Abstract
Cultivation of the marine cyanobacterium Moorea producens, collected from the Nabq Mangroves in the Gulf of Aqaba (Red Sea), led to the isolation of new apratoxin analogues, apratoxin H (1) and apratoxin A sulfoxide (2), together with the known apratoxins A-C, lyngbyabellin B and hectochlorin. The absolute configuration of these new potent cytotoxins was determined by chemical degradation, MS, NMR, and CD spectroscopy. Apratoxin H (1) contains pipecolic acid in place of the proline residue present in apratoxin A, expanding the known suite of naturally occurring analogues that display amino acid substitutions within the final module of the apratoxin biosynthetic pathway. The oxidation site of apratoxin A sulfoxide (2) was deduced from MS fragmentation patterns and IR data, and 2 could not be generated experimentally by oxidation of apratoxin A. The cytotoxicity of 1 and 2 to human NCI-H460 lung cancer cells (IC50 = 3.4 and 89.9 nM, respectively) provides further insight into the structure–activity relationships in the apratoxin series. Phylogenetic analysis of the apratoxin-producing cyanobacterial strains belonging to the genus Moorea, coupled with the recently annotated apratoxin biosynthetic pathway, supports the notion that apratoxin production and structural diversity may be specific to their geographical niche.
Marine cyanobacteria are capable of producing suites of highly bioactive secondary metabolites, often of mixed peptide and polyketide biosynthetic origin, with unique cancer cell toxicity profiles. A number of cyanobacterial depsipeptides have served as therapeutic lead compounds in cancer clinical trials.1 Adcetris® (SGN-35, brentuximab vedotin) is a synthetic dolastatin 10 analogue (monomethyl auristatin E), conjugated to an anti-CD30 monoclonal antibody,2 that has most recently gained accelerated FDA approval for Hodgkin's lymphoma (HL) and systemic anaplastic large cell lymphoma (ALCL) as an anti-tubulin agent. Examples of more recently reported cyanobacterial cyclodepsipeptides, with cancer-relevant mechanisms of action that are actively being pursued, include largazole,3 coibamide A,4 grassypeptolides5-7 and apratoxins.8 The depsipeptide scaffold of the apratoxins has become an attractive platform for synthetic efforts aimed at developing more potent and selective analogues, given their sub-nanomolar activity to several cancer cell lines and unique mechanism of action.8-10 This series of nine natural analogues to date is produced by several tropical marine cyanobacterial species of the Moorea genus (formerly classified as Lyngbya bouillonii, L. majuscula, and L. sordida).11-15 Apratoxin A is the first antitumor agent identified to inhibit the cellular secretory pathway by preventing the N-glycosylation and subsequent cotranslational translocation of several cancer-associated receptors (including receptor tyrosine kinases, RTKs) and secreted proteins (e.g., growth factors and cytokines) from the cytoplasm to the endoplasmic reticulum (ER), where they undergo rapid proteasomal degradation.9 Luesch and colleagues8 subsequently synthesized an apratoxin A/E hybrid with greater potency and efficacy. This synthetic analogue not only depleted cellular levels of the tyrosine kinase receptors VEGFR-2 (vascular endothelial growth factor receptor) and PDGFR-β (platelet-derived growth factor receptor, β polypeptide) in mouse colorectal carcinomas (HCT116 xenograft), but also displayed a marked reduction in toxicity compared to apratoxin A. This semi-synthetic hybrid is a second-generation lead that supports the identification of additional cytotoxic apratoxin analogues and medicinal chemistry studies to facilitate the development of a selective, efficacious clinical candidate.8
In the course of our studies to compare the biosynthetic capabilities of Red Sea cyanobacteria with those collected pantropically, we have isolated a new Moorea producens strain (RS05) that shares 98.9% sequence homology (SSU 16S rRNA gene) with the reported apratoxin-producing strains from Guam and Palau. Preliminary LC-MS profiling showed that this RS05 strain produced several compounds within the apratoxin series, as well as a diversity of unrelated depsipeptides. Thus, monophyletic cultures of Moorea producens RS05 were established, grown and harvested over a period of two years. Here we report the structure determination and biological activity of apratoxin H (1), and apratoxin A sulfoxide (2), as well as the isolation or detection of known apratoxins A-C, lyngbyabellin B16 and hectochlorin.17
RESULTS AND DISCUSSION
In 2007, a dark brown filamentous “Lyngbya” cyanobacterium was collected by hand (– 2-5 ft) while snorkeling in the Nabq Mangrove in the Gulf of Aqaba near Sharm el-Sheikh, Egypt. Microscopically, this cyanobacterium contained long, unbranched filaments with wide discoid cells that were contained within thick polysaccharide sheaths, which is consistent with the description of Moorea producens sp. nov.18 A unialgal culture, established through repetitive isolation and subculturing of individual trichomes, as previously described,6 was maintained in laboratory culture and used to propagate multiple large cultures over a period of two years. The CH2Cl2–MeOH (2:1) extract of the Red Sea Moorea producens RS05 was subjected to bioassay-guided fractionation via normal-phase vacuum-liquid chromatography (NPVLC) using a stepped solvent gradient of hexanes to EtOAc to MeOH. The fractions eluting with 80% and 100% EtOAc–hexanes were highly toxic to brine shrimp (0.1 μg/mL resulted in 100% kill). Reversed-phase C18 solid-phase extraction (RP18SPE) and exhaustive RPHPLC of these fractions yielded apratoxin H (1, 3.5 mg) and apratoxin A sulfoxide (2, 0.9 mg) as minor components, together with the known cytotoxic macrolide lyngbyabellin B (4.6 mg), and the highly cytotoxic depsipeptide apratoxin A11 (41.0 mg) as the major component. Furthermore, the 80% EtOAchexanes fraction also showed ion clusters for [M + H]+ at m/z 665/667/669 (100:75:20) and m/z 826.2, suggesting a trace amount of hectochlorin17 (a chlorinated lyngbyabellin analogue) and apratoxin B/C,12 respectively.
