Abstract
The liver is a tolerogenic environment exploited by persistent infections, such as hepatitis B (HBV) and C (HCV) viruses. In a murine model of intravenous hepatotropic adenovirus infection, liver-primed antiviral CD8+ T cells fail to produce proinflammatory cytokines and do not display cytolytic activity characteristic of effector CD8+ T cells generated by infection at an extrahepatic, that is, subcutaneous, site. Importantly, liver-generated CD8+ T cells also appear to have a T-regulatory (Treg) cell function exemplified by their ability to limit proliferation of antigen-specific T-effector (Teff) cells in vitro and in vivo via T-cell immunoglobulin and mucin 3 (Tim-3) expressed by the CD8+ Treg cells. Regulatory activity did not require recognition of the canonical Tim-3 ligand, galectin-9, but was dependent on CD8+ Treg cell-surface Tim-3 binding to the alarmin, high-mobility group box 1 (HMGB-1). Conclusion: Virus-specific Tim-3+CD8+ T cells operating through HMGB-1 recognition in the setting of acute and chronic viral infections of the liver may act to dampen hepatic T-cell responses in the liver microenvironment and, as a consequence, limit immune-mediated tissue injury or promote the establishment of persistent infections. (Hepatology 2014;59:1351-1365)
Immune cells within the liver are maintained in a state of active tolerance as a result of continuous exposure to microbe-derived pathogen-associated molecular patterns (PAMPs), toxins, and food-derived antigens.1 Hepatotropic viruses adapted to persist in the liver have evolved to take advantage of this environment and trigger dysfunctional adaptive immune responses during acute (adenovirus) and chronic (hepatitis B virus [HBV] and hepatitis C virus [HCV]) virus infections of the liver.2,3 The generation of antiviral CD8+ T-effector (Teff) cells is suboptimal in the liver, in part, because of defective antigen presentation and a skewed CD4+ Th1/Th2 cell balance resulting in diminished T-cell proliferation, decreased production of proinflammatory cytokines, and reduced expression of cytolytic effector molecules.4 Understanding the mechanisms that orchestrate liver tolerance is critical for antiviral immunotherapy target selection and development.
Another factor limiting intrahepatic CD8+ T-cell responses to infection is the generation of T-regulatory (Treg) cells.5 Treg cells suppress Teff cell responses through a variety of antigen-specific cell-contact–dependent and –independent mechanisms including interleukin (IL)-2 sequestration, release of anti-inflammatory cytokines (IL-10 and transforming growth factor beta),5 Cytotoxic T-lymphocyte antigen 4 (CTLA-4) costimulation,6 Fas/Fas ligand or perforin killing,7,8 granzyme B (GrB) negative feedback,9 and programmed death 1/PD-1 ligand (PD-1/PD-L1) signaling.10 Notably, intrahepatic CD8+ T cells responding to acute and chronic HCV infections are capable of producing IL-10 and coexpress PD-1/PD-L1 and T-cell immunoglobulin and mucin 3 (Tim-3). Compelling evidence suggests that simultaneous blockade of these regulatory elements improves intrahepatic immunity, prompting the development of novel therapeutics.11,12 CD8+ T-cell-derived IL-10 appears to reduce IL-12R, IL-2, interferon-gamma (IFN-γ), and Tbet expression through an autocrine mechanism.13 PD-1, upon engagement with PD-L1, suppresses CD8+ T-cell responses by direct small heterodimer partner 2-mediated dephosphorylation events and modulation of Ca++ fluxes downstream of T-cell receptor (TCR) signaling.14 Furthermore, galectin-9 (Gal-9) is abundant in the liver, is induced during viral infection by IFN-γ, and, upon engagement of Tim-3, dampens CD8+ T-cell IL-2, IFN-γ, and proliferation.15 Gal-9 recognizes N- and O-linked glycosylation sites of the Tim-3 mucin region. However, Escherichia coli–derived recombinant Tim-3 tetramers, inherently lacking oligosaccharides, can bind to B cells, T cells, dendritic cells (DCs), and macrophages, suggesting that other non-Gal-9 ligands for Tim-3 exist.16
