Abstract
Integration of data across spatial, temporal, and functional scales is a primary focus of biomedical engineering efforts. The advent of powerful computing platforms, coupled with quantitative data from high-throughput experimental platforms, has allowed multiscale modeling to expand as a means to more comprehensively investigate biological phenomena in experimentally relevant ways. This review aims to highlight recently published multiscale models of biological systems while using their successes to propose the best practices for future model development. We demonstrate that coupling continuous and discrete systems best captures biological information across spatial scales by selecting modeling techniques that are suited to the task. Further, we suggest how to best leverage these multiscale models to gain insight into biological systems using quantitative, biomedical engineering methods to analyze data in non-intuitive ways. These topics are discussed with a focus on the future of the field, the current challenges encountered, and opportunities yet to be realized.
Keywords: data integration, model validation, systems biology, bioinformatics, biochemical networks, model design
INTRODUCTION
Biological systems are inherently complex in nature; they are comprised of multiple functional networks that operate across diverse temporal and spatial domains to sustain an organism’s growth, development, and reproductive potential. These so-called “multiscale” systems extend from the most basic of amino acid substitutions that alter protein function to concerted multicellular signaling cascades regulating hormone release throughout an entire lifetime. Computational models are uniquely positioned to capture the connectivity between these divergent scales of biological function as they can bridge the gap in understanding between isolated in vitro experiments and whole-organism in vivo models.
While seemingly transparent, a careful definition of multiscale should be explored as it can very quickly spiral into the realm of catch-all scientific jargon. Fundamentally, a multiscale model must explicitly account for more than one level of resolution across measurable domains of time, space, and/or function. To clarify, many models of physical systems implicitly account for multiple spatial scales by simplifying their boundary conditions into “black boxes” where assumptions about other spatial or temporal domains are summarized by governing equations. Further, explicitly modeled tiers of resolution must also provide additional information that could not be obtained by independently exploring a single scale in isolation.
The classic engineering exercise of heat transfer through an insulated rod is an excellent case study in implicit multiscale modeling. Whether solved using continuous PDEs or a discrete finite element approach, all solutions to this problem rely on carefully defining spatial boundary conditions, the fundamental laws of thermodynamics of a closed system, and material properties such as a thermal conductivity coefficient. Using these tools, engineering students unwittingly wrangle molecular motion at the femtometer scale to reliably predict the distribution of temperatures across an idealized one-dimensional landscape measured in meters. However, were we to explicitly account for the motion of each molecule of metal in the rod would we gain any additional information about the system (assuming that this were not a computational intractable challenge)? In this case, the governing equations of thermodynamics sufficiently capture the probabilistic distributions of molecules without requiring explicit representation in the model.
Ultimately, this model system is explicitly analyzed at the scale of the rod while implicitly accessing information about molecular thermal motion using established equations of thermodynamics. However, as a “Law of Biological Systems” has yet to be codified into governing equations, the biomedical scientist lacks the means to accurately make assumptions across multiple tiers of measurable resolution. This challenge is further compounded by the complex nature of the system that is being investigated; that is to say, components of biological systems act differently in isolation than they do when integrated into the larger machinery of a living organism.
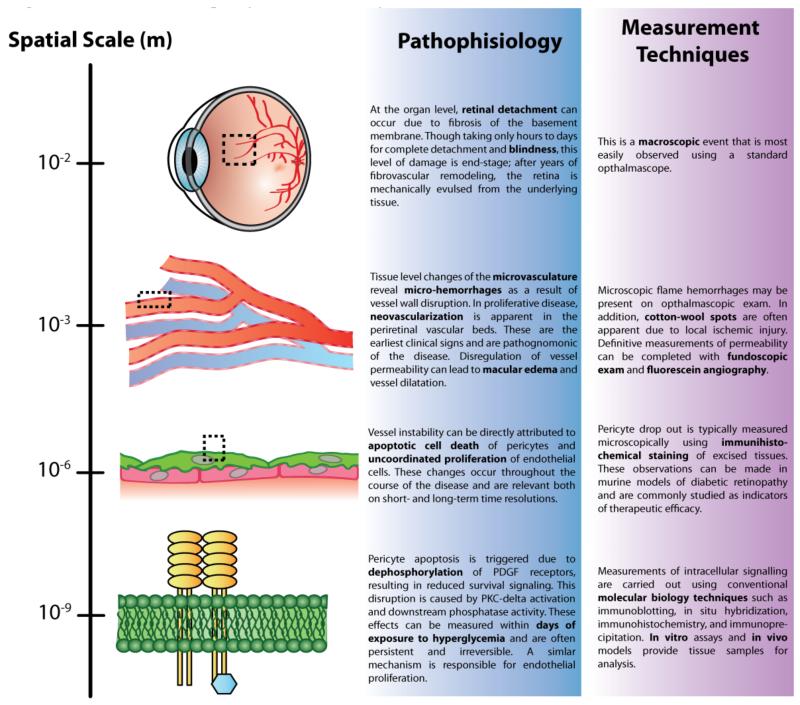
To further illustrate the need for explicit multiscale models in biology, let us consider the multiple levels of spatial, temporal, and functional scale that are known to operate in the pathophysiology of diabetic retinopathy (Figure 1). At its most advanced stage, proliferative diabetic retinopathy can result in blindness due to retinal detachment at the macroscopic level. This event, however, is preceded by years of tissue damage caused by microvascular hemorrhage and fibrovascular remodeling of the retinal basement membrane. These defects in the vessel wall are the result of pericyte (abluminal vascular support cell) apoptosis, leading to aberrant vessel growth and increased vessel permeability throughout the retina. Finally, pericyte apoptosis occurs due to reduction of PDGF receptor survival signaling mediated by activation of PKC-delta and downstream phosphatases in the setting of chronic hyperglycemia (1, 2).
Figure 1. Diabetic Retinopathy as a Case Study in Multiscale Pathophysiology.
A detailed look at how the pathogenesis of diabetic retinopathy is a function of multiple spatial scales across biology.
At what tier of resolution is the most information available for understanding the underlying mechanisms of this complex disease? Conversely, is there a tier of resolution that offers the least understanding of the disease? The debatable answers to these questions have driven model building for decades as investigators attempt to develop the highest information yield from their intellectual investments in computational modeling approaches. More recently however, investigators are turning to multiscale modeling techniques to generate detailed information about complex biological systems. In these multiscale models, perturbations to fine-grained parameters (e.g. protein modifications) can generate observable and measurable changes to coarse-grained outputs (e.g. tissue patterning), and vice versa. This integration across functional, spatial, and temporal scales in biological systems introduces a powerful tool for capturing and analyzing biological information that is inaccessible to other modeling and experimental techniques.
