-2A-Puro (Addgene plasmid ID: 48139), pSpCas9n(BB) (Addgene plasmid ID: 48873), pSpCas9n(BB)-2A-GFP (Addgene plasmid ID: 48140), pSpCas9n(BB)-2A-Puro (Addgene plasmid ID: 48141). Annotated GenBank files for the plasmids are available through Addgene and http://www.genome-engineering.org/
pUC19 (Invitrogen, cat. no. 15364-011) or any preferred cloning plasmid
PCR primers or oligos for sgRNA construction are listed in Table 1 and in supplementary Data 1. Primers longer than 60 bp can be ordered as 4-nmol ultramers (Integrated DNA Technologies)
UltraPure DNase/RNase-free distilled water (Life Technologies, cat. no. 10977-023)
Herculase II fusion polymerase with 5× reaction buffer (Agilent Technologies, cat. no. 600679) ▲ CRITICAL To minimize error in amplifying sgRNAs, it is important to use a high-fidelity polymerase. Other high-fidelity polymerases, such as PfuUltra (Agilent) or Kapa HiFi (Kapa Biosystems), may be used as substitutes.
Taq DNA polymerase with standard Taq buffer (NEB, cat. no. M0273S)
dNTP solution mix, 25 mM each (Enzymatics, cat. no. N205L)
MgCl2, 25 mM (Thermo Scientific, cat. no. R0971)
QIAquick gel extraction kit (Qiagen, cat. no. 28704)
QIAprep spin miniprep kit (Qiagen, cat. no. 27106)
UltraPure TBE buffer, 10× (Life Technologies, cat. no. 15581-028)
SeaKem LE agarose (Lonza, cat. no. 50004)
SYBR Safe DNA stain, 10,000× (Life Technologies, cat. no. S33102)
1-kb Plus DNA ladder (Life Technologies, cat. no. 10787-018)
TrackIt CyanOrange loading buffer (Life Technologies, cat. no. 10482-028)
FastDigest BbsI (BpiI) (Fermentas/Thermo Scientific, cat. no. FD1014)
Fermentas Tango buffer (Fermentas/Thermo Scientific, cat. no. BY5)
DTT (Fermentas/Thermo Scientific, cat. no. R0862)
T7 DNA ligase with 2× rapid ligation buffer (Enzymatics, cat. no. L602L). Alternative ligases, such as T4 DNA ligase, can also be used. If you are using other ligases, substitute with the compatible buffer
T4 polynucleotide kinase (New England BioLabs, cat. no. M0201S)
T4 DNA ligase reaction buffer, 10× (New England BioLabs, cat. no. B0202S)
Adenosine 5′-triphosphate, 10 mM (New England BioLabs, cat. no. P0756S)
PlasmidSafe ATP-dependent DNase (Epicentre, cat. no. E3101K)
One Shot Stbl3 chemically competent E. coli (Life Technologies, cat. no. C7373-03)
SOC medium (New England BioLabs, cat. no. B9020S)
LB medium (Sigma, cat. no. L3022)
LB agar medium (Sigma, cat. no. L2897)
Ampicillin, 100 mg ml–1, sterile filtered (Sigma, cat. no. A5354)
Mammalian cell culture
HEK 293FT cells (Life Technologies, cat. no. R700-07)
HUES 9 cell line (Harvard Stem Cell Science)
DMEM, high glucose (Life Technologies, cat. no. 10313-039)
DMEM, high glucose, no phenol red (Life Technologies, cat. no. 31053-028)
Dulbecco's PBS (DPBS; Life Technologies, cat. no. 14190-250)
FBS, qualified and heat inactivated (Life Technologies, cat. no. 10438-034)
Opti-MEM I reduced-serum medium (Life Technologies, cat. no. 11058-021)
Penicillin-streptomycin, 100× (Life Technologies, cat. no. 15140-163)
Puromycin dihydrochloride (Life Technologies, cat. no. A11138-03)
TrypLE Express, no phenol red (Life Technologies, cat. no. 12604-013)
Lipofectamine 2000 transfection reagent (Life Technologies, cat. no. 11668027)
Amaxa SF cell line 4D-Nucleofector X kit S, 32 RCT (Lonza, cat. no. V4XC-2032)
Geltrex LDEV-free reduced growth factor basement membrane matrix (Life Technologies, cat. no. A1413201)
mTeSR1 medium (Stemcell Technologies, cat. no. 05850)
Normocin (InvivoGen, cat. no. ant-nr-1)
Accutase cell detachment solution (Stemcell Technologies, cat. no. 07920)
Rho-associated protein kinase (ROCK) inhibitor (Y-27632; Millipore, cat. no. SCM075)
Amaxa P3 primary cell 4D-Nucleofector X kit S, 32 RCT (Lonza, cat. no. V4XP-3032)
Genotyping analysis
PCR primers for SURVEYOR, RFLP analysis or sequencing; see Table 1, supplementary Data 1 (alternatively, they can be custom made)
QuickExtract DNA extraction solution (Epicentre, cat. no. QE09050)
SURVEYOR mutation detection kit for standard gel electrophoresis (Transgenomic, cat. no. 706025)
TBE Gels, 4–20%, 1.0 mm, 15 well (Life Technologies, cat. no. C62255BOX)
Novex Hi-Density TBE sample buffer, 5× (Life Technologies, cat. no. LC6678)
SYBR Gold nucleic acid gel stain, 10,000× (Life Technologies, cat. no. S-11494)
FastDigest HindIII (Fermentas/Thermo Scientific, cat. no. FD0504)
FastDigest buffer, 10× (Fermentas/Thermo Scientific, supplied with FastDigest HindIII)
FastAP Antarctic phosphatase (Fermentas/Thermo Scientific, cat. no. EF0654)
Nextera XT index kit (Illumina, cat. no. FC-131-1001)
EQUIPMENT
Filtered sterile pipette tips (Corning)
Standard microcentrifuge tubes, 1.5 ml (Eppendorf, cat. no. 0030 125.150)
Axygen PCR plates, 96 well (VWR, cat. no. PCR-96M2-HSC)
Axygen 8-Strip PCR tubes (Fischer Scientific, cat. no. 14-222-250)
Falcon tubes, polypropylene, 15 ml (BD Falcon, cat. no. 352097)
Falcon tubes, polypropylene, 50 ml (BD Falcon, cat. no. 352070)
Round-bottom tube with cell strainer cap, 5 ml (BD Falcon, cat. no. 352235)
Petri dishes, 60 mm × 15 mm (BD Biosciences, cat. no. 351007)
Tissue culture plate, 24 wells (BD Falcon, cat. no. 353047)
Tissue culture plate, 96 wells flat bottom (BD Falcon, cat. no. 353075)
Tissue culture dish, 100 mm (BD Falcon, cat. no. 353003)
Nunc EasYFlask 225 cm2 (T225 flask), filter cap, 70-ml working volume (Thermo Scientific, cat. no. 159934)
Nunc EasYFlask 75 cm2 (T75 flask), filter cap, 25-ml working volume (Thermo Scientific, cat. no. 156499)
INCYTO C-Chip disposable hemocytometer (VWR, cat. no. 82030-468)
Steriflip-GP Filter Unit, 0.22 μM (Millipore, cat. no. SCGP00525)
Thermocycler with programmable temperature stepping functionality, 96 well (Applied Biosystems Veriti, cat. no. 4375786)
Desktop microcentrifuges (e.g., Eppendorf, cat. nos. 5424 and 5804)
Gel electrophoresis system (PowerPac basic power supply, Bio-Rad, cat. no. 164-5050), and Sub-Cell GT System gel tray (Bio-Rad, cat. no. 170-4401)
Novex XCell SureLock mini-cell (Life Technologies, cat. no. EI0001)
Digital gel imaging system (GelDoc EZ, Bio-Rad, cat. no. 170-8270), and blue sample tray (Bio-Rad, cat. no. 170-8273)
Blue-light transilluminator and orange filter goggles (SafeImager 2.0; Invitrogen, cat. no. G6600)
Gel quantification software (Bio-Rad, ImageLab or open-source ImageJ from the National Institutes of Health (NIH), USA, available at http://rsbweb.nih.gov/ij/)
UV spectrophotometer (NanoDrop 2000c, Thermo Scientific)
REAGENT SETUP
TBE electrophoresis solution
Dilute TBE buffer in distilled water to a 1× working solution, and store it at room temperature (18–22 °C) for up to 6 months.
ATP, 10 mM
Divide the solution into aliquots, and store them at –20 °C for up to 1 year; avoid repeated freeze-thaw cycles.
DTT, 10 mM
Prepare the solution in ddH2O, divide it into aliquots and store them at –70 °C for up to 2 years. Use a new aliquot for each reaction, as DTT is easily oxidized.
