Abstract
The role of protein phosphorylation for adjusting chloroplast functions to changing environmental needs is well established, whereas calcium signalling in the chloroplast is only recently becoming appreciated. The work presented here explores the potential cross-talk between calcium signalling and protein phosphorylation in chloroplasts and provides the first evidence for targets of calcium-dependent protein phosphorylation at the thylakoid membrane. Thylakoid proteins were screened for calcium-dependent phosphorylation by 2D gel electrophoresis combined with phospho-specific labelling and PsaN, CAS, and VAR1, among other proteins, were identified repeatedly by mass spectrometry. Subsequently their calcium-dependent phosphorylation was confirmed in kinase assays using the purified proteins and chloroplast extracts. This is the first report on the protein targets of calcium-dependent phosphorylation of thylakoid proteins and provides ground for further studies in this direction.
Keywords: CAS, calcium signalling, chloroplast, protein phosphorylation, thylakoid protein phosphorylation, Var1
Introduction
The chloroplast harbours many cellular processes that require tight regulation allowing plants to grow efficiently under fluctuating environmental conditions. Protein phosphorylation is important for the post-translational control of these processes and seems to be dominated in the chloroplast by three protein kinases: chloroplast casein kinase 2 (cpCK2) and the state transition kinases STN7 and STN8 (Bayer et al., 2012). cpCK2 localizes to the stroma and plays a role in chloroplast transcription and translation, as it was found to associate with the RNA polymerase complex and to phosphorylate parts of the transcription machinery and RNA-binding proteins (Baginsky et al., 1999; Ogrzewalla et al., 2002). The functional impact on plant growth of cpCK2 phosphorylation of the chloroplast transcription machinery was recently demonstrated by the use of phosphorylation site mutants of the Arabidopsis thaliana sigma factor 6 (AtSIG6) (Schweer et al., 2010). Furthermore, the discovery of a bacterial two-component-like sensor kinase (CSK for chloroplast sensor kinase) as the interaction partner of cpCK2 (Puthiyaveetil et al., 2008) provided a link between redox sensing and plastid transcription control (Puthiyaveetil et al., 2010). However, the action of cpCK2 is most likely not restricted to the control of chloroplastic gene regulation alone, as its preferred phosphorylation motive is strongly overrepresented in a diverse set of 174 identified chloroplast phosphoproteins (Reiland et al., 2009). This notion is further supported by the observation that cpCK2 seems to be responsible for the main protein kinase activity in the stroma. This conclusion was based on the observation that stromal protein extracts are equally well phosphorylated in the presence of GTP, a known and specific co-substrate of CK2 (Niefind et al., 1999), as by ATP (Bayer et al., 2012). STN7 and STN8 are integral membrane protein kinases of the thylakoid and their function is to optimize light harvesting for photosynthesis to the fluctuating light conditions and the repair of high-light photo-damaged photosynthetic complexes, respectively (Rochaix, 2007; Tikkanen and Aro, 2011). Accordingly, stunted growth of the stn7 and stn7/stn8 double mutants becomes paticularly visible under fluctuating light conditions (Tikkanen et al., 2010). STN7 mainly phosphorylates the light-harvesting complex (LHC) proteins, while STN8 specifically phosphorylates the subunits D1, D2, and CP43 of photosystem II (PSII). These proteins represent the majority of phosphorylated proteins in thylakoids and, recently, a hypothesis has been put forward that phosphorylation of PSII regulates the cation-dependent stacking of thylakoids because thylakoid stacking was found to be less dependent on Mg2+ in stn8 mutants (Fristedt et al., 2009b,2010). The authors reason that increased phosphorylation of the main thylakoid proteins in the thylakoid stack (grana) would provide a repulsion of negative phosphoryl groups between adjacent grana membranes and therefore provide space for the cycling of photo-damaged PSII proteins to the lamellae, where they are degraded and replaced (Tikkanen et al., 2008). STN7 and STN8 are also responsible for minor phosphorylations of other thylakoid proteins, respectively TSP9, a soluble protein involved in the regulation of light harvesting (Hansson et al., 2007; Fristedt et al., 2009a) and the calcium sensing protein (CaS), an integral membrane protein involved in the process of stomatal closure (Nomura et al., 2008; Vainonen et al., 2008; Weinl et al., 2008). Notably, in the stn7/stn8 double mutant, the remains of unknown phosphorylated thylakoid proteins could be detected, albeit to a much lower extent than the main phosphorylated proteins (Fristedt et al., 2009b). TAK (thylakoid associated kinase; Snyders and Kohorn, 1999), or an as yet unidentified protein kinase, could be responsible for this phosphorylation. Moreover, cross-phosphorylation of stromal protein kinases also cannot be excluded. In addition, the recently discovered chloroplast protein kinases, such as the ABC1 kinases (Ytterberg et al., 2006; Bayer et al., 2012) and plastid protein kinase (PPK) (Bayer et al., 2011) together with the variety of identified chloroplast phosphoproteins (Reiland et al., 2009) opens many new possibilities for future discoveries and broadens the potential impact of chloroplast phosphorylation on plant physiology.
