Abstract
Action potentials propagating along axons require the activation of voltage-gated Na+ (Nav) channels. How Nav channels are transported into axons is unknown. Here we show KIF5/kinesin-1 directly binds to ankyrin-G (AnkG) to transport Nav channels into axons. KIF5 and Nav1.2 channels bind to multiple sites in the AnkG N-terminal domain that contains 24 ankyrin repeats. Disrupting AnkG-KIF5 binding with siRNA or dominant-negative constructs markedly reduced Nav channel levels at the axon initial segment (AIS) and along entire axons, thereby decreasing action potential firing. Live-cell imaging showed that fluorescently-tagged AnkG or Nav1.2 co-transported with KIF5 along axons. Deleting AnkG in vivo or virus-mediated expression of a dominant-negative KIF5 construct specifically decreased the axonal level of Nav but not Kv1.2 channels in the mouse cerebellum. These results indicate AnkG functions as an adaptor to link Nav channels to KIF5 during axonal transport, before anchoring them to the AIS and nodes of Ranvier.
Introduction
Action potentials (APs) propagating along axons play a central role in cell-to-cell communication in the nervous system. AP firing minimally requires the sequential activation of two types of voltage-gated ion channels, Na+ and K+ (Kv) channels, as discovered by Hodgkin and Huxley 60 years ago (Hodgkin and Huxley, 1952). Activation of Nav channels initiates an AP, whereas activation of Kv channels helps terminate it. The Nav channel family contains ten Nav1 channel α-subunits with different channel biophysical properties, regulation, and expression and localization patterns in health and disease, and has been extensively studied (Armstrong and Hille, 1998; Boiko et al., 2003; Catterall, 2012; Hu et al., 2009; Vacher et al., 2008; Waxman, 2012; Payandeh et al., 2011; Payandeh et al., 2012). However, axonal transport of Nav channels is a long-standing mystery.
Crucial for efficient initiation and saltatory propagation of APs along myelinated axons of vertebrates, Nav channels are clustered at the AIS and nodes of Ranvier (Black et al., 1990; Clark et al., 2009; Stuart et al., 1997). The clustering of Nav channels, as well as some K+ channels and cell adhesion molecules at AISs and nodes, is mediated by AnkG (Bennett and Healy, 2009; Dzhashiashvili et al., 2007; Jenkins and Bennett, 2001; Pan et al., 2006; Salzer, 2003; Zhou et al., 1998). Vertebrate ankyrins are encoded by three genes, ankyrin-R, ankyrin-B, and AnkG. They are closely related in their ankyrin repeats in the N-terminus and spectrin-binding domains, but diverge in their C-terminal regulatory domains. Ankyrin repeats in AnkG associate with a variety of ion channels/pumps, calcium release channels, and cell adhesion molecules (Bennett and Baines, 2001; Bennett and Healy, 2009). AnkG links these key membrane proteins to the actin cytoskeleton via spectrins. AnkG is clustered at the AIS via a neuronal intrinsic mechanism, whereas it is recruited to nodes of Ranvier through an extrinsic mechanism via axonal neurofascin-186 guided by myelin membranes (Dzhashiashvili et al., 2007; Feinberg et al., 2011; Hedstrom et al., 2007; Sherman et al., 2005; Zonta et al., 2012). However, nothing is known about how AnkG itself is anterogradely transported into axons, particularly to the nodes of Ranvier far from the cell body (Barry and Gu, 2013).
The microtubule-based forward transport is mainly mediated by kinesin motors. The kinesin superfamily contains 45 members, which selectively transport many different cargos, including different ion channels (Goldstein, 2001; Hirokawa et al., 2010; Vale, 2003). Whereas PDZ- and coiled-coil- domain proteins function as adaptor proteins linking ionotropic glutamate and GABA receptors respectively to different kinesin motors, some voltage-gated ion channel/transporter proteins bind directly to kinesin motors during forward transport (Barry and Gu, 2013; Xu et al., 2010). Despite much progress in this research field, how most ion channels, including Nav channels, are linked to kinesin motors during intracellular forward transport is not known.
Conventional kinesin-1 is a major anterograde motor operating in axons, consisting of a heavy chain (KIF5A, KIF5B or KIF5C) dimer and two light chains (KLC) binding to the C-termini of the dimer. The heavy chains have an N-terminal motor domain, followed by a stalk domain responsible for dimerization through coiled-coil regions, and a C-terminal tail domain containing cargo-binding sites (Asbury et al., 2003; Gennerich and Vale, 2009; Hirokawa et al., 2010). The cargos of kinesin-1 can bind either to KLC or directly to the KIF5 C-terminal tail region (Barry and Gu, 2013; Glater et al., 2006; Hirokawa et al., 2010; Setou et al., 2002; Xu et al., 2010). In this study, we have identified a direct binding between AnkG and KIF5B. The binding is critical for axonal targeting of Nav channels and proper action potential firing. AnkG and Nav1.2 can co-transport with KIF5, revealed by live-cell timelapse imaging. Our finding is further supported by in vivo experiments using cerebellum-specific AnkG knockout mice and virus-mediated expression of a dominant-negative KIF5B construct. Taken together, our results show that AnkG functions as an adaptor to link Nav channels to KIF5 during axonal transport before anchoring them to the AIS and nodes of Ranvier.
Results
Interaction and colocalization of AnkG and KIF5
Using protein pulldown assays followed by tandem mass spectrometry analysis, we previously identified KIF5 as a binding protein of the Kv3.1 T1 domain (Xu et al., 2010). Tetramerized but not monomeric Kv3.1 T1 domains directly bind to the KIF5B tail domain, as the mechanism underlying channel-assembly-dependent axonal transport (Barry and Gu, 2013; Xu et al., 2010). The KIF5B tail domain, containing cargo-binding sites, directly binds to many other proteins (Xu et al., 2010). We used purified GST-tagged KIF5B tail as a bait to pulldown mouse brain lysates, followed by mass spectrometry analysis. We identified AnkG, among many other proteins, as a potential binding protein of KIF5B. However, in contrast to the clustering of AnkG at the AIS and nodes of Ranvier in mature neurons, KIF5 is not enriched at those sites.
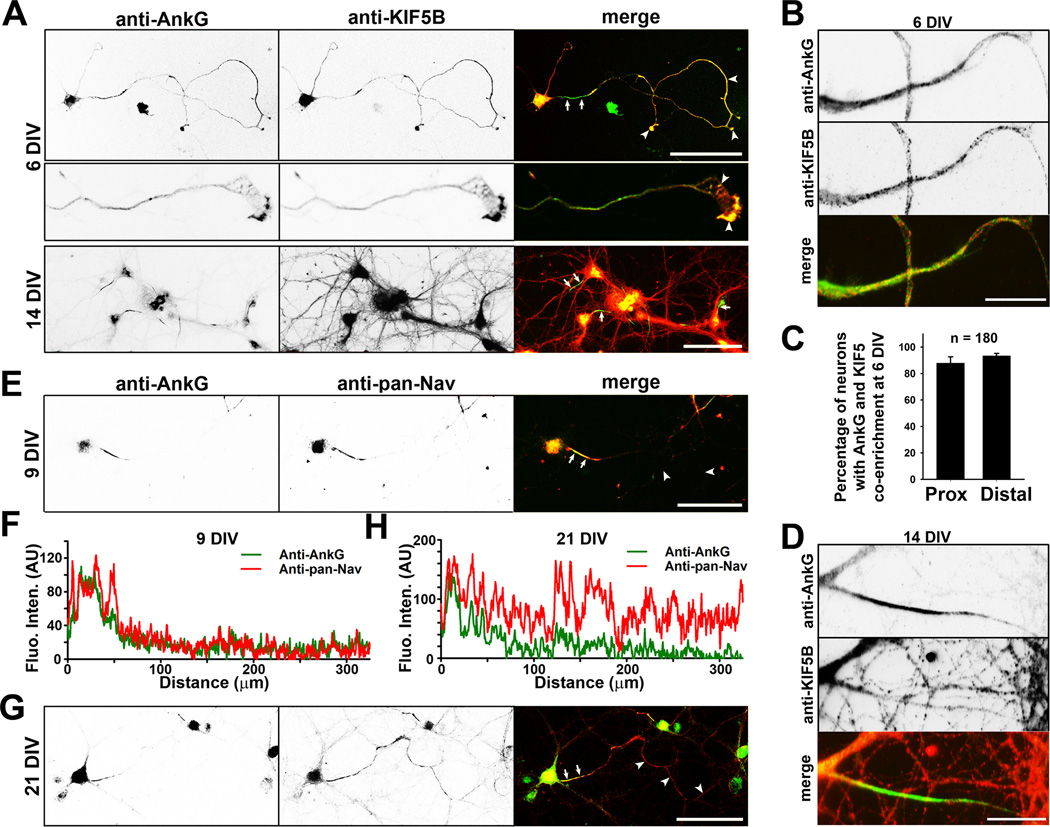
To determine whether AnkG and KIF5 can colocalize in young neurons, we used cultured rat hippocampal neurons, an established model for cell biological study of neurons, with well-characterized developmental stages (Dotti et al., 1988). Interestingly, there was much more colocalization of AnkG and KIF5 along axons in young neurons. At 6 days in vitro (DIV), when the axon-dendrite polarity is just established, AnkG was clearly observed both at the AIS and in distal axons, while KIF5B was colocalized with AnkG along axons, including axonal growth cones (Figure 1A–C). Despite being partially co-clustered, co-enrichment of KIF5B and AnkG was high in both proximal and distal axons (Fig. 1C). At 14 DIV, when synapses start to form, AnkG was highly concentrated at the AIS, while KIF5B was distributed throughout the whole neuron and present but not enriched at the AIS (Figure 1A,D).
Figure 1. Development-dependent colocalization of AnkG, KIF5B and Nav channels along axons.