High-mobility group box 1 (HMGB-1) was recently discovered to bind the FG cleft in the immunoglobulin variable region of Tim-3 within DC endosomes.17 HMGB-1, originally identified as a DNA-binding protein, is classified under the alarmins, which relay danger-associated molecular pattern signals to antigen presenting cells (APCs). Although passively released by necrotic cells, active secretion of HMGB-1 from monocytes, macrophages, DCs, and natural killer (NK) cells has been observed.18 Upon binding the receptor for advanced glycation end-products expressed by DCs and T cells, HMGB-1 can directly stimulate DC maturation and T-cell proliferation.19,20
During an analysis of CD8+ T-cell responses after infection with hepatotropic adenovirus, we hypothesized that antiviral intrahepatic CD8+ T cells had Treg cell properties likely dependent on production of IL-10, display of Gal-9, and/or expression of PD-L1. However, we observed that the PD-1/PD-L1+Tim-3+CD8+ T cells did not produce Gal-9; moreover, administration of blocking antibodies (Abs) to IL-10, PD-L1, and Gal-9 indicated that CD8+ Treg cell suppression was not dependent on these mechanisms. Surprisingly, we found that Tim-3, displayed on the surface of intrahepatic CD8+ Treg cells, sequesters HMGB-1, thereby limiting augmentation of CD8+ Teff cell proliferation. The capture of HMGB-1 by Tim-3 on the surface of CD8+ Treg cells offers an attractive explanation for the inability of CD8+ Teff cells to generate efficient antiviral responses in the liver.
Materials and Methods
Animals and Infections
Thy1.2+/+, Thy1.1+/−OT-I(Tcra/Tcrb)+/− (Taconic Farms, Hudson, NY), and IL-10 transcriptional reporter (Vert-X) mice, provided by Christopher Karp (University of Cincinnati College of Medicine, Cincinnati, OH), were used in these experiments. Animals were 6-10 weeks of age and housed in a pathogen-free facility under protocols approved by the institutional animal care and use committee at the University of Virginia (Charlottesville, VA).
Replication-deficient type 5 adenoviruses expressing ovalbumin (Ad-Ova) and beta-galactosidase (β-Gal; Ad-LacZ) were provided by Timothy L. Ratliff (University of Iowa, Iowa City, IA) and Gregory A. Helm (University of Virginia), respectively. Mouse cytomegalovirus expressing ovalbumin (MCMV-Ova) was provided by Ann B. Hill (Oregon Health and Science University, Portland, OR). Mice were infected with 2.5 × 107 IU Ad-Ova/LacZ or 1 × 104 IU MCMV-Ova by intravenous (IV) injection in the caudal vein or subcutaneous (SC) injection in the left flank.
Quantitative Polymerase Chain Reaction
Total RNA was isolated using the TRIzol method (Invitrogen, Carlsbad, CA) and reverse transcribed using High Capacity RNA-to-cDNA Master Mix (Applied Biosystems, Foster City, CA). Quantitative polymerase chain reaction (qPCR) was performed using Fast SYBR Green Master Mix (Applied Biosystems) on an AB StepOne Plus Real-Time PCR System. QuantiTect primers for Mus musculus Il10, Il2, Ifng, Tgfb, Lgals9, Havcr2, Pdl1, Pdc1, Ctla4, Klrc1, Foxp3, and Ikzf2 (Qiagen, Valencia, CA) and self-designed primers for Mus musculus hypoxanthine phosphoribosyltransferase (forward, 5′-CTCCGCCGGCTTCCTCCTCA-3′; reverse, 5′-ACCTGGTTCATCATCGCTAATC-3′) were used for detection.
Enzyme-Linked Immunosorbent Assay
IL-2, IL-10, and IFN-γ enzyme-linked immunosorbent assay (ELISAs) were performed according to the manufacturer's instructions (BD Biosciences, Franklin Lakes, NJ). Absorbance was read at 450 nm using a PowerWave XS Microplate Spectrophotometer (BioTek, Winooski, VT).