Calcium signalling in organelles is a relatively new topic (Stael et al., 2012). Compared with protein phosphorylation (Bennett, 1977), it was only recently appreciated that fluxes of free calcium ions (Ca2+) occur in the chloroplast and the topic has not yet been extensively studied (Johnson et al., 1995; Sai and Johnson, 2002). Ca2+ fluxes occur in the chloroplast stroma upon the transition from light to dark, as measured with a stromal targeted aequorin protein construct (a Ca2+ sensor protein) (Johnson et al., 1995; Sai and Johnson, 2002). The resting level of free Ca2+ was estimated at ~150 nM and increased 5 min after dark to a peak concentration of 5–10 μM in a time frame of 20–25 min. Interestingly, when the plants were exposed for several days to continuous light, the amount of Ca2+ release was found to be proportional to the light period. From earlier work it is known that chloroplasts take up Ca2+ from the surrounding media (cytosol) upon illumination (Kreimer, 1985; Roh et al., 1998). Given the characteristics of the Ca2+ flux, the chloroplast most likely takes up Ca2+ from the cytosol and stores it in the thylakoid membrane or a so far unknown store, which is then released on the transition from dark to light. The thylakoid lumen needs to take up Ca2+ in order to provide the oxygen evolving complex (OEC) with one of its necessary cofactors (Cinco et al., 2004). Accordingly, Ca2+ uptake into the lumen was found to be driven by the proton gradient through the activity of an unknown Ca2+/H+ exchanger (Ettinger et al., 1999). The increase of the Ca2+ concentration in the stroma was hypothesized to have a direct effect (inhibition) on the enzyme functions of the Calvin–Benson cycle, as it was observed in vitro (Racker and Schroeder, 1958; Portis and Heldt, 1976; Charles and Halliwell, 1980). More recently, the discovery of calmodulin-dependent processes, such as protein import (Chigri et al., 2005, 2006) and NAD kinase activity (Jarrett et al., 1982;Turner et al., 2004) suggest a role for Ca2+ as a secondary messenger in the chloroplast. Furthermore, two EF-hand (Ca2+-binding motive) containing proteins localize to the chloroplast: the Ca2+-activated RelA/SpoT homologue protein (CRSH) that resides in the chloroplast and is most likely involved in an evolutionary conserved signalling process called the ‘bacterial stringent response’ (Takahashi et al., 2004; Tozawa et al., 2007; Masuda et al., 2008) and a predicted S-adenosyl methionine transporter-like carrier (SAMTL) that localizes to the chloroplast envelope (Bayer et al., 2011; Stael et al., 2011b). The impact of chloroplast calcium handling on plant physiology is probably best illustrated by a rather unusual Ca2+ binding protein. CAS, for ‘calcium sensing’, binds Ca2+ with a low affinity and high capacity and down-regulation of its expression impairs the induction of [Ca2+]ext cytoplasmic Ca2+ oscillations in guard cells, thereby affecting stomatal movement (Han et al., 2003;Nomura et al., 2008; Weinl et al., 2008). First reported as a plasmamembrane-localized Ca2+-sensing receptor (Han et al., 2003), it was later unequivocally established as an integral thylakoid membrane protein (Friso et al., 2004;Nomura et al., 2008; Vainonen et al., 2008; Weinl et al., 2008). As mentioned earlier, CAS is increasingly phosphorylated under increasing light intensities in an STN8-dependent manner (Vainonen et al., 2008). It is still unclear exactly how CAS is able to ‘sense’ extracellular Ca2+ ([Ca2+]ext) variations and influence stomatal closure. To conclude, the chloroplast has all the prerequisites for calcium signalling, namely a low resting free Ca2+ concentration and the ability to produce Ca2+ fluxes as well as proteins to decode the calcium signal.
Materials and methods
Calcium-dependent phosphorylation assays of thylakoid membranes
Pisum sativum (pea) or Arabidopsis chloroplasts were isolated as previously described (Bayer et al., 2011). Plants were grown in the greenhouse (16/8 h light/dark) and harvested in the morning (1–2 h after lights on). For two reactions, Percoll-purified chloroplasts containing 140 μg of chlorophyll (chl) were lysed by adding five equal volumes of lysis buffer [25 mM TRIS-HCl pH 7.8, 75 mM NaCl, 10 mM MgCl2, 1 mM NaF, 0.5 mM NaVO3, 15 mM μ-glycerophosphate, 1 mM DTT, and a complete EDTA-free Protease Inhibitor Cocktail (Roche)], and incubating for 5 min on ice. Thylakoids were spun down for 2 min at 20 000 g at 4 °C and were washed with 5 volumes of lysis buffer. After centrifugation, the thylakoid pellet was re-suspended in 80 μl of lysis buffer and divided between two Eppendorf tubes (giving two reactions with 70 μg chl). To one tube, 1 μl of CaCl2 (of varying concentrations; 40, 10 or 1 mM) was added and to the other tube, 1 μl of the calcium-specific chelator EGTA (of varying concentrations; 40, 10 or 1 mM) was added. The tubes were gently mixed and incubated for 5 min at room temperature in the dark. The kinase reactions were started by adding 1 μl of an 8 mM ATP solution (final concentration of 0.2 mM ATP) and were incubated for 20 min at room temperature in the dark. The kinase reactions were stopped by the addition of 400 μl methanol, and thylakoid proteins were precipitated according to the chloroform/methanol technique (Wessel and Flugge, 1984).
2D protein separation by isoelectric focusing and SDS-PAGE gel electrophoresis
After chloroform/methanol precipitation, the protein pellets were solubilized in 125 μl of rehydration buffer [7 M urea, 2 M thiourea, 2% CHAPS, 2% IPG buffer (pH 3–11 NL, GE Healthcare), 0.002% bromphenol blue, and 50 mM DTT]. The protein solutions were applied to immobilized pH gradient strips (IPG strips Immobiline™ DryStrip gels pH 3–11 NL, 7 cm, GE Healthcare). After covering of the strips with mineral oil (Immobiline™ DryStrip Cover Fluid, GE Healthcare), they were left to rehydrate overnight. The day after, the strips were placed in a Multiphor II focusing unit (GE Healthcare) that was connected to an EPS 3501 XL power supply (GE Healthcare) and the strips were run according to the manufacturer’s protocol. Afterwards, the strips were equilibrated in SDS equilibration buffer (75 mM TRIS-HCl pH 8.8, 6 M urea, 29.3% glycerol, 2% SDS, and 0.002% bromphenol blue) containing DTT (100 mg per 10 ml buffer) for 20 min and subsequently in SDS equilibration buffer containing iodacetamide (250 mg per 10 ml buffer) for 20 min. Both strips, containing the phosphorylated proteins in the presence of Ca2+ and EGTA, were loaded on top of the same SDS-PAGE gel. The gel was run in a Protean II system (Bio-Rad) at a current of 10 mA gel−1 until the proteins from the strip had entered the SDS-PAGE. Subsequently, the current was increased to 30 mA gel−1.
Visualization of phosphorylated thylakoid proteins
To visualize phosphorylated thylakoid proteins, the 2D gels were stained by Pro-Q Diamond phosphoprotein gel stain according to the manufacturer’s protocol (Invitrogen). Alternatively, proteins of the 2D gels were transferred to polyvinylidene fluoride membranes (PVDF, Immobilon-P, Millipore) by Western blotting according to manufacturer’s protocol (Trans-Blot SD semi-dry electrophoretic transfer cell, Bio-Rad). The PVDF membranes were incubated with a phospho-threonine specific primary antibody (Cell Signalling No. 9381) and an ECL Rabbit IgG, HRP-linked secondary antibody (from donkey, GE Healthcare). Immunolabelled spots were detected with the ECL Plus Western Blotting Detection System (GE Healthcare) and films (Fujifilm) were developed in a CURIX60 table-top processor (AGFA). After completion of the phospho-specific stains, the gels and PVDF membranes were stained by silver stain or Coomassie Brilliant Blue (CBB) stain.