Herein we describe a meta-analysis of multiscale modeling, focusing foremost on recent publications from the biomedical engineering community. First, we will describe the tiers of biological resolution that have been modeled and the computational techniques leveraged to obtain insightful conclusions. Focus will shift to a discussion of best practices in model verification and validation as we discuss challenges unique to multiscale modeling. Once we have covered the questions and tools used to answer them, we will expand on how multiscale models capture biologically relevant data that may be inaccessible using conventional wet laboratory techniques. Finally, we will look to the future of the field and pose a set of specific landmarks that, if accomplished, may provide even greater insight into the form and function of complex biological systems.
CURRENT MULTISCALE MODELING EFFORTS
Scales of Biology: What Are We Modeling?
Multiscale models are pervasive in the biological sciences, covering many tiers of resolution and many disciplines. Using a selection of literature from the last decade we have highlighted and clustered broad biomedical disciplines based on the levels of spatial resolution they are investigating with multiscale models (Figure 2). A clear trend is shown in which metabolomics and genomics research are clustered separately as they are uniquely focused on sub-cellular resolutions (3-7). Further, studies of tissue mechanics and disciplines interested in cellular trafficking (i.e. cancer and immunology) display the most work at the organ and multisystem scales (8-18).
Figure 2. Clustergram of multiscale models as a function of biological discipline and spatial resolution.
Each publication was scored as containing (1) or not containing (0) a biological scale within the described multiscale model. For each discipline, the Boolean values were summed and then normalized to the total number of publications within that category, such that the weighted heatmap is scaled from 0 to 1. For example: of the 7 publications in Vascular Biology, 5 involved Whole Cell components, resulting in a weighted score of 5/7 = 0.71. A total of 39 publications were included in this analysis.
As might be expected, much effort is focused on the interrogation of biology at many resolutions, from signaling networks (i.e. subcellular simulations where proteins are not explicitly modeled) through to cell networks (i.e. tissue-level simulations comprising more than a single cell). In particular, the fields of cell biology, developmental biology, vascular biology, and cardiovascular research all share a very similar pattern of work at these tiers (19-42). A common theme among these fields is a desire to understand how subcellular networks may influence tissue-level patterning through the actions of individual cells.
Of course, it should be emphasized that these are trends from a subset of papers that have been broadly classified based on the field of biological research and the explicitly modeled tiers of resolution. This meta-analysis is also purely an evaluation of the quantity of publications in a given field and not the quality of the models being developed. Clearly, other disciplines are also using multiscale modeling to their advantage; even the disciplines shown contain researchers whose work does not neatly conform to the selected scales. This meta-analysis does, however, demonstrate a clear trend in the literature, which may allow us to glean some insight into current gaps in computational coverage within our disciplines of interest.
Most importantly, this analysis demonstrates that a major goal of the field is yet to be realized: no single comprehensive “gene-to-organism” multiscale model has been developed. Based on our observations there are many open avenues of research within each of the listed disciplines where multiscale efforts are either sparsely represented or completely nonexistent. This deficit is not a shortcoming, but rather an opportunity to push the boundaries of knowledge in these biomedical investigations using multiscale modeling as a platform for high-throughput, high-yield hypothesis generation and testing.
Models Within Models: That Which Comprises A Multiscale System
All modeling methodologies have strengths and weaknesses with regards to their ease and fidelity of capturing biological system dynamics. Typically, these techniques are broadly classified into continuous and discrete modeling strategies based on how the solution space is acquired. Additional classification into deterministic and stochastic models is an alternative method that divides systems based on whether they contain a degree of “randomness” that allows for multiple solutions to the same initial conditions. Importantly, while not an exhaustive list, the modeling techniques presented here are all taken from published multiscale models; these examples are already validated against experimental data and, therefore, serve as a foundation for future computational efforts.
Continuous modeling strategies include using systems of ordinary differential equations (ODEs) and partial differential equations (PDEs) to solve for steady state solutions. Solutions to these continuous systems are deterministic as they obey the Picard-Lindelöf Existence and Uniqueness Theorem (43). Because numerical tools for solving PDEs such as Finite Element and Finite Volume methods rely on reduction to a system of ODEs, the assumption of uniqueness still holds despite their ability to contain stochastic elements.
Generally, systems of ODEs using the law of Mass Action Kinetics are leveraged to represent chemical reactions within the cytosol and nucleus of the cell (13, 15, 18, 19, 44-46). As the kinetics of molecular binding, conformational switching, and diffusion are often occurring over very small time scales the assumption of steady state in the overall model architecture (which may be discretized into hours, days, weeks, etc.) is typically valid. Sun et al. (47) employed a system of ODEs executed with the COmplex PAthways SImulator (COMPASI) to explicitly model TGF-β1 function in a multiscale model of epidermal wound healing. Using this technique they expanded on a previous single-scale model and were able to decouple the promigratory and anti-proliferative effects of TGF-β1 on various cell types in an in silico skin wound closure model over time. Analogous reasoning and techniques are also used for analysis of metabolic and signaling networks in which a steady state flux is desired for informing higher tiers of function (3, 5, 6, 45, 48).
Models of reaction diffusion kinetics are also typically modeled in continuous time and are often used to represent intra- and extracellular molecular binding and diffusion (29, 38, 39, 41, 49). These models differ from previous diffusion/pathway models as they typically rely on systems of PDEs that are then solved using numerical approaches. Broadly speaking, finite element methods (and related finite volume methods) are also uniquely suited for monitoring geometrically-constrained properties such as cell surface interfaces and mechanical properties of tissues across all scales (17, 38, 50-54). Aguado-Sierra et al. (35) generated a patient-specific three-dimensional model of heart failure in which a finite element mesh was fitted to echocardiographs and mechanical parameters were directly estimated from a combination of MR and cardiac ultrasound. This work highlights the clinical value of computational models by using patient data to generate electrical conduction and mechanical contractility maps with the potential to inform interventional decisions as processing cost and time decrease. Note that these approaches are a hybrid of continuous and discrete strategies as finite element methods rely on discretization of continuous equations to generate numerical solutions for otherwise irreducible PDEs.