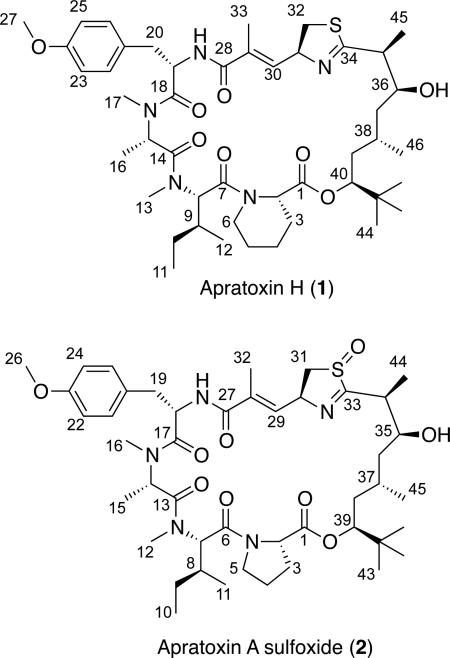
The 1H and 13C NMR spectra for apratoxin H (1) and apratoxin A sulfoxide (2) were very similar to those for apratoxin A11 from a Moorea sp. (previously reported as Lyngbya majuscula and Lyngbya bouillonii) collected from Apra Harbor, Guam. HRTOFMS data provided molecular formulas of C46H71N5O8S ([M + H]+ m/z 854.5074) for 1 and C45H69N5O9S ([M + H]+ m/z 856.4897) for 2, putatively differing from apratoxin A by a methylene and an oxygen atom, respectively. Furthermore, the HRMS data for 1 and 2 were not consistent with any of the known apratoxins (A–G), suggesting the isolation of two new apratoxin analogues from the Red Sea Moorea strain.
The 1H NMR spectrum for 1 showed two mutually coupled aromatic 2H doublets (δ 7.17, 6.82), indicating a para-disubstituted benzene, two 1H deshielded doublets (δ 6.36, 6.05), seven 1H multiplets characteristic of heteroatom-substituted methines or protons (δ 4.3-5.5), a putative methoxy 3H singlet (δ 3.79), two N-methyl 3H singlets (δ 2.71, 2.79) and a 9H methyl singlet (δ 0.89) characteristic of a tert-butyl group. Analysis of the 2D NMR spectra (HSQC, HMBC, COSY, ROESY) for compound 1 confirmed that it possesses identical spin systems and planar connectivities for the polyketide moiety (Dtena, 3,7-dihydroxy-2,5,8,8-tetramethyl-nonanoic acid) and four out of the five amino acid units of apratoxin A: N-Me-alanine (N-Me-Ala), modified cysteine (moCys), N-Me-isoleucine (N-Me-Ile) and O-methyl tyrosine (O-Me-Tyr). The HMBC and COSY experiments identified four contiguous methylene units within the remaining spin system of 1 (Table 1), attributable to the replacement of the proline residue in apratoxin A by pipecolic acid (Pip) in this new analogue, consequently designated as apratoxin H (1).
Table 1.
1H and 13C NMR Spectroscopic Data for Apratoxins H (1, 500 MHz) and A Sulfoxide (2, 700 MHz) in CDCl3.
| Apratoxin H (1) | Apratoxin A Sulfoxide (2) | ||||
|---|---|---|---|---|---|
| position | δC, type | δH (J in Hz)a | position | δC, type | δH (J in Hz)a |
| Pip | Pro | ||||
| 1 | 172.2, C | 1 | 172.6, C | ||
| 2 | 54.4, CH | 4.60, dd (7.0, 3.6) | 2 | 59.8, CH | 4.22, br t (7.4) |
| 3a | 25.7, CH2 | 1.82, m | 3a | 29.5, CH2 | 1.90, m |
| 3b | 2.03, m | 3b | 2.26, m | ||
| 4a | 19.2, CH2 | 1.36, m | 4a | 25.7, CH2 | 1.92, m |
| 4b | 1.62, m | 4b | 2.06, m | ||
| 5a | 24.3, CH2 | 1.56, m | 5a | 48.0, CH2 | 3.69, m |
| 5b | 1.82, m | 5b | 4.18, m | ||
| 6a | 43.1, CH2 | 3.50, m | |||
| 6b | 4.35, m | ||||
| N-Me-Ile | N-Me-Ile | ||||
| 7 | 169.7, C | 6 | 170.5, C | ||
| 8 | 54.4, CH | 5.48, d (11.5) | 7 | 57.3, CH | 5.23, d (11.5) |
| 9 | 31.8, CH | 2.29, m | 8 | 32.2, CH | 2.18, m |
| 10a | 24.6, CH2 | 0.99, m | 9a | 25.0, CH2 | 0.99, m |
| 10b | 1.33, m | 9b | 1.36, m | ||
| 11 | 9.0, CH3 | 0.91, ob | 10 | 9.6, CH3 | 0.95, d (7.5) |
| 12 | 14.5, CH3 | 0.92, ob | 11 | 14.5, CH3 | 0.97, d (7.3) |
| 13 | 30.4, CH3 | 2.71, s | 12 | 30.5, CH3 | 2.66, s |
| N-Me-Ala | N-Me-Ala | ||||
| 14 | 169.6b, C | 13 | 170.2, C | ||
| 15 | 60.8, CH | 3.31, br m | 14 | 60.8, CH | 3.31, m |
| 16 | 14.0, CH3 | 1.25, d (6.7) | 15 | 14.1, CH3 | 1.23, d (6.6) |
| 17 | 36.7, CH3 | 2.79, s | 16 | 36.9, CH3 | 2.82, s |
| O-Me-Tyr | O-Me-Tyr | ||||
| 18 | 170.5, C | 17 | 170.5, C | ||
| 19 | 50.6, CH | 5.06, ddd (10.9, 9.5, 4.8) | 18 | 50.8, CH | 5.05, m |
| 20a | 37.3, CH2 | 2.88, dd (−12.4, 4.8) | 19a | 37.2, CH2 | 2.88, dd (−12.5, 4.5) |
| 20b | 3.14, dd (−12.4, 4.8) | 19b | 3.14, m | ||
| 21 | 128.3, C | 20 | 128.4, C | ||
| 22/26 | 130.6, CH | 7.17, d (8.5) | 21/25 | 130.8, CH | 7.17, d (8.5) |
| 23/25 | 113.9, CH | 6.82, d (8.6) | 22/24 | 114.1, CH | 6.80, d (8.5) |
| 24 | 158.7, C | 23 | 158.9, C | ||
| 27 | 55.3, CH3 | 3.79, s | 26 | 55.5, CH3 | 3.79, s |
| NH | 6.05, d (9.4) | NH | 6.05, d (9.5) | ||
| moCys | moCys | ||||
| 28 | 169.6b, C | 27 | 169.5, C | ||
| 29 | 130.5, C | 28 | 133.8, C | ||
| 30 | 136.4, CH | 6.36, br d (9.6) | 29 | 135.0 | 5.90, d (9.9) |
| 31 | 72.6, CH | 5.23, ddd (9.4, 9.1, 4.5) | 30 | 71.8, CH | 5.70, m |
| 32a | 37.6, CH2 | 3.13, dd (−10.9, 4.7) | 31a | 57.0, CH2 | 2.78, dd (−13.9, 5.7) |
| 32b | 3.47, dd (−10.9, 8.7) | 31b | 3.32, dd (−14.0, 5.9) | ||
| 33 | 13.4, CH3 | 1.97, s | 32 | 13.6, CH3 | 2.0, s |
| Dtena | Dtena | ||||
| 34 | 177.1, C | 33 | 177.2, C | ||
| 35 | 48.8, CH | 2.65, ob | 34 | 48.7, CH | 2.84, dq (10.5, 7.0) |
| 36 | 71.7, CH | 3.56, dddd (10.9, 10.6, 10.4, 3.0) | 35 | 70.8, CH | 3.89, dddd (11.1, 10.8, 10.5, 3.5) |
| 37a | 38.5, CH2 | 1.11, ddd (−13.8, 10.9, 3.0) | 36a | 37.9, CH2 | 1.24, m |
| 37b | 1.46, ddd (−13.7, 11.1, 4.0) | 36b | 1.61, m | ||
| 38 | 24.5, CH | 2.11, br m | 37 | 24.7, CH | 2.12, m |
| 39a | 37.8, CH2 | 1.26, ob | 38a | 37.6, CH2 | 1.31, ob |
| 39b | 1.79, m | 38b | 1.82, m | ||
| 40 | 77.4, CH | 4.90, dd (12.7, 2.2) | 39 | 77.7, CH | 4.98, dd (12.6, 2.2) |
| 41 | 34.9, C | 40 | 35.1, C | ||
| 42/43/44 | 3 × 26.1, CH3 | 3 × 0.89, s | 41/42/43 | 3 × 26.3, CH3 | 3 × 0.89, s |
| 45 | 16.7, CH3 | 1.06, d (6.9) | 44 | 15.7, CH3 | 1.35, d (7.0) |
| 46 | 19.9, CH3 | 1.01, d (6.7) | 45 | 20.0, CH3 | 1.01, d (6.5) |
| OH | 4.40, d (10.9) | OH | 4.65, d (11.5) | ||
J values obtained from 1H spectrum recorded at 700 MHz.
