Abstract
Embryo quality is a critical parameter in assisted reproductive technologies. Although embryo quality can be evaluated morphologically, embryo morphology does not correlate perfectly with embryo viability. To improve this, it is important to understand which molecular mechanisms are involved in embryo quality control. Autophagy is an evolutionarily conserved catabolic process in which cytoplasmic materials sequestered by autophagosomes are degraded in lysosomes. We previously demonstrated that autophagy is highly activated after fertilization and is essential for further embryonic development. Here, we developed a simple fluorescence-based method for visualizing autophagic activity in live mouse embryos. Our method is based on imaging of the fluorescence intensity of GFP-LC3, a versatile marker for autophagy, which is microinjected into the embryos. Using this method, we show that embryonic autophagic activity declines with advancing maternal age, probably due to a decline in the activity of lysosomal hydrolases. We also demonstrate that embryonic autophagic activity is associated with the developmental viability of the embryo. Our results suggest that embryonic autophagic activity can be utilized as a novel indicator of embryo quality.
The recent development of assisted reproductive technologies (ART), such as in vitro fertilization (IVF), allows us to assess embryo quality prior to embryo transfer. In humans, it is generally recognized that multiple embryo transfer increases not only the chance of pregnancy but also the risk of multiple pregnancies1. Thus, selecting the most viable embryo is a critical step for ART. Embryo quality is usually classified according to embryo morphology2; however, these assessments do not always correlate perfectly with embryo viability. To overcome these issues, it is important to understand which molecular mechanism(s) can affect embryo viability.
In most animal species, the transformation from differentiated oocyte to totipotent embryo after fertilization, known as the oocyte-to-embryo transition, is critical for further embryonic development3. As this transition is completed during a short period, the embryo must activate bulk degradation systems in order to eliminate unnecessary maternal factors and recycle them for use in the synthesis of new zygotic products.
Macroautophagy (referred to hereafter simply as autophagy) is an evolutionarily conserved catabolic process in which portions of the cytoplasm sequestered by double-membrane structures (autophagosomes) are delivered to lysosomes, resulting in the degradation of cytoplasmic content by lysosomal proteases4. The basic roles of autophagy are the production of necessary amino acids during starvation and the maintenance of intracellular quality by eliminating misfolded proteins and abnormal organelles. In addition, autophagy is involved in many other processes, including cancer, aging, cell death, viral infection, and development5. Previously, we reported that autophagy is highly induced after fertilization, and that autophagy-deficient embryos die before implantation6; these observations highlight the importance of autophagy during preimplantation mammalian embryonic development. These findings led us to visualize autophagic activity in live embryos and to investigate the functional links between autophagic activity, maternal aging, and embryonic viability.
Results and discussion
Microtubule-associated protein 1 light chain 3 (LC3), a mammalian homolog of yeast Atg8, is a versatile marker of autophagy; phosphatidylethanolamine (PE)-conjugated LC3 (LC3-II: lipidated form) associates predominantly with autophagosomes until they undergo fusion with lysosomes7. Similar to endogenous LC3, green fluorescent protein fused to the N-terminus of LC3 (GFP-LC3) is commonly utilized to observe autophagy, both in cultured cells and whole organisms8. Although autophagy can be visualized by observing punctate GFP-LC3 structures (which primarily represent autophagosomes) under a fluorescence microscope, the entire autophagic process cannot be monitored9. However, because total cellular GFP-LC3 is degraded as an autophagic substrate in the lysosomes, turnover of this protein correlates well with overall autophagic activity. Therefore, quantitation of the total fluorescence intensity of GFP-LC3 has been used to monitor autophagic activity10. This method, generally accepted as an assay for autophagic flux, also allows autophagic activity to be monitored in living cells.