Discrete stochastic modeling techniques are a heterogeneous group of computational foundations that rely on non-deterministic solutions to generate constrained distributions of outputs. These techniques include methods such as Markov Chains, whose probabilistic transition matrices are suited to biological systems whose functions can be discretized into independent states. Along with the related class of discrete state-based Boolean Networks, these techniques have modeled receptor activation states (e.g. cardiomyocyte ion channels), compartmentalized signaling networks, and functional protein conformations (19, 20, 34, 37, 40, 55). Barua et al. (5) have recently developed an algorithm, GeneForce, to explore the Boolean rules in metabolic signaling networks and correct for inconsistencies between experimental results and model predictions. The model “forces” an optimized output by allowing for a set degree of rule violation; these perturbations to the original rule set revealed incorrectly silenced gene transcription which, when correct, allowed for agreement with experimental results. This approach generated as much as an 8% improvement in model predictive accuracy and was applied to well curated metabolome libraries for organisms such as E. coli.
Recently, Agent Based Modeling (ABM) has become a very popular and powerful tool for representing discrete stochastic biological processes as either compartmentalized or spatially-defined models. These models include geometries in one-, two-, and three-dimensional configurations and may be scaled such that each fundamental agent is as large (groups of organisms) or as small (sub-cellular membrane components) as is desired. Zahedmanesh et al. (52) incorporated a lattice-free ABM with a finite element approach to explore the effects of porosity, compliance, cyclic strain, and flow-induced shear stress on tissue engineered blood vessels. This investigation was able to explore how these complex and non-intuitive parameters combined to affect development of intimal hyperplasia over time with potential to make predictions across time scales that cannot be investigated using in vitro techniques. Owing to their diversity of scale, AMBs have been used to describe multicellular processes including tissue electrical conduction, cell trafficking, tissue mechanics, immunomodulation, arterial remodeling, inflammation and many others (10, 14, 18, 21, 37, 38, 44, 56, 57).
Selecting a Computational Method Based on Function and Spatial Resolution
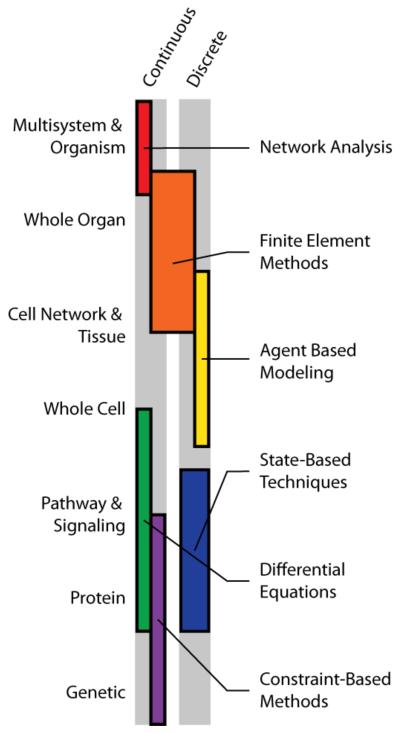
The computational techniques presented in the previous section were selected as examples currently being employed in multiscale models. We classified the techniques into continuous-deterministic and discrete-stochastic (with some exceptions and hybrids), while highlighting specific spatial and temporal domains that these models are suited to represent. This classification forms the basis for a discussion of how multiscale models can be designed by selecting the best computational techniques for the task rather than forcing a modeling technique to approximate a system for which it is poorly suited. To this end, we propose some guidelines for how these individual techniques can be combined across scales (Figure 3).
Figure 3. Map of Modeling Techniques by Scale.
Conceptual map of modeling techniques divided into continuous and discrete categories across spatial scales for which they are most suited.
As a class of modeling techniques, network analyses include discrete state-based techniques (e.g. Markov chains, Boolean networks) as well as continuous systems biology approaches (e.g. Flux Balance Analysis). These methods are well suited to modeling the smallest tiers of resolution: genomic, proteomic, and metabolomic. Recently demonstrated by Milne and colleagues (6), construction of a composite Gene-Protein-Reaction (GPR) model to simulate regulation of butanol production as a function of growth conditions (e.g. growth medium, atmosphere) supported the hypothesis that Clostridium beijerinckii was an ideal candidate for biofuel applications. The iCM925 model contained 925 genes coding for 938 reactions involving 881 metabolites – approximately 18% of the protein coding genome of C. beijerinckii. This level of detail and network annotation for a relatively understudied organism was captured and analyzed with linear algebraic equations defined by a homogenous ODE constrained with mass balance principals. Simply put, a vast amount of multiscale data was integrated using a single computational technique.
In the sub-cellular regime, continuous-deterministic systems of ODEs and PDEs are also ideal for monitoring concentrations of signaling molecules in both the intra- and extracellular domains. These systems are often less comprehensive than the previously described network analyses due to the paucity of relevant kinetic parameters; however, they excel at explicitly accounting for binding kinetics and monitoring rates of reactions as a function of time. Sample et al. (29) demonstrate the use of these continuum approaches to solve for gradients of morphogens within the developing Drosophila embryo. In their model, solving for department-dependent degradation rates was integral to understanding nuclear-cytoplasmic shuttling of morphogens, which are responsible for long-range patterning. Although only focusing on a single protein, this technique expands the resolution from purely intracellular reactions to subcellular components with intercellular interactions.
At this point, many of the internal cellular components (i.e. genome, proteome, and signaling networks) have been explicitly accounted for; the next tier of resolution, the whole cell, now requires additional consideration as the functions of interest are again interwoven with the scale of investigation. Here, the cell may be viewed as a mechanical entity with discretized membrane segments and interconnected cytoskeletal components or it may be viewed as itself being the smallest component of the system. This biological scale is a natural transition point where both continuum and discrete modeling approaches have been successful, and it falls to the investigator to make the final decision guided by the hypothesis to be tested. Practically, if the cell is the largest entity in the system (i.e. only a single cell is being modeled) a more fine-grained approach is necessary. The converse is also true: if the cell is part of a larger tissue network it must be more coarsely resolved to allow for observations to be feasible given limited computing resources.
For the sake of simplicity we will consider that the cell is itself a transition state between the sub-cellular and super-cellular domains (this notably excludes mechanical analyses of single cells which are often performed at the whole-cell level). Such a view favors a discrete-stochastic approach to cell behavior as this captures a degree of biological noise and allows for easy representation in physical space. ABMs are well suited to this task as they can be specifically adapted to represent cells as either single- or multi-agent entities within the system. Bentley et al. (57) chose the later approach and represented a capillary as a linear array of ten endothelial cells, each comprised of 1288 membrane agents. This representation was necessary as their analysis required discrete membrane localization of receptors, as well as detailed filopodial sprouting within a three dimensional extracellular space. Bailey et al. (24) opted on the former approach, representing each endothelial cell in the network as a single agent to generate a larger microvascular system. Again, this selection was reasonable based on the analysis at hand: leukocyte extravasation as a function of adhesion molecule expression in a tissue bed.