D10 culture medium
For culture of HEK 293FT cells, prepare D10 medium by supplementing DMEM with GlutaMAX and 10% (vol/vol) FBS. For routine cell line culture and maintenance, D10 medium can be further supplemented with 1× penicillin-streptomycin. Store the medium at 4 °C for up to 1 month.
mTeSR1 culture medium
For culture of human embryonic stem cells (hESCs), prepare mTeSR1 medium by supplementing it with the supplement solution supplied with the medium and 100 μg ml–1 Normocin. Prepared medium can be stored at 4 °C for up to 2 months.
PROCEDURE
Design of targeting components and the use of the CRISPR Design tool ● TIMING 1 d
Input target genomic DNA sequence. We provide an online CRISPR Design Tool (http://tools.genome-engineering.org) that takes an input sequence (for example, a 1-kb genomic fragment from the region of interest), identifies and ranks suitable target sites and computationally predicts off-target sites for each intended target. Alternatively, one can manually select guide sequences by identifying the 20-bp sequence directly upstream of any 5′-NGG.
Order necessary oligos and primers as specified by the online tool. If the cleavage site is chosen manually, the oligos or primers should be designed as described in Figure 4b,c.
Design of the ssODN template (optional) ● TIMING 1 h
-
3
Design and order custom ssODN. Purchase either the sense or antisense ssODN directly from IDT or the preferred supplier. We recommend manually designing homology arms of at least 40 nt on either side and preferably 90 nt for optimal HDR efficiency. It is not necessary to PAGE-purify the ssODN.
-
4
Resuspend and dilute ssODN ultramers to a final concentration of 10 μM. Do not combine or anneal the sense and antisense ssODNs. Store them at –20 °C.
Preparation of sgRNA expression construct
-
5
To generate the sgRNA expression construct, use either the PCR expression cassette (option A) or the plasmid-based procedure (option B).
(A) Generation of the sgRNA expression construct by PCR amplification ● TIMING 2 h
Preparation of diluted U6 PCR template. We recommend using pSpCas9(BB) or pSpCas9n(BB) (supplementary Data 2) as a PCR template, but any U6-containing plasmid can be used. Dilute the template with ddH2O to a concentration of 10 ng μl–1. Note that if a plasmid or cassette already containing a U6-driven sgRNA is used as a template, a gel extraction will need to be performed after PCR (Step 5A(iv)), using the QIAquick gel extraction kit according to the manufacturer's instructions, to ensure that the product contains only the intended sgRNA and no trace of sgRNA carryover from the template.
Preparation of diluted PCR primers. Dilute the U6-Fwd and U6-Rev (designed either using the CRISPR Design Tool or by hand and unique for each sgRNA, Step 1) primers (table 1) to a final concentration of 10 μM in ddH2O by adding 10 μl of the 100 μM primer stock to 90 μl of ddH2O.
- U6-sgRNA PCR. Set up the following reaction for each U6-Rev primer as follows:
Component Amount (μl) Final concentration Herculase II PCR buffer, 5× 10 1× dNTP, 100 mM (25 mM each) 0.5 1 mM U6 PCR template (pSpCas9(BB)) 1 0.2 ng μl−1 U6-Fwd primer (universal) 1 0.2 μM U6-Rev primer (sgRNA specific) 1 0.2 μM Herculase II fusion polymerase 0.5 Distilled water 36 Total 50
▲ CRITICAL STEP To minimize error in amplifying sgRNAs, it is important to use a high-fidelity polymerase. Other high-fidelity polymerases, such as PfuUltra II (Agilent) or Kapa HiFi (Kapa Biosystems), may be used as a substitute.
-
ivPerform a PCR by using the following cycling conditions:
Cycle number Denature Anneal Extend 1 95 °C, 2 m 2-31 95 °C, 20 s 60 °C, 20 s 72 °C, 20 s 32 72 °C, 3 min -
v
After the reaction is complete, run a sample of the product on a gel to verify successful amplification: cast a 2% (wt/vol) agarose gel in TBE buffer with SYBR Safe dye. Run 5 μl of the PCR product in the gel at 15 V cm–1 for 30 min. Successful reactions should yield a single 370-bp-long product, and the template should be invisible.
? TROUBLESHOOTING
-
vi
Purify the PCR product by using the QIAquick PCR purification kit according to the manufacturer's directions. Elute the DNA in 35 μl of EB buffer (part of the kit) or water.
■ PAUSE POINT Purified PCR products can be stored at –20 °C for up to several months.
(B) Cloning sgRNA into the pSpCas9(BB) vector for co-expression with Cas9 ● TIMING 3 d
- Preparation of the sgRNA oligos inserts. Resuspend the top and bottom strands of oligos for each sgRNA design (Step 1) to a final concentration of 100 μM. Prepare the following mixture for phosphorylating and annealing the sgRNA oligos (top and bottom strands):
Component Amount (μl) sgRNA top (100 μM) 1 sgRNA bottom (100 μM) 1 T4 ligation buffer, 10× 1 T4 PNK 1 ddH2O 6 Total 10 Phosphorylate and anneal the oligos in a thermocycler by using the following parameters: 37 °C for 30 min; 95 °C for 5 min; ramp down to 25 °C at 5 °C min–1.
Dilute phosphorylated and annealed oligos 1:200 by adding 1 μl of oligo to 199 μl of room temperature ddH2O.
- Cloning the sgRNA oligos into pSpCas9(BB). Set up a ligation reaction for each sgRNA, as described below. We recommend also setting up a no-insert, pSpCas9(BB)-only negative control for ligation. Note: if you are using the Cas9 D10A nickase mutant for subsequent applications, substitute pSpCas9(BB) with pSpCas9n(BB). Alternatively, if fluorescence-based screening or selection is needed, substitute with pSpCas9(BB)-2A-GFP, pSpCas9(BB)-2A-Puro, pSpCas9n(BB)-2A-GFP or pSpCas9n(BB)-2A-Puro. The following steps use pSpCas9(BB) as an example:
Components Amount (μl) pSpCas9(BB), 100 ng × Diluted oligo duplex from Step 5B(iii) 2 Tango buffer, 10× 2 DTT, 10 mM 1 ATP, 10 mM 1 FastDigest BbsI 1 T7 ligase 0.5 ddH2O to 20 Total 20 - Incubate the ligation reaction for a total of 1 h.
Cycle number Condition 1-6 37 °C for 5 min, 21 °C for 5 min - Treat the ligation reaction with PlasmidSafe exonuclease to digest any residual linearized DNA. This step is optional but highly recommended.
Component Amount (μl) Ligation reaction from Step 5B(v) 11 PlasmidSafe buffer, 10× 1.5 ATP, 10 mM 1.5 PlasmidSafe exonuclease 1 Total 15 Incubate the PlasmidSafe reaction at 37 °C for 30 min, followed by 70 °C for 30 min.
■ PAUSE POINT After PlasmidSafe treatment, the reaction can be stored at –20 °C for at least 1 week.
-
viii
Transformation. Transform the PlasmidSafe-treated plasmid into a competent E. coli strain, according to the protocol supplied with the cells. We recommend the Stbl3 strain for quick transformation. Briefly, add 2 μl of the product from Step 5B(vii) into 20 μl of ice-cold chemically competent Stbl3 cells, incubate the mixture on ice for 10 min, heat-shock it at 42 °C for 30 s and return it immediately to ice for 2 min. Add 100 μl of SOC medium and plate it onto an LB plate containing 100 μg ml–1 ampicillin. Incubate it overnight at 37 °C. Note that it is not necessary to incubate competent cells for the outgrowth period after heat shock when you are transforming ampicillin- resistant plasmids.
-
ix
Day 2: inspect the plates for colony growth. Typically, there are no colonies on the negative control plates (ligation of BbsI-digested pSpCas9(BB) alone without annealed sgRNA oligo insert), and there are tens to hundreds of colonies on the pSpCas9(sgRNA) (sgRNA inserted into pSpCas9(BB)) cloning plates.
? TROUBLESHOOTING
-
x
From each plate, pick two or three colonies to check for the correct insertion of sgRNA. Use a sterile pipette tip to inoculate a single colony into a 3-ml culture of LB medium with 100 μg ml–1 ampicillin. Incubate the culture and shake it at 37 °C overnight.
-
xi
Day 3: isolate the plasmid DNA from cultures by using a QIAprep spin miniprep kit according to the manufacturer's instructions.
-
xii
Sequence validation of CRISPR plasmid. Verify the sequence of each colony by sequencing from the U6 promoter using the U6-Fwd primer. Optional: sequence the Cas9 gene by using the Cbh-Fwd and SXRP002-007 primers listed in supplementary Data 1. Reference the sequencing results against the pSpCas9(BB) cloning vector sequence to check that the 20-nt guide sequence is inserted between the U6 promoter and the remainder of the sgRNA scaffold (Fig. 4c). Details and sequence of the pSpCas9(BB) map in GenBank vector map format (*.gb file) can be found at http://crispr.genome-engineering.org/.