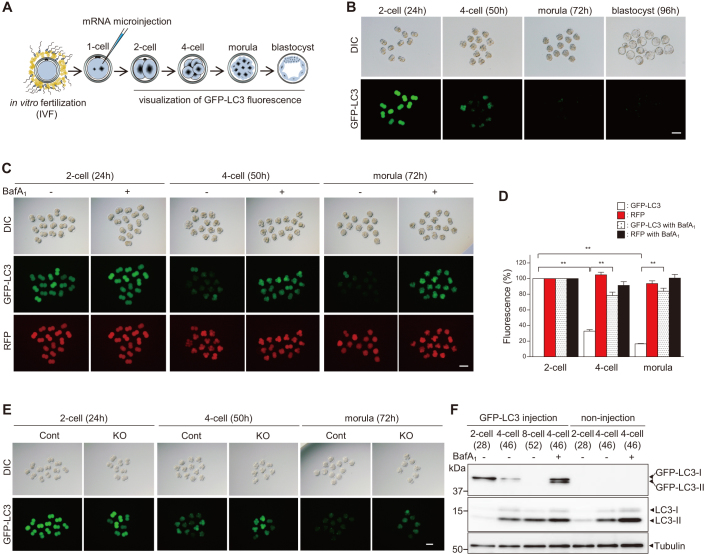
Taking advantage of the fact that total GFP-LC3 fluorescence decreases when autophagy is activated, we attempted to visualize autophagic activity in live mouse embryos. To this end, we microinjected mRNA encoding GFP-LC3 into 1-cell embryos, obtained 5 h after IVF, and visualized total fluorescence at the indicated time points under constant fluorescence settings (Fig. 1A). We detected bright GFP-LC3 fluorescence at the 2-cell stage (24 h after IVF), but this fluorescence rapidly disappeared at the 4-cell stage (50 h after IVF); no fluorescence was observed in the subsequent stages (Fig. 1B). This result is consistent with our previous study using GFP-LC3 transgenic mice8 in which the total level of GFP-LC3 fluorescence declined during early embryonic development6. Therefore, this experiment demonstrates the successful visualization of autophagic activity in live wild-type embryos.
Figure 1. Visualization of autophagic activity during preimplantation embryonic development.
(A) Experimental scheme for imaging autophagic activity in live embryos. (B) Visualization of total GFP-LC3 fluorescence during preimplantation embryonic development. Embryos injected with GFP-LC3 mRNA were observed from the 2-cell to morula stage under constant fluorescence settings. (C) Comparison of GFP-LC3 and control RFP fluorescence intensity in developing embryos cultured with or without BafA1. (D) Graph showing total fluorescence intensities of GFP-LC3 and control RFP during embryonic development; values are expressed relative to the fluorescence at the 2-cell stage. Values are the mean ± s.e.m. of three different experiments, with at least 10 embryos analyzed per group. **P < 0.01 (Student's t-test). (E) GFP-LC3 mRNA was injected into 1-cell embryos collected either from Atg5flox/+;Zp3-Cre (control; Cont) or Atg5flox/−;Zp3-Cre (oocyte-specific Atg5 knockout; KO) females mated with Atg5+/+ (wild-type) males, and total fluorescence was observed during embryonic development. (F) Western blotting with GFP and LC3 antibodies shows GFP-LC3 turnover and endogenous autophagic activity during preimplantation development. Forty embryos were loaded in each lane, and tubulin was used as the loading control. DIC, differential interference contrast. Parentheses, time after IVF (B, C, E, F). Scale bars, 100 μm (B), (C), (E).
To validate our visualization of GFP-LC3 degradation in developing embryos, we first investigated whether the reduction in GFP-LC3 fluorescence occurs in an autophagy-dependent manner. We injected embryos with a mixture of GFP-LC3 and control red fluorescent protein (RFP) mRNAs, and incubated them with bafilomycin A1 (BafA1), an inhibitor of the lysosomal vacuolar proton pump that increases lysosomal pH, thereby blocking lysosomal protease activity. Previously, we demonstrated that BafA1 treatment does not affect embryonic development until the 8-cell stage11. The fluorescence intensity of GFP-LC3 remained high in the presence of BafA1, whereas the fluorescence decayed rapidly in non-treated embryos (Fig. 1C and Supplementary Videos S1, 2). By contrast, RFP fluorescence remained stable over the period monitored (Fig. 1C). At the 4-cell stage, BafA1 treatment resulted in a 45% reduction in GFP-LC3 degradation compared with non-treated embryos, whereas RFP was unaffected (Fig. 1D). Confocal microscopic analysis revealed a massive accumulation of GFP-LC3 dots (which represent autophagosomes) in the BafA1-treated embryos (Supplementary Fig. S1). We observed similar trends in GFP-LC3 fluorescence when we treated embryos with various lysosomal or autophagic inhibitors (Supplementary Fig. S2A, B): E64d and pepstatin A, which inhibit lysosomal proteases; and wortmannin, which inhibits the Class III phosphoinositide 3-kinase pathway, thereby preventing autophagosome formation12.