Immunoprecipitation and Western Blotting
We added 5μg of recombinant (r) mouse Tim-3 human immunoglobulin G (IgG)1 chimeric protein (rTim-3Fc; R&D Systems, Minneapolis, MN) to 500 μL of supernatant and immunoprecipitated with Protein A/G PLUS-Agarose (Santa Cruz Biotechnology, Dallas, TX). Proteins were resolved, western blotted, and incubated with rabbit anti-HMG1/2/3 (pAb; Santa Cruz Biotechnology), biotinylated anti-human IgG (pAb; SouthernBiotech, Birmingham, AL), horseradish peroxidase (HRP)-linked anti-rabbit IgG (pAb; Cell Signaling Technology, Danvers, MA), and streptavidin-HRP (R&D Systems), followed by visualization with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Rochester, NY).
Liver and Spleen Mononuclear Cell Isolation
Mononuclear cells (MNCs) were isolated from livers by Histodenz (Sigma-Aldrich, St. Louis, MO) gradient centrifugation and spleens over a Ficoll (Atlanta Biologicals, Lawrenceville, GA) gradient, according to previous work.2
In Vitro Suppression Assay
Bone-marrow–derived dendritic cells (BMDCs) were matured for 1 week in RPMI 1640 medium containing 10% HyClone fetal bovine serum, 15 mM of HEPES buffer, 50 μM of beta-mercaptoethanol, 20 ng/mL of rIL-4, and 20 ng/mL of recombinant granulocyte macrophage colony-stimulating factor (eBioscience, San Diego, CA). BMDCs (5 × 103) were pulsed for 5 hours with 10 ng/mL of SIINFEKL or ICPMYARV peptides (AnaSpec, Fremont, CA), then cultured with 5 × 104 carboxyfluorescein succinimidyl ester (CFSE)-labeled (Invitrogen) naïve Thy1.1+CD8+ OT-I T cells. CD8+ T cells from SC- or IV-infected C57BL/6 mice were then added at the appropriate ratio. CD8+ T cells were positively sorted using anti-CD8α magnetic beads (Miltenyi Biotec, Auburn, CA).
In Vivo Suppression Assay
For in vivo liver responses studied, 5 × 105 CFSE-labeled naïve Thy1.1+CD8+ OT-I T cells were transferred into naïve, day 7 Ad-Ova-infected, or day 7 Ad-LacZ-infected mice before IV MCMV-Ova infection. For in vivo lymph node responses, 3 × 106 CD8+ T cells from SC- or IV-infected C57BL/6 mice were cotransferred with 1.5 × 106 CFSE-labeled naïve Thy1.1+CD8+ OT-I T cells into SC-infected C57BL/6 mice at day 0.
Ab Blockade and Cell Treatments
In vivo whole-animal blockade of HMGB-1, PD-L1, and Tim-3 was conducted by intraperitoneal (IP) injection of 300 μg of anti-HMGB-1 (pAb; Shino-Test Corporation, Kanagawa, Japan), anti-PD-L1 (10F.9G2), or anti-Tim-3 (RMT3-23; BioXCell, West Lebanon, NH). For in vitro and in vivo lymph node blockade, CD8+ Treg cells were precoated with 20 μg/mL of anti-PD-L1 and/or anti-Tim-3 for 1 hour at 37°C. Recombinant mouse Gal-9 (rGal-9; 1.0 μg/mL; R&D Systems), 20 μg/mL of anti-Gal-9 (RG9-1), 20 μg/mL of anti-IL-10R (1B1.3A; BioXCell), and 0.5 μg/mL of anti-HMGB-1 (pAb; eBioscience) were added to culture media in relevant experiments.
Flow Cytometry
Antibodies from BD Biosciences, BioLegend (San Diego, CA), eBioscience, and R&D Systems were used for detection. H2-Kb Ova-tetramer APCs (Baylor College of Medicine, Houston, TX) and H2-Kb β-Gal-tetramer phycoerythrin (NIAID, Atlanta, GA) identified antigen-specific CD8+ T cells. MNCs (1.5 × 106) were blocked with anti-CD16/CD32 (2.4G2; University of Virginia), followed by specific Ab labeling. Nuclear transcription factor staining was achieved using the forkhead box protein 3 (FoxP3) Staining Set (eBioscience). For intracellular cytokine detection, cells were restimulated with 5 ng/mL of phorbol myristate acetate (PMA) and 500 ng/mL of ionomycin (Sigma-Aldrich) or 2 μg/mL of SIINFEKL peptide (AnaSpec) and blocked with 1 μL/mL of GolgiPlug/GolgiStop (BD Biosciences). Data were collected on a BD fluorescence-activated cell sorting (FACS) Canto II (BD Immunocytometry Systems, San Jose, CA) and analyzed using FlowJo software (8.8.6; Tree Star Inc., Ashland, OR). Cells were FACS sorted on an iCyt Reflection Cell Sorter (iCyt Mission Technology, Champaign, IL).