? TROUBLESHOOTING
Functional validation of sgRNAs: HEK 293FT cell culture and transfections ● TIMING 3–4 d
▲ CRITICAL The CRISPR-Cas system has been used in a number of mammalian cell lines. Conditions may vary for each cell line. Below we detail transfection conditions for HEK 293FT cells. Note that ssODN-mediated HDR transfections, performed with Amaxa SF cell line Nucleofector kit, are described in Steps 14–29. For hESC (HUES9) culturing and transfection, follow Steps 30–53.
-
6
HEK 293FT maintenance. Cells are maintained according to the manufacturer's recommendations. Cells are cultured in D10 medium supplemented with 10% (vol/vol) FBS at 37 °C and 5% CO2.
-
7
To passage, remove the medium and rinse the cells once by gently adding DPBS to the side of the vessel, so as not to dislodge the cells. Add 2 ml of TrypLE to a T75 flask, and incubate the mixture for 5 min at 37 °C. Add 10 ml of warm D10 medium to inactivate the trypsin, and transfer the cells to a 50-ml Falcon tube. Dissociate the cells by pipetting them up and down gently, and then reseed them into new flasks as necessary.
▲ CRITICAL STEP We typically passage cells every 2–3 d at a split ratio of 1:4 or 1:8, never allowing cells to reach more than 70% confluency. Cells are discarded upon reaching passage number 15.
-
8
Preparation of cells for transfection. Plate the well-dissociated cells onto 24-well plates in D10 medium without antibiotics 16–24 h before transfection. Seed the cells at a density of 1.3 × 105 cells per well in a total volume of 500 μl. Scale up or down according to the cell line supplier's manual as needed.
▲ CRITICAL STEP Do not plate more cells than the recommended density, as doing so may reduce transfection efficiency.
-
9On the day of transfection, cells are optimal at 70–90% confluency. Cells can be transfected with Lipofectamine 2000 or the Amaxa SF cell line 4D-Nucleofector X kit according to the manufacturers’ instructions. Transfections should be performed as follows: for sgRNAs cloned into pSpCas9(BB), transfect 500 ng of sequence-verified CRISPR plasmid (pSpCas9(sgRNA)); if you are transfecting more than one plasmid (Box 2), mix them at equimolar ratios and use no more than 500 ng of total DNA. For sgRNA amplified by PCR, mix the following:
pSpCas9 (Cas9 only) 400 ng sgRNA amplicon from Step 5A (each) 20 ng pUC19 (carrier DNA) Fill up total DNA to 500 ng
▲ CRITICAL STEP We recommend transfecting in technical triplicates for reliable quantification, and including transfection controls (e.g., GFP plasmid) to monitor transfection efficiency. pSpCas9(sgRNA)-2A-GFP or pSpCas9(sgRNA)-2A-Puro may be used in place of pSpCas9 if fluorescence sorting or drug selection, respectively, is desired. In addition, the pSpCas9(BB) cloning plasmid and/or the sgRNA amplicon may be transfected alone as a negative control for downstream functional assays.
-
10
Add Lipofectamine complex to the cells gently, as HEK 293FT cells can detach easily from the plate, which will result in a lower transfection efficiency.
-
11
Check cells after 24 h for transfection efficiency. The percentage of fluorescent cells in the transfection control (e.g., GFP) can be estimated by using a fluorescence microscope. Typically, more than 70% of cells are transfected.
? TROUBLESHOOTING
-
12
Supplement the culture medium with an additional 500 μl of warm D10 medium.
▲ CRITICAL STEP Add D10 very slowly to the side of the well, and do not use cold medium, as cells can detach easily. Puromycin selection can be applied at a concentration of 1–3 μg ml–1 for HEK 293FT cells (may vary depending on the cell line).
-
13
Incubate the cells for a total of 48–72 h after transfection before passaging them for downstream applications or harvesting for indel analysis.
Co-transfection of CRISPR plasmids and HDR templates into HEK 293FT cells (optional) ● TIMING 3–4 d
-
14
Linearize 1–2 μg of targeting vector if possible by cutting once at a restriction site in the vector backbone near one of the homology arms or at the distal end of either homology arm.
Alternatively, if you are using ssODNs, simply resuspend them to a final concentration of 10 μM (see Step 4) and skip Steps 15 and 16.
-
15
Run a small amount of the linearized plasmid alongside uncut plasmid on a 0.8–1% (wt/vol) agarose gel to check for successful linearization. Linearized plasmids should run above the supercoiled plasmid.
-
16
Purify the linearized plasmid with the QIAQuick PCR Purification kit, and elute in 35 μl of EB buffer.
-
17
Preparation of cells for transfection. Culture HEK 293FT in T75 or T225 flasks. Plan ahead to have sufficient cells for the day of transfection (2 × 105 cells per transfection if you are using the Amaxa SF cell line 4D-Nucleofector X kit S).
-
18
Prewarming plates for transfection. Add 1 ml of warm D10 medium into each well of a 12-well plate. Place the plates in the incubator to keep the medium warm.
-
19
Use option A in the table below for preparing the co-transfection of the HDR targeting plasmid with the Cas9 plasmid or option B for the co-transfection of ssODN with the Cas9 plasmid. To prepare transfection controls, see Step 9. If an sgRNA is cloned into pSpCas9(BB)-2A-GFP, cells may also be sorted by fluorescence. If you are using Cas9 nickase to mediate HDR, substitute pSpCas9(sgRNA) with pSpCas9n(sgRNA) from Step 5B(v).
▲ CRITICAL STEP For HDR applications, we recommend cloning sgRNA guides into one of the sgRNA expression plasmids described in Step 5B, rather than using the PCR-based expression approach.
| (A) For the cotransfection of the HDR-targeting plasmid with the Cas9 plasmid: | |
| (i) Pre-mix the following DNA in PCR tubes: | |
| Cas9 plasmid (pSpCas9(sgRNA)) | 500 ng |
| Linearized targeting plasmid | 500 ng |
| (B) For the cotransfection of ss0DN and with the Cas9 plasmid: | |
| (i) Pre-mix the following DNA in PCR tubes: | |
| Cas9 plasmid (pSpCas9(sgRNA)) | 500 ng |
| ssODN template (10 μM) | 1 μl |
-
20
Dissociation of cells for transfection. Remove the medium and rinse the cells once gently with DPBS, taking care not to dislodge cells. Add 2 ml of TrypLE to a T75 flask and incubate it for 5 min at 37 °C, and then add 10 ml of warm D10 medium and triturate gently in a 50-ml Falcon tube.
▲ CRITICAL STEP Ensure that the cells are triturated gently and dissociated to single cells. Large clumps will reduce transfection efficiency.
-
21
Take a 10-μl aliquot from the cell suspension and dilute it into 90 μl of D10 medium for counting. Count the cells and calculate the number of cells and the volume of suspension needed for transfection. We typically transfect 2 × 105 cells per condition with the Amaxa SF cell line 4D-Nucleofector X kit S, and we recommend calculating for 20% more cells than required to adjust for volume loss in subsequent pipetting steps. Transfer the volume needed (20 μl per transfection plus waste volume) into a new Falcon tube.
-
22
Spin down the cells from Step 21 at 200g for 5 min at room temperature.
-
23
Prepare the transfection solution by mixing the SF solution and S1 supplement supplied in the Amaxa SF cell line 4D-Nucleofector X kit S; a total of 20 μl of supplemented SF solution is used per transfection. Likewise, we recommend calculating for 20% more volume than required.
-
24
Remove the medium completely from the pelleted cells from Step 22, and gently resuspend the cells in an appropriate volume (20 μl per 2 × 105 cells) of S1-supplemented SF solution. Do not leave the cells in SF solution for extended periods of time.
-
25
Pipette 20 μl of resuspended cells into each DNA premix from Step 19. Pipette gently to mix and transfer to a Nucleocuvette strip chamber. Repeat this step for each transfection condition.
-
26
Electroporate the cells by using the Nucleofector 4D program recommended by Amaxa, CM-130.
-
27
Gently and slowly pipette 100 μl of warm D10 medium into each Nucleocuvette strip chamber, and transfer all the volume into a well with the prewarmed medium from Step 18.
▲ CRITICAL STEP Cells are very fragile at this stage, and harsh pipetting can cause cell death.
-
28
Incubate the mixture for 24 h. At this point, transfection efficiency can be estimated from the fraction of fluorescent cells in the positive transfection control. Nucleofection typically results in >70–80% transfection efficiency.
? TROUBLESHOOTING
-
29
Slowly add 1 ml of warm D10 medium to each well without dislodging the cells. Puromycin selection can be applied at a concentration of 1–3 μg ml–1 for HEK 293FT cells (may vary depending on the cell line). Incubate the cells with puromycin for at least 72 h. Cells can then be cultured in regular medium for downstream experiments or harvested for genotyping.
hESC (HUES 9) culture and transfection ● TIMING 3–4 d
▲ CRITICAL hESCs and human induced pluripotent stem cells can vary widely in their transfection efficiency, tolerance of single-cell dissociation and maintenance conditions. For a given cell line of interest, relevant literature or the distributor should be consulted.