To further confirm that the reduction in GFP-LC3 fluorescence is dependent on autophagy, we microinjected GFP-LC3 mRNA into autophagy-deficient embryos, collected from oocyte-specific Atg5-knockout females mated with wild-type males, and monitored fluorescence under the same conditions described above. As expected, the total fluorescence of GFP-LC3 was not reduced in autophagy-deficient embryos even in the morula stage (Fig. 1E). In addition, the fluorescence level of neither GFP alone nor GFP-rer1 (GFP targeted to the Golgi membrane13) changed during the monitored period (data not shown). Taken together, these results indicate that the reduction in GFP-LC3 fluorescence is mainly due to increased autophagic degradation, rather than intrinsic GFP instability.
We next analyzed the association between endogenous autophagic activity and the timing of GFP-LC3 degradation during embryo development. To achieve this, we microinjected embryos with GFP-LC3 mRNA and cultured them in the presence or absence of BafA1. At the indicated times, the embryos were subjected to western blot analysis using an anti-GFP antibody and an anti-LC3 antibody that recognizes endogenous LC3-I (cytoplasmic form, apparent mobility: MW = 17 kDa) and LC3-II (membrane-bound form, apparent mobility: MW = 15 kDa). The analysis revealed that GFP-LC3 was present at the 2-cell stage, but degraded rapidly during the 4- and 8-cell stages (Fig. 1F). By contrast, endogenous LC3-II content significantly increased at the 4- and 8-cell stages, a period that corresponds to the massive decay in GFP-LC3 fluorescence that we observed in the developing embryos. Moreover, BafA1 treatment completely blocked GFP-LC3 degradation and increased both GFP-LC3-II and endogenous LC3-II content (Fig. 1F), indicating that GFP-LC3 degradation was due to increased autophagic flux. These results also suggest that the increase in autophagic activity during the 4- and 8-cell stages is required for the massive degradation of GFP-LC3. In addition, because the LC3-I and LC3-II content in embryos microinjected with GFP-LC3 was similar to that in non-injected embryos, it is unlikely that the overexpression of GFP-LC3 interfered with endogenous autophagic activity.
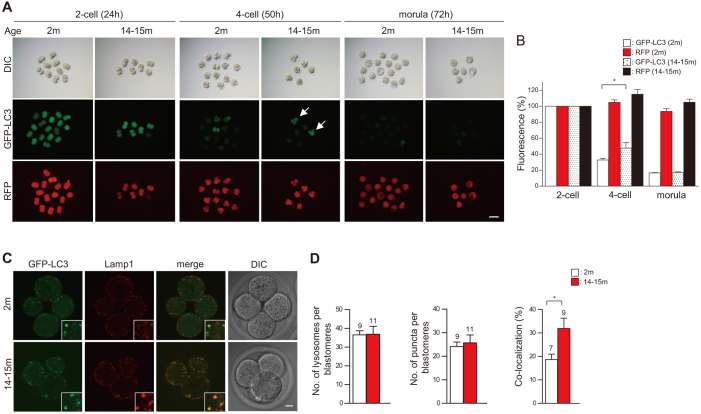
Aging in several model organisms and tissues reflects, at least in part, a decrease in autophagic activity14, leading us to predict that autophagic activity might be reduced in mouse embryos from aged mothers. To test this hypothesis, we used in vitro fertilized embryos derived from 14–15-month-old females, in which the number of ovulated oocytes was significantly decreased (Supplementary Fig. S3A), a well-known hallmark of maternal aging15. Conversely, the rates of IVF and the subsequent development of embryos from aged mothers were similar to those of embryos from young mothers (Supplementary Fig. S3B). To determine whether embryonic autophagic activity declines during aging, we microinjected GFP-LC3 mRNA into 1-cell embryos from either young (2-month-old) or aged (14–15-month-old) females and visualized autophagic activity during embryonic development. The total fluorescence levels were higher in some embryos from aged mothers than in embryos from young mothers (Fig. 2A, B), although there was no significant difference in the developmental process itself throughout the 4-cell stage (50–51 h after IVF) (Supplementary Fig. S3C). Quantitation revealed that the numbers of autophagosomes (GFP-LC3 dots) and lysosomes (Lamp1-positive dots) were not significantly different between embryos from young and aged mothers (Fig. 2C, D). However, the extent of the co-localization of GFP-LC3 and Lamp1 (i.e., the number of dots positive for both GFP-LC3 and Lamp1, shown as yellow dots in Fig. 2C) was significantly higher in embryos from aged mothers (Fig. 2C, D). The high levels of co-localization of GFP-LC3 with Lamp1 reflect a decrease in lysosomal clearance during aging. To test this speculation, we stained embryos with DQ-Red BSA, a derivative of BSA conjugated to a self-quenching red fluorescent dye that is de-quenched by lysosomal proteases, and observed its fluorescence signals at the 4-cell stage. Confocal imaging analysis showed that the number of DQ-Red BSA dots was significantly lower in embryos from aged mothers (Supplementary Fig. S4A, B), supporting our speculation that aging is associated with a decline in lysosomal hydrolase activity, resulting in the decreased clearance of GFP-LC3 in lysosomes.