Microscopy
Human hepatocellular carcinoma (HCC) and HCV liver biopsies, kindly provided by the Biorepository and Tissue Research Facility and Valeria R. Mas (University of Virginia), respectively, were fixed in chilled acetone for 2 minutes. Mouse tissues were flushed with 1× phosphate-buffered saline and periodate-lysine-paraformaldehyde (PLP) fixative, excised, incubated in PLP for 3 hours at 4°C, and passed over a sucrose gradient. Tissues were frozen in optimal cutting temperature (OCT) medium, sectioned at 5 μm thickness, blocked with 2.4G2 solution (2.4G2 media containing anti-CD16/32, 30% chicken/donkey/horse serum, and 0.1% NaN3), and stained with Abs from BioLegend, eBioscience, Santa Cruz Biotechnology, and Sino Biological (Beijing, China). In select experiments, cells were also labeled with CFSE and Vybrant CM-DiI (Invitrogen). Confocal microscopy was performed on a Zeiss LSM-700, and data were analyzed using Zen 2009 Light Edition software (Carl Zeiss MicroImaging GmbH, Jena, Germany).
Statistical Analysis
Significant differences between experimental groups were calculated using the two-tailed Student t test or one-way analysis of variance (ANOVA; with group comparisons ≥3). Data analysis was performed using Prism software (5.0a; GraphPad Software Inc., La Jolla, CA). Values of P < 0.05 were regarded as being statistically significant and noted as P < 0.05, P < 0.01, and P < 0.001.
Results
Liver-Primed CD8+ T Cells Exert a Treg Suppressor Function on the Priming of Naïve CD8+ T Cells In Vitro by Linked Suppression
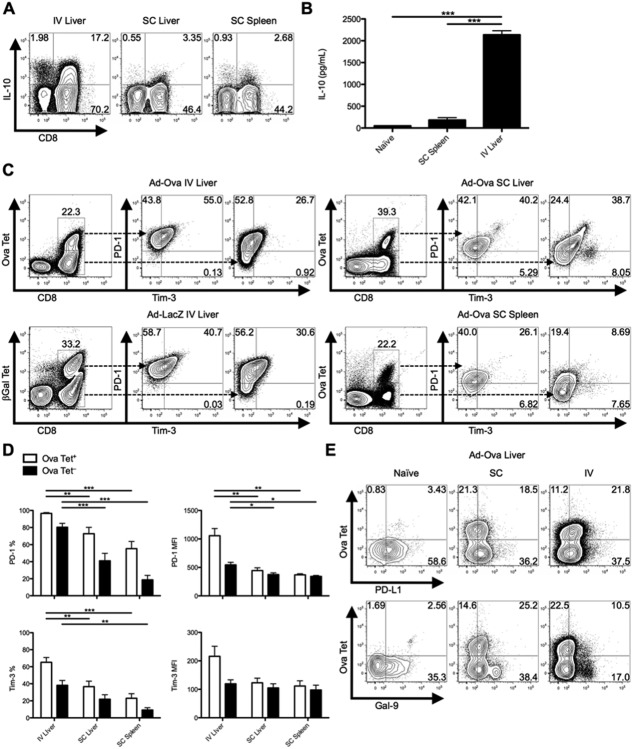
IV adenovirus infection of mice is characterized by a decrease in target cell lysis from day 7 to 14, exemplified by a drop in serum alanine aminotransferase. Antigen-specific CD8+ T cells primed directly in the liver after IV infection peak in absolute number at day 7, but fail to produce tumor necrosis factor alpha (TNF-α), IFN-γ, and GrB, compared to CD8+ T cells that have trafficked to the liver from the inguinal lymph node (Ig LN) in response to SC adenovirus inoculation (Supporting Fig. 1).2 To expand our studies upon the potential roles of anti-inflammatory mediators utilized by liver-primed CD8+ T cells, we examined cellular production of IL-10 and expression of PD-1/PD-L1 and Tim-3. After IV infection with Ad-Ova, liver CD8+ T cells had elevated intracellular IL-10 at day 7, compared to SC-generated CD8+ T cells (Fig. 1A). Secretion of IL-10 was also detectable