Excision of proteins spots and identification by mass spectrometry
Protein spots that showed differences in intensity between the Ca2+ and EGTA phosphorylation assays on the phospho-specific stained gel were manually mapped to the protein spots of the corresponding silver-stained or CBB-stained gel. The protein spots were excised and prepared for mass spectrometry analysis (MS) as previously described by Bayer et al. (2011). For the pea samples, MS/MS analysis was carried out as previously described (Bayer et al., 2011) and proteins were identified from a recently created pea EST database (Brautigam et al., 2008). For the Arabidopsis samples, peptides were separated on an UltiMate 3000 HPLC system (Dionex), by loading the sample digests on a trapping column (PepMap C18, 5 μM particle size, 300 μM i.d.×5 mm) equilibrated with 0.1% TFA (trifluoric acetic acid) and separating on an analytical column (PepMap C18, 3 μM, 75 μM i.d.×150 mm), applying a 90 min linear gradient from 2.5% up to 40% ACN with 0.1% formic acid. The HPLC was directly coupled to a LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) equipped with a nanoelectrospray ionization source (Proxeon), for which the electrospray voltage was set to 1500 V. The mass spectrometer was operated in the data-dependent mode: 1 full scan (m/z: 300–1800, resolution 60 000) with lock mass enabled was followed by maximal 20 MS/MS scans. The lock mass was set at the signal of polydimethylcyclosiloxane at m/z 445.120025. Screening of the charge state was on, singly charged signals and ions with no charge state assigned were excluded from fragmentation. The collision energy was set at 35%, Q-value at 0.25, and the activation time at 10 ms. Fragmented ions were set onto an exclusion list for 90 s. Raw spectra were interpreted by Mascot 2.2.04 (Matrix Science) using Mascot Daemon 2.2.2. The peptide tolerance was set to 2 ppm, MS/MS tolerance was set to 0.8 Da. Proteins were identified from the full genome sequence of TAIR9 (ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR9_genome_release/, on www.arabidopsis.org). Carbamidomethylcysteine was set as a static modification and oxidation of methionine as a variable modification. Trypsin was selected as the protease and two missed cleavages were allowed. MASCOT results were loaded into Scaffold (Ver. 3.00.02; Proteome Software) for an X! Tandem Search. Peptide identifications were accepted at a probability greater than 95% and protein identifications at a probability greater than 99%, as assigned by the Protein Prophet algorithm (Keller et al., 2002; Nesvizhskii et al., 2003). Mostly, multiple proteins were identified per spot. Therefore, peptide count was used as a relative measure of protein abundance and this was combined with information on predicted pI and mass to single out the most likely protein per spot. All identified proteins per spot are summarized in Supplementary Table 2.
Preparation of chloroplast soluble and extrinsic membrane proteins for the recombinant phosphorylation assay
Arabidopsis chloroplasts were hypotonically disrupted in lysis buffer (20 mM Tricine pH 7.6, 10% glycerol, and 0.5% DTT), supplemented with protease inhibitors (complete EDTA-free Protease Inhibitor Cocktail, Roche), phosphatase inhibitors (Phospho-Stop, Roche), and 5 mM EGTA. After incubation on ice for 15 min, membranes and soluble components were separated by centrifugation at 60 000 g for 10 min. To solubilize extrinsic membrane proteins, the membrane fraction was washed with lysis buffer containing 0.8 M NaCl. The 60 000 g supernatants of the first and second centrifugation were combined, desalted, and concentrated using Vivaspin 500 spin columns (3 kDa cutoff, GE Healthcare). All operations were carried out either on ice or at 4 °C.
Calcium-dependent phosphorylation assays of selected recombinant proteins
PsaN, lacking the N-terminal 81 amino acids (forward primer: 5′-TTATTATCCATGGCTGCTTCTGCTAATGCTGGCGTCAT-3′ and reverse primer: 5′-AATATAGCGGCCGCATATAAGAATAGATGAAAAC-3′), the C-terminus of CAS (amino acids 216–387; forward primer: 5′-GATCGGGCCCATGGGTTACAAAGGTGATCTTACGCC-3′ and reverse primer: 5′-ACTAGCGGCCGCAGTCGGAGCTAGGAAGGAAC-3′), the C-terminus of VAR1 (amino acids 221–704, forward primer: 5′-GATCGGGCCCATGGGTGGTCCTGGAGGTTTAGG-3′ and reverse primer: 5′-ACTAGCGGCCGCAAGAAACATATAACTCGGCT-3′) and the C-terminus of VAR2 (amino acids 198–695; forward primer: 5′-GATCGGGCCCATGGGTGGACCTGGTGGTCC-3′ and reverse primer: 5′-ACTAGCGGCCGCAGACAGCAGCTGGTGTTGGT-3′) were obtained by PCR from Arabidopsis cDNA using primers containing the restriction sites for NcoI (or ApaI) and NotI. The PCR products of PsaN and CAS were cloned in-frame with an N-terminal intein tag into pTWIN (New England Biolabs) and the PCR products of VAR1 and VAR2 were cloned in-frame with a gluthathione-S-transferase (GST) tag into pGEX4T-3 (GE Healthcare). Proteins were expressed according to the manufacturer’s protocol, which resulted in the removal of the intein tag from PsaN and CAS. Protein phosphorylation assays were carried out in kinase buffer [20 mM Tricine pH 7.6, 10 mM MgCl2, 5 μM cold ATP, and 2–5 μCi (γ-32P) ATP (3000 Ci mmol−1, Perkin Elmer), 10% glycerol, and 0.5% DTT] supplemented with either 2 mM EGTA or 5 mM CaCl2. In a total reaction mix of 50 μl was added 5 μl of chloroplast soluble and extrinsic membrane proteins. Reactions were stopped after 25 min at room temperature by the addition of 12 μl of 4× SDS-sample buffer and were separated by SDS-PAGE gel electrophoresis and stained with CBB. Radiolabelled proteins were detected by exposure for 3 d on a X-ray film (FUJI) at −80 °C.
Results
Several thylakoid proteins are phosphorylated in a calcium-dependent manner in pea and Arabidopsis
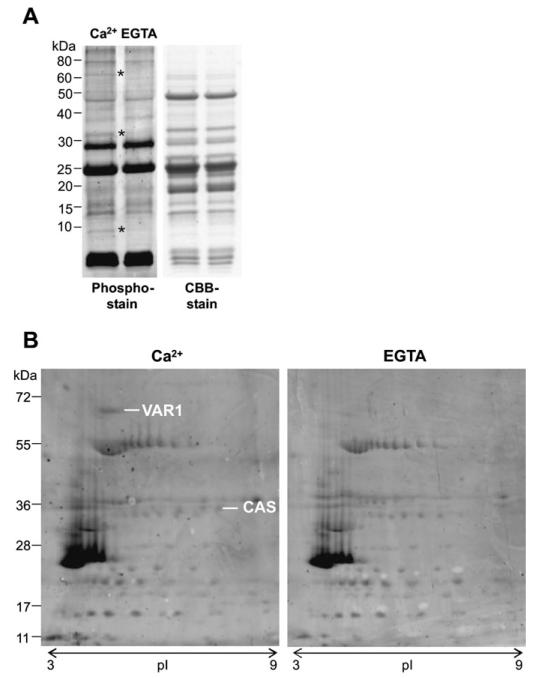
In order to identify targets of Ca2+ dependent phosphorylation in thylakoids, Percoll-purified chloroplasts of pea were subjected to in vitro phosphorylation assays in the presence of Ca2+. Control experiments were performed in the presence of EGTA to chelate any contaminating Ca2+. The higher specificity of EGTA towards Ca2+ combined with the saturating amount of Mg2+ in the kinase buffer ensured that all differences in phosphorylation were Ca2+ specific. Initial experiments were performed with pea in the presence of 1 mM Ca2+, and the concentration of Ca2+ was lowered to 25 μM in the subsequent experiments using Arabidopsis thylakoids. After loading the two kinase reactions (+/− Ca2+) on a denaturing SDS-PAGE gel, phosphorylated bands were visualized with Pro-Q Diamond phosphoprotein gel stain. Protein bands at approximately 65, 35, and 10 kDa, respectively, showed a clear dependence on Ca2+ for phosphorylation as displayed for pea thylakoids in Fig. 1A.