Figure 2. Decline of autophagic activity with advancing maternal age.
(A) Comparisons of GFP-LC3 and control RFP fluorescence during the development of embryos from young (2 months) and aged (14–15 months) mothers. Embryos that retained GFP-LC3 fluorescence are indicated by arrows. (B) Graph showing the total fluorescence intensities of GFP-LC3 and control RFP during embryonic development; values are expressed relative to the fluorescence at the 2-cell stage. Values are the mean ± s.e.m. of three different experiments, with more than 5 embryos analyzed per group. (C) Comparison of the localization of GFP-LC3 (green) and Lamp1 (red) between embryos from young and aged mothers. The insets show higher magnification images. (D) Comparisons of the number of GFP-LC3 puncta and lysosomes per blastomere between embryos from young and aged mothers, and of the percentage of co-localization of GFP-LC3 with lysosomes. Numbers of embryos analyzed are shown above the bars. Scale bars, 100 μm (A) and 10 μm (C). Error bars, s.e.m.; *P < 0.05 (Student's t-test).
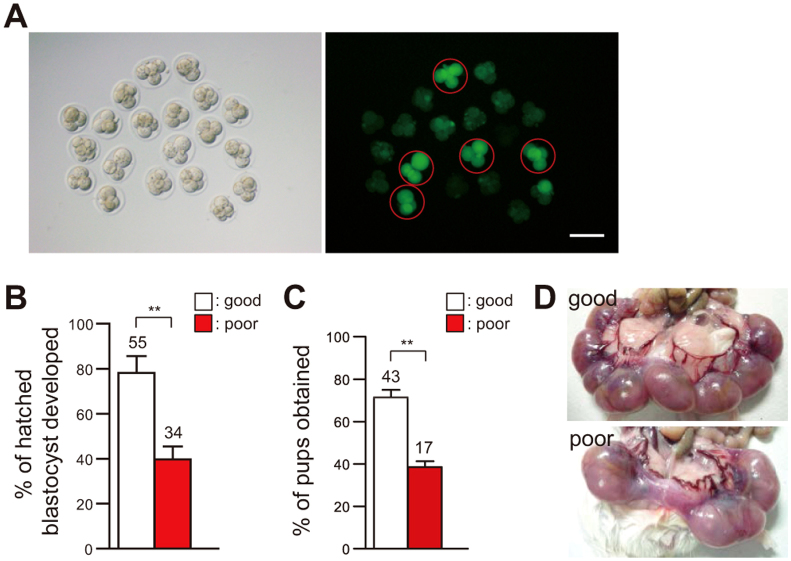
Our successful visualization of autophagic activity in live embryos prompted us to test whether our method could be applied to the assessment of embryonic viability. To this end, we imaged autophagic activity at the 4-cell stage and categorized embryos into two groups according to their GFP-LC3 fluorescence level: embryos with low or high fluorescence were classified as good or poor, respectively (Fig. 3A and Supplementary Table S1). These embryos were further cultured until the hatched blastocyst stage or transferred to a pseudo-pregnant foster mother. Approximately 80% of the good embryos developed to the hatched blastocyst stage, whereas only around 50% of the poor embryos did (Fig. 3B). In addition, the ability to implant and undergo full-term embryonic development was significantly higher in the good embryos than in the poor embryos (Fig. 3C, D, and Supplementary Table S1), suggesting that autophagic activity is related, at least in part, to the ability of an embryo to undergo normal development.
Figure 3. Autophagic activity is related to the developmental ability of the embryo.