in culture supernatants of TCR-stimulated CD8+ T cells sorted from the IV-infected liver (Fig. 1B). PD-1 and Tim-3 coexpression peaked at day 7 (Supporting Fig. 2) and was significantly enriched on Ova Tet+CD8+ T cells in livers of IV-infected mice (Fig. 1C,D). In spite of the lower absolute number of CD8+ T cells infiltrating livers and spleens of SC-infected mice, the frequency of Ova-specific CD8+ T cells was comparable between IV and SC infections. IV infection with adenovirus expressing β-Gal (Ad-LacZ) yielded comparable results (Fig. 1C). Route of infection had no effect on expression of PD-L1 in the liver. Expression of the Tim-3 ligand, Gal-9, was readily detectable on naïve and SC CD8+ T cells, but only low expression was observed on liver-primed CD8+ T cells (Fig. 1E).
Fig 1.
IL10, PD-1, and Tim-3 are coexpressed on liver-primed CD8+ T cells after IV Ad infection. C57BL/6 mice were SC or IV infected with 2.5 × 107 IU of Ad-Ova. (A) Endogenous day 7 liver and spleen T-cell IL-10 production was assessed after a 5-hour restimulation with 5 ng/mL of PMA and 500 ng/mL of ionomycin (n = 3 per group). (B) CD8+ T cells sorted from naïve liver, day 7 SC spleen, and day 7 IV liver were restimulated in vitro with plate-bound anti-CD3ε Ab and soluble anti-CD28 Ab. The concentration of IL-10 was measured in the day 2 culture supernatant by ELISA (one-way ANOVA/Tukey's posttest; n = 3 per group). (C and D) Coexpression of PD-1 and Tim-3 was determined directly ex vivo for Ova Tet+ and Ova Tet− populations of CD8+ T cells at day 7 postinfection. Ad-Ova IV infection in the liver was also compared to day 7 livers from Ad-LacZ IV-infected C57BL/6 mice (one-way ANOVA/Tukey's posttest; n = 4-9 per group). (E) PD-L1 and Gal-9 ligand expression was characterized in livers of naïve, SC, and IV Ad-Ova-infected animals (n = 3-6 per group). Numbers in the scatter plots represent percentages (mean ± standard error of the mean). *P < 0.05; **P < 0.01; ***P < 0.001.
Fig 2.
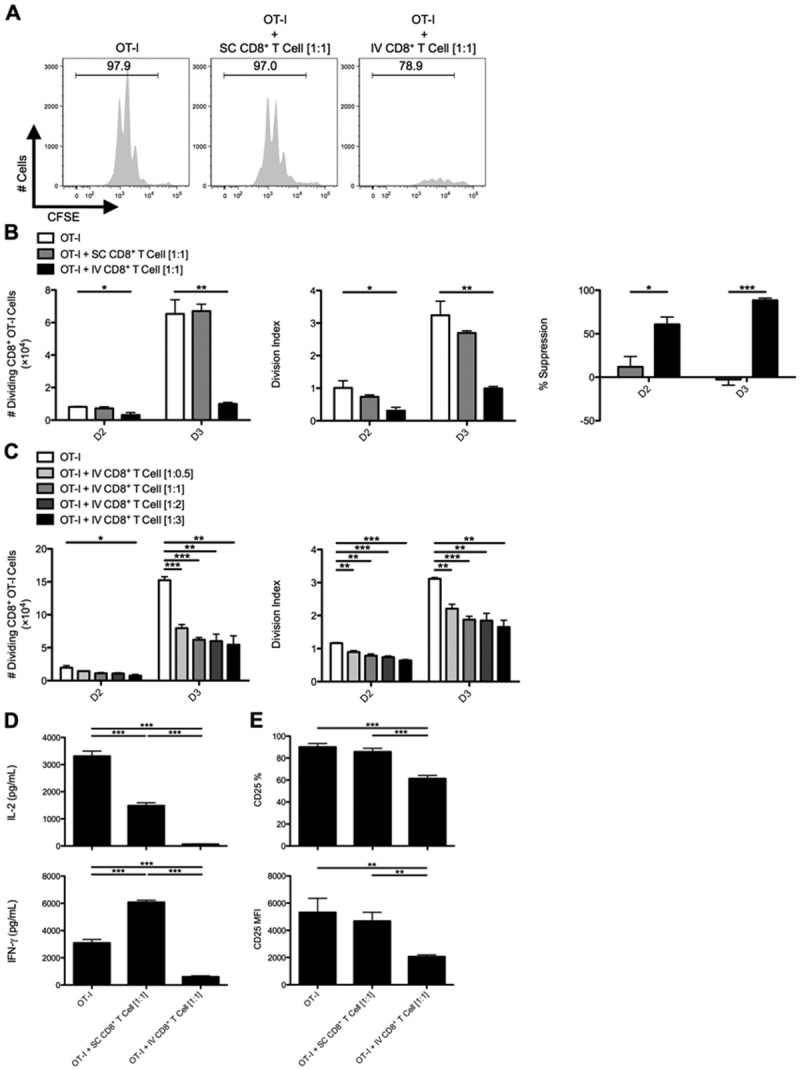
CD8+ T cells from livers of IV-infected animals suppress the activation and expansion of naïve CD8+ OT-I T cells in vitro. C57BL/6 mice were SC or IV infected with 2.5 × 107 IU of Ad-Ova, and (A) bulk CD8+ T cells were isolated from day 7 SC spleens and IV livers, then cultured with CFSE-labeled naïve Thy1.1+CD8+ OT-I T cells at a 1:1 ratio. SIINFEKL-pulsed BMDCs were used as the source of antigen, and cocultures were analyzed at