Fig. 1. Calcium-dependent phosphorylation of pea thylakoid proteins.
(A) The left panel displays a Pro-Q Diamond phosphoprotein gel stain that reveals three differentially phosphorylated protein bands (indicated with an asterisk) upon the addition of 1 mM Ca2+, compared with 1 mM EGTA. The right panel is the protein loading control (CBB=Coomassie Brilliant Blue). (B) 2D protein separation according to isoelectric point (pI) and size (kDa) of the same Ca2+-dependent phosphorylation assay followed by a Pro-Q Diamond phosphoprotein gel stain (experiment 1). Two protein spots were identified as the FtsH protease, Variegated 1 (VAR1) and ‘Calcium sensing’ protein (CAS).
2D gel analysis identifies VAR1 and CAS as targets of Ca2+-dependent phosphorylation
To identify the Ca2+-dependent phosphorylated proteins, the proteins from the two phosphorylation assays of pea thylakoids (1 mM Ca2+ and 1 mM EGTA) were separated in a 2D approach. In the first dimension, the proteins were separated according to their pI on a non-linear pH gradient from pH 3–11 and, in the second dimension, on a 12% SDS-PAGE gel. Phosphorylated proteins were again visualized with Pro-Q Diamond phosphoprotein stain and the gel was subsequently stained with Coomassie Brilliant Blue (CBB). Protein spots, corresponding to a molecular mass of 65 kDa and 35 kDa, respectively, showed differences in phosphorylation (Fig. 1B) confirming the first observation from the initial experiment (Fig. 1A). The protein spots from the phosphostain were manually mapped to the CBB-stained gel, cut out, and analysed by tandem mass spectrometry. After parsing of all identified proteins (see Materials and methods), the protein spots at 65 kDa and 35 kDa corresponded, respectively, to Variegated 1 (VAR1) and CAS.
Repeated 2D gel analysis of Arabidopsis thylakoids confirms results obtained in pea and uncovers additional targets of Ca2+-dependent phosphorylation
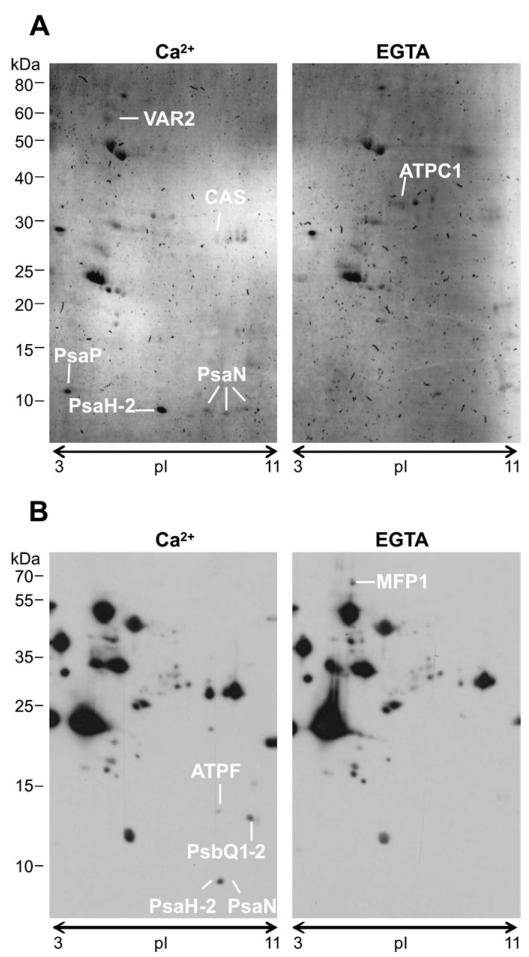
In subsequent experiments, the Ca2+-dependent phosphorylation assays were repeated with thylakoids from Percoll-purified chloroplasts of Arabidopsis. In order to get closer to physiological relevant Ca2+ concentrations, the experiment was repeated three times with 250 μM Ca2+/EGTA (Fig. 2) and three times with 25 μM Ca2+/EGTA. Furthermore, two complementary approaches were used to detect phosphorylated proteins. In four of the six experiments, a Western blot with a phosphothreonine-specific antibody was used, and for the remaining two experiments the Pro-Q Diamond phosphoprotein stain was used. When phosphorylated proteins were detected by Western blot, a reference gel was run under identical conditions and the protein spots were mapped to this reference gel with the help of the CBB stain of the Western blot. The results of these seven independent experiments are summarized in Supplementary Table 1. Those proteins, which were identified multiple times in independent experiments, are presented in Table 1. From these proteins, PsaN (subunit N of photosystem 1) was identified most frequently and it probably represents the phosphorylated band appearing at a molecular mass of 10 kDa in the initial pea experiment (Fig. 1A). Following in number of identifications were CAS, PsbP (subunit P of photosystem 2), and PsaH-2 (subunit H-2 of photosystem 1). VAR1 and VAR2 were both included in this table, because protein identification ambiguity existed between these closely homologous proteins.
Fig. 2. Calcium-dependent phosphorylation of Arabidopsis thylakoid proteins visualized by Pro-Q Diamond phosphoprotein gel stain (A) and phospho-threonine specific antibody (B) after 2D-gel separation according to isoelectric point (pI) and size (kDa).
(A) Protein spots identified in experiment 4 (250 μM Ca2+/EGTA) are the FtsH protease Variegated 2 (VAR2), ‘Calcium sensing’ protein (CAS), Photosystem I subunit P (PsaP), Photosystem I subunit H-2 (PsaH-2), and Photosystem I subunit N (PsaN; three times identified). (B) Protein spots identified in experiment 3 (250 μM Ca2+/EGTA) are the ATPase subunit F (ATPF), the Photosystem II subunits Q1 and Q2 (PsbQ1-2), PsaH-2, and PsaN. Stronger phosphorylation of spots were also observed in the EGTA samples, as it is the case in (A) for the ATPase C1 subunit (ATPC1) and in (B) for the MAR binding filament-like protein 1 (MFP1).