(A) Categorization of 4-cell embryos into good or poor embryos based on GFP-LC3 fluorescence intensity. Embryos judged as poor are shown as red circles. Scale bars, 100 μm (B) Hatched blastocyst developmental rates of good and poor embryos. Numbers of embryos analyzed are shown above the bars. Error bars, s.e.m.; **P < 0.01 (Student's t-test). (C) Full-term developmental rates of good and poor embryos. Numbers of pups analyzed are shown above the bars. Error bars, s.e.m.; **P < 0.01 (Student's t-test). (D) Photograph showing embryonic development in Day 19 uteri of foster mothers that had received the same number of embryos categorized as good (upper) or poor (bottom).
In this study we developed a simple fluorescence-based method for visualizing autophagic activity in live embryos. Using this method, we showed that autophagic activity declined in embryos from aged mothers, probably due to a decline in lysosomal protease activity. We also demonstrated that autophagic activity is associated with embryo viability. Clinically, for ART including IVF, it is critical to select embryos with high developmental potential to increase the chance of pregnancy after transfer to the uterus. To date, however, few reliable methods are available for evaluating the competence of individual embryos. Although the monitoring of autophagic activity must be improved for our method to be applied to human embryos, we believe that in combination with other methods, such as the evaluation of embryo morphology2 and cytoplasmic flow16, our method could facilitate more reliable predictions of embryonic viability. Our findings may also have therapeutic potential, in that embryonic quality might be improved or maintained if autophagic activity could be regulated artificially. To achieve this, further analysis would be required on how to control fertilization-induced autophagy precisely at the molecular level.
Methods
Mice and embryo culture
All animal experiments were performed in accordance with the relevant guidelines and regulations and were approved by the Animal Care and Use Committee of the National Institute of Radiological Sciences. The mice used in this study were C57BL/6 and MCH (ICR) supplied by SLC and Charles River, respectively. Oocyte-specific Atg5-knockout mice were generated as described previously6,17. For IVF, females were superovulated by intra-peritoneal injections of 5 IU pregnant mare's serum gonadotropin (PMSG; Asuka Pharmaceutical) and 5 IU human chorionic gonadotropin (hCG; Asuka Pharmaceutical) at an interval of 46–48 h. Metaphase II (MII) oocytes were collected from females 14–16 h after hCG injection, cultured in mHTF medium, and inseminated with spermatozoa collected from the cauda epididymis of a >12-week-old C57BL/6 male. Fertilized embryos (1-cell embryos), exhibiting 2 pronuclei and a second polar body, were collected 5 h after insemination and used for microinjection. One-cell embryos were further cultured in KSOM-AA medium, except for those used for live-cell imaging, which were cultured in CZB medium. In some experiments, the embryos were co-cultured with the following reagents, diluted in KSOM-AA medium: BafA1 (200 nM; Wako), E64d and pepstatin A (10 μM and 10 μg/ml, respectively; Peptide Institute, Inc.), and wortmannin (200 nM; Sigma). All the media used in this study were routinely made in our laboratory18. Embryo culture was performed under mineral oil (Sigma) in an atmosphere of 5% CO2 in air at 37°C. In some experiments, 4-cell embryos were transferred into the oviducts of pseudo-pregnant ICR females on Day 1 after mating with vasectomized ICR males. These females were sacrificed at Day 19, and implantation sites and pups were counted.
RNA synthesis and microinjection
For in vitro mRNA synthesis, the cDNA fragment of rat LC3 was cloned into the eukaryotic expression vector pcDNA3.1 (Invitrogen), which contained a GFP-epitope tag, to yield GFP-LC3. For transcription of EGFP and RFP mRNAs, the EGFP or RFP cDNA alone was cloned into the pcDNA3.1 expression vector. After validating the sequence, these constructs were linearized, and capped mRNAs with poly(A) tails were synthesized using the mMESSAGE mMACHINE Ultra Kit (Ambion) according to the manufacturer's instructions. The synthesized mRNA was purified using a MegaClear Kit (Ambion), and the mRNA concentration was determined using NanoDrop (Thermo Fisher) or e-Spect (Malcom). Before microinjection, the mRNA was diluted in TE buffer (pH 7.4) to 100 ng/μl and then filtered using Ultrafree-MC (Merck Millipore) to remove insoluble materials. We titrated the mRNA concentration without a detrimental effect on embryonic development (>80% of the injected embryos developed normally into blastocysts), and all injection experiments were carried out using 100 ng/μl of the mRNA in TE buffer. Microinjection was performed as previously described11.