day 3. (B) The number of dividing OT-I T cells, OT-I T cell division index, and percent suppression displayed by either SC CD8+ T cells or IV CD8+ T cells was calculated at days 2 and 3 of culture (one-way ANOVA/Tukey's posttest; n = 12 per group). (C) Naïve Thy1.1+CD8+ OT-I T cells were also cultured at various ratios relative to CD8+ T cells collected from livers of IV-infected mice (1:0.5, 1:1, 1:2, and 1:3; one-way ANOVA/Tukey's posttest; n = 3 per group). (D) Concentrations of IL-2 and IFN-γ were measured in the day 3 culture supernatants by ELISA, and (E) CD25 expression was determined directly ex vivo on day 3 Thy1.1+CD8+ OT-I T cells (one-way ANOVA/Tukey's posttest; n = 5-6 per group). Numbers in histograms represent percentages (mean ± standard error of the mean). *P < 0.05; **P < 0.01; and ***P < 0.001.
Because IL-10-producing, PD-1/PD-L1+Tim-3+CD8+ T cells appeared at a late time point postadenovirus infection (day 7), we hypothesized that liver-primed CD8+ T cells, rather than acting as effector cells, contribute to limiting local inflammatory responses by suppressing Teff cells migrating to the hepatic microenvironment. To test this, we utilized an in vitro system employing TCR transgenic (Tg) OT-I T cells, where Thy1 congenically mismatched CFSE-labeled naïve CD8+ OT-I T cells were placed in coculture with CD8+ T cells isolated from infected day 7 SC spleens or IV livers. BMDCs pulsed with ovalbumin octapeptide (Ova257-264/SIINFEKL) served as the APC. CD8+ T cells isolated from day 7 livers of IV Ad-infected mice had markedly suppressed OT-I T cell proliferative expansion by days 2-3 of culture, as evident from the absolute number and division index of dividing OT-I T cells, whereas CD8+ T cells isolated from the day 7 SC spleens had no suppressive effect (Fig. 2A,B). Suppression was observed over a range of OT-I/IV CD8+ T ratios (Fig. 2C). Significantly less IL-2 and IFN-γ could be detected in supernatants collected from OT-I/IV CD8+ T cell cocultures (Fig. 2D), along with diminished CD25 expression on OT-I T cells exposed to liver-primed CD8+ T cells (Fig. 2E). The regulatory activity mediated by these liver-primed CD8+ Treg cells was also dependent on TCR recognition of specific antigen (Supporting Fig. 3), suggesting that liver-primed CD8+ Treg cells mediate suppression in an antigen-specific manner. Liver-primed CD8+ Treg cells did not appear to induce cell death in responder OT-I T cells (Supporting Fig. 4).