Hippocampal neurons cultured from rat E18 embryos were fixed and stained at different developmental stages. A, Development-dependent colocalization of KIF5B (red in merged) and AnkG (green in merged) in neurons. Signals are inverted in gray scale images. KIF5B and AnkG colocalized in an axonal growth cone (middle, 3-fold higher magnification compared to the top). B, High magnification images show colocalization of AnkG and KIF5B in proximal axons of neurons at 6 DIV. C, Summary of percentage of neurons with the co-enrichment of AnkG and KIF5B in the proximal (left) and distal (right) axons. D, High magnification images show the AnkG and KIF5B staining in the proximal axon from a neuron at 14 DIV. E, Colocalization of AnkG (green in merged) and pan-Nav channels (red in merged) in hippocampal neurons at 9 DIV. The anti-pan-Nav channel antibodies were used throughout this study. F, Fluorescence intensities of AnkG (green) and pan-Nav channels (red) along the axon in E. G, Colocalization of AnkG (green) and pan-Nav channels (red) in hippocampal neurons at 21 DIV. H, Fluorescence intensities along the axon in G. Arrows, proximal axons including the AIS; Arrowheads, distal axons. Scale bars, 100 µm in A,E,G; 25 µm in B,D. See also Figure S1.
We next examined Nav channels at different developmental stages. At the early stage (6 DIV), both AnkG and KIF5B colocalized with Nav channels along axons and at growth cones (Figure S1A,B). At 9 DIV, AnkG and Nav channels highly clustered at the AIS with much lower levels in distal axons (Figure 1E,F). In older neurons (> 21 DIV), whereas AnkG was highly concentrated at the AIS, Nav channels were present not only at the AIS, but in distal axons as well (Figures 1G,H and S1C). These results raised two interesting questions: Does KIF5 transport both AnkG and Nav channels into axons? Does Nav channel axonal transport require AnkG?
Multiple binding sites mediate the physical interaction between AnkG and KIF5
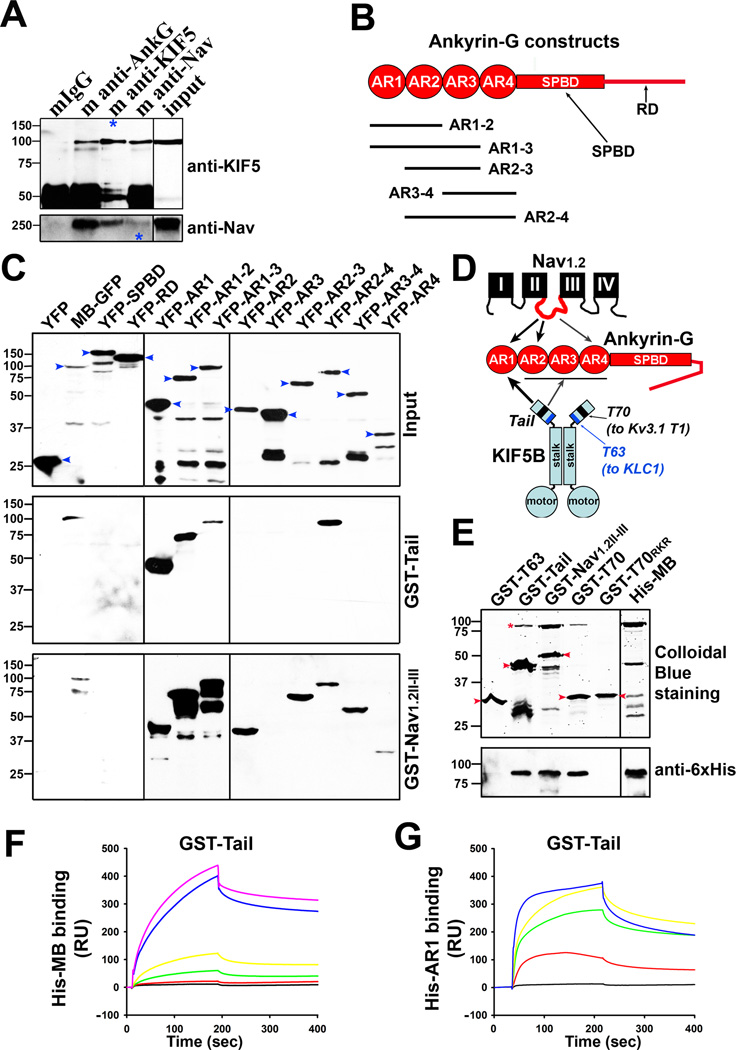
To determine whether AnkG and KIF5 physically interact, we first performed the immunoprecipitation (IP) assay from brain lysates of mouse pups. Both anti-AnkG and anti-pan-Nav channel antibodies precipitated KIF5 (H2) (Figure 2A). Under the same condition, the AnkG antibody did not pull down KIF3A but caused a faint band for KIF4 (Figure S2A,B). The faint band above the KIF4 bands is most likely a nonspecific band, which can be observed sometimes in control IgG as well. On the other hand, though unlikely, it could be interpreted as a real KIF4 band with post-translational modification, which resulted in the shift (Figure S2B). The Ankyrin B (AnkB) antibody did not pull down KIF5 (Figure S2C). Conversely, the KIF5 antibody, but not the KIF3A or KIF4 antibody, pulled down AnkG (Figure S2D). The pull down of AnkG by KIF5 was not very efficient (Figure S2D), which may result from AnkG’s large size and many binding proteins. While AnkG and Nav channel interactions are well known, the binding between AnkG and KIF5 is new. Next, to determine the binding sites mediating the AnkG-KIF5 interaction, we used bacterially expressed GST-Tail (KIF5B tail, aa 758–963), GST as a negative control, and GST-Nav1.2II–III (containing the AnkG-binding site (aa 1080–1203) in Nav1.2 channel as a positive control (Garrido et al., 2003)) to pull down YFP-tagged AnkG fragments (Figures 2B and S2E). Both GST fusion proteins precipitated MB-GFP (the N-terminal membrane-binding domain of AnkG that contains 24 ankyrin repeats) but not YFP-SPBD and YFP-RD expressed in HEK293 cells (Figure 2C). Interestingly, both precipitated the first 6-ankyrin-repeat cluster YFP-AR1 (Figure 2C,D). GST-Nav1.2II–III but not GST-Tail pulled down the second cluster YFP-AR2 (Figure 2C,D). GST-Nav1.2II–III also precipitated the 4th (YFP-AR4) but not the 3rd cluster (YFP-AR3) (Figure 2C,D). Thus, our results surprisingly show that there are multiple binding sites for Nav1.2 in the AnkG N-terminus. In contrast, GST-Tail failed to bind either YFP-AR3 or YFP-AR4, but bound to a larger fragment including the 2nd, 3rd and 4th clusters (YFP-AR2-4) (Figure 2C,D). Therefore, our results show that KIF5 and Nav1.2 have multiple binding sites but distinct binding patterns in the AnkG MB domain (Figure 2D). This raised an intriguing possibility that AnkG can function as an adaptor protein to link Nav channels to KIF5 motors (Figure 2D).
Figure 2. Novel direct binding between AnkG and KIF5.
A,, AnkG and Nav channel interacted with KIF5 in brain lysates from mouse pups (postnatal day 7 to 21). Blue asterisks, 10% of KIF5 (H2) pulldown was loaded (top); 5% of Nav channel pulldown was loaded (bottom). 5% was loaded for both inputs. B, Diagram of AnkG domains and YFP fusion constructs. AR, ankyrin repeat; SPBD, spectrin-binding domain; RD, regulatory domain. C, Identifying the KIF5- and Nav-channel-binding sites within AnkG. Both GST-Nav1.2II–III and GST-Tail pulled down the N-terminal MB domain (membrane-binding domain containing 24 ankyrin repeats), but not SPBD and C-terminal RD. GST-Tail and GST-Nav1.2II–III differ in binding to different ankyrin repeat clusters within the MB domain. YFP-fusion proteins expressed in HEK293 cells (inputs on the top) were pulled down by purified GST-Tail (middle) or GST-Nav1.2II–III (bottom), revealed in Western blotting with an anti-GFP antibody. Molecular weights are indicated on the left. Blue arrowheads, full-length YFP-fusion proteins. D, Diagram shows different binding sites of KIF5 and Nav1.2 in the MB domain of AnkG. The T63 (aa 758–820) and T70 (aa 865–934) regions in KIF5B tail bind to KLC1 and Kv3.1 T1 domain, respectively. E, The T70 region binds to the AnkG N-terminus. The pulldown assay was performed with purified proteins and shown with Colloidal Blue staining for proteins (top) and confirmed with Western blotting with an anti-6×His antibody (bottom). The bands of full-length GST-fusion proteins are indicated with red arrowheads. The full-length His-MB bands are indicated with a red asterisk. There were smaller degradation bands for some proteins. GST-Nav1.2II–III had a weak band near the position of His-MB. F, SPR response curves in measuring the binding affinity between GST-Tail and His-MB. His-MB concentrations (µM): 0, 0.001, 0.005, 0.01, 0.05, and 0.1. G, SPR response curves in measuring the binding affinity between GST-Tail and His-AR1. His-AR1 concentrations (µM): 0, 0.053, 0.265, 0.53, and 1.59. See also Figure S2.
To confirm whether the AnkG-KIF5 binding is direct, we purified His-MB and several GST-tagged KIF5 tail fragments. Indeed, purified GST-Tail and GST-Nav1.2II–III pulled down purified His-MB shown by the Colloidal Blue staining (Figure 2E). To determine whether KIF5 can bind to Nav channels through AnkG, we performed the following pull down assay. We used GST or GST-Tail coated beads to incubate with or without purified His-MB, and further incubated the beads with HEK293 cell lysates containing expressed full-length Nav1.2-GFP channel. GST-Tail but not GST pulled down His-MB and Nav1.2-GFP (Figure S2F left). In the absence of His-MB, GST-Tail did not pull down Nav1.2-GFP (Figure S2F right). Therefore, this result shows that AnkG is required for the interaction between Nav channels and KIF5 motors, and these three proteins can form a complex.
Interestingly, GST-T70 (aa 865–934 in KIF5B tail, containing the Kv3.1 T1-binding site), but not GST-T63 (aa 758–820 in KIF5B tail, containing the KLC1-binding site), nor GST-T70RKR (with the three basic residues (R892K893R894) mutated to three Ds, no longer binding to Kv3.1 T1) (Barry et al., 2013), pulled down His-MB (Figure 2E). Thus, the AnkG MB domain binds to the Kv3.1 T1-binding site in KIF5B tail. To further pinpoint the binding site, we tested three smaller fragments covering the entire T70 region. None of them (GST-Tail865–891, GST-Tail892–913 and GST-Tail913–934) pulled down His-MB efficiently (Figure S2G). Therefore, the binding is not mediated by a small region and requires most of the residues within the T70 region.