Fluorescence microscopy
To visualize GFP-LC3 fluorescence in the developing embryos, 1-cell embryos microinjected with the indicated RNAs (described above) were placed in 1–2 μl of PB1 medium under mineral oil in a glass-bottomed dish (Matsunami Glass) and observed at the following stages and time points: 2-cell, 4-cell, morula, and blastocyst stages were observed 24, 50, 72, and 96 h after IVF, respectively. We used an Olympus IX70 inverted microscope equipped with a DP72 CCD camera (Olympus), a 10× objective lens (NA0.3; Olympus), and an ND6 neutral density filter (Olympus), which allowed the transmission of only 6% of the total fluorescence to the embryos. Fluorescence images of live embryos were captured in 5-sec exposures and analyzed using LuminaVision software (Mitani Corporation). The fluorescence intensity in all embryos was measured at the indicated stages using ImageJ software (National Institutes of Health). The imaging procedure for each sample was completed within 5 min. The imaged embryos were further cultured in KSOM-AA.
To categorize GFP-LC3 injected embryos at the 4-cell stage into either good or poor embryos (see Fig. 3A), the whole fluorescence intensity of individual embryos, including the nucleus and cytoplasm, was measured using ImageJ and compared to that obtained from the embryos co-cultured with the lysosomal inhibitor BafA1. Embryos with similar fluorescence levels of the BafA1-treated embryos (an average of 26.5, s.d. = 5.0, n = 55) were predicted as poor, while the embryos with lower fluorescence levels than the BafA1-treated embryos (an average of 12.5, s.d. = 3.7, n = 87) were predicted as good. The average fluorescence intensities of the good and poor embryos are shown in Supplementary Table 1. To exclude the possibility that differences in GFP-LC3 expression levels affected the fluorescence decay, only those embryos with highly similar GFP-LC3 fluorescence levels at the 2-cell stage were used in this study. The two groups of embryos were either cultured separately or transferred immediately to a foster mother.
For time-lapse imaging, GFP-LC3 injected embryos that had developed normally to the 2-cell stage were transferred into 5 μl of CZB medium in a glass-bottomed dish. Fluorescence images were recorded at 1-h intervals over 48 hours using a BioStation (Nikon) with the following settings: objective lens, 20× NA0.5; filter, FITC (488 nm); light, 3%; exposure time, 1 s; gain, 46; resolution, 1280 × 960. The internal temperature was maintained at 37°C in an atmosphere containing 5% CO2. We confirmed that the imaged embryos developed normally from the 2-cell to morula stage under these settings.
Embryo staining
Embryos were fixed in 3.7% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min. After blocking and permeabilization in PBS containing 0.4% Triton X-100, 10% goat serum, and 3% bovine serum albumin (BSA; Sigma) at 4°C overnight, the embryos were stained with antibodies against Lamp1 (1:200; Abcam ab25245) overnight at 4°C. After washing with 3% BSA in PBS, the embryos were stained with anti-rat IgG DyLight™ 549 (1:750; Rockland) for 50 min at room temperature. After further washing, the embryos were placed in glass-bottomed dishes and observed with an FV1000 (Olympus) laser scanning confocal system. For DQ-Red BSA staining, 2-cell embryos were washed in BSA-free PB1 containing 3 mg/ml polyvinylpyrrolidone (PVP; Sigma) and cultured with 50 μg/ml DQ-Red BSA (self-quenching red BODIPY dye conjugated to BSA; Molecular Probes) for 30 min at 37°C on a warm plate. After staining, the embryos were washed three times with PB1 and further cultured in KSOM-AA until use. DQ-Red BSA-stained embryos were fixed in 3.7% paraformaldehyde as described above and observed using an FV10i (Olympus). Fluorescence measurements and dot counts were performed using ImageJ software.
Western blotting
Forty embryos injected with or without GFP-LC3 mRNA were collected at the indicated stages, washed more than four times in PBS containing PVP instead of BSA, and then lysed in 10 μl of 4× Laemmli sample buffer (Sigma). These samples were separated by SDS-PAGE (13.5% gel) and transferred to PVDF membranes. The membranes were probed with anti-GFP (1:1,000; Wako mFX75), anti-LC3 (1:1,000; Sigma L7543), and anti-tubulin (1:1,000; Cell Signaling #2148) antibodies, followed by horseradish peroxidase-conjugated secondary antibodies (1:10,000; Cell Signaling #7076 for GFP, #7074 for LC3 and tubulin, respectively). When necessary, the antibodies were diluted in XL-Enhancer solution (APRO Science) to enhance signal intensity. Western Lightning Ultra (PerkinElmer) was used for the chemiluminescence reaction. The signals were obtained using a ChemiDoc-It BioChemi (UVP).