-
30
Maintaining HUES9 cells. We routinely maintain HUES9 cells (a hESC cell line) in feeder-free conditions with mTesR1 medium. Prepare mTeSR1 medium by adding the 5× supplement included with the basal medium and 100 μg ml–1 Normocin.
-
31
Prepare a 10-ml aliquot of mTeSR1 medium supplemented further with 10 μM ROCK inhibitor.
-
32
Coating a tissue culture plate. Dilute cold GelTrex 1:100 in cold DMEM and coat the entire surface of a 100-mm tissue culture plate.
-
33
Place the plate in an incubator for at least 30 min at 37 °C.
-
34
Thaw a vial of cells at 37 °C, transfer the cells to a 15-ml Falcon tube, add 5 ml of mTeSR1 medium and pellet at 200g for 5 min at room temperature.
-
35
Aspirate the GelTrex coating (Step 32) and seed ~1 × 106 cells with 10 ml of mTeSR1 medium containing ROCK inhibitor from Step 31.
-
36
Replace with mTeSR1 medium without ROCK inhibitor after 24 h and refeed daily.
-
37
Passaging cells. Passage the cells before they reach 70% confluency.
-
38
Aspirate the mTeSR1 medium and wash the cells once with DPBS.
-
39
Dissociate the cells by adding 2 ml of Accutase and incubating them at 37 °C for 3–5 min.
-
40
Add 10 ml of mTeSR1 medium to the detached cells, transfer the mixture to a 15-ml Falcon tube and resuspend gently.
-
41
Replate the cells onto GelTrex-coated plates in mTeSR1 medium with 10 μM ROCK inhibitor.
-
42
Replace with normal mTeSR1 medium 24 h after plating.
-
43
Transfection. We recommend culturing cells for at least 1 week after thawing and before transfecting by using the Amaxa P3 primary cell 4D Nucleofector kit.
-
44
Refeed log-phase growing cells (50–70% confluency) with fresh medium 2 h before transfection.
-
45
Dissociate to single cells or small clusters of no more than ten cells (as viewed under the microscope) with Accutase and gentle resuspension.
-
46
Count the number of cells needed for nucleofection (200,000 cells per transfection) and spin down at 200g for 5 min at room temperature.
-
47
Remove the medium completely and resuspend it in 20 μl of S1-supplemented P3 nucleofection solution, per 2 × 105 cells.
-
48
Pipette the resuspended cells with added DNA (Steps 9 and 19) into electroporation cuvettes and electroporate according to the suggested program. For 2 × 105 cells, we typically use 1 μg of total DNA.
-
49
Gently plate the electroporated cells onto coated 100-mm plates supplemented with 10 μM ROCK inhibitor.
-
50
Check transfection success (Steps 11 and 28) and refeed the cells daily with regular mTeSR1 medium beginning 24 h after nucleofection. Puromycin selection can be applied at a concentration of 0.5 μg ml–1 (may vary depending on the cell line). Typically, we observe >70% transfection efficiency with Amaxa nucleofection.
? TROUBLESHOOTING
-
51
At 48–72 h post transfection, dissociate the cells with Accutase and resuspend them gently in a 5× volume of mTeSR1. Reserve a fraction of the resuspended cells at this stage for replating (Steps 41 and 42; make sure to add ROCK inhibitor for each passaging), for downstream applications or clonal isolation (Steps 54–70), and use the remaining cells for genotyping (Steps 71–126).
▲ CRITICAL STEP Do not dissociate the cells mechanically without Accutase.
-
52
Spin the cells down at 200g for 5 min at room temperature.
▲ CRITICAL STEP Do not spin the cells without inactivating the Accutase first, or above the recommended speed; doing so may cause cells to lyse.
-
53
Process pelleted cells directly for DNA extraction with the QuickExtract solution (Steps 71–74).
Isolation of clonal cell lines by FACS ● TIMING 2–3 h hands-on; 2–3 weeks expansion
▲ CRITICAL Isolation of clonal cell populations from the transfected cells (Step 51) can be performed 24 h after transfection by FACS (Steps 54–65) or by serial dilutions (Steps 66–70). Given that cell types can vary greatly in their response to FACS, clonal-density dilution or other isolation procedures, literature specific to the cell type of interest should be consulted.
-
54
Preparation of FACS media. Cells can be sorted in regular D10 medium supplemented with penicillin-streptomycin. The antibiotics-containing medium should be filtered through a 0.22-μM Steriflip filter. If fluorescence sorting is also required, phenol red–free DMEM or DPBS is substituted for normal DMEM.
-
55
To 96-well plates, add 100 μl of D10 medium supplemented with penicillin-streptomycin per well.
▲ CRITICAL STEP Not all sorted cells may survive the FACS process or the subsequent outgrowth; therefore, the number of plates prepared and cells sorted may need to be adjusted to ensure an adequate number of clonal lines derived.
-
56
Preparation of cells for FACS. Dissociate the cells (from Steps 11 or 28) by aspirating the medium completely and adding enough TrypLE to thinly cover the adherent layer of transfected cells. Incubate the mixture for 5 min at 37 °C and add 400 μl of warm D10 medium.
-
57
Transfer the resuspended cells into a 15-ml Falcon tube and gently triturate 20 times.
▲ CRITICAL STEP Check under the microscope to ensure dissociation to single cells.
-
58
Spin down the cells at 200g for 5 min at room temperature.
-
59
Aspirate the medium, and resuspend it in 200 μl of FACS medium.
-
60
Filter the cells into the cell strainer tube through its mesh cap. Place the cells on ice until sorting.
-
61
Sort single cells into the 96-well plates prepared from Step 55. If sgRNA is cloned into pSpCas9(BB)-2A-GFP, fluorescence may be used to enrich for transfected cells. After sorting, examine the plate under a microscope and determine the presence of a single cell in most of the wells on the plate.
? TROUBLESHOOTING
-
62
Return the cells to the incubator and allow them to expand for 2–3 weeks. Add 100 μl of warm D10 medium 5 d after sorting. Change 100 μl of the medium every 3–5 d as necessary.
-
63
Inspect the colonies for “clonal” appearance 1 week after sorting: rounded colonies radiating from a central point. Mark off the wells that are empty or that may have been seeded more than a single cell.
-
64
When the cells are more than 60% confluent, prepare replica plates for passaging (one well for each clone) by adding 100 μl of D10 medium to each well in the replica plates. Dissociate the cells directly by pipetting up and down vigorously 20 times, and plate 20% of each of the resuspended volumes into the replica wells to keep the clonal lines. Change the medium every 2–3 d thereafter and passage accordingly.
-
65
Use the remaining 80% of cells for DNA isolation and genotyping (Steps 71–74).
Isolation of clonal cell lines by dilution ● TIMING 2–3 h hands-on; 2–3 weeks expansion
▲ CRITICAL As cell types can vary greatly in their response to FACS, clonal-density dilution or other isolation procedures, literature specific to the cell type of interest should be consulted.
-
66
Dissociate the cells from the transfected wells (Steps 11 or 28) 48 h after transfection. Take care to dissociate to single cells. A cell strainer (Step 60) can be used to prevent clumping of cells.
-
67
Count the number of cells from each 24-well plate, and serially dilute them in D10 medium to a final concentration of 0.5 cells per 100 μl to reduce the likelihood of having multiple cells per well. We recommend using 60 cells in 12 ml of D10 medium for each 96-well plate, and plating at least two 96-well plates for each transfected population.
▲ CRITICAL STEP Single-cell dissociation and accurate count of cell number are critical for clonal dilution. We recommend examining the dissociated cells under a microscope to ensure successful dissociation and recounting cells at an intermediate serial dilution stage to ensure accuracy.
? TROUBLESHOOTING
-
68
Multichannel-pipette 100 μl of diluted cells to each well of a 96-well plate. The remaining cell suspension can be kept and used for genotyping at the population level to gauge overall modification efficiency.
-
69
Inspect the colonies for a clonal appearance ~1 week after plating (rounded colonies radiating from a central point). Mark off the wells that may have been seeded with multiple cells.
-
70
Return the cells to the incubator and allow them to expand for 2–3 weeks. Refeed and replica-plate the cells as needed and as detailed in Steps 64 and 65.
Functional testing: detection of indel mutations by the SURVEYOR nuclease assay ● TIMING 5–6 h
▲ CRITICAL Before assaying the cleavage efficiency of transfected cells, we recommend testing each new SURVEYOR primer on negative (untransfected) control samples for the intended cell type by SURVEYOR nuclease digestion.
-
71
Harvesting cells for DNA extraction. Dissociate all transfected cells (from Steps 13, 29, 53, 65 or 70) and spin them down at 200g for 5 min at room temperature. Keep the replica plates as needed to maintain transfected cell lines in culture.