Fig 3.
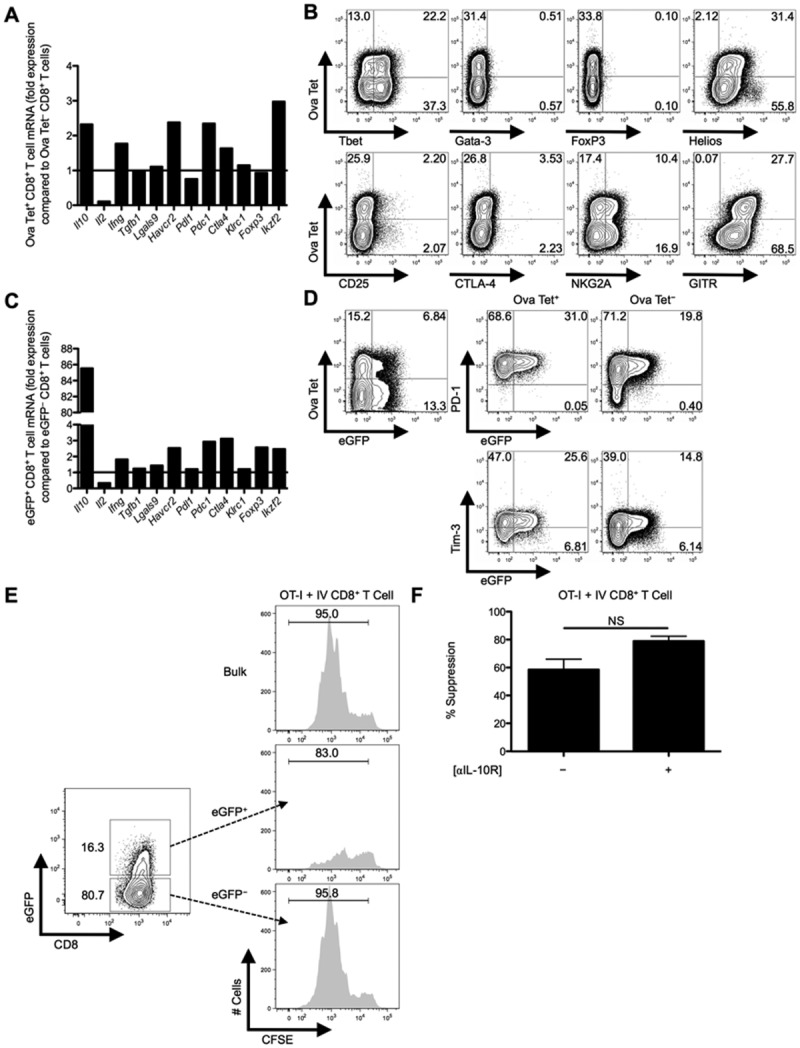
eGFP+CD8+ Treg cells are more potent suppressors compared to eGFP−CD8+ Treg cells from IL-10 transcriptional reporter mice and display some canonical Treg markers. C57BL/6 mice were IV infected with 2.5 × 107 IU of Ad-Ova, day 7 liver MNCs were isolated, and total RNA was collected from FACS-sorted Ova Tet+ and Ova Tet−CD8+ Treg cells. (A) Il10 (IL-10), Il2 (IL-2), Ifng (IFN-γ), Tgfb (TGF-β), Lgals9 (Gal-9), Havcr2 (Tim-3), Pdl1 (PD-L1), Pdc1 (PD-1), Ctla4 (CTLA-4), Klrc1 (NKG2A), Foxp3 (FoxP3), and Ikzf2 (Helios) mRNA was measured by qPCR. (B) Expression of intranuclear/cellular Tbet/Gata-3/FoxP3/Helios and surface CD25/CTLA-4/NKG2A/GITR was determined on CD8+ Treg cells from livers of IV-infected mice. (C) Similarly, transcript from FACS-sorted eGFP+CD8+ Treg cells was compared to that of eGFP−CD8+ Treg cells from day 7 IV-infected Vert-X mice. (D) PD-1 and Tim-3 surface expression along the eGFP profile was separately assessed on Ova Tet+ and Ova Tet−CD8+ Treg cells (n = 3 per group). (E) Representative in vitro suppression assays from bulk CD8+ Treg cells and FACS-sorted eGFP+CD8+ Treg cells or eGFP−CD8+ Treg cells from day 7 Ad-Ova