These carbons have the same chemical shift.
ob = obscured.
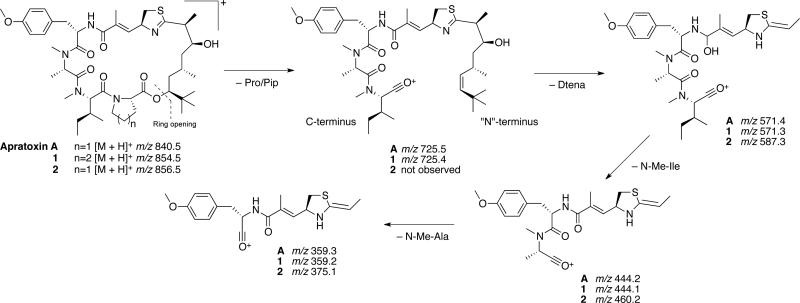
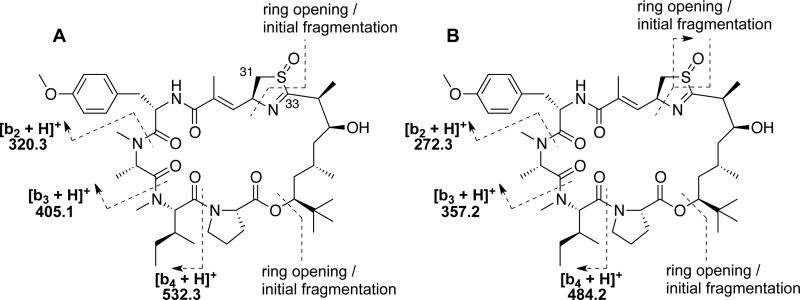
The 1D NMR data for 2 (Table 1) resembled those for apratoxin A most closely. However, an additional midfield resonance (δC 57.0, C-31) was apparent in the 13C NMR spectrum for 2, which also showed one less methylene resonance in the crowded region between δC 36-38. Examination of the 2D NMR spectra (HSQC, HMBC, COSY, ROESY) for compound 2 confirmed that, while the more shielded thiazoline methylene 13C resonance present in the spectrum for apratoxin A (δC 37.6, C-31)11 was absent, compound 2 comprised the same residues and planar connectivity as apratoxin A. Thus, the midfield shift of C-31 in 2, together with an increase of 16 mass units relative to apratoxin A, was consistent with an oxidized nitrogen or sulfur atom within the thiazoline moiety of 2. An IR absorption at 1031 cm−1 was more consistent with an S=O than an N-O (930-980 cm−1) stretching vibration, suggesting the assignment of 2 as apratoxin A sulfoxide. To gain further support for this unusual oxidation, ESIMS data were examined. The mass fragmentation patterns observed for compounds 1 and 2 relative to apratoxin A (Scheme 1) support a pseudo c ion (m/z 725), resulting from ring opening at the ester linkage and loss of Pip/Pro, for compound 1 and apratoxin A. The subsequent loss of the Dtena subunit to leave a fragment of m/z 571 from compound 1 and apratoxin A, is followed by detection of a series of b ions due to loss of N-Me-Ile (m/z 444) and N-Me-Ala (m/z 359), which is consistent with previously reported cyclic peptide fragmentations that exhibit mostly b ions.19 Compound 2 yielded a minor fragmentation pattern that was identical to that of both 1 and apratoxin A. However, the pseudo c ion for apratoxin A and 1 (Scheme 1) was not observed for compound 2. Instead, two major fragmentation pathways were observed, resulting from ring opening and fragmentation of the thiazoline (Figure 1; Supporting Information, Figure S19). A difference of 48 mass units (SO) between the two b ion series is consistent with the presence of a sulfoxide group in 2. For most cyclic peptides, fragmentation occurs following protonation of a peptide bond to open the macrocycle and give a linear form of the molecule with an acylium ion representing the C-terminus.20 However, in compound 2, it appears that oxidation of the thiazoline sulfur atom may lead to an increase in thiazoline ring strain and weakening of the carbon sulfur bond at C-33.
Scheme 1.
Mass fragmentation pattern for compounds 1 and 2 relative to the major MS2 fragmentation pattern observed for apratoxin A.
Figure 1.
Assignment of MS2 fragmentation data for apratoxin A sulfoxide (2).