-
72
Aspirate the medium completely.
-
73
For DNA extraction, use the QuickExtract solution according to the manufacturer's instructions. We typically use 50 μl or 10 μl of the solution for each well of a 24-well or 96-well plate, respectively.
-
74
Normalize the extracted DNA to a final concentration of 100–200 ng μl–1 with ddH2O.
■ PAUSE POINT Extracted DNA can be stored at –20 °C for several months.
-
75Setting up the SURVEYOR PCR. Master-mix the following using the SURVEYOR primers provided by the CRISPR Design Tool (Step 1):
Component Amount (μl) Final concentration Herculase II PCR buffer, 5× 10 1× dNTP, 100 mM (25 mM each) 1 2 mM SURVEYOR-Fwd primer, 10 μM 1 0.2 μM SURVEYOR-Rev primer, 10 μM 1 0.2 μM Herculase II fusion polymerase 1 MgCl2, 25 mM 2 1 mM DNA template 1 2 ng μl−1 ddH2O 33 Total 50
▲ CRITICAL STEP SURVEYOR assays rely on the detection of single-base mismatches; therefore, it is crucial to use a high-fidelity polymerase. Other high-fidelity polymerases, such as PfuUltra (Agilent) or Kapa HiFi (Kapa Biosystems), may be used as a substitute. In addition, because SURVEYOR cleavage results can detect naturally occurring single-nucleotide polymorphisms, it is important to run negative control samples of untransfected or otherwise unmodified cells.
-
76Perform a PCR with the following cycling conditions, for no more than 30 amplification cycles:
Cycle number Denature Anneal Extend 1 95 °C, 2 min 2-31 95 °C, 20 s 60 °C, 20 s 72 °C, 30 s 32 72 °C, 3 min -
77
Run 2–5 μl of the PCR products on a 1% (wt/vol) agarose gel to check for single-band products. Although these PCR conditions are designed to work with most pairs of SURVEYOR primers, some primers may need additional optimization by adjusting the template concentration, MgCl2 concentration and/or the annealing temperature.
? TROUBLESHOOTING
-
78
Purify the PCRs with the QIAQuick PCR purification kit, and normalize the eluted product to 20 ng μl–1.
■ PAUSE POINT Purified PCR products can be stored at –20 °C for up to several months.
-
79DNA heteroduplex formation. Set up the annealing reaction as follows:
Component Amount (μl) Taq PCR buffer, 10× 2 Normalized PCR product, 20 ng μl−1 18 Total volume 20 -
80Anneal the reaction by using the following conditions:
Cycle number Condition 1 95 °C, 10 min 2 95-85 °C, −2 °C s−1 3 85 °C, 1 min 4 85-75 °C, −0.3 °C s−1 5 75 °C, 1 min 6 75-65 °C, −0.3 °C s−1 7 65 °C, 1 min 8 65-55 °C, −0.3 °C s−1 9 55 °C, 1 min 10 55-45 °C, −0.3 °C s−1 11 45 °C, 1 min 12 45-35 °C, −0.3 °C s−1 13 35 °C, 1 min 14 35-25 °C, −0.3 °C s−1 15 25 °C, 1 min 16 25-4 °C, −0.3 °C s−1 17 4 °C, hold -
81SURVEYOR nuclease S digestion. Master-mix and add the following components on ice to the annealed heteroduplexes from Step 80, to a final volume of 25 μl:
Component Amount (μl) Final concentration Annealed heteroduplex 20 MgCl2 stock solution supplied with kit, 0.15 M 2.5 15 mM ddH2O 0.5 SURVEYOR nuclease S 1 1× SURVEYOR enhancer S 1 1× Total 25
▲ CRITICAL STEP Note that the MgCl2 used for SURVEYOR nuclease digestion (included in the SURVEYOR mutation detection kit) is a higher concentration from that used for SURVEYOR PCR.
-
82
Vortex the mixture well and spin it down briefly. Incubate the reaction at 42 °C for 30 min.
-
83
(Optional) Add 2 μl of the Stop Solution from the SURVEYOR kit if you do not intend to visualize the reaction products (next step) immediately.
■ PAUSE POINT The digested products with Stop Solution can be stored at –20 °C for at least 2 d at this point for later analysis.
-
84
Visualizing the SURVEYOR reaction. SURVEYOR nuclease digestion products can be visualized on a 2% (wt/vol) agarose gel. For better resolution, products can be run on a 4–20% gradient polyacrylamide TBE gel. Load 10 μl of the product with the recommended loading buffer and run the gel according to the manufacturer's instructions. Typically, we run the gel until the bromophenol blue dye has migrated to the bottom of the gel. Include the DNA ladder and negative (untransfected) controls on the same gel.
-
85
Stain the gel with SYBR Gold dye diluted 1:10,000 in TBE (20 μl of stock in 200 ml of TBE). Rock the gel gently for 15 min. Be sure to shield the staining solution from light to avoid photobleaching of the dye.
-
86
Image the gel by using a quantitative imaging system without overexposing the bands. The negative controls should have only one band corresponding to the size of the PCR product, but they may have occasional nonspecific cleavage bands of other sizes. These will not interfere with analysis if they are distinct in size from the target cleavage bands. The sum of target cleavage band sizes, provided by the CRISPR Design Tool, should be equal to the size of the PCR product.
? TROUBLESHOOTING
-
87
Estimation of the cleavage intensity. Measure the integrated intensity of the PCR amplicon and cleaved bands by using ImageLab, ImageJ or other gel quantification software.
-
88
For each lane, calculate the fraction of the PCR product cleaved (fcut) by using the following formula: fcut = (b + c)/(a + b + c), where a is the integrated intensity of the undigested PCR product and b and c are the integrated intensities of each cleavage product. A sample gel is shown in Figure 6.
-
89Indel occurrence can be estimated with the following formula, based on the binomial probability distribution of duplex formation:
Functional testing: detection of genomic microdeletions by PCR ● TIMING 3–4 h hands-on; 2–3 weeks expansion
-
90
Transfect the cells as described in Steps 8–13 or Steps 43–51 with a pair of sgRNAs flanking the region to be deleted.
-
91
At 24 h after transfection, isolate the clones by FACS or by serial dilution as described above (Steps 54–70).
-
92
Expand the cells for 2–3 weeks.
-
93
Extract the DNA from clonal lines as described above (Steps 71–74) by using 10 μl of QuickExtract solution, and normalize the genomic DNA with ddH2O to a final concentration of 100 ng μl–1.
-
94
PCR amplification and analysis of the modified region. For analysis of (micro)deletions, follow option A; for analysis of inversions, follow option B.
(A) Deletion or microdeletion analysis
For the analysis of microdeletions, use the Out-Fwd and Out-Rev primers, both of which are designed to anneal outside of the deleted region, to verify the successful deletion by product size analysis. If the deletion size is more than 1 kb, set up a parallel set of PCRs with In-Fwd and In-Rev primers to screen for the presence of the WT allele (Fig. 5c).
As with SURVEYOR assays, include a negative (untransfected sample) control. Set up the PCR as follows:
| Component | Amount (μl) | Final concentration |
|---|---|---|
| Herculase II PCR buffer, 5× | 10 | 1× |
| dNTP, 100 mM (25 mM each) | 1 | 2 mM |
| Out-Fwd primer, 10 μM | 1 | 0.2 μM |
| Out-Rev primer, 10 μM | 1 | 0.2 μM |
| Herculase II fusion polymerase | 1 | |
| MgCl2, 25 mM | 2 | 1 mM |
| DNA template | 1 | 2 ng μl−1 |
| ddH2O | 33 | |
| Total | 50 |
(B) Inversion analysis
- To screen for inversions, set up the PCR (Fig. 5c) as described below. Note that primers are paired either as Out-Fwd + In-Fwd or Out-Rev + In-Rev. Include a negative control.
Component Amount (μl) Final concentration Herculase II PCR buffer, 5× 10 1× dNTP, 100 mM (25 mM each) 1 2 mM Out-Fwd or Out-Rev primer, 10 μM 1 0.2 μM In-Fwd or In-Rev primer, 10 μM 1 0.2 μM Herculase II fusion polymerase 1 MgCl2, 25 mM 2 1 mM DNA template 1 2 ng μl−1 ddH2O 33 Total 50
-
95Perform a PCR by using the following cycling conditions:
Cycle number Denature Anneal Extend 1 95 °C, 2 min 2-31 95 °C, 20 s 60 °C, 20 s 72 °C, 30 s 32 72 °C, 3 min -
96
Run 2–5 μl of PCR product on a 1–2% (wt/vol) agarose gel to check for size of the products in the case of deletions, or for the presence or absence of PCR products in the case of inversions. Although these PCR conditions are designed to work with most primers, some primers may need additional optimization by adjusting the template concentration, MgCl2 concentration and/or the annealing temperature.