Table 1. Overview of most frequently identified calcium-dependent phosphorylated proteins.
Indicated in the table is, in which species the proteins were identified (At=Arabidopsis thaliana; Ps=Pisum sativum), which stain was used to reveal the phosphorylated proteins (Pro-Q=Pro-Q Diamond phosphoprotein gel stain; pThr=phospho-threonine specific antibody), if the protein is included in the phospho-peptide database PhosPhat 3.0 (Heazlewood et al., 2008; Durek et al, 2010), and in which experiment the protein was identified. Images to the seven experiments are included in the supplementary figures.
| ID | AGI code | Description | Species | Pro-Q | p-Thr | PhosPhat | Experiment |
|---|---|---|---|---|---|---|---|
| PsaN | At5g64040 | Photosystem I subunit N | At | 4 | 4 | y | (3)(4)(5) (6) (7) |
| CAS | At5g23060 | ‘Calcium sensing’ protein | Ps/At | 2 | 1 | y | (1)(2)(4) |
| VAR1/VAR2 | At5g42270/At2g30950 | Variegated 1 and 2, FtsH proteases | Ps/At | 2 | 0 | y/n | (1)(4) |
| PsbP-1 | At1g06680 | Photosystem II subunit P-1 | At | 1 | 1 | y | (2)(7) |
| PsaH-2 | At1g52230 | Photosystem I subunit H-2 | At | 1 | 1 | n | (3)(4) |
Kinase assays on purified proteins confirm the calcium-dependent phosphorylation of VAR1, CAS, and PsaN
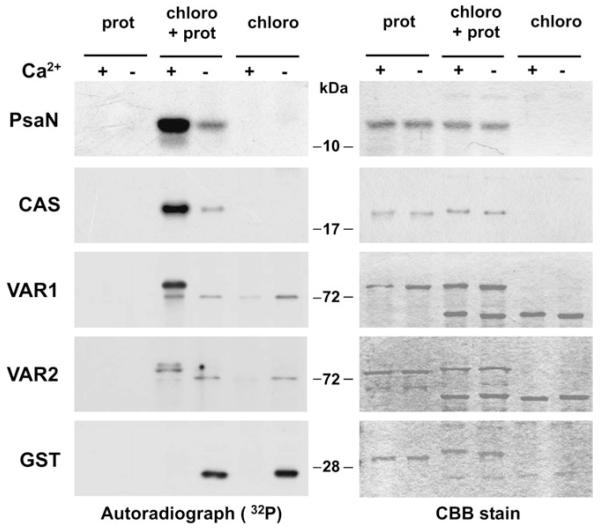
The four proteins PsaN, CAS, VAR1, and VAR2, which were identified with the highest confidence as targets of Ca2+-dependent phosphorylation at thylakoid membranes (Table 1) were recombinantly expressed and purified from E. coli to test the Ca2+ dependency of their phosphorylation in an independent approach. In the case of CAS, VAR1, and VAR2, only the C-termini of the proteins without the transmembrane domains were expressed, in order to avoid protein solubility problems. The C-termini were chosen as they contain the known phosphorylation sites of these proteins (Vainonen et al., 2008; Reiland et al., 2009). PsaN and CAS were purified without a protein tag, whereas the addition of a gluthathione-S-transferase (GST) protein tag to VAR1 and VAR2 seemed to increase their protein stability (data not shown). For the kinase assays, soluble and extrinsic proteins of Percoll-purified Arabidopsis chloroplasts were used to phosphorylate the purified proteins in the presence of Ca2+ or EGTA (two middle lanes of Fig. 3). No phosphorylation was detected when only the recombinant proteins were incubated with ATP (two left lanes of Fig. 3). Phosphorylation assays without purified proteins were included for comparison to the background phosphorylation of the chloroplast extract (two right lanes of Fig. 3). As a control, kinase assays were performed on GST. A clear calcium-dependent phosphorylation was visible for PsaN, CAS, and VAR1. VAR2, on the other hand, was notably less phosphorylated than VAR1, if compared with the background phosphorylation. Taken together, these data confirm the previously reported phosphorylation of those proteins from an unbiased phosphoproteomics study (Reiland et al., 2009).
Fig. 3. Calcium-dependent phosphorylation assays on recombinant PsaN, CAS, VAR1, VAR2, and GST protein.
The two middle lanes of the left panel display the autoradiograph of 32P-labelled proteins of a Ca2+-dependent phosphorylation assay of recombinant protein (prot) by an Arabidopsis chloroplast protein extract (chloro). The two left lanes are a control of protein only, whereas the two right lanes are a control for the background phosphorylation in the chloroplast protein extract. Note that GST is not being phosphorylated, instead a strong phosphorylation is visible in the EGTA sample of the chloroplast protein extract. The right panel is the protein loading control (CBB=Coomassie Brilliant Blue).
Discussion
The exploration of potential cross-talk between calcium signalling and protein phosphorylation at the chloroplast thylakoid membranes has delivered the first evidence for the calcium-dependent phosphorylation of PsaN, CAS, and VAR1, among other proteins. This led to the question as to what might be the function of this calcium-dependent phosphorylation? While this is difficult to answer for PsaN, one can speculate about the impact of calcium-dependent phosphorylation on VAR1 and CAS. VAR1, which is also called FTSH5, is a conserved thylakoid localized protease that, together with its close homologue VAR2 (or FTSH2), is responsible for the protein turnover of photo-damaged D1 subunits of photosystem II (Lindahl et al., 2000; Aro et al., 2005; Kato et al., 2009). Subunit D1 seems to be the main ‘victim’ of the reactive photochemical reactions of the photosystem II and constantly needs to be recycled, even under low light, to prevent photo-inhibition of the photosystems (Aro et al., 1993). The important function of VAR1 and VAR2 for the plant becomes obvious in their loss-of-function mutants, which exhibit variegated leaf phenotypes (green and white sectoring of the leaf) (Sakamoto et al., 2003). Phosphorylation of VAR1 might be a means to altering its proteolytic activity, as it is known for another important group of proteases, the caspases (Kurokawa and Kornbluth, 2009). Accordingly, we hypothesize that an increase in free stromal Ca2+ on the transition from light to dark (Sai and Johnson, 2002) could mediate the phosphorylation of VAR1, a protease that degrades photo-damaged D1 protein in the light, to switch off its activity when it is no longer needed in the dark. From our experiments it seems that VAR1 is mainly subjected to post-translational modification, since VAR2 was not phosphorylated significantly above background levels (Fig. 3). As both VAR1 and VAR2 are operating in heterocomplexes (Sakamoto et al., 2003;Zhang et al., 2010), the post-translational modification of VAR1 could have an influence on the activity of VAR2. In this respect, phosphorylation of VAR1 could affect complex formation, as it is known for the photosynthetic complexes (Tikkanen and Aro, 2011). For CAS it is tempting to speculate that the observed increase of phosphorylation under increased light intensities (Vainonen et al., 2008) is mediated by an increase of Ca2+ uptake into the chloroplast under light (Kreimer, 1985; Roh et al., 1998). How this behaviour then relates to the observed function in ‘calcium sensing’ of CAS for stomatal movement remains an open question. CAS was shown to bind Ca2+ with low affinity/high capacity to a part of the protein N-terminal of the transmembrane region (Han et al., 2003). Interestingly, for the phosphorylation assay of Fig. 3, the C-terminal part of CAS was used because it had been proven to be phosphorylated (Vainonen et al., 2008; Reiland et al., 2011). How the binding of Ca2+ to the N-terminus of CAS relates to the Ca2+-dependent phosphorylation of the C-terminus adds to question on the function of CAS. It should be noted that CAS was found in this study through a naïve search for calcium-dependent phosphorylated proteins. Nevertheless, its prior connection to chloroplast calcium signalling fits nicely with the finding of calcium-dependent protein phosphorylations in the thylakoid. Further detailed studies are necessary to elucidate the functional impact of calcium-dependent phosphorylation on VAR1 and CAS.