Statistical analysis
Data were analyzed using GraphPad Prism 5 (GraphPad Software, Inc.), and are expressed as means ± s.e.m., except those in Supplementary Table 1. Two-tailed Student's t tests were used to evaluate significance and calculate P values. P values less than 0.05 were considered to represent statistically significant differences.
Author Contributions
S.T. conceived and designed the experiments. S.T., T.H. and A.Y. performed the experiments, and S.K., N.M., T. Kubota, K.S. and T. Kokubo helped interpret the data. S.T. wrote the initial manuscript, and T.H., N.M. and K.S. edited the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Supplementary Information
Supplementary Videos S1
Supplementary Videos S2
Acknowledgments
We thank Dr. Noboru Mizushima (The University of Tokyo) for providing Atg5+/− and Atg5flox mice, and for valuable comments. We acknowledge Ayako Wada and Megumi Ibayashi for assistance with embryo manipulation. This research was supported in part by the Takeda Science Foundation (Visionary Research), MEXT/JSPS KAKENHI grant 25111005, the joint research program of the Institute for Molecular and Cellular Regulation, Gunma University 11022 (S.T.), by MEXT/JSPS KAKENHI grants 24116707 and 24590341 (T.H.), and by the Funding Program for Next Generation World-leading Researchers (NEXT program), The Sumitomo Foundation, The Naito Foundation, and The Mochida Memorial Foundation (K.S.).
References
- Katz P., Nachtigall R. & Showstack J. The economic impact of the assisted reproductive technologies. Nat Cell Biol 4 Suppl, s29–32 (2002). [DOI] [PubMed] [Google Scholar]
- Scott L., Alvero R., Leondires M. & Miller B. The morphology of human pronuclear embryos is positively related to blastocyst development and implantation. Hum Reprod 15, 2394–2403 (2000). [DOI] [PubMed] [Google Scholar]
- Telford N. A., Watson A. J. & Schultz G. A. Transition from maternal to embryonic control in early mammalian development: a comparison of several species. Mol Reprod Dev 26, 90–100 (1990). [DOI] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev 21, 2861–2873 (2007). [DOI] [PubMed] [Google Scholar]
- Mizushima N. & Komatsu M. Autophagy: renovation of cells and tissues. Cell 147, 728–741 (2011). [DOI] [PubMed] [Google Scholar]
- Tsukamoto S. et al. Autophagy is essential for preimplantation development of mouse embryos. Science 321, 117–120 (2008). [DOI] [PubMed] [Google Scholar]
- Kabeya Y. et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19, 5720–5728 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Matsui M., Yoshimori T. & Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15, 1101–1111 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T. & Levine B. Methods in mammalian autophagy research. Cell 140, 313–326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvets E., Fass E. & Elazar Z. Utilizing flow cytometry to monitor autophagy in living mammalian cells. Autophagy 4, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
- Tsukamoto S. et al. Functional analysis of lysosomes during mouse preimplantation embryo development. J Reprod Dev 59, 33–39 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E. & Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 6, 764–776 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Nishikawa S. & Nakano A. Membrane protein retrieval from the Golgi apparatus to the endoplasmic reticulum (ER): characterization of the RER1 gene product as a component involved in ER localization of Sec12p. Mol Biol Cell 6, 1459–1477 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo A. M. Autophagy and aging: keeping that old broom working. Trends Genet 24, 604–612 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister L. M. et al. Age-related meiotic segregation errors in mammalian oocytes are preceded by depletion of cohesin and Sgo2. Curr Biol 20, 1511–1521 (2010). [DOI] [PubMed] [Google Scholar]
- Ajduk A. et al. Rhythmic actomyosin-driven contractions induced by sperm entry predict mammalian embryo viability. Nat Commun 2, 417 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T. et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 (2006). [DOI] [PubMed] [Google Scholar]
- Kito S. et al. Improved in vitro fertilization and development by use of modified human tubal fluid and applicability of pronucleate embryos for cryopreservation by rapid freezing in inbred mice. Comp Med 54, 564–570 (2004). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Videos S1
Supplementary Videos S2