? TROUBLESHOOTING
Functional testing: genotyping of HDR-mediated targeted modifications by RFLP analysis ● TIMING 3–4 h
-
97
Extract the DNA as described in Steps 71–74 by using the QuickExtract solution, and normalize the genomic DNA with water to a final concentration of 100–200 ng μl–1.
-
98PCR amplification of the modified region. The HDR-Fwd and HDR-Rev primers are designed to anneal outside of the region of homology between the ssODN and targeted genomic region, to verify successful sequence conversion. Include a negative (untransfected) control sample. Set up the PCR as follows:
Component Amount (μl) Final concentration Herculase II PCR buffer, 5× 10 1× dNTP, 100 mM (25 mM each) 1 2 mM HDR-Fwd primer, 10 μM 1 0.2 μM HDR-Rev primer, 10 μM 1 0.2 μM Herculase II fusion polymerase 1 MgCl2, 25 mM 2 1 mM DNA template 1 2 ng μl−1 ddH2O 33 Total 50 -
99Run the following program:
Cycle number Denature Anneal Extend 1 95 °C, 2 min 2-31 95 °C, 20 s 60 °C, 20 s 72 °C, 30-60 s per kb 32 72 °C, 3 min -
100
Run 5 μl of PCR product on a 0.8–1% (wt/vol) agarose gel to check for a single band of product. Primers may need additional optimization by adjusting the template concentration, MgCl2 concentration and/or the annealing temperature.
? TROUBLESHOOTING
-
101
Purify the PCRs by using the QIAQuick PCR purification kit.
-
102In the HDR example provided in Figure 6, a HindIII restriction site was inserted into the EMX1 gene. These are detected by an RFLP analysis of the PCR amplicon:
Component Amount (μl) Purified PCR amplicon x (200-300 ng) FastDigest buffer 1 Hindlll (or other enzyme as necessary) 0.5 ddH2O Up to 10 Total 10 -
103
Digest the DNA for 10 min at 37 °C.
-
104
Run 10 μl of the digested product with loading dye on a 4–20% gradient polyacrylamide TBE gel until the xylene cyanol band has migrated to the bottom of the gel.
-
105
Stain the gel with SYBR Gold dye while rocking for 15 min. Be sure to shield the staining solution from light to avoid photobleaching of the dye.
-
106
Image and quantify the cleavage products as described above for the SURVEYOR assay section (Steps 86–89).
-
107
HDR efficiency is estimated by using the following formula: (b + c)/(a + b + c), where a is the integrated intensity for the undigested HDR PCR product, and b and c are the integrated intensities for the HindIII-cut fragments.
-
108
Alternatively, clone the genotype-purified PCR amplicons from Step 101 via Sanger sequencing (Steps 109–117) or deep sequencing (Steps 118–126).
Assessment of Cas9 cleavage or HDR-mediated target modification efficiency by Sanger sequencing ● TIMING 3 d
▲ CRITICAL Instead of the SURVEYOR or RFLP assays, genomic amplicons of the target region (produced in Step 78 or 101) from transfected cells (Steps 8–13, and 14–28 for HEK 293FT cells, or steps 43–51 for HUES9 cells) can be cloned into a plasmid, and a set of clones can be prepared for Sanger sequencing to assess Cas9 cleavage or HDR-mediated target modification efficiency. SURVEYOR or HDR primers can be used for Sanger sequencing if appropriate restriction sites are appended to the forward and reverse primers. For cloning into the recommended pUC19 backbone, EcoRI can be used for the Fwd primer and HindIII for the Rev primer.
-
109Target-site amplicon digestion. Set up the digestion reaction as follows:
Component Amount (μl) FastDigest buffer, 10× 3 FastDigest EcoRI 1 FastDigest HindIII 1 Purified PCR product, 20 ng μl−1 10 (Step 78 or 101) ddH2O 15 Total volume 30 -
110pUC19 backbone digestion. Set up the digestion reaction as follows and incubate it at 37 °C for 15 min:
Component Amount (μl) FastDigest buffer, 10× 3 FastDigest EcoRI 1 FastDigest HindIII 1 FastAP alkaline phosphatase 1 pUC19 vector (200 ng μl−1) 5 ddH2O 20 Total volume 30 -
111
Purify the digestion reactions with the QIAQuick PCR purification kit.
■ PAUSE POINT Purified PCR product can be stored at –20 °C overnight.
-
112Ligate the digested pUC19 backbone and PCR product at a 1:3 vector:insert ratio and incubate it at room temperature for 15 min. As always, it is important to include a vector-only ligation control.
Component Amount (μl) Digested pUC19 x (50 ng) Digested PCR product (insert) x (1:3 vector to insert molar ratio) T7 Ligase 1 Rapid Ligation buffer, 2× 10 ddH2O Up to 20 Total volume 20 -
113Treat the ligation reaction with PlasmidSafe exonuclease to digest any residual linearized DNA. This step is optional but highly recommended.
Component Amount (μl) Ligation reaction from Step 112 11 PlasmidSafe buffer, 10× 1.5 ATP, 10 mM 1.5 PlasmidSafe exonuclease 1 Total 15 -
114
Transformation of bacteria. Transform the PlasmidSafe-treated plasmid into a competent E. coli strain, according to the protocol supplied with the cells. We recommend Stbl3 for quick transformation. Briefly, add 5 μl of the product from Step 113 into 20 μl of ice-cold chemically competent Stbl3 cells; incubate the mixture on ice for 10 min, heat-shock it at 42 °C for 30 s, return it immediately to ice for 2 min, add 100 μl of SOC medium and plate it onto LB plates containing 100 μg ml–1 ampicillin. Incubate the mixture overnight at 37 °C.
-
115
Day 2: inspect the plates for colony growth. Typically, there are no colonies on the negative control plates (ligation of vector only, with no Sanger amplicon insert), and tens to hundreds of colonies on the experimental plates. Pick a minimum of 48 clones to inoculate in 3 ml of LB-ampicillin culture.
? TROUBLESHOOTING
-
116
Day 3: isolate the plasmid DNA from overnight cultures by using a QIAprep spin miniprep kit.
-
117
Sanger sequencing. Verify the sequence of each colony by sequencing from the pUC19 backbone using the pUC19-Fwd or pUC19-Rev primer. Reference the sequencing results against the expected genomic sequence to check for the presence of Cas9-induced NHEJ or HDR modifications. Calculate the percentage of editing efficiency as (no. of modified clones)/ (no. of total clones). It is important to pick a reasonable number of clones (>24) to generate an accurate approximation of modification efficiencies.
Deep sequencing and off-target analysis ● TIMING 2–3 d
-
118
Designing deep-sequencing primers. Primers for deep sequencing are designed to produce short PCR amplicons, typically in the 100- to 200-bp size range. You can manually design primers by using the NCBI Primer-Blast or generate them with the CRISPR Design Tool (http://tools.genome-engineering.org).
-
119
Extract genomic DNA from Cas9-targeted cells (Steps 71–74). Normalize QuickExtract genomic DNA to 100–200 ng μl–1 with ddH2O.
-
120Initial library preparation-PCR. By using the primers from Step 118, prepare the initial library preparation PCR (include untransfected sample as negative control):
Component Amount (μl) Final concentration Herculase II PCR buffer, 5× 10 1× dNTP, 100 mM (25 mM each) 1 2 mM Fwd primer (10 μM) 1 0.2 μM Rev primer (10 μM) 1 0.2 μM Herculase II fusion polymerase 1 MgCl2 (25 mM) 2 1 mM DNA template 1 2 ng μl−1 ddH2O 33 Total 50 -
121Perform the PCR with the following cycling conditions, for no more than 20 amplification cycles:
Cycle number Denature Anneal Extend 1 95 °C, 2 min 2-21 95 °C, 20 s 60 °C, 20 s 72 °C, 15 s 22 72 °C, 3 min -
122
Run 2–5 μl of PCR product on a 1% (wt/vol) agarose gel to check for single-band product. As with all genomic DNA PCRs, the primers may require additional optimization by adjusting the template concentration, MgCl2 concentration and/or the annealing temperature.
? TROUBLESHOOTING
-
123
Purify the PCRs by using the QIAQuick PCR purification kit and normalize the eluants to 20 ng μl–1.
■ PAUSE POINT Purified PCR product can be stored at –20 °C overnight or longer.
-
124
Nextera XT DNA sample preparation kit. According to the manufacturer's protocol, generate Miseq sequencing-ready libraries with unique bar codes for each sample.
-
125
Sequence the samples prepared in Step 124 on the Illumina Miseq according to the Illumina user manual.
-
126
Analyze sequencing data. By using the expected reference genome sequence, perform indel analysis with read alignment programs such as ClustalW (http://www.clustal.org/), Geneious (http://www.geneious.com/) or by simple sequence analysis scripts.
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 2.
TABLE 2.