To determine the binding affinity between AnkG MB and KIF5B tail, we performed a Surface Plasmon Resonance (SPR) experiment using purified GST-Tail and His-MB or His-AR1 with the Biacore T100 instrument. His-MB bound to GST-Tail with high affinity (KD: 2.4 ± 0.9 ×10−9 M; KON: 2.9 ± 0.6 ×105 M−1s−1; KOFF: 5.9 ± 2.1 ×10−4 s−1; n = 6) (Figure 2F). The binding affinity for His-AR1 was approximately 20 fold less (KD: 4.8 ± 0.8 ×10−8 M; KON: 3.2 ± 0.6 ×104 M−1s−1; KOFF: 1.4 ± 0.2 ×10−3 s−1; n = 6) (Figure 2G). Both the association and disassociation rates for His-AR1 appear faster. It is important to note the binding between AnkG and KIF5 in vivo may differ from purified domains due to different microenvironment. Nonetheless, this result confirms the direct binding between AnkG and KIF5B.
Disrupting AnkG function reduces Nav channel axonal targeting and AP firing
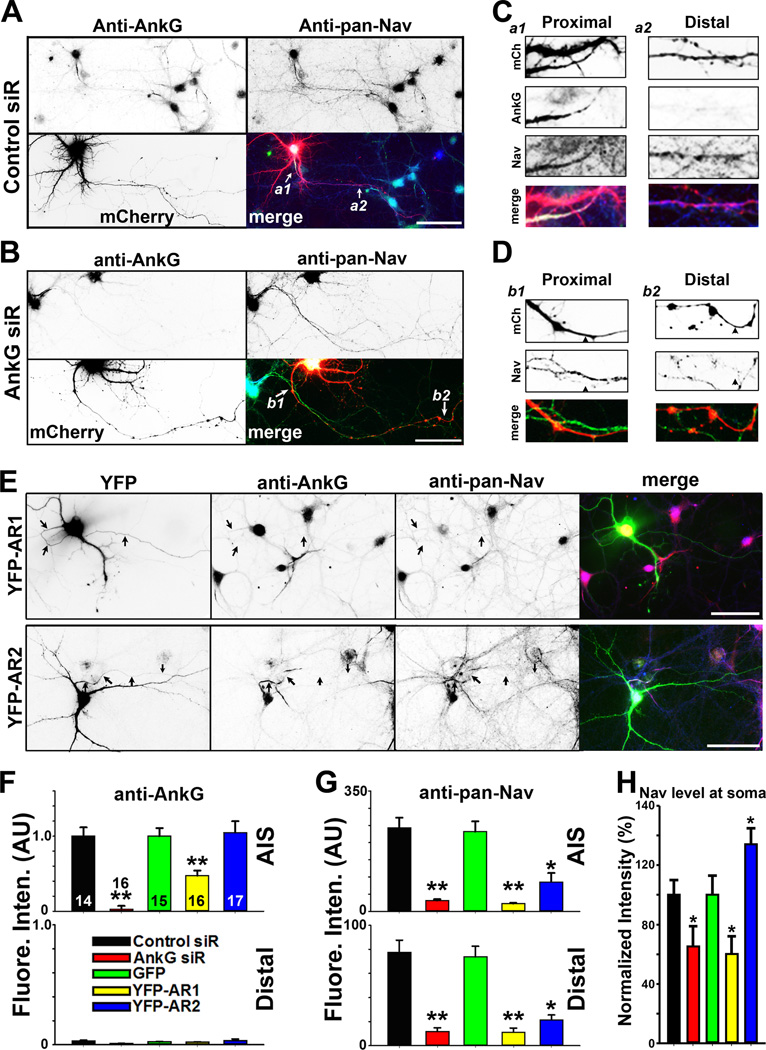
We next examined whether AnkG mediates axonal localization of Nav channels in neurons. Suppressing AnkG expression with siRNA in mature neurons significantly reduced the level of Nav channels not only at the AIS, but also in distal axons (Figures 3A–D,F,G). Expression of AnkG dominant-negative mutants to interfere with AnkG-KIF5 binding also reduced Nav channel levels in axons. YFP-AR1 significantly reduced AnkG at the AIS and overall Nav channel levels (including its level at the soma), whereas YFP-AR2 did not change the AnkG level but reduced Nav channel axonal levels (Figures 3E–G and S3A,B). Interestingly, whereas YFP-AR1 expression reduced Nav channel level at soma, YFP-AR2 expression increased it (Figure 3E,H). This appears to be consistent with our binding data that YFP-AR1 binds to both KIF5 tail and Nav1.2II–III, whereas YFP-AR2 only binds to Nav1.2II–III (Figures 2C,D and 3E). YFP-AR2 may only disrupt the AnkG-Nav binding to prevent Nav channels from entering the axon, resulting in accumulation of Nav channels at soma. On the other hand, since YFP-AR1 can disrupt both AnkG-KIF5 and AnkG-Nav binding, it reduces general forward trafficking of both AnkG and Nav channels, which may stimulate channel degradation at the same time. This also appeared to be the case for AnkG siRNA knockdown (Figure 3H). The exact mechanism underlying interactions among protein synthesis, forward trafficking and degradation of Nav channels remains to be determined in future studies. To confirm that YFP-AR1 disrupts AnkGKIF5 binding, we performed a competition assay using purified proteins and showed that an increased amount of His-AR1 reduced the amount of His-MB binding to GST-Tail (Figure S3C,D). Although it is not clear how many different proteins can bind to AR1, we show that expression of YFP-AR1 did not reduce the axonal levels of CFP-KLC1, CFP-VAMP2, or CFP-SNAP25 (Figure S3E). Taken together, the siRNA knockdown and dominant-negative results support the hypothesis that AnkG-KIF5 binding are required for axonal localization of Nav channels.
Figure 3. Disrupting AnkG-KIF5 interaction suppresses axonal targeting of Nav channels.
A, Control siRNA had no effect on endogenous AnkG and Nav channels along axons. Hippocampal neurons were transfected with control siRNA at 5 DIV, fixed and stained at 21 DIV. Signals are inverted in gray-scale images. In merged image: anti-AnkG, blue; anti-pan-Nav, green; mCherry from the siRNA vector, red. White arrows, proximal and distal axons. B, Knocking down AnkG with siRNA reduced the overall Nav channel level along the entire axon. C, Two enlarged areas (3 fold) in A. a1, proximal axon; a2, middle-distal axon. D, Two enlarged areas (3 fold) in B. b1, proximal axons; b2, distal axons. Black arrowheads, axons with AnkG siRNA. E, YFP-AR1, but not YFP-AR2, reduced AnkG (red in merged) and Nav channel (blue in merged) levels at the AIS. Hippocampal neurons were transfected at 12 DIV, fixed and stained at 17 DIV. In single images, signals are inverted. Black arrows, proximal axons. Scale bars, 100 µm. F, Summary of the effect of siRNA knock down of endogenous AnkG and over-expression of YFP-AR1 and YFP-AR2 (transfected on 12 DIV and fixed on 17 DIV) on endogenous AnkG along axons. Fluorescence intensities were normalized with Control siRNA or GFP. G, Summary of the effect on endogenous Nav channels along axons. H, Normalized fluorescence intensity of the anti-pan-Nav staining at the soma of the neurons. The "n" number is indicated in each column in the upper panel of F. Unpaired t-test was used for the siRNA experiments and One-Way ANOVA followed by Dunnett's test was used for the experiments of YFP-AR1 and YFPAR2. *, p < 0.05; **, p < 0.01. See also Figure S3.
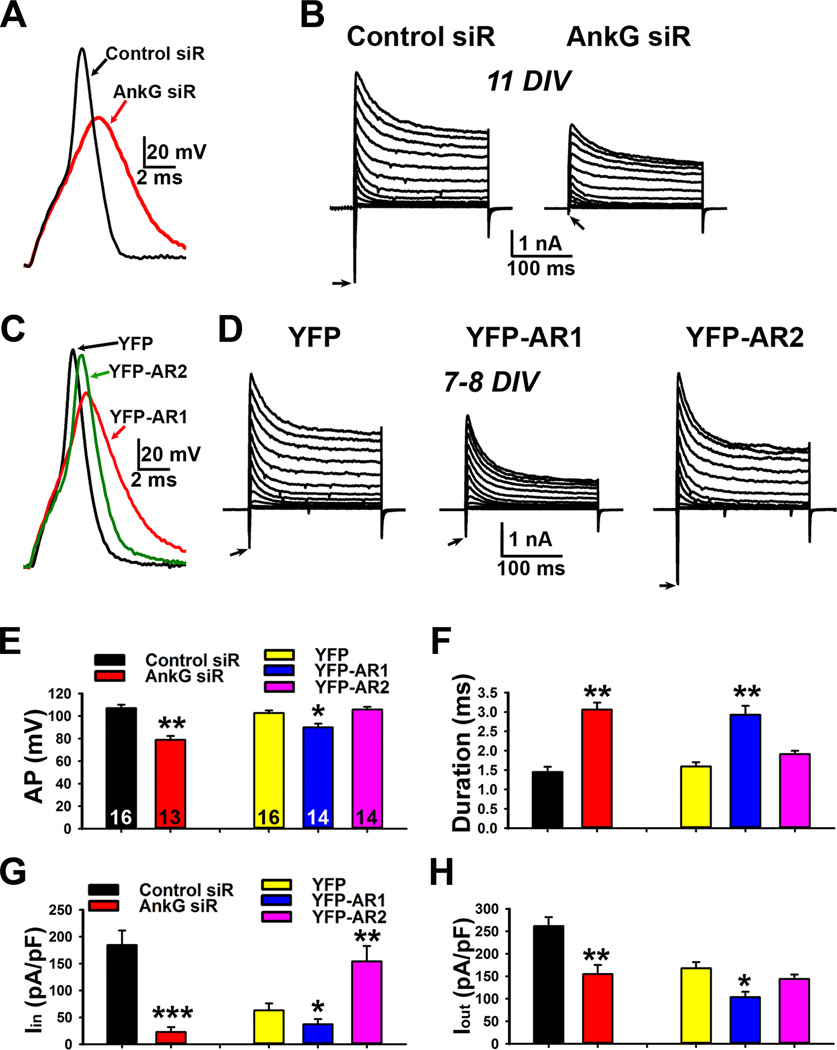
To determine how axonal targeting of Nav channels regulates AP firing, we performed current-and voltage-clamp recording on cultured neurons transfected with either AnkG siRNA or dominant-negative mutants. In current clamp recording, a current pulse was injected into the soma via electrode, which picked up back-propagated APs as previously described (Gu et al., 2012). Suppressing AnkG markedly reduced AP amplitude and increased AP duration, which is consistent with the elimination of inward current density (Figures 4A,B,E–H and S4A–C). On the other hand, YFPAR1 but not YFP-AR2 significantly reduced AP amplitude and increased AP duration (Figures 4C–H and S4D–F). Whereas YFP-AR1 significantly reduced both inward and outward current densities, YFP-AR2 surprisingly increased the inward current without affecting the outward current (Figure 4D,G,H). In YFP-AR2 expressing neurons, Nav channels accumulating at the soma (Figure 3E,H), can give rise to larger inward current (Figure 4D right) and support AP firing. These results are consistent with both binding and staining data (Figures 2C,D and 3).