The presence of a thiazoline sulfoxide moiety in a natural product is unprecedented in the literature. However, prenylated quinones containing a rare 1,1-dioxo-1,4-thiazine ring (e.g., ascidiathiazone A and B) have been reported from marine ascidians.21 Additionally, there are numerous reports of methionine sulfoxide or sulfone residues of putative biosynthetic origin in natural products such as leinamycin22 from a Streptomyces sp., dendroamide C23 from the epilithic cyanobacterium Stigonema dendroideum Fremy, tenuecyclamide D24 isolated from a terrestrial cyanobacterium Nostoc spongiaeforme var. tenue, waiakeamide25 from the Indonesian sponge Ircinia dendroides, haligramide B26 from a Haliclona nigra sponge collected in Papua New Guinea and carriebowmide27 from the marine cyanobacterium “Lyngbya” polychroa. Interestingly, Rashid et al.26 performed the oxidative conversion of haligramide A to waiakeamide in order to compare the overall stereostructures of the two compounds, which presents the possibility that waiakeamide may be an artifact rather than a natural product. However, Sera et al.28 later reported the isolation of a waiakeamide derivative containing both a methionine sulfoxide moiety and a methionine sulfone unit, while differential chemical oxidation of the two methionine units within the haligramides was not observed by Rashid et al.26 Baumann and colleagues29 reported that the cyclic octapeptide planktocyclin undergoes oxidation of its methionine residue upon acidification of the HPLC eluents and during long-term storage under atmospheric oxygen. Therefore, to examine the possibility that apratoxin A was oxidized to sulfoxide 2 during the isolation process, apratoxin A was treated with 30% aqueous hydrogen peroxide with and without acetic acid according to the methods of Rashid et al.26 Analysis of the apratoxin A reaction products by RP18HPLC-MS revealed the presence of apratoxin A and the cysteic acid-containing derivative (m/z [M + H]+ 906.4; [M + Na]+ 928.4; [M – H + 2Na]+ 950.4; 1:1:1) in the ratio of 2.9:1 and 1.7:1 for treatments with and without acetic acid, respectively. An S-dioxo- or S-oxo thiazoline-containing analogue was not detected in any of the LC-MS profiles, suggesting that the sulfur atom within the apratoxin A thiazoline is fully oxidized to cysteic acid under these reaction conditions. Noteworthy is that the haligramides contain a thiazole ring within their core structure that does not appear to undergo oxidation following treatment with acetic acid and 30% aqueous hydrogen peroxide for 6 h.26 Although apratoxin A could not be oxidized to sulfoxide 2 under these reaction conditions, it is still possible that 2 was formed during the isolation process given that aqueous solutions of chlorinated solvents (CH2Cl2–MeOH, 2:1) become weakly acidic over time. E-Dehydroapratoxin A is a derivative that is presumed to form from the acid-catalyzed decomposition of apratoxin A in CDCl3.12 However, E-dehydroapratoxin A was not detected here within the fraction containing compound 2. Alternatively, apratoxin A could undergo posttranslational modification within the cyanobacterium or through the actions of associated heterotrophic bacteria. The selective oxidation of a thiazoline moiety is intriguing and there is no evidence that 2 is an isolation artifact, particularly in the absence of any doubling of NMR chemical shifts that would suggest the presence of diastereomers generated by non-enzymatic oxidation. Further investigation of the function of some of the uncharacterized proteins within biosynthetic pathway of the apratoxins30 may shed light on the possible biosynthetic origin of apratoxin A sulfoxide (2).
The absolute configurations of the amino acid units within apratoxin H (1) and apratoxin A sulfoxide (2) were determined by RP18HPLC-MS of the ozonolysis and acid hydrolysis products of 1 and 2 derivatized with N-α-(5-fluoro-2,4-dinitrophenyl-)-l-leucinamide (advanced Marfey's reagent). Marfey's analyses indicated the presence of N-Me-l-Ala, d-Cya for an S configuration at the α-carbon, N-Me-l-Ile and O-Me-l-Tyr for both 1 and 2. Additionally, l-Pip was present in the hydrolysate of 1, while 2 contained a corresponding l-Pro residue. The relative configuration of the polyketide Dtena moiety (C-33–C-45) of apratoxin H (1) was assigned as in apratoxin A based on little to no variation in the 13C NMR chemical shifts, which is consistent with the closely matching 1H NMR shifts and coupling constants11 (Table 1). Interestingly, the oxidized analogue (2) showed minor 13C chemical shift variations (< 1 ppm) relative to apratoxin A for the C-35 chiral center and CH3-44 within the Dtena subunit (S20), suggesting that oxidation of the sulfur atom alters the conformation of the thiazoline and adjacent carbon skeleton relative to apratoxin A. However, these chemical shift differences, coupled with similar 1H coupling constants11 (Table 1) do not suggest a change in configuration at the C-34 and C-35 chiral centers. The overall stereostructures of apratoxin H (1) and apratoxin A were further compared by circular dichroism (CD), which produced nearly identical negative and positive Cotton effects (CEs) near 236 and 210 nm, respectively (Figure S21). In contrast, sulfoxide 2 displayed a negative CE (ca. 236 nm) nearly 3–fold smaller than for apratoxins A and H (1). This effect is likely caused by a slight change in the overall conformation of the oxidized structure.
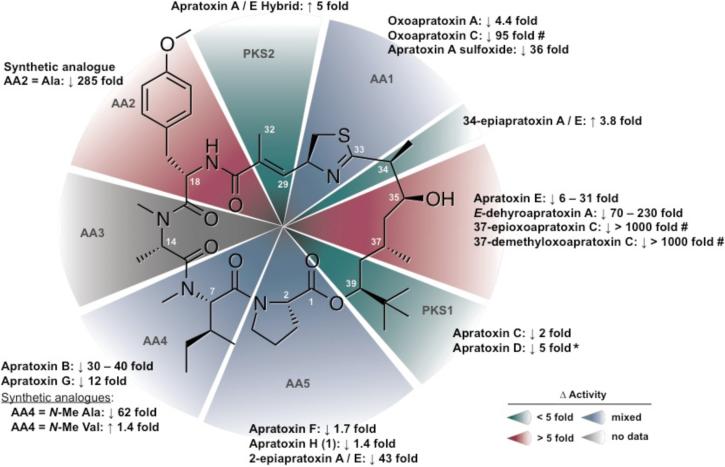
Apratoxin H (1) showed significant cytotoxicity to human NCI-H460 lung cancer cells (IC50 3.4 nM), which is comparable to the results obtained for apratoxin A in this study (IC50 2.5 nM; Supporting Information, Figure S22). The small difference (~1.4-fold) in cytotoxicity between 1 and apratoxin A is consistent with the results obtained for the Pro to N-Me-Ala substitution present in apratoxin F,15 indicating that changes in AA5 do not alter the cytotoxic potential of the molecule (Figure 2). However, a Pro to N-Me-Ala substitution at AA5 in combination with an N-Me-Ile to N-Me-Val exchange (apratoxin G) at the neighboring position (AA4) results in a 7–fold reduction in activity.15 Furthermore, in the apratoxin A/E hybrid, a change in the C-2 configuration of Pro resulted in a 226–fold loss of cytotoxicity, suggesting that the overall stereostructure in this region of the molecule is critical for potent cytotoxicity.8N-Me-Ala substitution at AA48 in the apratoxin A scaffold during the course of chemical mutagenesis studies resulted in a 62–fold decrease in activity against HCT116 colorectal carcinoma cells. In contrast, incorporation of N-Me-Val at AA4 in the apratoxin A scaffold does not affect cytotoxicity,8 (Figure 2) suggesting that a more bulky hydrophobic group (i.e. Ile or Val) is required at this position.