What is the nature of the kinase responsible for calcium-dependent protein phosphorylation? Most likely, several protein kinases are involved, since protein substrates from both sides of the thylakoid membrane have been identified. For example, the C-terminal part of VAR1 faces the stromal side of the thylakoid (Sakamoto et al., 2003), whereas PsaN and the oxygen-evolving complex subunit PsbP are attached to the luminal side of the thylakoid membrane. Another clue comes from the confirmation experiments (Fig. 3) in which the recombinantly purified proteins PsaN, VAR1, and CAS were phosphorylated by a salt-washed soluble chloroplast extract. This would rather exclude the membrane-intrinsic thylakoid protein kinases STN7 and STN8 as responsible kinases for this phosphorylation. Therefore, even though CAS phosphorylation was found to be dependent on STN8 (Vainonen et al., 2008), a direct phosphorylation has not been demonstrated and leaves space for additional soluble kinases downstream of STN8. A possible candidate kinase for calcium-dependent phosphorylation could be CIPK13, which is supposed to be soluble and was reported to reside in the chloroplast (Schliebner et al., 2008). CIPKs are CBL interacting protein kinases that are known to be regulated in a calcium-dependent manner through the interaction with the EF-hand containing calcineurin B-like proteins (CBL) (Weinl and Kudla, 2009). However, CIPK13 has not been characterized in detail so far and the absence of CBL proteins in the chloroplast casts doubt on the function of CIPK13 in the chloroplast. Also several other calcium-dependent protein kinases are predicted to be localized to plastids. However, it turned out that they are membrane-localized due to N-acylation, which prevents their chloroplast import (Mehlmer et al., 2010; Bayer et al., 2012; Stael et al., 2011a). It should be noted that, through the use of phosphatase inhibitors (NaF, NaVO3, and β-Glycerophosphate) the influence of phosphatases can be excluded in this experimental set-up. Of course, it cannot be excluded that in vivo protein phosphatases could play a role in calcium-dependent protein phosphorylations of the thylakoid. On the other hand, instead of Ca2+ influencing the kinase, Ca2+ might also bind the substrate protein first leading to a conformational change or enhancing the interaction with a protein kinase which, in turn, could phosphorylate the substrate. This could be the case for CAS and PsbP, which are known to bind calcium. Similarly, conformational changes, which are induced by the depletion of Ca2+ (upon addition of EGTA), could be the reason for observed increases in phosphorylation, which is visible for ATPC1 and MFP1 in Fig. 2A and B, respectively. This effect was also observed in other experiments (see Supplementary Figs S1–S7).
In the future, the functional impact of calcium-dependent phosphorylation on plant physiology can be estimated through targeted mutations of the phosphorylation sites of VAR1 and CAS. Quantitative phosphoproteomics have been proved to work for chloroplasts (Reiland et al., 2011) and could be used to obtain an even more comprehensive overview of calcium-dependent phosphorylated proteins of the thylakoid, including the mapping of exact phosphorylation sites.
Supplementary Material
Excel table of all the protein spots identified from different 2D gels.
Acknowledgements
This work has been funded by the Austrian GEN-AU program in the ERA-PG project CROPP (Project No. 818514), the Austrian Science Foundation (FWF) (P19825-B12), and by the EU in the Marie-Curie ITN COSI (ITN 2008 GA 215-174).
References
- Aro EM, McCaffery S, Anderson JM. Photoinhibition and D1 protein degradation in peas acclimated to different growth irradiances. Plant Physiology. 1993;103:835–843. doi: 10.1104/pp.103.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro EM, Suorsa M, Rokka A, Allahverdiyeva Y, Paakkarinen V, Saleem A, Battchikova N, Rintamaki E. Dynamics of photosystem II: a proteomic approach to thylakoid protein complexes. Journal of Experimental Botany. 2005;56:347–356. doi: 10.1093/jxb/eri041. [DOI] [PubMed] [Google Scholar]
- Baginsky S, Tiller K, Pfannschmidt T, Link G. PTK, the chloroplast RNA polymerase-associated protein kinase from mustard (Sinapis alba), mediates redox control of plastid in vitro transcription. Plant Molecular Biology. 1999;39:1013–1023. doi: 10.1023/a:1006177807844. [DOI] [PubMed] [Google Scholar]
- Bayer RG, Stael S, Csaszar E, Teige M. Mining the soluble chloroplast proteome by affinity chromatography. Proteomics. 2011;11:1287–1299. doi: 10.1002/pmic.201000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer RG, Stael S, Rocha AG, Mair A, Vothknecht UC, Teige M. Chloroplast localized protein kinases: a step forward towards a complete inventory. Journal of Experimental Botany. 2012;63:1713–1723. doi: 10.1093/jxb/err377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett J. Phosphorylation of chloroplast membrane polypeptides. Nature. 1977;269:344–346. [Google Scholar]
- Brautigam A, Shrestha RP, Whitten D, Wilkerson CG, Carr KM, Froehlich JE, Weber AP. Low-coverage massively parallel pyrosequencing of cDNAs enables proteomics in non-model species: comparison of a species-specific database generated by pyrosequencing with databases from related species for proteome analysis of pea chloroplast envelopes. Journal of Biotechnology. 2008;136:44–53. doi: 10.1016/j.jbiotec.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Charles SA, Halliwell B. Action of calcium ions on spinach (Spinacia oleracea) chloroplast fructose bisphosphatase and other enzymes of the Calvin cycle. Biochemical Journal. 1980;188:775–779. doi: 10.1042/bj1880775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chigri F, Hormann F, Stamp A, Stammers DK, Bolter B, Soll J, Vothknecht UC. Calcium regulation of chloroplast protein translocation is mediated by calmodulin binding to Tic32. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:16051–16056. doi: 10.1073/pnas.0607150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chigri F, Soll J, Vothknecht UC. Calcium regulation of chloroplast protein import. The Plant Journal. 2005;42:821–831. doi: 10.1111/j.1365-313X.2005.02414.x. [DOI] [PubMed] [Google Scholar]