Figure 4. Knocking down AnkG or over-expression of AnkG dominant-negative fragments differentially changes action potential firing by altering inward and outward currents.
A, Effects of knocking down AnkG on the action potential waveform recorded with current clamp. B, Effects of knocking down AnkG on inward (downward) and outward currents recorded with voltage clamp. Neurons were transfected with siRNA plasmids at 5 DIV and recorded at 11 DIV. C, Differential effects of expressing YFP-AR1 and YFP-AR2 on action potential firing recorded with current clamp. Neurons were transfected at 5 DIV and recorded at 7–8 DIV. D, Effects of YFP-AR1 and YFP-AR2 on inward and outward currents recorded with voltage clamp. E, Summary of the effects on AP amplitude (AP in mV). F, Summary of the effects on AP duration (ms). G, Summary of the effects on inward current density (Iin). H, Summary of the effects on outward current density (Iout). The "n" number is indicated in each column in E. An unpaired t-test was used for the siRNA experiments and One-Way ANOVA followed by Dunnett's test was used for the experiments of YFP-AR1 and YFP-AR2. *, p < 0.05; **, p < 0.01; ***, p < 0.001. See also Figure S4.
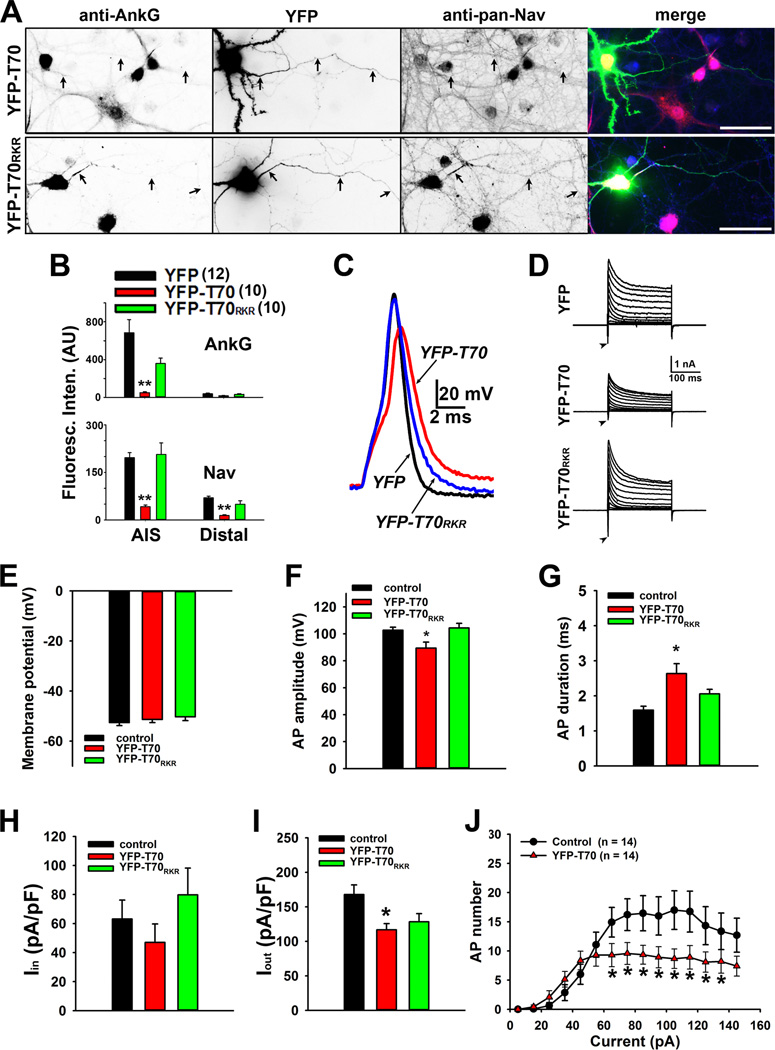
Expressing the AnkG-binding site in KIF5B tail, YFP-T70, reduces axonal levels of AnkG and Nav channels
We further examined how expressing the AnkG-binding site in KIF5B tail alters localization and function of AnkG and Nav channels. YFP-T70 (aa 865–934 within KIF5B tail), but not YFP and YFPT70RKR (aa 865–934 with R892K893R894 mutated to DDD), significantly reduced the levels of AnkG and Nav channels at both the AIS and distal axons (Figures 5A,B and S5A). Since T70 is highly conserved in KIF5A, KIF5B and KIF5C (Xu et al., 2010), YFP-T70 can be used as a dominant-negative mutant for all KIF5 motors eliminating potential compensatory effects. Importantly, YFP-T70 expression significantly reduced AP firing probably via reducing the inward current (Figures 5C–J and S5B), consistent with its inhibitory effect on Nav channel axonal targeting. Taken together, our results support the model that KIF5 transports Nav channels into axons through direct binding to AnkG.
Figure 5. Disruption of AnkG-KIF5 binding by expression of the KIF5 tail fragment reduces axonal targeting of Nav channels and action potential firing.
A, YFP-T70 but not YFP-T70RKR reduces the endogenous levels of AnkG and Nav channels along axons. Black arrows, proximal and distal axons. In merged images: YFP in green, anti-AnkG in red, and anti-pan-Nav in blue. Scale bars, 100 µm. B, Summary of fluorescence intensities of AnkG (top) and Nav channel (bottom) in the AIS and distal axons of neurons expressing either YFP, YFP-T70, or YFP-T70RKR. The "n"s numbers are provided on the top. C, A single action potential induced in the presence of YFP (black), YFP-T70 (red) and YFP-T70RKR (blue) with expanded time scale. D, Voltage-clamp recording of inward and outward currents corresponding to the action potential traces in C. E, The resting membrane potential did not change. F, YFP-T70, but not YFP and YFP-T70RKR, reduced AP amplitude. G, YFP-T70 significantly increased AP duration. H, The inward current remained same. I, YFP-T70 reduced the outward current. J, The input-output relationship significantly reduced in YFP-T70- expressing neurons. 1000-ms duration currents of increasing amplitude (from 5 to 145 pA with increments of 10 pA) were injected into the soma to induce APs. The "n" numbers for electrophysiology experiments are provided in J. One way ANOVA followed by Dunnett's test was used in B, E–I, and an unpaired t-test was used in J. *, p < 0.01; **, p < 0.001. See also Figure S5.
Axonal transport of AnkG and Nav1.2 channels by KIF5B motors
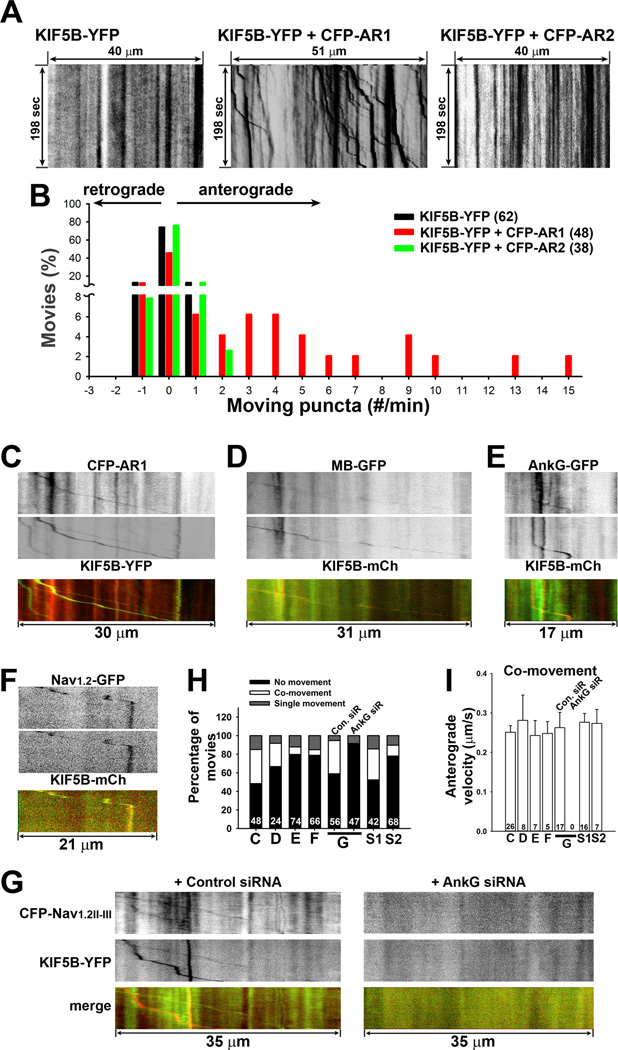
To directly visualize axonal transport of AnkG and Nav channels by KIF5B, we performed live-cell timelapse imaging. First, we examined the effects of CFP-AR1 and CFP-AR2 on the motility of KIF5BYFP. Consistent with the binding result that YFP-AR1 but not YFP-AR2 binds to the KIF5 tail domain (Figure 2C,D), CFP-AR1 but not CFP-AR2 markedly increased the anterograde transport frequency of KIF5B-YFP (Figure 6A,B). Binding of CFP-AR1 activated KIF5B-YFP likely by releasing autoinhibition of the motor protein, similar to other KIF5-binding proteins (Barry et al., 2013), but unlike Kv3.1, CFP-AR1 did not induce clusters of KIF5B-YFP. We further performed the two-color timelapse imaging. Indeed, CFP-AR1 and KIF5B-YFP co-transported along axons (Figure 6C,H and Movie S1). The whole N-terminal of AnkG, MB-GFP was also found to co-transport with KIF5B-mCh (Figure 6D,H). However, the co-movement of full-length AnkG-GFP and KIF5B-mCh was difficult to find under normal conditions, which may result from many other binding proteins of AnkG including the spectrin/actin cytoskeleton. Therefore, we added Latrunculin A (2.5 µM) to depolymerize actin filaments and subsequently performed live-cell imaging. We indeed observed co-transport of AnkGGFP and KIF5B-mCh (Figures 6E,H and S6A,F). Importantly, we observed co-transport of the full-length Nav1.2-GFP and the AnkG-binding site of Nav1.2 (CFP-Nav1.2II–III) with KIF5B constructs (Figure 6F,G). Nav1.2-GFP was highly concentrated at the AIS and present in distal axons at a much lower level (Figure S6B), as previously described (Lee and Goldin, 2009). Knocking down AnkG with siRNA markedly reduced the axonal level of Nav1.2-GFP (Figure S6C). Consistently, no co-movement was observed between CFP-Nav1.2II–III and KIF5B-YFP when endogenous AnkG was knocked down by siRNA (Figure 6G,H). Interestingly, the moving percentage is much higher with co-expression of CFP-Nav1.2II–III, KIF5B-YFP and control siRNA, compared to KIF5B-YFP alone (Figure 6G,H). Although Nav1.2II–III does not directly bind to KIF5B, it may enhance the recruitment of endogenous AnkG to KIF5 and activate KIF5. In the absence of endogenous AnkG, Nav1.2II–III may not activate KIF5, and KIF5 puncta moving frequency becomes comparable to KIF5b-YFP alone (Figure 6G,H).