Figure 2.
Structure-activity relationships profile of the apratoxins modified from Tidgewell et al.15 to include cytotoxicity data for 1 and 2. Fold change in activity is relative to apratoxin A in the same cell line by standard MTT assay unless otherwise stated. (* Indicates an indirect comparison across different cell lines; # indicates cytotoxicity relative to oxoapratoxin A).
Apratoxin A sulfoxide (2) showed a nearly 36–fold reduction in cytotoxicity against human NCI-H460 lung cancer cells (IC50 89.9 nM) relative to apratoxin A, while a synthetic oxazoline analogue of apratoxin A was only 4.4–fold less cytotoxic to HeLa cervical carcinoma cells than apratoxin A.10 Together, these results suggest that apratoxin cytotoxicity is sensitive to certain modifications of the moCys thiazoline unit (C-27–C-32). The synthetic apratoxin A/E hybrid, which lacks the allylic methyl group (C-32) and α,β-unsaturation (Δ28), shows a 5.2–fold increase in potency against HCT116 colorectal carcinoma cells and less nonspecific, off-target effects.8 The latter is evidenced by greater in vivo tumor selectivity and a significant reduction of toxicity, which may stem from the inability to form conjugates with cellular nucleophiles following reduction of the Δ28 olefin. The C-34 epimer of the apratoxin A/E hybrid was equipotent,8 which suggests that the conformational flexibility observed in the upper portion of the polyketide moiety in compound 2 does not affect the activity. Thus, it is likely that a loss of pi bond characteristics in the thiazoline ring through resonance between the imino unsaturation and the sulfoxide group may play a more critical role in the loss of activity for 2. Further medicinal chemistry efforts focused on additional modifications of the thiazoline and proline ring systems flanking the polyketide portion of the apratoxin scaffold may fine tune apratoxin selectivity and potency and greatly enhance the SAR with respect to the overall geometry required for this highly cytotoxic molecule.
Apratoxins F, G and H (1) are the only naturally occurring apratoxin analogues containing amino acid substitutions that may stem from relaxed substrate specificity within the adenylation domains of their respective biosynthetic enzymes. Coincidentally, the enzyme responsible for conversion of l-Lys to l-Pip in the biosynthesis of the immunosuppressants rapamycin and FK506 bears a close resemblance to ornithine cyclodeaminases, which convert l-Orn to l-Pro.31 The ability of the final module of the apratoxin biosynthetic pathway to activate and incorporate l-Lys and l-Pip is therefore plausible. However, the incorporation of N-Me-Ala at this terminal position in apratoxin F and G likely results from changes to several of the critical binding pocket residues that mediate substrate specificity or the presence of multiple, cryptic adenylation (A) domains.32 Consistent with these observations, the recently annotated apratoxin biosynthetic gene cluster from an apratoxin-producing Moorea bouillonii cyanobacterium revealed that the final module contained two A-domains and a methyl transferase (MT).30 The presence of an MT in this module explains the incorporation of an N-methyl group in apratoxin F and G, but not the final alanine residue. Furthermore, this MT is likely not functional in the apratoxin A pathway due to the specificity for Pro of the neighboring A-domain. Although the functionality of the second domain could not be predicted accurately due to the absence of the highly conserved active site residue Lys,30 it is likely that such a deletion accounts for the relaxed substrate specificity observed within this domain.
In an effort to characterize the evolutionary relationship of the apratoxin-producing strains of Moorea sp., which have been identified historically as Lyngbya bouillonii, Lyngbya majuscula,and Lyngbya sordida,18 a phylogenetic analysis was performed using the 16S rRNA gene (Figure S23). Noteworthy is that Apratoxins A–C were isolated from collections of Moorea sp. made from Guam and Palau,11,12 while collections of Moorea sp. inhabiting Papua New Guinea13 and Palmyra Atoll15 have afforded apratoxins A–D and apratoxins F and G, respectively (Figure S23). Collectively, these strains, coupled with our apratoxin-producing Red Sea Moorea sp., share 98.9% sequence homology within their SSU 16S rRNA gene, suggesting that these organisms are closely related species. Furthermore, this distinct cluster of Moorea spp. seemingly share a nearly identical biosynthetic pathway for the production of apratoxins, which is likely due to the presence of putative transposases flanking the boundaries of this biosynthetic cluster,30 leading to its mobility and horizontal gene transfer. Remarkably, however, the apratoxins have only been reported from M. bouillonii, and M. producens, suggesting that the successful horizontal gene transfer of this relatively large (57.4 kb) biosynthetic gene cluster has either not occurred outside of this clade, or has thus far evaded detection. Additionally, transposase genes were not detected between any of the genes within the biosynthetic cluster.30 Thus, many of the apratoxin analogues may have resulted from the activation of several cryptic genes within the apratoxin biosynthetic pathway, which are likely regulated by environmental and epigenetic cues present within each organism's unique niche. Furthermore, Luesch and co-workers14 reported the differential production of apratoxin A and a new analogue, apratoxin E, based on the depth from which a Guamanian Moorea bouillonii cyanobacterium was collected, and the presence or absence of the closely associated red alpheid shrimp, Alpheus frontalis. In agreement with these observations, the Red Sea Moorea producens strain was grown in BG-11 medium that had been modified to closely resemble the relatively extreme environmental conditions of the cyanobacterium's natural environment (pH 8.4 and salinity 41‰), which may have contributed to the production of the previously unreported apratoxin analogue, apratoxin H (1). Notably, additional apratoxin analogues were detected in our extracts of the cultured Moorea producens RS05 cyanobacterium. However, these minor components could not be purified in sufficient quantity. Thus, a more meticulous examination of extract components from this and other apratoxin-producing cyanobacteria collected with temporal and/or environmental variations in mind may reveal additional, more diverse apratoxin analogues.