Tiers of resolution beyond the cell network and tissue level, as demonstrated in Figure 2, remain largely unexplored as components of multiscale models. This focus may be due to technical limitations such that computational power is not yet available to track discretized agents throughout an entire organism. Larger, whole-organ models do exist and typically adopt a finite element approach where each cell is represented as part of the discretized mesh. Moreno and colleagues (20) were very successful with this technique, using a finite element approach to analyze the effects of antiarrhythmic pharmaceuticals on cardiac conduction through a fully rendered three-dimensional human heart. This model stands out particularly, as the smallest explicitly resolved element was the well-studied cardiomyocyte sodium channel. This voltage gated channel was modeled using Markov states that were altered in the presence of various inhibitors. Most notably, cardiotoxic concentrations of antiarrhythmics could not be predicted at the single-cell scale; however, when cells operating with the same parameters were linked into a network (and ultimately a complete tissue), the model very closely matched clinically observed data.
To summarize: function and spatial resolution beget modeling technique. Based on our current understanding and computational limitations it is necessary to view some biological processes as continuous equations and others as discrete states. As we ascend from sub- to super-cellular resolutions, continuous models that were once exceptionally accurate begin to lose resolving power. Conversely, discrete models are often computationally expensive and become most useful at lower resolution for cell networks and tissues where cells are easily viewed as individual modules. Yet larger systems may require a return to network approaches to account for spatial distances and boundaries between organ systems that are too large to be explicitly modeled at the cellular level.
VERIFICATION AND VALIDATION OF MULTISCALE MODELS
Validating Across Multiple Scales
As with all computational models, multiscale approaches must be rigorously tested against independent data sets for proper validation prior to use as experimental constructs. Recently, Qu and colleagues (58) have reviewed how information is translated between scales of models and highlighted several of the challenges associated with validation across tiers of resolution. A key observation is that inherently noisy stochastic systems and noiseless deterministic systems can generate dramatically different outputs when used to model the same biological phenomena (Keizer’s Paradox). Furthermore, adding noise to a previously noiseless system by combining deterministic and stochastic models may increase the likelihood of phase transitions, increasing the number of stable solutions. These additional solutions may be biologically relevant; however, they may also become problematic as their addition could be viewed as incongruence between continuous and discrete systems.
Ultimately, this challenge reduces to the simple fact that we lack the computational resources to explicitly model every protein in a living organism simultaneously. Multiscale models must rely on techniques such as those mentioned above (selecting appropriately resolvable approaches based on function and spatial scale; using integrative systems biology) to capture accurate and robust information from each tier of resolution. It stands to reason that by linking potentially divergent modeling techniques we may introduce inconsistencies into our multiscale systems. To reach model agreement (both inter-model agreement and agreement with biological experiments), we must decide on a validation strategy that is both theoretically sound and computationally practical.
Individual Verification vs. Complete Multiscale Verification
Multiscale models often originate by linking individual models from two different scales to generate a composite system. In the cases where each tier of a model has been independently published they must, by definition, be validated at the single-scale level before validating at the multiscale level. Our lab has, in collaboration with others, followed this strategy to generate high quality multiscale models from successfully implemented single-scale models (21, 59, 60). In this particular example, the multiscale model captured continuum elements (extracellular matrix composition, fluid dynamics, etc.) as well as discrete elements (mechanical properties as determined by cell number and orientation) to generate a blood vessel wall for measuring adaptation to chronic hypertension.
As explored by Hayenga and colleagues (61), prior to generating a comprehensive model, the continuum and discrete systems shared common outputs that were independently validated. Importantly, despite sharing independently validated outputs, the models were not in complete agreement as they drew on data from different scales. The discrete agent based model was generated from cell-level data acquired primarily from reduced in vitro systems that no longer maintained systems-level responses. Conversely, the continuous constrained mixture model was based on tissue-level data from studies of tissue parameters in which different systems-level responses were potentially still intact. Disagreement between the models presented a significant challenge, as neither was, strictly speaking, incorrect.
Ultimately, to reconcile these differences between scales and allow for comprehensive model validation, agreement on shared variables was required. As such, each model was deemed equally “unreliable” for the purposes of weighting a Genetic Algorithm approach to parameter estimation. Agreement between the continuous and the discrete models was achieved for shared parameters, allowing for validation of independent terms using a shared data set.
Fedosov et al. (62) describe a multiscale model of erythrocyte membrane mechanics in the context of malaria infection and how changes in material properties and cell geometry impact bulk blood viscosity. In this example, validation was performed at the cellular level using optical-tweezer and optical magnetic twisting cytometry to measure deformability of erythrocytes during different stages of malaria parasite development. Bulk blood viscosity was validated against a separate data set to demonstrate that each tier of model resolution independently achieved agreement with biologically relevant data sets. In order to perform these validations, previously dimensionless particle models had to be scaled using erythrocyte diameter as a reference length. This example highlights how careful selection of units and appropriate parameter selection is necessary to achieve multiscale validation.
Multiscale models are subject to scrutiny at both individual and integrated tiers of resolution. To appropriately parameterize a model and achieve validation, it is necessary to ensure that each module or computational technique is itself in agreement with biological data before advancing to a complete multiscale simulation. Further validation of the multiscale model is required to test the reliability of data transfer between computational scales such that crosstalk between continuous and discrete systems does not introduce artifacts or discrepancies. As with all modeling efforts, thorough and thoughtful validation is key to achieving acceptance in the biological community; the predictive power of a model is dependent on the rigor of this validation.
BIOLOGICAL INSIGHT FROM MODELS
Measuring the Unmeasurable
Most modeling endeavors begin with a hypothesis that cannot be easily tested using even the most cutting edge experimental assays. Tracking individual macrophages in real time in vivo, measuring chemokine concentration gradients throughout an entire tissue region, determining frequency responses to mechanical stimuli in the human ear, observing capillary and lymphatic filling as a function of muscle contraction, quantifying the effects of drug therapy on granuloma formation over the course of 300 days with receptor-level resolution – these are just a few examples of recent investigations that would not otherwise be possible without multiscale modeling approaches (24, 39, 42, 44, 51). Multiscale models are capable of quantifying any explicitly implemented variable as an output across all tiers of resolution.