Figure 6. Co-transport of AnkG, Nav1.2 and KIF5B along axons.
Hippocampal neurons were transfected at 5 DIV and imaged 2–3 days later. A, CFP-AR1 but not CFP-AR2 increased the anterograde transport of KIF5B-YFP, indicated by kymographs of KIF5B-YFP puncta along axonal segments alone (left), or in the presence of CFP-AR1 (middle) or CFP-AR2 (right). B, Frequency of axonal transport of KIF5B-YFP puncta under 3 different conditions in A. C, Kymograph of anterograde co-transport of CFP-AR1 (red in merged) and KIF5B-YFP (green in merged). D, Kymograph of anterograde co-transport of MB-GFP (green in merged) and KIF5B-mCh (red in merged). E, Kymograph of anterograde co-transport of AnkG-GFP (green in merged) and KIF5B-mCh (red in merged), preincubated with 2.5 µM Latrunculin A at 37°C for 2 hrs. F, Kymograph of anterograde co-transport of Nav1.2-GFP (green in merged) and KIF5B-mCh (red in merged). G, Kymographs of co-movement of CFP-Nav1.2II–III (green in merged) and KIF5B-YFP (red in merged) in the presence of control siRNA (left) or AnkG siRNA (right). H, Percentage of anterograde and retrograde co-moving puncta (C: CFP-AR1 + KIF5B-YFP; D: MB-GFP + KIF5B-mCh; E: AnkG-GFP + KIF5B-mCh; F: Nav1.2-GFP + KIF5B-mCh; G: CFP-Nav1.2II–III + KIF5B-YFP; S1: CFP-Nav1.2II–III + YFP-AR1; S2: CFP-AR1 + Nav1.2-YFP). The movie number is provided for each bar. The time length was 198 sec for all kymographs and the distance is provided for each one. I, Average anterograde velocity of co-moving puncta in different conditions. The puncta number is provided for each bar. See also Figure S6.
Furthermore, we visualized co-transport of Nav1.2-YFP and CFP-AR1, and CFP-Nav1.2II–III and YFP-AR1 (Figures 6H and S6D,E and Movie S2). Most co-moving puncta in each condition were anterograde ones (Figure S6G). The average velocities of anterograde co-moving puncta were similar (Figure 6I). We did not detect a clear difference between single-moving and co-moving puncta in terms of transport velocity. Therefore, our live-cell imaging experiments directly showed the anterograde co-transport of AnkG and KIF5B. We also found that the co-transport of Nav1.2 and KIF5B requires endogenous AnkG.
Deleting AnkG in vivo decreases the axonal level of Nav but not Kv1.2 channels
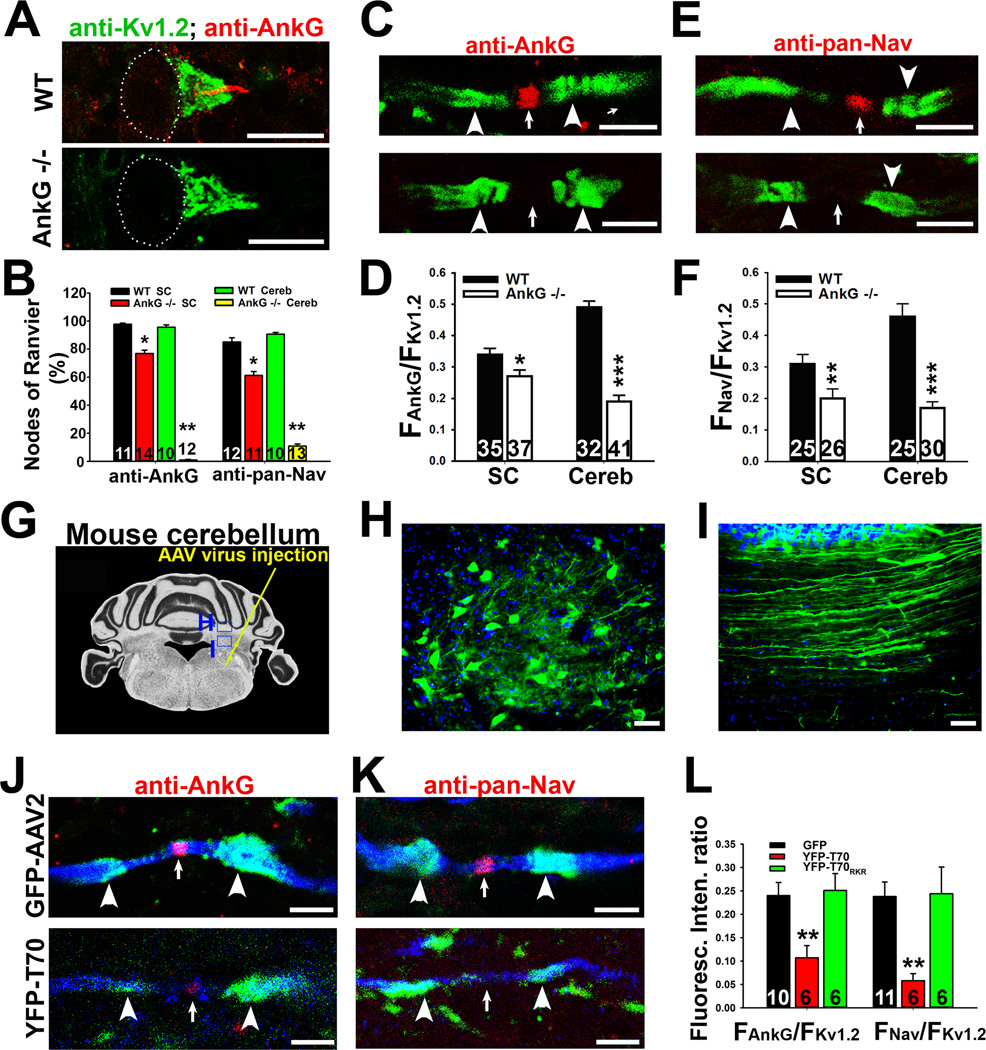
We further tested our model in vivo. In the cerebellum of wildtype (WT) mice, AnkG and Nav channels colocalized at the AIS and nodes of Ranvier, whereas Kv1.2 channels are clustered both in basket cell terminals and at the juxtaparanodal (JXP) regions flanking the nodes of myelinated axons (Jenkins and Bennett, 2001; Wang et al., 1993). In the cerebellum of AnkG KO mice, both AnkG and Nav channel staining was largely eliminated, but the JXP Kv1.2 staining was not altered (Figure 7A–F). In this KO mouse, the AnkG splice variant mainly concentrated in the cerebellum is deleted, while AnkG in other brain regions remains unchanged (Zhou et al., 1998). In spinal cord white matter of AnkG KO mice, AnkG and Nav channels were also missing from some nodes, while the JXP Kv1.2 persisted (Figures 7B and S7A–D). Importantly, when AnkG is absent, the axonal Nav channel was not just dispersed along axons, instead its overall axonal levels were markedly reduced (Figures 7C–F and S7E,F). These in vivo data confirm that AnkG is a critical adaptor protein to mediate axonal transport of Nav channels.
Figure 7. AnkG deletion or disruption of AnkG-KIF5 binding by expressing a KIF5B tail fragment reduces the axonal level of Nav channels in vivo.
Costaining of Kv1.2 and AnkG (or Nav channels) was performed on coronal sections of mouse cerebellum. A, Confocal images of AnkG (red) and Kv1.2 (green) co-staining at the AIS of Purkinje neurons in WT (top) and AnkG −/− (bottom) mice. White dots indicate the soma of Purkinje neurons. B, Summary of the percentage of nodes of Ranvier with AnkG and Nav channel staining in WT and AnkG −/− mice in the cerebellum (Cereb) and spinal cord (SC). The nodal regions were defined by a pair of JXP Kv1.2 clusters. C, High magnification confocal images of AnkG (red) and Kv1.2 (green) staining in cerebellar white matter myelinated axons of WT (top) and AnkG −/− (bottom) mice. D, Summary of the ratio of AnkG and Kv1.2 staining fluorescence along the axonal segment. E, High magnification confocal images of pan-Nav (red) and Kv1.2 (green) staining in cerebellar white matter myelinated axons of WT (top) and AnkG −/− (bottom) mice. F, Summary of the ratio of pan-Nav and Kv1.2 staining fluorescence intensities. G, Diagram for AAV virus injection into mouse cerebellum. H, Neurons in cerebellar nuclei expressing AAV-GFP. I, Axons expressing AAV-GFP in cerebellum white matter. J, Axons expressing either GFP (top, blue) or YFP-T70 (bottom, blue) were co-stained for endogenous AnkG (red) and Kv1.2 (green). K, Axons expressing either GFP (top, blue) or YFP-T70 (bottom, blue) were co-stained for endogenous pan-Nav (red) and Kv1.2 (green) channels. L, Summary of the effect of AAV-mediated expression of dominant-negative construct of KIF5B tail domain. White arrowheads, JXP region; White arrows, nodes of Ranvier. Scale bars, 200 µm in H,I; 50 µm in A; 10 µm in C,E,J,K. An unpaired t-test was used in B,D,F; One way ANOVA followed by Dunnett's test was used in L. *, p < 0.05; **, p < 0.01; ***, p < 0.001. See also Figure S7.