- Cinco RM, Robblee JH, Messinger J, Fernandez C, McFarlane Holman KL, Sauer K, Yachandra VK. Orientation of calcium in the Mn4Ca cluster of the oxygen-evolving complex determined using polarized strontium EXAFS of photosystem II membranes. Biochemistry. 2004;43:13271–13282. doi: 10.1021/bi036308v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durek P, Schmidt R, Heazlewood JL, Jones A, MacLean D, Nagel A, Kersten B, Schulze WX. PhosPhAt: the Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Research. 2010;38:D828–D834. doi: 10.1093/nar/gkp810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger WF, Clear AM, Fanning KJ, Peck ML. Identification of a Ca2+/H+ antiport in the plant chloroplast thylakoid membrane. Plant Physiology. 1999;119:1379–1386. doi: 10.1104/pp.119.4.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso G, Giacomelli L, Ytterberg AJ, Peltier JB, Rudella A, Sun Q, Wijk KJ. In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. The Plant Cell. 2004;16:478–499. doi: 10.1105/tpc.017814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R, Carlberg I, Zygadlo A, Piippo M, Nurmi M, Aro EM, Scheller HV, Vener AV. Intrinsically unstructured phosphoprotein TSP9 regulates light harvesting in Arabidopsis thaliana. Biochemistry. 2009a;48:499–509. doi: 10.1021/bi8016334. [DOI] [PubMed] [Google Scholar]
- Fristedt R, Granath P, Vener AV. A protein phosphorylation threshold for functional stacking of plant photosynthetic membranes. PLoS One. 2010;5:e10963. doi: 10.1371/journal.pone.0010963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R, Willig A, Granath P, Crevecoeur M, Rochaix JD, Vener AV. Phosphorylation of photosystem II controls functional macroscopic folding of photosynthetic membranes in Arabidopsis. The Plant Cell. 2009b;21:3950–3964. doi: 10.1105/tpc.109.069435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Tang R, Anderson LK, Woerner TE, Pei ZM. A cell surface receptor mediates extracellular Ca2+ sensing in guard cells. Nature. 2003;425:196–200. doi: 10.1038/nature01932. [DOI] [PubMed] [Google Scholar]
- Hansson M, Dupuis T, Stromquist R, Andersson B, Vener AV, Carlberg I. The mobile thylakoid phosphoprotein TSP9 interacts with the light-harvesting complex II and the peripheries of both photosystems. Journal of Biological Chemistry. 2007;282:16214–16222. doi: 10.1074/jbc.M605833200. [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Durek P, Hummel J, Selbig J, Weckwerth W, Walther D, Schulze WX. PhosPhAt: a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Research. 2008;36:D1015–D1021. doi: 10.1093/nar/gkm812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett HW, Brown CJ, Black CC, Cormier MJ. Evidence that calmodulin is in the chloroplast of peas and serves a regulatory role in photosynthesis. Journal of Biological Chemistry. 1982;257:13795–13804. [PubMed] [Google Scholar]
- Johnson CH, Knight MR, Kondo T, Masson P, Sedbrook J, Haley A, Trewavas A. Circadian oscillations of cytosolic and chloroplastic free calcium in plants. Science. 1995;269:1863–1865. doi: 10.1126/science.7569925. [DOI] [PubMed] [Google Scholar]
- Kato Y, Miura E, Ido K, Ifuku K, Sakamoto W. The variegated mutants lacking chloroplastic FtsHs are defective in D1 degradation and accumulate reactive oxygen species. Plant Physiology. 2009;151:1790–1801. doi: 10.1104/pp.109.146589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Analytical Chemistry. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Kreimer G, Melkonian M, Holtum JAM, Latzko E. Characterization of calcium fluxes across the envelope of intact spinach chloroplasts. Planta. 1985;166:515–523. doi: 10.1007/BF00391276. [DOI] [PubMed] [Google Scholar]
- Kurokawa M, Kornbluth S. Caspases and kinases in a death grip. Cell. 2009;138:838–854. doi: 10.1016/j.cell.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl M, Spetea C, Hundal T, Oppenheim AB, Adam Z, Andersson B. The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. The Plant Cell. 2000;12:419–431. doi: 10.1105/tpc.12.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Mizusawa K, Narisawa T, Tozawa Y, Ohta H, Takamiya K. The bacterial stringent response, conserved in chloroplasts, controls plant fertilization. Plant and Cell Physiology. 2008;49:135–141. doi: 10.1093/pcp/pcm177. [DOI] [PubMed] [Google Scholar]
- Mehlmer N, Wurzinger B, Stael S, Hofmann-Rodrigues D, Csaszar E, Pfister B, Bayer R, Teige M. The Ca2+-dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. The Plant Journal. 2010;63:484–498. doi: 10.1111/j.1365-313X.2010.04257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Analytical Chemistry. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Niefind K, Putter M, Guerra B, Issinger OG, Schomburg D. GTP plus water mimic ATP in the active site of protein kinase CK2. Nature Structural Biology. 1999;6:1100–1103. doi: 10.1038/70033. [DOI] [PubMed] [Google Scholar]
- Nomura H, Komori T, Kobori M, Nakahira Y, Shiina T. Evidence for chloroplast control of external Ca2+-induced cytosolic Ca2+ transients and stomatal closure. The Plant Journal. 2008;53:988–998. doi: 10.1111/j.1365-313X.2007.03390.x. [DOI] [PubMed] [Google Scholar]
- Ogrzewalla K, Piotrowski M, Reinbothe S, Link G. The plastid transcription kinase from mustard (Sinapis alba L.). A nuclear-encoded CK2-type chloroplast enzyme with redox-sensitive function. European Journal of Biochemistry. 2002;269:3329–3337. [PubMed] [Google Scholar]
- Portis AR, Jr, Heldt HW. Light-dependent changes of the Mg2+ concentration in the stroma in relation to the Mg2+ dependency of CO2 fixation in intact chloroplasts. Biochimica et Biophysica Acta. 1976;449:434–436. doi: 10.1016/0005-2728(76)90154-7. [DOI] [PubMed] [Google Scholar]
- Puthiyaveetil S, Ibrahim IM, Jelicic B, Tomasic A, Fulgosi H, Allen JF. Transcriptional control of photosynthesis genes: the evolutionarily conserved regulatory mechanism in plastid genome function. Genome Biology and Evolution. 2010;2:888–896. doi: 10.1093/gbe/evq073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthiyaveetil S, Kavanagh TA, Cain P, Sullivan JA, Newell CA, Gray JC, Robinson C, van der Giezen M, Rogers MB, Allen JF. The ancestral symbiont sensor kinase CSK links photosynthesis with gene expression in chloroplasts. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10061–10066. doi: 10.1073/pnas.0803928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racker E, Schroeder EA. The reductive pentose phosphate cycle. II. Specific C-1 phosphatases for fructose 1,6-diphosphate and sedoheptulose 1,7-diphosphate. Archives of Biochemistry and Biophysics. 1958;74:326–344. doi: 10.1016/0003-9861(58)90004-3. [DOI] [PubMed] [Google Scholar]