In addition to quantifying individual variables with relative ease, multiscale models also allow for simultaneous observation of multiple parameters across resolution domains. Tracking multiple variables across a range of parameter values allows for construction of valuable phase planes to describe systems-level behaviors (13, 20, 29, 57). Bifurcations in these phase plane analyses offer insights into system stability and potential interventional targets that may yield higher likelihoods of maintaining transitions from one equilibrium state of a biological system to another. For example, Kim et al. (63) explored reorientation of individual CD8+ T-killer lymphocytes in the two-dimensional parameter space defined by microtubule length and initial centrosome orientation relative to a target cell. This analysis reveals complex relationships between the parameters, suggesting certain combinations that would render the T-killer unable to properly orient itself for productive cytolytic activity. Such incompatible orientations could not by predicted by either parameter alone, emphasizing the need for more rigorous analysis.
Similarly, as in Holland et al. (22), acquiring large sums of data across multiple scales of resolution allows for more informed selection of reducible components within complex systems. Using a graphical approach in a normalized phase plane to study the kinetics of β-adrenergic signaling, this investigation demonstrates a method to identify reactions that can reasonably be reduced to steady-state when evaluating system dynamics. Each reaction trajectory in the system was compared relative to steady-state values: trajectories in the phase plane with greater deviations from steady-state had larger hysteresis loops and could be identified as necessary for capturing dynamics of system behaviors.
These examples highlight how traditional engineering approaches to capturing system behaviors can be applied to biological systems. However, as these approaches generally require large sums of quantitative data to be useful, traditional wet lab experiments are not easily translated to these analysis techniques. Computational models, in particular multiscale models, offer an alternative source of data that can be acquired across many parameter values in a high throughput manner. As experimental methodologies develop, these predictions can themselves be independently validated or they may provide insight into unexplored hypotheses that can be tested immediately.
The Virtual Bench: in silico Perturbations
Beyond simply capturing otherwise inaccessible measurements with high resolution across large changes in scale, multiscale modeling allows for precise manipulation of network variables to isolate true effects from experimental artifacts. Even the most precise RNA interference strategies (including small interfering, short hairpin, and micro RNA) are capable of producing off-target effects either directly (e.g. silencing alternative binding sites) or indirectly (e.g. diminution of native RNA translation), resulting in confounding or erroneous observations (64). This statement does not imply that models are error-free; models simply are capable of isolating perturbations as defined by the user’s constraints without concern of unknown interactions.
Thus, at first glance multiscale modeling offers the unique advantage of being able to make perturbations across any tier of resolution. This is potentially very powerful as it allows for not only knockdown and overexpression experiments, but also very precise changes to the degree of expression of a single gene or set of genes. This control, quite simply, is not possible with current microbiology techniques. While note a replacement for experimental investigation, these approaches can serve to contextualized data and reconcile discrepancies that may be caused by off-target effects. Further, computational methods may also refine experimental approaches by surveying all possible perturbations to narrow the scope of experimental interrogation.
As an example, Fallahi-Sichani and colleagues (13, 44) have described the effect of modifying NF-κB signaling mechanisms at the level of transcript stability with implications of temporal variables (e.g. degradation rate, activation rate) affecting the outcomes of Mycobacterium tuberculosis infection. In this work, pharmaceutical therapies were applied using a system of ODEs to capture intracellular signaling pathways while cellular behaviors were executed as a discrete probabilistic agent based model at the tissue level scale. This union of subcellular pathway manipulation and multicellular function allows for direct investigation of pharmaceutical intervention on a relevant pathophysiological outcome that would otherwise be unobtainable by modeling an individual tier of resolution.
Alternatively, for systems being modeled with a top-down approach, more general questions can be answered by completely removing subsystems from multiscale models. In these cases functional impairments are evaluated as opposed to specific physiological interventions. Using this approach, Shirinifard and colleagues (9) demonstrated unique growth patterns in avascular tumors by removing the capability for angiogenic growth from their multiscale model of solid tumors. Insights from such a broad phenotype perturbation (i.e. complete abrogation of angiogenesis rather than impairment of a single component in the pathway with downstream effects) allow for investigations into the minimal functions necessary for individual system behaviors.
Further, the concept of in silico perturbations can be extended as a direct analogy to bench work – with the exception that it can be executed at high throughput with low resource allocation. More so than modeling at a single resolution, multiscale models can be directly mapped to biological assays for both experimental validation and hypothesis testing. For some of these cases, multiscale models have proven predictive for optimizing biopolymer scaffolds based on altering material properties to investigate extracellular matrix mechanotransduction and cell seeding (16, 52, 56). Here, small parameter changes that could easily be completed with computational iteration would require extensive material cost and time commitment to generate comparable data sets.
LOOKING FORWARD
Throughout this manuscript we have highlighted the currently available computational tools for multiscale modeling and the best practices for their implementation. As shown in Figure 2, many disciplines of biological research have yet to fully leverage the power of multiscale modeling across more than a few tiers of resolution. That being said, examples do exist that span the spectrum from the most fundamental genetic modifications to organ-level perturbations. Combining these tools across all of these scales simultaneously may seem at this point an intractably difficult problem; however, some preliminary efforts are already emerging.
The Physiome Projects are a collection of biological databases, mathematical models, and utilities being gathered with a singular purpose: integration (65, 66). Models from every spatial, temporal, and functional scale are being curated as individual modules such that they can be preserved for integration into larger, multiscale simulations. The efforts of this project are ongoing as it recognizes that, primarily due to computational limitations, a single, whole organism model that explicitly incorporates all tiers of biological resolution has yet to be realized. As we have noted earlier, the majority of information is concentrated near the cellular level with decreasing availability of models and data at the genetic and whole organism levels. This ongoing effort shows much promise as a means to begin generating larger multiscale models from validated, optimized modules that have been assembled with integration in mind.
Beyond implementing better and more comprehensive multiscale models, the future of the field also holds potential to advance other recently accelerating fields of biomedical engineering. In particular, efforts in synthetic biology are using multiscale data and analysis to inform design optimization and control systems theory of novel biological systems. In a recent publication, Nawroth and colleagues (67) describe the design, development, and implementation of a synthetic jellyfish capable of self-propulsion dubbed the “Medusoid.” They describe the reverse engineering process as occurring over several orders of space and time in order to capture the necessary information to generate synthetic muscle fibers capable of productive, concerted contraction. Callura et al. (68) are similarly beginning to use multiscale approaches as they scale up from a single gene to a composite “genetic switchboard.” Capable of regulating four metabolic genes in E. coli, this synthetic regulatory system reliably shunted flux through different carbon-utilizing pathways as measured by mRNA levels and direct quantification of metabolites. This effort demonstrates in a strictly in vitro sense how multiscale theory can be applied to better understand and engineer biological systems.