EXPERIMENTAL SECTION
General Experimental Procedures
Optical rotations were measured on a JASCO P-1010 polarimeter. UV spectra and CD measurements were recorded using a JASCO J-815 spectropolarimeter. IR spectra were recorded on a Thermo Scientific Nicolet IR100 FT-IR spectrometer. NMR data were acquired in CDCl3 referenced to residual CHCl3 chemical shifts (δC 77.2, δH 7.26) on a Bruker Avance III 700 MHz spectrometer equipped with a 5mm 13C cryogenic probe for compound 2. NMR data for 1 were acquired in CDCl3 on a Bruker Avance III 500 MHz spectrometer equipped with a 5 mm TXI probe. High-resolution mass spectrometry was performed in positive ion mode on an AB SCIEX Triple TOF 5600 mass spectrometer. Low resolution LC-ESIMS3 data were obtained on an AB SCIEX 3200 Q TRAP mass spectrometer. HPLC was performed using a Shimadzu dual LC-20AD solvent delivery system with a Shimadzu SPD-M20A UV/VIS photodiode array detector.
Collection, Isolation and Culture of the Red Sea Moorea producens strain RS05
A dark brown filamentous assemblage of cyanobacteria was collected by hand while snorkeling from the Nabq Mangroves (1–5 ft) in the Gulf of Aqaba near Sharm el-Sheikh, Egypt (N 27° 21.146′ E 33° 54.472′) on May 31, 2007 (collection code ENq-05/31/07-3, extraction code Nabq-4). Cyanobacterial isolation, morphological characterization and culturing was performed as described previously.6
DNA Extraction, Amplification of Cyanobacterial 16S rRNA
Genomic DNA was extracted and amplified by polymerase chain reactions (PCR) as previously described.6 The 16S rRNA partial gene sequences were inspected visually and assembled using CAP3.33 The resulting contig was analyzed for chimeric sequences using Pintail34 and compared to sequences in the Ribosomal Database Project database (http://rdp.cme.msu.edu) and GenBank (http://www.ncbi.nlm.nih.gov). The consensus sequence was deposited in GenBank under accession number JX470179.
Phylogenetic Analysis
A 16S rRNA gene library of 60 marine and freshwater cyanobacteria was produced from in house SSU (16S) rRNA sequence data for Red Sea cyanobacterial isolates and from sequences collected from the GenBank sequence database. The library was screened for chimeric sequences using the computer program Mallard,35 aligned using ClustalW in Geneious 5.6,36 and the resulting alignment edited to exclude gaps and missing data, for a total of 1275 positions (90.3% identity) in the final dataset covering the V2 to V8 hypervariable regions within the 16S rRNA gene. Prior to phylogenetic predictions, a statistical selection of best-fit models of nucleotide substitutions for the SSU (16S) rRNA data set was selected using Akaike and Bayesian information criteria (AIC and BIC) in jModelTest 0.1.1.37 Phylogenetic trees were calculated using the Bayesian (MrBayes)38 and phylogenetic maximum likelihood (PhyML v3.0)39 algorithms in Geneious 5.6.36 The PhyML analysis was performed with 500 bootstrap replicates using the GTR+I+G model (selected by AIC and BIC in jModelTest; proportion of invariable sites (pINV) = 0.450, shape parameter (α) = 0.387, number of rate categories = 4). Bayesian analysis was performed with the GTR substitution model (number of rate categories = 4). The Markov chain length (three heated) was set to 3 million with sampling performed every 100 generations (25% burn-in).
Extraction and Isolation of Compounds 1, 2, Apratoxin A and Lyngbyabellin B
Culture collections of Moorea producens RS05 (30 × 1.5 L cultures) grown over a span of two years yielded 1.32 g organic extract (CH2Cl2-MeOH, 2:1). The organic extract was subjected to bioassay-guided fractionation via NPVLC using a stepped solvent gradient of hexanes to EtOAc to MeOH to produce nine fractions (A–I). The fractions eluting with 80% EtOAc–hexanes (fraction F) and 100% EtOAc (fraction G) showed potent toxicity to brine shrimp and were further separated C18 reversed-phase (RP18) solid-phase extraction (SPE) using a stepped solvent gradient from 60% MeOH-H2O to 100% MeOH, followed by 100% CH2Cl2. Isocratic RPHPLC (column: Synergi Fusion-RP, 10 × 250 mm, 90% MeOH-H2O, 3.5 mL/min) of the SPE fractions eluting in 80% MeOH-H2O for fraction F and G yielded lyngbyabellin B (4.6 mg, tR = 5.9 min.) and three impure compounds from each fraction with matching UV profiles and retention times. Corresponding peaks were combined from each fraction and targeted for further purification by RPHPLC (Synergi Max, 4.6 × 250 mm) with 80% MeOH-H2O (0.8 mL/min), yielding 2 (0.9 mg, tR = 14.1 min.), or 90% MeOH-H2O (0.7 mL/min), yielding apratoxin A (41.0 mg, tR = 9.3 min.) and 1 (3.5 mg, tR = 10.8 min). LC-MS profiling (Synergi Fusion-RP, 2 × 100 mm, 0.2 mL/min, linear gradient of 65 to 100% MeCN in 0.1% (v/v) aqueous TFA) of fraction F also showed an [M + H]+ m/z 665/667/669 (100:75:20) ion cluster and [M + H]+ m/z 826.2, suggesting a trace amount of hectochlorin and apratoxin B/C, respectively.
Apratoxin H (1): white, amorphous solid; [α]30D–207 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 195 (4.85), 226 (4.45), 284 (3.23) nm; FTIR (neat) νmax 3428 (br), 2962, 2934, 2874, 1738, 1626, 1512, 1456, 1385, 1280, 1247, 1180, 1074 cm−1; 1H and 13C NMR data, see Table 1; HRTOFMS m/z 876.5041 [M + Na]+ (calcd for C46H71N5O8SNa, 876.4921), m/z 854.5074 [M + H]+ (calcd for C46H72N5O8S, 854.5102).
Apratoxin A sulfoxide (2): white, amorphous solid; [α]30D–94 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 193 (4.67), 226 (4.26), 284 (3.14) nm; FTIR (neat) νmax 3402 (br), 2964, 2932, 2872, 1740, 1626, 1512, 1453, 1384, 1277, 1247, 1178, 1072, 1031 cm−1; 1H and 13C NMR data, see Table 1; HRTOFMS m/z 856.4897 [M + H]+ (calcd for C45H70N5O9S, 856.4894).
Apratoxin A: white, amorphous solid; [α]30D–198 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 193 (4.97), 226 (4.41), 269 (3.92) nm; FTIR (neat) νmax 3422 (br), 2965, 2933, 2876, 1742, 1625, 1512, 1456, 1385, 1277, 1248, 1180, 1074 cm−1; 1H and 13C NMR data, see Supporting Information (Figures S15 and S16); HRTOFMS m/z 862.4666 [M + Na]+ (calcd for C45H69N5O8SNa, 862.4764), m/z 840.4927 [M + H]+ (calcd for C45H70N5O8S, 840.4945).