Troubleshooting table.
| Step | Problem | Possible reason | Possible solution |
|---|---|---|---|
| 5A(v) | No amplification of sgRNA | Incorrect template or primer. Incorrect template or primer concentration | Titrate U6-template concentration to 10-50 ng for a 50-μl reaction. Titrate primer concentration to a final concentration of 0.1-0.5 μM |
| 5B(ix), 115 | Colonies growing on negative control plate | Incomplete digestion of pSpCas9(BB) or pUC19 plasmid | Increase the amount of restriction enzymes; add phosphatase treatment to the plasmid digestions to reduce self-ligation of empty vector |
| 5B(xii) | No sgRNA sequences or wrong sequences | Ligation failure, incomplete digestion of cloning plasmid | Screen additional colonies; reanneal sgRNA oligos; titrate sgRNA oligo concentration during ligation; redigest pSpCas9(BB) or pUC19 |
| 11 | Low Lipofectamine transfection efficiency | Incorrect amount or poor quality of DNA used for transfection; poorly or unevenly seeded cells | Use low-passage-number cells (passage number <20) and do not let cells reach >90% confluence; titrate DNA (200 to 500 ng for 200,000 cells); add GFP transfection control; reseed cells evenly at recommended density; prepare new DNA for transfection |
| 28, 50 | Low nucleofection transfection efficiency | Incorrect amount or poor quality of DNA used for transfection; clumpy cells | Use low-passage-number cells (passage number <20); check cell density; titrate DNA; gently dissociate cells to single-cell suspension |
| 61 | Clumps or no cells after FACS | Cells not dissociated well in Step 56 | Filter cells before FACS; dissociate to single cells; re-suspend in no more than 107 cells per ml |
| 67 | Clumps or no cells in serial dilution | Cells not dissociated well; incorrect cell number or concentration | Dissociate to single cells and filter through strainer; recount cells; check serial dilution to make sure cell density is at an average of 0.5 cells per well |
| 77, 96, 100, 122 | Multiple bands or no amplification in SURVEYOR, microdeletion, RFLP analysis, or deep-sequencing PCR | Inefficient or nonspecific priming; incorrect template concentration | Titrate MgCl2 (0-1 mM final concentration); normalize and titrate template concentration (50-200 ng total); titrate annealing temperature (58-62 °C); re-design primers to minimize primer dimer formation or nonspecific amplification |
| 86 | High SURVEYOR background in negative sample; nonspecific or smeared bands on SURVEYOR gel; no SURVEYOR cleavage | Polymorphism within the examined region or co-amplification of highly homologous region; impure input DNA; input DNA too low or too high; improper annealing; overdigestion by SURVEYOR nuclease; sgRNA for a particular locus does not mediate cleavage | Redesign primers to different locations in the locus examined; purify PCR product; normalize total input DNA to 200-400 ng; reanneal heteroduplex (Step 80); incubate SURVEYOR digestion at 42 °C for <60 min; re-design sgRNAs; sequence verify sgRNA or Cas9 on pSpCas9(BB) backbone |
● TIMING
Steps 1–4, design of targeting components (sgRNA and ssODN) and use of the CRISPR Design Tool: 1 d
Step 5A, PCR-based generation of sgRNA expression cassette: 2 h
Step 5B, cloning of sgRNA expression vector: 3 d
Steps 6–13, functional validation of sgRNAs: HEK 293FT cell culture and transfections: 3–4 d
Steps 14–29, co-transfection of CRISPR plasmids and HDR templates into HEK 293FT cells (optional): 3–4 d
Steps 30–53, hESC (HUES 9) culture and transfection: 3–4 d
Steps 54–65, isolation of clonal cell lines by FACS: 2–3 h hands-on; 2–3 weeks expansion
Steps 66–70, isolation of clonal cell lines by dilution: 2–3 h hands-on; 2–3 weeks expansion
Steps 71–89, SURVEYOR assay for the assessment of CRISPR cleavage efficiency: 5–6 h
Steps 90–96, detection of genomic microdeletion by PCR: 3–4 h hands-on; 2–3 weeks expansion
Steps 97–108, genotyping of HDR-mediated targeted modifications by RFLP analysis: 3–4 h
Steps 109–117, genotyping by Sanger sequencing: 3 d
Steps 118–126, genotyping by next-generation deep sequencing: 2–3 d
ANTICIPATED RESULTS
We have compiled a list of most-frequently asked questions from our web-based CRISPR discussion forum (discuss.genome-engineering.org) to clarify points of confusion when applying the CRISPR system (Box 3). CRISPR-Cas can be easily multiplexed to facilitate high-efficiency genome editing in mammalian cells: by using two sgRNAs, we were able to demonstrate simultaneous targeting of the human DYRK1A and GRIN2B loci at efficiencies of 65–68% for each locus in HEK 293FT cells (Fig. 5b). Likewise, a pair of sgRNAs can be used to mediate microdeletions, such as excision of EMX1 exon 3, which we genotyped by PCR at a clonal level (Fig. 5c). Note that the precise location of exon junctions can vary. We also demonstrate here the use of ssODNs and targeting vector to mediate HDR (Fig. 6a,b) with both WT and the D10A nickase mutant of Cas9 in HEK 293FT and HUES9 cells (Fig. 6c). Note that we have not been able to detect HDR in HUES9 cells by using Cas9n with a sgRNA, which may be due to low efficiency or a potential difference in repair activities in HUES9 cells. The state of ploidy of the cell type of interest may also affect modification efficiency. However, Cas9n may be paired with two sgRNAs to mediate a DSB and stimulate HDR in HUES9 cells.
Supplementary Material
Box 1 | Considerations for off-target cleavage activities.
Similarly to other nucleases, Cas9 can cleave off-target DNA targets in the genome at reduced frequencies51,52,61. The extent to which a given guide sequence exhibits off-target activity depends on a combination of factors including enzyme concentration and the abundance of similar sequences in the target genome. For routine application of Cas9, it is important to consider ways to minimize the degree of off-target cleavage and also to be able to detect the presence of off-target cleavage51,52,61.
Minimizing off-target activity. For application in cell lines, we recommend following two steps to reduce the degree of off-target genome modification. First, by using our online CRISPR Design Tool, it is possible to computationally assess the likelihood of a given guide sequence to have off-target sites. These analyses are performed through an exhaustive search in the genome for off-target sequences that are similar to the guide sequence. Comprehensive experimental investigation of the effect of mismatching bases between the sgRNA and its target DNA revealed that mismatch tolerance is (i) position dependent: the 8–14 bp on the 3′ end of the guide sequence is less tolerant of mismatches than the 5′ bases; (ii) quantity dependent: in general, more than three mismatches are not tolerated; (iii) guide sequence dependent: some guide sequences are less tolerant of mismatches than others; and (iv) concentration dependent: off-target cleavage is highly sensitive to the transfected amounts, as well as relative ratios of Cas9 and sgRNA51. As shown in the illustration (adapted with permission from ref. 51; error bars show s.e.m. (n = 3)), Cas9 can exhibit off-target cleavage in the genome, which may be minimized by carefully titrating the amount of pSpCas9 DNA transfected. The CRISPR Design Tool integrates these criteria to provide predictions for likely off-target sites in the target genome. We also recommend titrating the amount of Cas9 and sgRNA expression plasmid to minimize off-target activity.
Detection of off-target activities. We have found experimentally that Cas9 can cleave at genomic off-target sites with either 5′-NGG or 5′-NAG PAMs. By using our CRISPR-targeting web tool, it is possible to generate a list of the most likely off-target sites, as well as primers for performing SURVEYOR or sequencing analysis of those sites. For isogenic clones generated using Cas9, we strongly recommend sequencing candidate off-target sites to check for any undesired mutations. It is worth noting that there may be off-target modifications in sites that are not included in the predicted candidate list, and full genome sequencing should be performed to completely verify the absence of off-target sites. Furthermore, in multiplex assays where several DSBs are induced within the same genome, there may be low rates of translocation events and they can be evaluated by using a variety of techniques such as deep sequencing62.
Box 2 | Double-nicking strategy for minimizing off-target mutagenesis.
To minimize off-target activity, a double nicking strategy can be used to introduce DSBs at the target site43. Whereas the WT Cas9 nuclease is guided by an sgRNA to mediate a DSB at the target locus, the D10A mutant Cas9 nickase (Cas9n) can be specified by a pair of appropriately spaced and oriented sgRNAs to simultaneously introduce single-stranded nicks on both strands of the target DNA. The DSBs from double nicking are then repaired via NHEJ and result in indel formation with similar levels of efficiency to that of WT Cas9. As single-stranded nicks are repaired without indel formation, DSBs would only occur if both sgRNAs are able to locate target sequences within a defined space. Thus, this strategy effectively doubles the number of bases that need to be specifically recognized at the target site and significantly increases the specificity of genome editing.