Multiscale modeling, above all, strives to better understand the fundamental processes that sustain biological life. Unquestionably, effects at the genetic level are responsible for both subtle and dramatic phenotypic expression of an entire organism. We are only just starting to begin to construct computational models that can explicitly demonstrate this same degree of emergent pattern phenomena through appropriate inter-scale connectivity. It is our hope that the techniques and practices presented here are able to guide future efforts in this field towards high quality multiscale model implementation.
SUMMARY POINTS.
Multiscale models are explicitly executed simulations of complex biological systems that have been integrated across temporal, spatial, and functional domains. Through simultaneous evaluation of multiple tiers of resolution, multiscale models provide access to systems behaviors that are not observable using single-scale techniques.
A combination of multiple computational techniques, including both continuous and discrete systems, is optimal for efficiently capturing information across biological scales. Each spatial scale can be summarized by the biological functions occupying that tier of resolution, allowing for modeling techniques to be implemented based on how well they represent these functions.
Multiscale models more closely recapitulate traditional bench top experimentation while allowing for high throughput hypothesis generation and testing, quantitation of values that cannot be measured, and translation to in vivo systems. Perturbations to high-resolution parameters (e.g. protein binding constants) can generate low-resolution outputs that are biologically relevant (e.g. tissue developmental patterning), allowing for simultaneous access to quantifiable values across all scales of biology.
FUTURE ISSUES.
Fundamental to the model building process, sensitivity analyses are performed to explore the parameter space for potentially interesting and useful “tuning” variables on which system outputs are strongly dependent. Multiscale models must be thoroughly investigated to determine whether sensitivities are truly a function of the system behavior or an artifact of coarse-graining lower resolution outputs. This area will require further investigation through the continued development of multiscale and complex systems models.
Appropriate parameter selection remains a concern in the computational modeling community, as many of the parameter values required to develop multiscale models are either difficult or impossible to measure. Values obtained from in vitro data may not be suitable for multiscale models operating at a tissue network or larger spatial scale. As such, exploration of parameter estimation techniques may be required to better parameterize multiscale models. Alternatively, emerging in vivo molecular imaging techniques may grant access to previously unobtainable parameter values.
ACKNOWLEDGEMENTS
The authors wish to acknowledge funding from the National Institutes of Health (R01 GM088244 to Jason A. Papin and NIH-HL082838 to Shayn M. Peirce) and the Cystic Fibrosis Research Foundation (grant 1060 to Jason A. Papin).
References
- 1.Frank RN. Diabetic retinopathy. The New England journal of medicine. 2004;350:48–58. doi: 10.1056/NEJMra021678. [DOI] [PubMed] [Google Scholar]
- 2.Geraldes P, Hiraoka-Yamamoto J, Matsumoto M, Clermont A, Leitges M, et al. Activation of PKC-delta and SHP-1 by hyperglycemia causes vascular cell apoptosis and diabetic retinopathy. Nature medicine. 2009;15:1298–306. doi: 10.1038/nm.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Modi SR, Camacho DM, Kohanski MA, Walker GC, Collins JJ. Functional characterization of bacterial sRNAs using a network biology approach. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15522–7. doi: 10.1073/pnas.1104318108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seok J, Xiao W, Moldawer LL, Davis RW, Covert MW. A dynamic network of transcription in LPS-treated human subjects. BMC systems biology. 2009;3:78. doi: 10.1186/1752-0509-3-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barua D, Kim J, Reed JL. An automated phenotype-driven approach (GeneForce) for refining metabolic and regulatory models. PLoS computational biology. 2010;6:e1000970. doi: 10.1371/journal.pcbi.1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milne CB, Eddy JA, Raju R, Ardekani S, Kim PJ, et al. Metabolic network reconstruction and genome-scale model of butanol-producing strain Clostridium beijerinckii NCIMB 8052. BMC systems biology. 2011;5:130. doi: 10.1186/1752-0509-5-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosgrove BD, Alexopoulos LG, Saez-Rodriguez J, Griffith LG, Lauffenburger DA. A multipathway phosphoproteomic signaling network model of idiosyncratic drug- and inflammatory cytokine-induced toxicity in human hepatocytes. Conference proceedings: … Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Conference. 2009;2009:5452–5. doi: 10.1109/IEMBS.2009.5334019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eissing T, Kuepfer L, Becker C, Block M, Coboeken K, et al. A computational systems biology software platform for multiscale modeling and simulation: integrating whole-body physiology, disease biology, and molecular reaction networks. Frontiers in physiology. 2011;2:4. doi: 10.3389/fphys.2011.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shirinifard A, Gens JS, Zaitlen BL, Poplawski NJ, Swat M, Glazier JA. 3D multi-cell simulation of tumor growth and angiogenesis. PloS one. 2009;4:e7190. doi: 10.1371/journal.pone.0007190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Folcik VA, Broderick G, Mohan S, Block B, Ekbote C, et al. Using an agent-based model to analyze the dynamic communication network of the immune response. Theoretical biology & medical modelling. 2011;8:1. doi: 10.1186/1742-4682-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caputo KE, Hammer DA. Adhesive dynamics simulation of G-protein-mediated chemokine-activated neutrophil adhesion. Biophysical journal. 2009;96:2989–3004. doi: 10.1016/j.bpj.2008.12.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pospieszalska MK, Zarbock A, Pickard JE, Ley K. Event-tracking model of adhesion identifies load-bearing bonds in rolling leukocytes. Microcirculation. 2009;16:115–30. doi: 10.1080/10739680802462792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fallahi-Sichani M, Kirschner DE, Linderman JJ. NF-kappaB Signaling Dynamics Play a Key Role in Infection Control in Tuberculosis. Frontiers in physiology. 2012;3:170. doi: 10.3389/fphys.2012.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown BN, Price IM, Toapanta FR, DeAlmeida DR, Wiley CA, et al. An agent-based model of inflammation and fibrosis following particulate exposure in the lung. Mathematical biosciences. 2011;231:186–96. doi: 10.1016/j.mbs.