Lyngbyabellin B: white, amorphous solid; [α]30D–135 (c 0.10, MeOH); UV (MeOH) λmax (log ε) 196 (4.48), 240 (3.98) nm; HRTOFMS m/z 679.1775 [M + H]+ (calcd for C28H41Cl2N4O7S2, 679.1794).
Absolute Configuration of Apratoxin H (1) and Apratoxin A Sulfoxide (2)
The amino acid standards relevant to compounds 1 and 2 were obtained commercially and prepared as 50 mM solutions in H2O. Each standard was then derivatized for Marfey's analysis by adding 1 M NaHCO3 (10 μL) and N-α-(5-fluoro-2,4-dinitrophenyl-)-l-leucinamide (l-FDLA or d-FDLA, 1% w/v in acetone, 50 μL) to 25 μL of each standard solution. The mixture was heated at 40 °C for 1 h with continuous stirring, cooled to room temperature, acidified with 2 N HCl (5 μL), evaporated to dryness and resuspended in 1:1 MeCN-H2O (250 μL).
Approximately 0.4 mg of 1 and 0.2 mg of 2 were dissolved separately in 3 mL CH2Cl2 (−78 °C). Ozone was then bubbled through each solution for 15 min. The solution was dried under a stream of N2 gas, followed by an oxidative workup of the residue (0.6 mL of H2O2-HCOOH 1:2 at 70 °C for 20 min). The oxidation product was concentrated under vacuum and hydrolyzed with 6 N HCl (1 mL) in an Ace high-pressure tube, 1200 W microwave for 50 s and immediately cooled to 0 °C and evaporated to dryness. The hydrolyzed products were resuspended in 50 μL MeCN-H2O (1:10) and derivatized for Marfey's analysis in a similar manner to the derivatized chromatographic standards. The Marfey's products of 1 and 2 were resuspended in 1:1 MeCNH2O (100 μL) and analyzed by LC-MS (Kinetex XB-C18 110A, 4.6 x 100 mm, 2.6 μm, 1.5 mL/min, UV and ESIMS detection, 340 nm and negative ion mode, respectively) using a linear gradient of 30 to 70% MeCN (containing 0.1% [v/v] formic acid) and 0.1% (v/v) formic acid in H2O over 15 min. The retention time (tR min, base peak m/z) of the residues in the hydrolysate of 1 matched standards for N-Me-l-Ala (5.29, 396.2), d-Cya (2.12, 462.1), N-Me-l-Ile (7.88, 438.2), l-Pip (7.10, 422.3) and O-Me-l-Tyr (7.20, 488.3). The retention times for the residues in the hydrolysate of 2 were consistent with the results for 1, with the exception of an l-Pro (4.54, 408.2) standard rather than l-Pip matching the corresponding residue in the hydrolysate of 2. The retention times (tR min, base peak m/z) of the other amino acid standards analyzed were: NMe-d-Ala (5.49, 396.2), l-Cya (2.22, 462.1), N-Me-d-Ile (9.48, 438.2), N-Me-l-allo-Ile (8.03, 438.2), N-Me-d-allo-Ile (9.58, 438.2), d-Pip (6.56, 422.3), d-Pro (5.92, 408.2) and O-Me-d-Tyr (8.78, 488.3).
Biological Assays
Lethality of the organic extract and crude fractions was evaluated against brine shrimp (Artemia salina) as previously described40 with some modifications. Samples were added to 24-well plates containing newly hatched (24 h) brine shrimp (10 to 15 per well) in artificial seawater at final concentrations ranging from 1 to 100 ppm. Brine shrimp toxicity was determined after a 24 h incubation period (28 °C) by counting the number of dead shrimp versus total number in each well.
Cytotoxicity of the purified compounds was evaluated in human NCI-H460 lung cancer cells (ATCC, Manassas, VA) as previously described,6 with minor modifications. Cells were seeded into 96-well plates (6,000 cells per well) in 50 μL of medium 12 h before treatment. Approximately 2 h before treatment, test samples were generated from a stock solution (6 mg/mL, 100% DMSO) that was serially diluted in serum-free medium. All purified apratoxins were added to the culture medium at final concentrations ranging from 0.00595 nM to 17.85 μM. Each 96-well plate also contained untreated and vehicle-treated control cells. Cell viability was determined after 48 h treatment using a standard 3-(4,5-dimethylthiazol-2-yl)-2,5,diphenyltetrazolium bromide (MTT) assay, as done previously.6 The cytotoxic potential of each purified compound was assessed in at least three independent cultures with the viability of vehicle-treated control cells defined as 100% in all experiments. Dose response curves were plotted using GraphPad Prism® (v5.0) and IC50 values were derived from nonlinear regression analysis.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Red Sea Protectorate for permission to make collections of Red Sea cyanobacteria, and Jeff Morré of the Environmental Health Sciences Center at OSU for mass spectrometric data acquisition (NIEHS P30 ES00210). The National Science Foundation (CHE-0722319) and the Murdock Charitable Trust (2005265) are acknowledged for their support of the OSU Natural Products and Small Molecule Nuclear Magnetic Resonance. Funding was provided by the OSU College of Pharmacy.