- Reiland S, Finazzi G, Endler A, et al. Comparative phosphoproteome profiling reveals a function of the STN8 kinase in fine-tuning of cyclic electron flow (CEF) Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12955–12960. doi: 10.1073/pnas.1104734108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S. Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiology. 2009;150:889–903. doi: 10.1104/pp.109.138677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix JD. Role of thylakoid protein kinases in photosynthetic acclimation. FEBS Letters. 2007;581:2768–2775. doi: 10.1016/j.febslet.2007.04.038. [DOI] [PubMed] [Google Scholar]
- Roh MH, Shingles R, Cleveland MJ, McCarty RE. Direct measurement of calcium transport across chloroplast inner-envelope vesicles. Plant Physiology. 1998;118:1447–1454. doi: 10.1104/pp.118.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai J, Johnson CH. Dark-stimulated calcium ion fluxes in the chloroplast stroma and cytosol. The Plant Cell. 2002;14:1279–1291. doi: 10.1105/tpc.000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W, Zaltsman A, Adam Z, Takahashi Y. Coordinated regulation and complex formation of yellow variegated1 and yellow variegated2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. The Plant Cell. 2003;15:2843–2855. doi: 10.1105/tpc.017319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliebner I, Pribil M, Zuhlke J, Dietzmann A, Leister D. A survey of chloroplast protein kinases and phosphatases in Arabidopsis thaliana. Current Genomics. 2008;9:184–190. doi: 10.2174/138920208784340740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweer J, Turkeri H, Link B, Link G. AtSIG6, a plastid sigma factor from Arabidopsis, reveals functional impact of cpCK2 phosphorylation. The Plant Journal. 2010;62:192–202. doi: 10.1111/j.1365-313X.2010.04138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyders S, Kohorn BD. TAKs, thylakoid membrane protein kinases associated with energy transduction. Journal of Biological Chemistry. 1999;274:9137–9140. doi: 10.1074/jbc.274.14.9137. [DOI] [PubMed] [Google Scholar]
- Stael S, Bayer RG, Mehlmer N, Teige M. Protein N-acylation overrides differing targeting signals. FEBS Letters. 2011a;585:517–522. doi: 10.1016/j.febslet.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stael S, Rocha AG, Robinson AJ, Kmiecik P, Vothknecht UC, Teige M. Arabidopsis calcium-binding mitochondrial carrier proteins as potential facilitators of mitochondrial ATP-import and plastid SAM-import. FEBS Letters. 2011b;585:3935–3940. doi: 10.1016/j.febslet.2011.10.039. [DOI] [PubMed] [Google Scholar]
- Stael S, Wurzinger B, Mair A, Mehlmer N, Vothknecht UC, Teige M. Plant organellar calcium signalling: an emerging field. Journal of Experimental Botany. 2012c;63:1525–1542. doi: 10.1093/jxb/err394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Kasai K, Ochi K. Identification of the bacterial alarmone guanosine 5′-diphosphate 3′-diphosphate (ppGpp) in plants. Procedeings of the National Academy of Sciences, USA. 2004;101:4320–4324. doi: 10.1073/pnas.0308555101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen M, Aro EM. Thylakoid protein phosphorylation in dynamic regulation of photosystem II in higher plants. Biochimica et Biophysica Acta. 2011 doi: 10.1016/j.bbabio.2011.05.005. doi:10.1016/j.bbabio.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Tikkanen M, Grieco M, Kangasjarvi S, Aro EM. Thylakoid protein phosphorylation in higher plant chloroplasts optimizes electron transfer under fluctuating light. Plant Physiology. 2010;152:723–735. doi: 10.1104/pp.109.150250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen M, Nurmi M, Kangasjarvi S, Aro EM. Core protein phosphorylation facilitates the repair of photodamaged photosystem II at high light. Biochimica et Biophysica Acta. 2008;1777:1432–1437. doi: 10.1016/j.bbabio.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Tozawa Y, Nozawa A, Kanno T, Narisawa T, Masuda S, Kasai K, Nanamiya H. Calcium-activated (p)ppGpp synthetase in chloroplasts of land plants. Journal of Biological Chemistry. 2007;282:35536–35545. doi: 10.1074/jbc.M703820200. [DOI] [PubMed] [Google Scholar]
- Turner WL, Waller JC, Vanderbeld B, Snedden WA. Cloning and characterization of two NAD kinases from Arabidopsis. Identification of a calmodulin binding isoform. Plant Physiology. 2004;135:1243–1255. doi: 10.1104/pp.104.040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainonen JP, Sakuragi Y, Stael S, et al. Light regulation of CaS, a novel phosphoprotein in the thylakoid membrane of Arabidopsis thaliana. FEBS Journal. 2008;275:1767–1777. doi: 10.1111/j.1742-4658.2008.06335.x. [DOI] [PubMed] [Google Scholar]
- Weinl S, Held K, Schlucking K, Steinhorst L, Kuhlgert S, Hippler M, Kudla J. A plastid protein crucial for Ca2+-regulated stomatal responses. New Phytologist. 2008;179:675–686. doi: 10.1111/j.1469-8137.2008.02492.x. [DOI] [PubMed] [Google Scholar]
- Weinl S, Kudla J. The CBL-CIPK Ca2+-decoding signaling network: function and perspectives. New Phytologist. 2009;184:517–528. doi: 10.1111/j.1469-8137.2009.02938.x. [DOI] [PubMed] [Google Scholar]
- Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Analytical Biochemistry. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Ytterberg AJ, Peltier JB, van Wijk KJ. Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiology. 2006;140:984–997. doi: 10.1104/pp.105.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Kato Y, Zhang L, Fujimoto M, Tsutsumi N, Sodmergen, Sakamoto W. The FtsH protease heterocomplex in Arabidopsis: dispensability of type-B protease activity for proper chloroplast development. The Plant Cell. 2010;22:3710–3725. doi: 10.1105/tpc.110.079202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Excel table of all the protein spots identified from different 2D gels.