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheff JD, Mavroudis PD, Calvano SE, Lowry SF, Androulakis IP. Modeling autonomic regulation of cardiac function and heart rate variability in human endotoxemia. Physiological genomics. 2011;43:951–64. doi: 10.1152/physiolgenomics.00040.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sander EA, Stylianopoulos T, Tranquillo RT, Barocas VH. Image-based multiscale modeling predicts tissue-level and network-level fiber reorganization in stretched cell-compacted collagen gels. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17675–80. doi: 10.1073/pnas.0903716106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharafi B, Ames EG, Holmes JW, Blemker SS. Strains at the myotendinous junction predicted by a micromechanical model. Journal of biomechanics. 2011 doi: 10.1016/j.jbiomech.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adra S, Sun T, MacNeil S, Holcombe M, Smallwood R. Development of a three dimensional multiscale computational model of the human epidermis. PloS one. 2010;5:e8511. doi: 10.1371/journal.pone.0008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenstein JL, Winslow RL. Integrative systems models of cardiac excitation-contraction coupling. Circulation research. 2011;108:70–84. doi: 10.1161/CIRCRESAHA.110.223578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreno JD, Zhu ZI, Yang PC, Bankston JR, Jeng MT, et al. A computational model to predict the effects of class I anti-arrhythmic drugs on ventricular rhythms. Science translational medicine. 2011;3:98ra83. doi: 10.1126/scitranslmed.3002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thorne BC, Hayenga HN, Humphrey JD, Peirce SM. Toward a multi-scale computational model of arterial adaptation in hypertension: verification of a multi-cell agent based model. Frontiers in physiology. 2011;2:20. doi: 10.3389/fphys.2011.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holland DO, Krainak NC, Saucerman JJ. Graphical approach to model reduction for nonlinear biochemical networks. PloS one. 2011;6:e23795. doi: 10.1371/journal.pone.0023795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt BJ, Papin JA, Lawrence MB. Nano-motion dynamics are determined by surface-tethered selectin mechanokinetics and bond formation. PLoS computational biology. 2009;5:e1000612. doi: 10.1371/journal.pcbi.1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bailey AM, Thorne BC, Peirce SM. Multi-cell agent-based simulation of the microvasculature to study the dynamics of circulating inflammatory cell trafficking. Annals of biomedical engineering. 2007;35:916–36. doi: 10.1007/s10439-007-9266-1. [DOI] [PubMed] [Google Scholar]
- 25.Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. The Journal of cell biology. 2010;188:11–9. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grima R. Multiscale modeling of biological pattern formation. Current topics in developmental biology. 2008;81:435–60. doi: 10.1016/S0070-2153(07)81015-5. [DOI] [PubMed] [Google Scholar]
- 27.N’Dri NA, Shyy W, Tran-Son-Tay R. Computational modeling of cell adhesion and movement using a continuum-kinetics approach. Biophysical journal. 2003;85:2273–86. doi: 10.1016/s0006-3495(03)74652-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brodland GW, Chen X, Lee P, Marsden M. From genes to neural tube defects (NTDs): insights from multiscale computational modeling. HFSP journal. 2010;4:142–52. doi: 10.2976/1.3338713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sample C, Shvartsman SY. Multiscale modeling of diffusion in the early Drosophila embryo. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10092–6. doi: 10.1073/pnas.1001139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomlin CJ, Axelrod JD. Biology by numbers: mathematical modelling in developmental biology. Nature reviews. Genetics. 2007;8:331–40. doi: 10.1038/nrg2098. [DOI] [PubMed] [Google Scholar]
- 31.von Dassow G, Meir E, Munro EM, Odell GM. The segment polarity network is a robust developmental module. Nature. 2000;406:188–92. doi: 10.1038/35018085. [DOI] [PubMed] [Google Scholar]
- 32.Meir E, Munro EM, Odell GM, Von Dassow G. Ingeneue: a versatile tool for reconstituting genetic networks, with examples from the segment polarity network. The Journal of experimental zoology. 2002;294:216–51. doi: 10.1002/jez.10187. [DOI] [PubMed] [Google Scholar]
- 33.Longo D, Peirce SM, Skalak TC, Davidson L, Marsden M, et al. Multicellular computer simulation of morphogenesis: blastocoel roof thinning and matrix assembly in Xenopus laevis. Developmental biology. 2004;271:210–22. doi: 10.1016/j.ydbio.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 34.Silva JR, Rudy Y. Multi-scale electrophysiology modeling: from atom to organ. The Journal of general physiology. 2010;135:575–81. doi: 10.1085/jgp.200910358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aguado-Sierra J, Krishnamurthy A, Villongco C, Chuang J, Howard E, et al. Patient-specific modeling of dyssynchronous heart failure: A case study. Progress in biophysics and molecular biology. 2011 doi: 10.1016/j.pbiomolbio.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newman SA, Christley S, Glimm T, Hentschel HG, Kazmierczak B, et al. Multiscale models for vertebrate limb development. Current topics in developmental biology. 2008;81:311–40. doi: 10.1016/S0070-2153(07)81011-8. [DOI] [PubMed] [Google Scholar]
- 37.Das A, Lauffenburger D, Asada H, Kamm RD. A hybrid continuum-discrete modelling approach to predict and control angiogenesis: analysis of combinatorial growth factor and matrix effects on vessel-sprouting morphology. Philosophical transactions. Series A, Mathematical, physical, and engineering sciences. 2010;368:2937–60. doi: 10.1098/rsta.2010.0085. [DOI] [PubMed] [Google Scholar]
- 38.Liu G, Qutub AA, Vempati P, Mac Gabhann F, Popel AS. Module-based multiscale simulation of angiogenesis in skeletal muscle. Theoretical biology & medical modelling. 2011;8:6. doi: 10.1186/1742-4682-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vempati P, Popel AS, Mac Gabhann F. Formation of VEGF isoform-specific spatial distributions governing angiogenesis: computational analysis. BMC systems biology. 2011;5:59. doi: 10.1186/1752-0509-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bauer AL, Jackson TL, Jiang Y, Rohlf T. Receptor cross-talk in angiogenesis: mapping environmental cues to cell phenotype using a stochastic, Boolean signaling network model. Journal of theoretical biology. 2010;264:838–46. doi: 10.1016/j.jtbi.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 41.Hashambhoy YL, Chappell JC, Peirce SM, Bautch VL, Mac Gabhann F. Computational modeling of interacting VEGF and soluble VEGF receptor concentration gradients. Frontiers in physiology. 2011;2:62. doi: 10.3389/fphys.2011.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Causey L, Cowin SC, Weinbaum S. Quantitative model for predicting lymph formation and muscle compressibility in skeletal muscle during contraction and stretch. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9185–90. doi: 10.1073/pnas.1206398109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coddington EA, Levinson N. Theory of ordinary differential equations. McGraw-Hill; New York: 1955. p. 429. [Google Scholar]