Reducing nodal Nav channels in vivo by AAV-mediated expression of YFP-T70
Finally, we examined the effect of YFP-T70 in mouse cerebellar neurons. We made adeno-associated virus type 2 (AAV2) expressing YFP, YFP-T70, or YFP-T70RKR. The virus was injected into the right side of mouse cerebellum (Figure 7G). Three weeks after injection, we fixed and stained the mouse cerebellar sections. Many neurons of cerebellar nuclei were successfully infected. The soma and axons of those neurons contained GFP or YFP fluorescence (Figure 7H,I). In myelinated axons in cerebellar white matter, expression of YFP-T70 did not alter the Kv1.2 JXP clusters, but significantly decreased the nodal AnkG and Nav channel levels (FAnkG/FKv1.2: GFP, 0.24 ± 0.03; YFP-T70, 0.11 ± 0.03; YFP-T70RKR, 0.25 ± 0.04; FNav/FKv1.2: GFP, 0.24 ± 0.03; YFP-T70, 0.06 ± 0.02; YFP-T70RKR, 0.24 ± 0.06) (Figures 7J–L and S7E,F). Expression of YFP-T70RKR did not alter AnkG, Nav and Kv1.2 channels, similar to the effect of GFP expression. Therefore, disrupting KIF5-AnkG binding eliminated axonal transport of AnkG/Nav channels at the nodes, but not axonal transport of JXP Kv1.2 channels. The result that Kv1.2 JXP clusters were not affected by either deleting AnkG or expressing YFP-T70 is consistent with our previous finding that Kv1.2 is likely transported by kinesin-2 motors (Gu et al., 2006).
Discussion
In this study, we have identified a direct binding between AnkG and KIF5. The binding is mediated by the KIF5 tail domain and the MB domain of AnkG. Importantly, there are multiple binding sites for both KIF5 and Nav1.2 within this MB domain. Thus, AnkG links Nav channels to the KIF5 motor. Indeed, AnkG-KIF5 binding is critical for axonal targeting of pan Nav channels and proper action potential firing. The axonal level of Nav channels was also significantly reduced in cerebellar white matter of both AnkG knockout mice and WT mice expressing YFP-T70, YFP-tagged KIF5 tail fragment containing the AnkG-binding site. The anterograde co-transport of fluorescently-tagged AnkG (or Nav1.2) and KIF5 was observed with two-color live-cell imaging. Therefore, our results show that AnkG functions as an adaptor to link Nav channels to KIF5 during axonal transport.
Multiple binding sites in the MB domain of AnkG
Multiple binding sites were identified for both KIF5 and Nav1.2 in the AnkG MB domain (Figure 2C,D). While Nav1.2 and AnkG binding is well known, the direct binding between KIF5 and AnkG is novel. Our study further shows that both interactions involve multiple regions in the AnkG MB domain, although their binding patterns are different. Our findings are actually consistent with previous studies. Anion exchanger binds to two sites in repeats 7–12 and 13–24 of erythrocyte ankyrin, while neurofascin binds to repeats 7–18 and 13–24 (Michaely and Bennett, 1995a, b). Multiple binding sites raise an interesting possibility that AnkG can bind to ion channels and KIF5 motors simultaneously, and function as an adaptor protein.
Within the KIF5B tail region containing about 200 residues, there are different domains. For instance, the T63 region (aa 758–820) binds to KLC and the T70 region (aa 865–934) binds to Kv3.1 T1 domain, which we identified recently (Barry et al., 2013; Xu et al., 2010). Interestingly, the AnkG binding site in the KIF5 motor is the T70 region (Figure 2E), the exact same binding site for Kv3.1 T1 domain. Since Kv3.1 directly binds to KIF5, AnkG is not required for axonal transport of Kv3.1 channels. In fact, Kv3.1 channels may compete with AnkG for binding to KIF5 motors. The binding affinities of His-31T1 (KD: 6.0 ± 1.4 ×10−8 M) and His-MB (KD: 2.4 ± 0.9 ×10−9 M) to GST-Tail are comparable (Figure 2F) (Barry et al., 2013). It is important to note that although the binding affinities were measured using purified domains that can independently fold and function, these values might be different from those of full-length proteins. Nonetheless, our findings have demonstrated direct binding among these proteins. They also explain why Kv3.1 channels are not concentrated at the AIS in some neurons (Devaux et al., 2003; Johnston et al., 2011; Xu et al., 2007).
AnkG is involved in KIF5-mediated transport of some axonal proteins
Recent studies show that AnkG is localized at the AIS to regulate the passage of axonal proteins and cargos (Maniar et al., 2011). An AnkG and F-actin-dependent cytoplasmic filter at the AIS emerges within two days after axon-dendrite differentiation, allowing axonal entry of small diffusible (10 kDa) dextrans but not large (70 kDa) ones (Song et al., 2009). In mature neurons, AnkG at the AIS plays an important role in axon-dendrite polarity maintenance (Sobotzik et al., 2009). AnkG-based AIS formation is regulated by distal axonal cytoskeleton components, including AnkB, αII and βII spectrins (Galiano et al., 2012). Despite the functional significance of AnkG, how AnkG is transported down the axon, especially at the nodes of Ranvier far from the soma along myelinated axons, was not known. In the present study, our in vitro and in vivo experimental results show that KIF5 directly binds to AnkG to transport Nav channels into axons, including the AIS. This represents an important way for large protein complexes and organelles to be transported through the AIS. Suppressing endogenous AnkG by siRNA knockdown reduced the pan Nav channel level not only at the AIS but also in distal axons (Figures 3A–D,F,G). Consistent with the immunostaining results, the AP amplitude decreased and its duration increased, due to significant reduction of inward currents (Figure 4A,B,E,F). In AnkG KO mice, Nav channel levels reduced both at nodes and along entire axons (Figures 7A–H and S7). Thus, deleting AnkG seems not just to disperse the Nav channels clustered at the nodes, but to reduce the overall channel protein levels. The result supports the hypothesis that AnkG functions as an adaptor to transport Nav channels.
Using dominant-negative mutants (YFP-AR1 and YFP-T70) to disrupt AnkG-KIF5 binding not only reduced AnkG at the AIS, but also decreased pan Nav channel level both at the AIS and along distal axons (Figures 3E–G and 5A,B). The reduction of Nav channels at the AIS is consistent with reduced AP firing (Figures 4C–H and 5C–J). Similar to the binding to Kv3.1 T1 domain (Barry et al., 2013), mutating R892K893R894 in KIF5 tail also eliminated the AnkG-KIF5 binding (Figure 2E), so YFP-T70RKR did not affect Nav channel targeting and AP firing (Figure 5). These results are consistent with the hypothesis that AnkG-KIF5 binding is required in axonal targeting of Nav channels.
Our hypothesis that AnkG links Nav channels to KIF5 motors during axonal transport is further supported by two-color live cell imaging results. Both fragments and the full-length of AnkG co-transported with KIF5B along axons of cultured hippocampal neurons (Figure 6). However, the co-transport of AnkG-GFP and KIF5B-mCh was difficult to find under normal conditions. The co-transport events significantly increased when actin filaments were disrupted in the presence of Latrunculin A (Figure 6). It is possible that normally a few AnkG molecules are sufficient to mediate the loading of a Nav-channel-containing vesicle to KIF5 motors. Consistent with this idea, we did observe co-transport of Nav1.2-GFP and KIF5-mCh, which requires endogenous AnkG (Figures 6 and S6). Taken together, our studies indicate that AnkG can participate in selective transport of some axonal cargos, particularly those AnkG-binding proteins, as a key adaptor protein of KIF5 motor proteins.
Selective transport of nodal and JXP proteins along myelinated axons
Our results have demonstrated the causal relationship between KIF5 motor and nodal proteins, including AnkG and Nav channels, consistent with a recent work showing the slow replenishment of components during node maintenance depends on transport (Zhang et al., 2012). Cell adhesion molecules rely on diffusion to cluster to the nodes of Ranvier, whereas the channel proteins and cytoskeleton proteins rely on axonal transport to cluster into the nodes of Ranvier. Interestingly, either deleting AnkG or disrupting AnkG-KIF5 binding by expression of the KIF5 tail fragment did not affect the JXP clustering of Kv1.2 channels (Figure 7). This result is consistent with our previous findings that Kv1.2 channels are likely transported by KIF3/kinesin-2 motors (Gu et al., 2006; Gu and Gu, 2010). However, it is not known whether other JXP proteins, such as transient axonal glycoprotein-1 and contactin-associated protein-like 2 (Gu and Gu, 2011), are also transported by kinesin-2. Similarly, although AnkG binds to many membrane proteins at nodes of Ranvier, it remains to be determined whether all nodal proteins and all 10 Nav channel α subunits are exclusively transported by KIF5 through AnkG. Furthermore, after reaching the proper axonal site (such as the AIS and node of Ranvier), AnkG will disassociate from KIF5 to unload the cargo, including Nav channels. AnkG can remain associated with Nav channels for local retention. Some AnkG molecules, especially those in unmyelinated axons, may be transported back to soma and/or degraded, which remains to be investigated. The regulation of disassociation of AnkG and KIF5 may be mediated by posttranslational modifications, such as phosphorylation, which has been implicated in regulation of AnkG binding to other proteins and of KIF5 binding to cargo proteins (Barry and Gu, 2013; Cianci et al., 1988; Ghosh et al., 2002; Woroniecki et al., 2003). These are interesting research topics for future studies.
In the present study, our experimental data lead to a model that AnkG functions as an adaptor protein to link multiple types of Nav channels to KIF5 motors during axonal transport to the AIS and nodes of Ranvier, and subsequently clusters these channel proteins at these sites. This set of machinery for axonal transport and local retention is not shared by axonal Kv1.2 channels. The precise targeting of Nav channels is vital for efficient initiation and saltatory conduction of APs.