Footnotes
Supporting Information
Tables of 1D and 2D NMR data and NMR spectra for compounds 1 and 2 and apratoxin A in CDCl3. MS/MS and CD spectra for compounds 1 and 2 and apratoxin A. Dose response curves for apratoxins H and A sulfoxide (1 and 2) and A in NCI-H460 cells. Phylogenetic analysis of apratoxin-producing cyanobacteria and photographs and photomicrographs of the laboratory-cultured cyanobacterium. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Tan LT. J. Appl. Phycol. 2010;22:659–676. [Google Scholar]
- 2.Francisco JA, Cerveny CG, Meyer DL, Mixan BJ, Klussman K, Chace DF, Rejniak SX, Gordon KA, DeBlanc R, Toki BE, Law CL, Doronina SO, Siegall CB, Senter PD, Wahl AF. Blood. 2003;102:1458–1465. doi: 10.1182/blood-2003-01-0039. [DOI] [PubMed] [Google Scholar]
- 3.Hong J, Luesch H. Nat. Prod. Rep. 2012;29:449–456. doi: 10.1039/c2np00066k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hau AM, Greenwood JA, Löhr CV, Serrill JD, Proteau PJ, Ganley IG, McPhail KL, Ishmael JE. PLoS ONE. 2013;8:e65250. doi: 10.1371/journal.pone.0065250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwan JC, Ratnayake R, Abboud KA, Paul VJ, Luesch H. J. Org. Chem. 2010;75:8012–8023. doi: 10.1021/jo1013564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thornburg CC, Thimmaiah M, Shaala LA, Hau AM, Malmo JM, Ishmael JE, Youssef DT, McPhail KL. J. Nat. Prod. 2011;74:1677–1685. doi: 10.1021/np200270d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu H, Liu Y, Wang Z, Xing X, Maguire AR, Luesch H, Zhang H, Xu Z, Ye T. Chem. Eur. J. 2013;19:6774–6784. doi: 10.1002/chem.201203667. [DOI] [PubMed] [Google Scholar]
- 8.Chen QY, Liu Y, Luesch H. ACS Med. Chem. Lett. 2011;2:861–865. doi: 10.1021/ml200176m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Law BK, Luesch H. Mol. Pharmacol. 2009;76:91–104. doi: 10.1124/mol.109.056085. [DOI] [PubMed] [Google Scholar]
- 10.Ma D, Zou B, Cai G, Hu X, Liu JO. Chem. Eur. J. 2006;12:7615–7626. doi: 10.1002/chem.200600599. [DOI] [PubMed] [Google Scholar]
- 11.Luesch H, Yoshida WY, Moore RE, Paul VJ, Corbett TH. J. Am. Chem. Soc. 2001;123:5418–5423. doi: 10.1021/ja010453j. [DOI] [PubMed] [Google Scholar]
- 12.Luesch H, Yoshida WY, Moore RE, Paul VJ. Biorg. Med. Chem. 2002;10:1973–1978. doi: 10.1016/s0968-0896(02)00014-7. [DOI] [PubMed] [Google Scholar]
- 13.Gutiérrez M, Suyama TL, Engene N, Wingerd JS, Matainaho T, Gerwick WH. J. Nat. Prod. 2008;71:1099–1103. doi: 10.1021/np800121a. [DOI] [PubMed] [Google Scholar]
- 14.Matthew S, Schupp PJ, Luesch H. J. Nat. Prod. 2008;71:1113–1116. doi: 10.1021/np700717s. [DOI] [PubMed] [Google Scholar]
- 15.Tidgewell K, Engene N, Byrum T, Media J, Doi T, Valeriote FA, Gerwick WH. ChemBioChem. 2010;11:1458–1466. doi: 10.1002/cbic.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milligan KE, Marquez BL, Williamson RT, Gerwick WH. J. Nat. Prod. 2000;63:1440–1443. doi: 10.1021/np000133y. [DOI] [PubMed] [Google Scholar]
- 17.Marquez BL, Watts KS, Yokochi A, Roberts MA, Verdier-Pinard P, Jimenez JI, Hamel E, Scheuer PJ, Gerwick WH. J. Nat. Prod. 2002;65:866–871. doi: 10.1021/np0106283. [DOI] [PubMed] [Google Scholar]
- 18.Engene N, Rottacker EC, Kaštovský J, Byrum T, Choi H, Ellisman MH, Komárek J, Gerwick WH. Int. J. Syst. Evol. Microbiol. 2012;62:1171–1178. doi: 10.1099/ijs.0.033761-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu W-T, Ng J, Meluzzi D, Bandeira N, Gutierrez M, Simmons TL, Schultz AW, Linington RG, Moore BS, Gerwick WH, Pevzner PA, Dorrestein PC. Anal. Chem. 2009;81:4200–4209. doi: 10.1021/ac900114t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ngoka L, Gross M. J. Am. Soc. Mass Spectrom. 1999;10:360–363. doi: 10.1016/S1044-0305(99)00006-9. [DOI] [PubMed] [Google Scholar]
- 21.Pearce AN, Chia EW, Berridge MV, Clark GR, Harper JL, Larsen L, Maas EW, Page MJ, Perry NB, Webb VL, Copp BR. J. Nat. Prod. 2007;70:936–940. doi: 10.1021/np060626o. [DOI] [PubMed] [Google Scholar]
- 22.Hara M, Asano K, Kawamoto I, Takiguchi T, Katsumata S, Takahashi K, Nakano H. J. Antibiot. 1989;42:1768–1774. doi: 10.7164/antibiotics.42.1768. [DOI] [PubMed] [Google Scholar]
- 23.Ogino J, Moore RE, Patterson GML, Smith CD. J. Nat. Prod. 1996;59:581–586. doi: 10.1021/np960178s. [DOI] [PubMed] [Google Scholar]
- 24.Banker R, Carmeli S. J. Nat. Prod. 1998;61:1248–1251. doi: 10.1021/np980138j. [DOI] [PubMed] [Google Scholar]
- 25.Mau CMS, Nakao Y, Yoshida WY, Scheuer PJ, Kelly-Borges M. J. Org. Chem. 1996;61:6302–6304. doi: 10.1021/jo960771e. [DOI] [PubMed] [Google Scholar]
- 26.Rashid MA, Gustafson KR, Boswell JL, Boyd MR. J. Nat. Prod. 2000;63:956–959. doi: 10.1021/np000051+. [DOI] [PubMed] [Google Scholar]
- 27.Gunasekera SP, Ritson-Williams R, Paul VJ. J. Nat. Prod. 2008;71:2060–2063. doi: 10.1021/np800453t. [DOI] [PubMed] [Google Scholar]
- 28.Sera Y, Adachi K, Fujii K, Shizuri Y. J. Nat. Prod. 2003;66:719–721. doi: 10.1021/np020271i. [DOI] [PubMed] [Google Scholar]
- 29.Baumann HI, Keller S, Wolter FE, Nicholson GJ, Jung G, Süssmuth RD, Jüttner F. J. Nat. Prod. 2007;70:1611–1615. doi: 10.1021/np0700873. [DOI] [PubMed] [Google Scholar]
- 30.Grindberg RV, Ishoey T, Brinza D, Esquenazi E, Coates RC, Liu W.-t., Gerwick L, Dorrestein PC, Pevzner P, Lasken R, Gerwick WH. PLoS ONE. 2011;6:e18565. doi: 10.1371/journal.pone.0018565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gatto GJ, Boyne MT, Kelleher NL, Walsh CT. J. Am. Chem. Soc. 2006;128:3838–3847. doi: 10.1021/ja0587603. [DOI] [PubMed] [Google Scholar]
- 32.Challis GL, Ravel J, Townsend CA. Chem. Biol. 2000;7:211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 33.Huang X, Madan A. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. Appl. Environ. Microbiol. 2005;71:7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. Appl. Environ. Microbiol. 2006;72:5734–5741. doi: 10.1128/AEM.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drummond A, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A. 2010.
- 37.Posada D. Mol. Biol. Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 38.Huelsenbeck JP, Ronquist F. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 39.Guindon S, Gascuel O. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 40.Carballo J, Hernandez-Inda Z, Perez P, Garcia-Gravalos M. BMC Biotechnol. 2002;2:17. doi: 10.1186/1472-6750-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.