- 44.Fallahi-Sichani M, Flynn JL, Linderman JJ, Kirschner DE. Differential risk of tuberculosis reactivation among anti-TNF therapies is due to drug binding kinetics and permeability. Journal of immunology. 2012;188:3169–78. doi: 10.4049/jimmunol.1103298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quo CF, Moffitt RA, Merrill AH, Wang MD. Adaptive control model reveals systematic feedback and key molecules in metabolic pathway regulation. Journal of computational biology: a journal of computational molecular cell biology. 2011;18:169–82. doi: 10.1089/cmb.2010.0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laise P, Di Patti F, Fanelli D, Masselli M, Arcangeli A. Deterministic and stochastic aspects of VEGF-A production and the cooperative behavior of tumoral cell colony. Journal of theoretical biology. 2011;272:55–63. doi: 10.1016/j.jtbi.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 47.Sun T, Adra S, Smallwood R, Holcombe M, MacNeil S. Exploring hypotheses of the actions of TGF-beta1 in epidermal wound healing using a 3D computational multiscale model of the human epidermis. PloS one. 2009;4:e8515. doi: 10.1371/journal.pone.0008515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haggart CR, Bartell JA, Saucerman JJ, Papin JA. Whole-genome metabolic network reconstruction and constraint-based modeling. Methods in enzymology. 2011;500:411–33. doi: 10.1016/B978-0-12-385118-5.00021-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tveito A, Lines GT, Edwards AG, Maleckar MM, Michailova A, et al. Slow Calcium-Depolarization-Calcium waves may initiate fast local depolarization waves in ventricular tissue. Progress in biophysics and molecular biology. 2012 doi: 10.1016/j.pbiomolbio.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson AB, Gayzik FS, Moreno DP, Rhyne AC, Vavalle NA, Stitzel JD. A paradigm for human body finite element model integration from a set of regional models. Biomedical sciences instrumentation. 2012;48:423–30. [PubMed] [Google Scholar]
- 51.Zhang X, Gan RZ. A comprehensive model of human ear for analysis of implantable hearing devices. IEEE transactions on bio-medical engineering. 2011;58:3024–7. doi: 10.1109/TBME.2011.2159714. [DOI] [PubMed] [Google Scholar]
- 52.Zahedmanesh H, Lally C. A multiscale mechanobiological modelling framework using agent-based models and finite element analysis: application to vascular tissue engineering. Biomechanics and modeling in mechanobiology. 2011 doi: 10.1007/s10237-011-0316-0. [DOI] [PubMed] [Google Scholar]
- 53.Goktepe S, Abilez OJ, Parker KK, Kuhl E. A multiscale model for eccentric and concentric cardiac growth through sarcomerogenesis. Journal of theoretical biology. 2010;265:433–42. doi: 10.1016/j.jtbi.2010.04.023. [DOI] [PubMed] [Google Scholar]
- 54.Du P, O’Grady G, Davidson JB, Cheng LK, Pullan AJ. Multiscale modeling of gastrointestinal electrophysiology and experimental validation. Critical reviews in biomedical engineering. 2010;38:225–54. doi: 10.1615/critrevbiomedeng.v38.i3.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trayanova NA, Rice JJ. Cardiac electromechanical models: from cell to organ. Frontiers in physiology. 2011;2:43. doi: 10.3389/fphys.2011.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Artel A, Mehdizadeh H, Chiu YC, Brey EM, Cinar A. An agent-based model for the investigation of neovascularization within porous scaffolds. Tissue engineering. Part A. 2011;17:2133–41. doi: 10.1089/ten.tea.2010.0571. [DOI] [PubMed] [Google Scholar]
- 57.Bentley K, Gerhardt H, Bates PA. Agent-based simulation of notch-mediated tip cell selection in angiogenic sprout initialisation. Journal of theoretical biology. 2008;250:25–36. doi: 10.1016/j.jtbi.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 58.Qu Z, Garfinkel A, Weiss JN, Nivala M. Multi-scale modeling in biology: How to bridge the gaps between scales? Progress in biophysics and molecular biology. 2011 doi: 10.1016/j.pbiomolbio.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Valentin A, Humphrey JD. Evaluation of fundamental hypotheses underlying constrained mixture models of arterial growth and remodelling. Philosophical transactions. Series A, Mathematical, physical, and engineering sciences. 2009;367:3585–606. doi: 10.1098/rsta.2009.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valentin A, Humphrey JD. Parameter sensitivity study of a constrained mixture model of arterial growth and remodeling. Journal of biomechanical engineering. 2009;131:101006. doi: 10.1115/1.3192144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hayenga HN, Thorne BC, Peirce SM, Humphrey JD. Ensuring Congruency in Multiscale Modeling: Towards Linking Agent Based and Continuum Biomechanical Models of Arterial Adaptation. Annals of biomedical engineering. 2011 doi: 10.1007/s10439-011-0363-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fedosov DA, Lei H, Caswell B, Suresh S, Karniadakis GE. Multiscale modeling of red blood cell mechanics and blood flow in malaria. PLoS computational biology. 2011;7:e1002270. doi: 10.1371/journal.pcbi.1002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim MJ, Maly IV. Deterministic mechanical model of T-killer cell polarization reproduces the wandering of aim between simultaneously engaged targets. PLoS computational biology. 2009;5:e1000260. doi: 10.1371/journal.pcbi.1000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Singh S, Narang AS, Mahato RI. Subcellular fate and off-target effects of siRNA, shRNA, and miRNA. Pharmaceutical research. 2011;28:2996–3015. doi: 10.1007/s11095-011-0608-1. [DOI] [PubMed] [Google Scholar]
- 65.Bassingthwaighte JB. Predictive Modeling and Integrative Physiology: The Physiome Projects. The open pacing, electrophysiology & therapy journal. 2010;3:66–74. doi: 10.2174/1876536X01003010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bassingthwaighte JB, Chizeck HJ. The Physiome Projects and Multiscale Modeling. IEEE signal processing magazine. 2008;25:121–44. doi: 10.1109/MSP.2007.914723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nawroth JC, Lee H, Feinberg AW, Ripplinger CM, McCain ML, et al. A tissue-engineered jellyfish with biomimetic propulsion. Nature biotechnology. 2012 doi: 10.1038/nbt.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Callura JM, Cantor CR, Collins JJ. Genetic switchboard for synthetic biology applications. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5850–5. doi: 10.1073/pnas.1203808109. [DOI] [PMC free article] [PubMed] [Google Scholar]