Experimental Procedures
cDNA constructs
AnkG-GFP (270 kDa), MB-GFP, GST-31aC, GST-Tail, GST-T63, YFP-T63 and YFP-T70 were described previously (Gu et al., 2003; Xu et al., 2007; Xu et al., 2010). YFP-T70RKR was made by using the Quickchange method to mutate residues R892K893R894 to DDD. GST-Nav1.2II–III was made by fusing the intracellular loop region between II and III membrane segment clusters (aa 1080–1203) of Nav1.2 to the glutathione S-transferase (GST) C-terminus in pGEX4T-2. YFP-SPBD (aa 792–1871), YFP-RD (aa 1782–2622), YFP-AR1 (aa 2–236), YFP-AR2 (aa 237–467), YFP-AR3 (aa 468–665), YFPAR4 (aa 666–791), YFP-AR1-2 (aa 2–467), YFP-AR1-3 (aa 2–665), YFP-AR2-3 (aa 237–665), YFPAR2-4 (aa 237–791), and YFP-AR3-4 (aa 468–791) were made by fusing the regions, indicated by residual number of AnkG (270 kDa) to the YFP C-terminus in pEYFP-C1. His-MB (aa 2–791) and His- AR1 (aa 2–236) were made by inserting the regions into the pRSET-B vector. All constructs were confirmed by sequencing. Nav1.2-GFP was kindly provided by Dr. Alan Goldin.
Hippocampal neuron cultures and transfection
Hippocampal neuron culture was prepared as previously described from embryonic day 18 (E18) embryos (Gu et al., 2006). See supplementary materials for details.
Short interfering RNA knockdown of endogenous AnkG
Construction and validation of vector-based short interfering RNA (siRNA) strategy to suppress the levels of endogenous proteins in rat hippocampal neurons were previously described (Xu et al., 2007).
Fluorescence microscopy and quantification
Fluorescence images were captured with a Spot CCD camera RT slider (Diagnostic Instrument Inc., Sterling Heights, MI, USA) in a Zeiss upright microscope, Axiophot, using Plan Apo objectives 20×/0.75 and 100×/1.4 oil, saved as 16-bit TIFF files, and analyzed with NIH ImageJ and SigmaPlot 12.0 for fluorescence intensity quantification.
Surface Plasmon Resonance (SPR) experiments
SPR experiments were performed on a Biacore T100 instrument (GE Healthcare, Piscataway, NJ, USA) with CM5 sensor chips using IP buffer (50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 1% Triton X-100) as the running buffer. 0.6 µg of purified GST, GST-Tail, and GST-Nav1.2II–III were immobilized onto different flow cells using the GST capture kit (Biacore). Purified His-MB or His-AR1 (0 – 1590 nM) were injected at a flow rate of 20 µl/min for 3 min. All sensorgram data shows the total signal (GST fusion proteins) that was subtracted by the nonspecific signal (GST). The KD was calculated using kinetics fitting.
Whole-cell patch-clamp recording
Both current clamp and voltage clamp recording procedures were previous described (Gu et al., 2012).
Two-color live-cell imaging
Both the procedure and quantification of two-color live-cell imaging were previously described (Barry et al., 2013; Gu and Gu, 2010; Xu et al., 2010).
AnkG KO mice, cardiac perfusion, tissue fixation and sectioning
AnkG KO mice were previously published (Jenkins and Bennett, 2001). The procedure of cardiac perfusion, tissue fixation and sectioning were previously described (Jukkola et al., 2012; Jukkola et al., 2013).
Immunofluorescent staining of sections of brain and spinal cord
Sections were incubated in PBS/0.3% Triton X-100 for 1 hr at RT to permeabilize the tissue, then blocked with 2.5% normal goat or donkey serum (matched with the host species of the secondary antibody) for 1 h at RT. The primary antibodies were then added in blocking solution, and the sections were incubated for 3 hr at RT, then overnight at 4°C. The next day, the sections were rinsed 10 × 5 min at RT, the appropriate secondary antibody was added in blocking solution, and the sections were incubated for 3 hr at RT. Sections were rinsed 10 × 5 min at RT and coverslipped using tris-buffered Fluoro-Gel mounting media (Electron Microscopy Sciences, Hatfield, PA, USA).
Confocal microscopy
High-magnification confocal images were captured with a Leica TCS SL confocal imaging system (Leica Microsystems, Mannheim, Germany), using a 100× HCX Plan Apo CS oil immersion objective (numerical aperture = 1.40). Multiple channels were acquired simultaneously, and the signal was averaged over six scans. Channel crosstalk was eliminated through optimization of the laser line intensity by acousto-optical tunable excitation filters, and by spectral detectors allowing precisely-defined bandwidth adjustment. Images were saved as 8-bit TIFF files and adjusted for brightness and contrast using Adobe Photoshop 7.0.
Adeno-associate virus type 2 (AAV2) construction and production
YFP, YFP-T70 and YFP-T70RKR were subcloned into the AAV vector (kindly provided by the Viral Core at The Research Institute at Nationwide Childrens' Hospital) using NotI restriction site. AAV2 viruses expressing YFP (titer: 2.1 ×1013), YFP-T70 (titer: 1.9 × 1013), or YFP-T70RKR (titer: 1.0 × 1013) were made at the Viral Core facility at The Research Institute at Nationwide Childrens’ Hospital.
Surgeries and microinjection of viral vectors
YFP, YFP-T70, or YFP-T70RKR was overexpressed in mouse cerebellum by stereotaxic injection of recombinant AAV2 virus. Each mouse was unilaterally injected on the right side, leaving the uninjected left side a within-subject control. Cohorts of mice injected with AAV-YFP (n = 6), AAV-YFP-T70 (n = 6), or AAV-YFP-T70RKR (n = 6) were produced for between-subject comparisons. After injection, the mice were sutured and administered post-operative care for one week. Mouse cerebellum was fixed and sectioned 2 to 3 weeks after injection for immunohistochemistry studies.
Statistical analysis
We performed One-Way ANOVA when comparing 2 or 3 experimental groups to one control group, and unpaired t-tests when comparing two groups, using Sigmaplot 12.0. Results are provided as mean ± SEM.
Supplementary Material
Highlights.
AnkG and KIF5 are highly colocalized along axons of young but not mature neurons.
KIF5 and Nav1.2 channels bind to multiple sites in the AnkG N-terminus.
AnkG mediates axonal Nav channel transport by KIF5.
AnkG and KIF5 binding is required for targeting of Nav but not Kv1.2 channels.
Acknowledgement
We thank University of California, Davis-NIH NeuroMab facility (a nonprofit supplier) for monoclonal anti-Kv1.2 channel, anti-GFP and anti-AnkG antibodies, Drs. Michael Murphy and Andrew Dangel for comments on the SPR experimental results, Plant-Microbe Genomics Facility for providing the Biacore T100 instrument, Dr. Alan Goldin for the Nav1.2-GFP cDNA construct, Dr. Vann Bennett for comments on the manuscript, and the Viral Core facility at The Research Institute at Nationwide Children’s Hospital for AAV2 virus production. This work was supported by a grant from the US National Institute of Neurological Disorders and Stroke/National Institutes of Health (R01NS062720) to CG, and NIH R01DA014610 to HG and NIH HL084583, HL083422, HL114383 to PJM. All animal experiments have been conducted in accordance with the NIH Animal Use Guidelines.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Figures S1–S7
Supplemental Movies S1 and S2
Supplemental Experimental Procedures
Supplemental References
Author Contribution
C.G. designed and supervised the research; J.B, Y.G., P.J. and C.G. performed experiments. B.O., K.R. and H.G. performed viral vector injection. P.J.M. provided mouse anti-AnkG antibody and AnkG −/− mice. J.B., Y.G. and C.G. analyzed the data, made figures and wrote the paper. H.G. and P.J.M. edited and commented on the manuscript.