To facilitate efficient double nicking, the pair of sgRNAs must be designed such that 5′ overhangs are generated upon nicking. The target loci for the sgRNA pairs must also be offset with an optimal gap of 0–20 bp (see illustration: target DNA loci, blue N′s; PAM, pink; predicted cleavage sites on each strand, red triangles)43. The expression constructs for sgRNA pairs can be prepared by the PCR-based method as described for sgRNAs (Step 5A). The sgRNAs can then be combined at 1:1 ratio and introduced along with Cas9n by using identical procedures as for WT Cas9 and sgRNAs (Step 9, 20 ng for each sgRNA). Editing achieved by using this double-nicking strategy can be similarly detected using SURVEYOR assay or DNA sequencing. In addition to facilitating DSB- and NHEJ-mediated mutagenesis, double nicking can also be used to promote HDR with comparable efficiency as WT Cas9.
A web tool to help with the identification of suitable sgRNA pairs for double nicking can be accessed at http://tools.genome-engineering.org.
Box 3 | Frequently asked questions from the CRISPR Discussion Forum.
The following questions are selected from the CRISPR Discussion Forum (discuss.genome-engineering.org).
Q1: Should I use crRNA + tracrRNA or sgRNA?
Although we initially reported that sgRNAs with a truncated tracrRNA 3′ tail, sgRNA(+48), are less effective for gene targeting in mammalian cells than using crRNA and tracrRNA pairs22, we have optimized the sgRNA architecture by extending its tracrRNA tail. sgRNA(+85), used in pSpCas9 vectors, is able to mediate the highest efficiency of genome modification57.
Q2: My 20-bp target site does not start with G; can I replace the first base with G?
For efficient U6 transcription of sgRNA, a G is preferred at the 5′ position, which corresponds with the first base of the 20-bp guide sequence. For guide sequences that do not begin with a G, we recommend adding an additional G to the 5′ of the guide sequence, resulting in a 21-bp guide sequence (5′-GNNNNNNNNNNNNNNNNNNNN-3′, where the 20 Ns correspond to the 20-bp genomic target sequence immediately upstream of the 5′-NGG PAM). The addition of a 5′ G does not alter the specificity of the sgRNA or affect the efficiency of Cas9 cleavage.
Q3: Should I include the PAM sequence in my sgRNA?
No, the PAM sequence should not be included as a part of the sgRNA. The PAM sequence is located in the genomic target site, immediately following the 20-bp target sequence, but it is not a part of the sgRNA. Only the 20-bp target sequence is used to construct the sgRNA. For S. pyogenes Cas9, the PAM sequence is 5′-NGG.
Q4: Which one is more efficient for mediating HDR, WT Cas9 or Cas9 nickase?
WT Cas9 is more efficient at mediating homologous recombination than Cas9 nickase. The efficiency of HDR mediated by Cas9 nickase is highly dependent on the cell type. Although Cas9 nickase can reduce the risk for off-target activity, we suggest testing both WT Cas9 and Cas9 nickase when performing gene targeting experiments.
Q5: Does the CBh promoter in pSpCas9 work in my cell line?
The CBh promoter is an altered version of the CAG promoter69, and we have validated it in the following cell lines: HEK 293FT, human ESCs, mouse ESCs, HepG2, HeLa and Neuro 2a. For other cell lines, we recommend testing Cas9 expression by using pSpCas9(BB)-2A-GFP and checking for green fluorescence, or staining for Cas9 expression using antibodies against the 3× FLAG epitope that has been fused to the N-term of Cas9.
Q6: Which method of sgRNA expression is more effective, PCR-generated U6-sgRNA expression cassette or pSpCas9 plasmid carrying the appropriate sgRNA?
We have found that the use of pSpCas9 plasmid often results in higher cleavage efficiency. The use of PCR-generated U6-sgRNA expression cassettes allows easy and rapid comparison of sgRNA efficiencies so that the most optimal sgRNA, in terms of both efficiency and specificity, can be identified before subsequent cloning into pSpCas9.
Q7: Can the HDR template (plasmid or ssODN) contain the sgRNA target site?
We have found that it is possible to achieve HDR by using both plasmids and ssODNs that contain the target site. However, single bases can be silently mutated to prevent cleavage of the templates.
ACKNOWLEDGMENTS
We thank B. Holmes for help with computational tools. P.D.H. is a James Mills Pierce Fellow and D.A.S. is a National Science Foundation (NSF) pre-doctoral fellow. V.A. is supported by NIH Training Grants T32GM007753 and T32GM008313. This work was supported by an NIH Director's Pioneer Award (1DP1-MH100706); an NIH Transformative R01 grant (1R01-DK097768); the Keck, McKnight, Damon Runyon, Searle Scholars, Vallee, Merkin, Klingenstein and Simons Foundations; Bob Metcalfe; and Jane Pauley. Reagents are available to the academic community through Addgene and associated protocols; support forums and computational tools are available via the Zhang lab website (http://www.genome-engineering.org/).
Footnotes
AUTHOR CONTRIBUTIONS F.A.R., P.D.H., J.W., D.A.S. and F.Z. designed and performed the experiments. V.A. contributed to the online tool. F.A.R., P.D.H. and F.Z. wrote the manuscript with help from all authors.
Any Supplementary Information and Source Data files are available in the online version of the paper.
COMPETING FINANCIAL INTERESTS The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Ding Q, et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12:238–251. doi: 10.1016/j.stem.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soldner F, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlson DF, et al. Efficient TALEN-mediated gene knockout in livestock. Proc. Natl. Acad. Sci. USA. 2012;109:17382–17387. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geurts AM, et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433–433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takasu Y, et al. Targeted mutagenesis in the silkworm Bombyx mori using zinc finger nuclease mRNA injection. Insect Biochem. Molec. 2010;40:759–765. doi: 10.1016/j.ibmb.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Watanabe T, et al. Non-transgenic genome modifications in a hemimetabolous insect using zinc-finger and TAL effector nucleases. Nat. Commun. 2012;3:1017. doi: 10.1038/ncomms2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 8.Miller JC, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 9.Sander JD, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat. Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood AJ, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christian M, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang F, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hockemeyer D, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reyon D, et al. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 16.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 17.Sanjana NE, et al. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 2010;64:475–493. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 19.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 20.Makarova KS, et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 22.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jinek M, et al. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 26.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 27.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc-finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 30.Hsu PD, Zhang F. Dissecting neural function using targeted genome engineering technologies. ACS Chem. Neurosci. 2012;3:603–610. doi: 10.1021/cn300089k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez EE, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen F, et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saleh-Gohari N, Helleday T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004;32:3683–3688. doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 37.Sapranauskas R, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magadan AH, Dupuis ME, Villion M, Moineau S. Cleavage of phage DNA by the Streptococcus thermophilus CRISPR3-Cas system. PLoS ONE. 2012;7:e40913. doi: 10.1371/journal.pone.0040913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, et al. Processing-Independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol. Cell. 2013;50:488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen B, et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013;23:720–723. doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang N, et al. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gratz SJ, et al. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;4:1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedland AE, et al. Heritable genome editing in C. elegans via a CRISPRCas9 system. Nat. Methods. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmid-Burgk JL, Schmidt T, Kaiser V, Honing K, Hornung V. A ligation-independent cloning technique for high-throughput assembly of transcription activator-like effector genes. Nat. Biotechnol. 2013;31:76–81. doi: 10.1038/nbt.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 51.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tuschl T. Expanding small RNA interference. Nat. Biotechnol. 2002;20:446–448. doi: 10.1038/nbt0502-446. [DOI] [PubMed] [Google Scholar]
- 54.Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS. Insertion of DNA sequences into the human chromosomal -globin locus by homologous recombination. Nature. 1985;317:230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- 55.Thomas KR, Folger KR, Capecchi MR. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986;44:419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- 56.Hasty P, Rivera-Perez J, Bradley A. The length of homology required for gene targeting in embryonic stem cells. Mol. Cell Biol. 1991;11:5586–5591. doi: 10.1128/mcb.11.11.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu S, Ying GX, Wu Q, Capecchi MR. A protocol for constructing gene targeting vectors: generating knockout mice for the cadherin family and beyond. Nat. Protoc. 2008;3:1056–1076. doi: 10.1038/nprot.2008.70. [DOI] [PubMed] [Google Scholar]
- 58.Elliott B, Richardson C, Winderbaum J, Nickoloff JA, Jasin M. Gene conversion tracts from double-strand break repair in mammalian cells. Mol. Cellular Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guschin DY, et al. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 60.Loman NJ, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat. Biotechnol. 2012;30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 61.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oliveira TY, et al. Translocation capture sequencing: a method for high throughput mapping of chromosomal rearrangements. J. Immunol. Methods. 2012;375:176–181. doi: 10.1016/j.jim.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gray SJ, et al. Optimizing promoters for recombinant adeno-associated virus-mediated gene expression in the peripheral and central nervous system using self-complementary vectors. Human Gene Ther. 2011;22:1143–1153. doi: 10.1089/hum.2010.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.