References
- Armstrong CM, Hille B. Voltage-gated ion channels and electrical excitability. Neuron. 1998;20:371–380. doi: 10.1016/s0896-6273(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Asbury CL, Fehr AN, Block SM. Kinesin moves by an asymmetric hand-over-hand mechanism. Science. 2003;302:2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry J, Xu M, Gu Y, Dangel W, Jukkola P, Shrestha C, Gu C. Activation of Conventional Kinesin Motors in Clusters by Shaw Voltage-Gated Potassium Channels. J Cell Sci. 2013;126:2027–2041. doi: 10.1242/jcs.122234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry J, Gu C. Coupling mechanical forces to electrical signaling: molecular motors and intracellular transport of ion channels. Neuroscientist. 2013;19:145–159. doi: 10.1177/1073858412456088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett V, Baines AJ. Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol Rev. 2001;81:1353–1392. doi: 10.1152/physrev.2001.81.3.1353. [DOI] [PubMed] [Google Scholar]
- Bennett V, Healy J. Membrane domains based on ankyrin and spectrin associated with cell-cell interactions. Cold Spr Harb Persp in Biol. 2009;1:a003012. doi: 10.1101/cshperspect.a003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JA, Kocsis JD, Waxman SG. Ion channel organization of the myelinated fiber. Trends Neurosci. 1990;13:48–54. doi: 10.1016/0166-2236(90)90068-l. [DOI] [PubMed] [Google Scholar]
- Boiko T, Van Wart A, Caldwell JH, Levinson SR, Trimmer JS, Matthews G. Functional specialization of the axon initial segment by isoform-specific sodium channel targeting. J Neurosci. 2003;23:2306–2313. doi: 10.1523/JNEUROSCI.23-06-02306.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA. Voltage-gated sodium channels at 60: structure, function and pathophysiology. J Physiol. 2012;590:2577–2589. doi: 10.1113/jphysiol.2011.224204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianci CD, Giorgi M, Morrow JS. Phosphorylation of ankyrin down-regulates its cooperative interaction with spectrin and protein 3. J Cell Biochem. 1988;37:301–315. doi: 10.1002/jcb.240370305. [DOI] [PubMed] [Google Scholar]
- Clark BD, Goldberg EM, Rudy B. Electrogenic tuning of the axon initial segment. Neuroscientist. 2009;15:651–668. doi: 10.1177/1073858409341973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux J, Alcaraz G, Grinspan J, Bennett V, Joho R, Crest M, Scherer SS. Kv3.1b is a novel component of CNS nodes. J Neurosci. 2003;23:4509–4518. doi: 10.1523/JNEUROSCI.23-11-04509.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. J Neurosci. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhashiashvili Y, Zhang Y, Galinska J, Lam I, Grumet M, Salzer JL. Nodes of Ranvier and axon initial segments are ankyrin G-dependent domains that assemble by distinct mechanisms. J Cell Biol. 2007;177:857–870. doi: 10.1083/jcb.200612012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg K, Eshed-Eisenbach Y, Frechter S, Amor V, Salomon D, Sabanay H, Dupree JL, Grumet M, Brophy PJ, Shrager P, Peles E. A glial signal consisting of gliomedin and NrCAM clusters axonal Na+ channels during the formation of nodes of Ranvier. Neuron. 2011;65:490–502. doi: 10.1016/j.neuron.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiano MR, Jha S, Ho TS, Zhang C, Ogawa Y, Chang KJ, Stankewich MC, Mohler PJ, Rasband MN. A distal axonal cytoskeleton forms an intra-axonal boundary that controls axon initial segment assembly. Cell. 2012;149:1125–1139. doi: 10.1016/j.cell.2012.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido JJ, Giraud P, Carlier E, Fernandes F, Moussif A, Fache MP, Debanne D, Dargent B. A targeting motif involved in sodium channel clustering at the axonal initial segment. Science. 2003;300:2091–2094. doi: 10.1126/science.1085167. [DOI] [PubMed] [Google Scholar]
- Gennerich A, Vale RD. Walking the walk: how kinesin and dynein coordinate their steps. Curr Opin Cell Biol. 2009;21:59–67. doi: 10.1016/j.ceb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Dorsey FC, Cox JV. CK2 constitutively associates with and phosphorylates chicken erythroid ankyrin and regulates its ability to bind to spectrin. J Cell Sci. 2002;115:4107–4115. doi: 10.1242/jcs.00102. [DOI] [PubMed] [Google Scholar]
- Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol. 2006;173:545–557. doi: 10.1083/jcb.200601067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein LS. Kinesin molecular motors: transport pathways, receptors, and human disease. Proc Natl Acad Sci U S A. 2001;98:6999–7003. doi: 10.1073/pnas.111145298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Gu Y. Clustering and activity tuning of Kv1 channels in myelinated hippocampal axons. J Biol Chem. 2011;286:25835–25847. doi: 10.1074/jbc.M111.219113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Jan YN, Jan LY. A conserved domain in axonal targeting of Kv1 (Shaker) voltage-gated potassium channels. Science. 2003;301:646–649. doi: 10.1126/science.1086998. [DOI] [PubMed] [Google Scholar]
- Gu C, Zhou W, Puthenveedu MA, Xu M, Jan YN, Jan LY. The microtubule plusend tracking protein EB1 is required for Kv1 voltage-gated K+ channel axonal targeting. Neuron. 2006;52:803–816. doi: 10.1016/j.neuron.2006.10.022. [DOI] [PubMed] [Google Scholar]
- Gu Y, Barry J, McDougel R, Terman D, Gu C. Alternative splicing regulates kv3.1 polarized targeting to adjust maximal spiking frequency. J Biol Chem. 2012;287:1755–1769. doi: 10.1074/jbc.M111.299305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Gu C. Dynamics of Kv1 channel transport in axons. PLoS One. 2010;5:e11931. doi: 10.1371/journal.pone.0011931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedstrom KL, Xu X, Ogawa Y, Frischknecht R, Seidenbecher CI, Shrager P, Rasband MN. Neurofascin assembles a specialized extracellular matrix at the axon initial segment. J Cell Biol. 2007;178:875–886. doi: 10.1083/jcb.200705119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Niwa S, Tanaka Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 2010;68:610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF. Currents carried by sodium and potassium ions through the membrane of the giant axon of Loligo. J Physiol. 1952;116:449–472. doi: 10.1113/jphysiol.1952.sp004717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Tian C, Li T, Yang M, Hou H, Shu Y. Distinct contributions of Na(v)1.6 and Na(v)1.2 in action potential initiation and backpropagation. Nat Neurosci. 2009;12:996–1002. doi: 10.1038/nn.2359. [DOI] [PubMed] [Google Scholar]
- Jenkins SM, Bennett V. Ankyrin-G coordinates assembly of the spectrin-based membrane skeleton, voltage-gated sodium channels, and L1 CAMs at Purkinje neuron initial segments. J Cell Biol. 2001;155:739–746. doi: 10.1083/jcb.200109026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J, Forsythe ID, Kopp-Scheinpflug C. Going native: voltage-gated potassium channels controlling neuronal excitability. J Physiol. 2011;588:3187–3200. doi: 10.1113/jphysiol.2010.191973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukkola P, Lovett-Racke A, Zamvil S, Gu C. Alterations of K+ channels in the progression of EAE. Neurobiol Dis. 2012;47:280–293. doi: 10.1016/j.nbd.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukkola P, Guerrero T, Gray V, Gu C. Astrocytes mediating neurovascular coupling differentially react to inflammatory autoimmune insults and imbalances of neural activity. Acta Neuropathol Comm. 2013;1:70. doi: 10.1186/2051-5960-1-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Goldin AL. Role of the terminal domains in sodium channel localization. Channels. 2009;3:171–180. doi: 10.4161/chan.3.3.8854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniar TA, Kaplan M, Wang GJ, Shen K, Wei L, Shaw JE, Koushika SP, Bargmann CI. UNC-33 (CRMP) and ankyrin organize microtubules and localize kinesin to polarize axon-dendrite sorting. Nat Neurosci. 2011;15:48–56. doi: 10.1038/nn.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaely P, Bennett V. The ANK repeats of erythrocyte ankyrin form two distinct but cooperative binding sites for the erythrocyte anion exchanger. J Biol Chem. 1995a;270:22050–22057. doi: 10.1074/jbc.270.37.22050. [DOI] [PubMed] [Google Scholar]
- Michaely P, Bennett V. Mechanism for binding site diversity on ankyrin. Comparison of binding sites on ankyrin for neurofascin and the Cl-/HCO3- anion exchanger. J Biol Chem. 1995b;270:31298–31302. doi: 10.1074/jbc.270.52.31298. [DOI] [PubMed] [Google Scholar]
- Pan Z, Kao T, Horvath Z, Lemos J, Sul JY, Cranstoun SD, Bennett V, Scherer SS, Cooper EC. A common ankyrin-G-based mechanism retains KCNQ and NaV channels at electrically active domains of the axon. J Neurosci. 2006;26:2599–2613. doi: 10.1523/JNEUROSCI.4314-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J, Gamal El-Din TM, Scheuer T, Zheng N, Catterall WA. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 2011;486:135–139. doi: 10.1038/nature11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2012;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer JL. Polarized domains of myelinated axons. Neuron. 2003;40:297–318. doi: 10.1016/s0896-6273(03)00628-7. [DOI] [PubMed] [Google Scholar]
- Setou M, Seog DH, Tanaka Y, Kanai Y, Takei Y, Kawagishi M, Hirokawa N. Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature. 2002;417:83–87. doi: 10.1038/nature743. [DOI] [PubMed] [Google Scholar]
- Sherman DL, Tait S, Melrose S, Johnson R, Zonta B, Court FA, Macklin WB, Meek S, Smith AJ, Cottrell DF, Brophy PJ. Neurofascins are required to establish axonal domains for saltatory conduction. Neuron. 2005;48:737–742. doi: 10.1016/j.neuron.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Sobotzik JM, Sie JM, Politi C, Del Turco D, Bennett V, Deller T, Schultz C. AnkyrinG is required to maintain axo-dendritic polarity in vivo. Proc Natl Acad Sci U S A. 2009;106:17564–17569. doi: 10.1073/pnas.0909267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song AH, Wang D, Chen G, Li Y, Luo J, Duan S, Poo MM. A selective filter for cytoplasmic transport at the axon initial segment. Cell. 2009;136:1148–1160. doi: 10.1016/j.cell.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Stuart G, Spruston N, Sakmann B, Hausser M. Action potential initiation and backpropagation in neurons of the mammalian CNS. Trends Neurosci. 1997;20:125–131. doi: 10.1016/s0166-2236(96)10075-8. [DOI] [PubMed] [Google Scholar]
- Vacher H, Mohapatra DP, Trimmer JS. Localization and targeting of voltage-dependent ion channels in mammalian central neurons. Physiol Rev. 2008;88:1407–1447. doi: 10.1152/physrev.00002.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- Wang H, Kunkel DD, Martin TM, Schwartzkroin PA, Tempel BL. Heteromultimeric K+ channels in terminal and juxtaparanodal regions of neurons. Nature. 1993;365:75–79. doi: 10.1038/365075a0. [DOI] [PubMed] [Google Scholar]
- Waxman SG. Sodium channels, the electrogenisome and the electrogenistat: lessons and questions from the clinic. J Physiol. 2012;590:2601–2612. doi: 10.1113/jphysiol.2012.228460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woroniecki R, Ferdinand JR, Morrow JS, Devarajan P. Dissociation of spectrin-ankyrin complex as a basis for loss of Na-K-ATPase polarity after ischemia. American J Physiol - Renal Physiol. 2003;284:F358–F364. doi: 10.1152/ajprenal.00100.2002. [DOI] [PubMed] [Google Scholar]
- Xu M, Cao R, Xiao R, Zhu MX, Gu C. The axon-dendrite targeting of Kv3 (Shaw) channels is determined by a targeting motif that associates with the T1 domain and ankyrin G. J Neurosci. 2007;27:14158–14170. doi: 10.1523/JNEUROSCI.3675-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Gu Y, Barry J, Gu C. Kinesin I transports tetramerized Kv3 channels through the axon initial segment via direct binding. J Neurosci. 2010;30:15987–16001. doi: 10.1523/JNEUROSCI.3565-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Bekku Y, Dzhashiashvili Y, Armenti S, Meng X, Sasaki Y, Milbrandt J, Salzer JL. Assembly and maintenance of nodes of Ranvier rely on distinct sources of proteins and targeting mechanisms. Neuron. 2012;73:92–107. doi: 10.1016/j.neuron.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Lambert S, Malen PL, Carpenter S, Boland LM, Bennett V. AnkyrinG is required for clustering of voltage-gated Na channels at axon initial segments and for normal action potential firing. J Cell Biol. 1998;143:1295–1304. doi: 10.1083/jcb.143.5.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonta B, Desmazieres A, Rinaldi A, Tait S, Sherman DL, Nolan MF, Brophy PJ. A critical role for Neurofascin in regulating action potential initiation through maintenance of the axon initial segment. Neuron. 2011;69:945–956. doi: 10.1016/j.neuron.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.