IV-infected C57BL/6 and Vert-X mice, respectively, cocultured with CFSE-labeled naïve Thy1.1+CD8+ OT-I T cells at a 1:1 ratio are depicted (n = 3 per group). (F) Day 3 percent suppression by day 7 Ad-Ova liver-primed CD8+ Treg cells during coculture with CFSE-labeled naïve Thy1.1+CD8+ OT-I T cells at a 1:1 ratio and SIINKFEKL-pulsed BMDCs are displayed. Select wells also contained anti-IL-10R Ab in media during culture (n = 3 per group). Numbers in histograms and scatter plots represent percentages.
Fig 4.
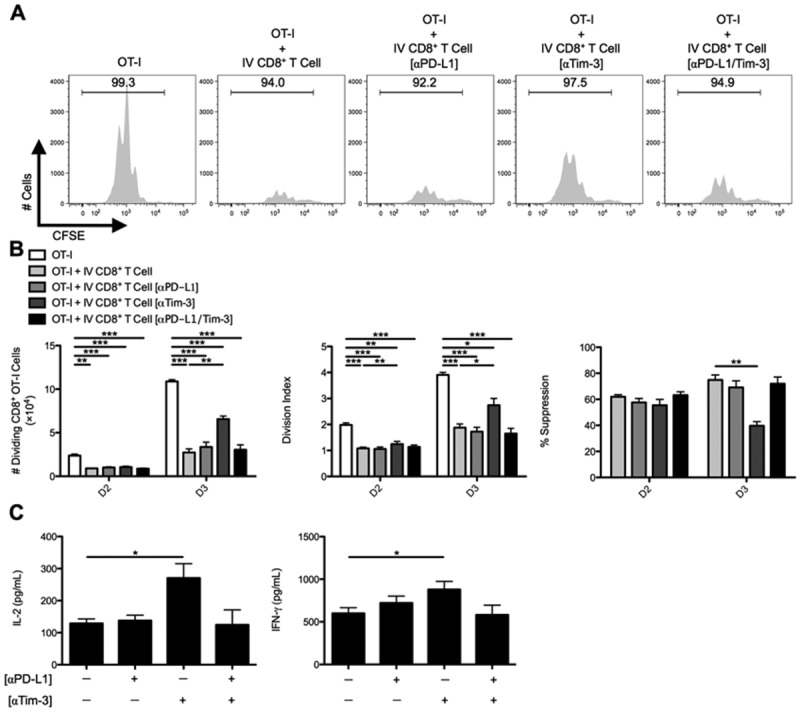
In vitro CD8+ Treg cell suppression is dictated by the Tim-3 inhibitory pathway. C57BL/6 mice were IV infected with 2.5 × 107 IU of Ad-Ova, and (A) bulk CD8+ Treg cells were isolated from day 7 IV livers. Before coculture with CFSE-labeled naïve Thy1.1+CD8+ OT-I T cells at a 1:1 ratio and SIINKFEKL-pulsed BMDCs, CD8+ Treg cells were left alone or precoated with anti-PD-L1 Ab, anti-Tim-3 Ab, or both. Cell cultures were analyzed at day 3 for CFSE dilution. (B) The number of dividing OT-I T cells, OT-I T-cell division index, and percent suppression displayed by IV CD8+ Treg cells was calculated at days 2 and 3 of culture. (C) Concentrations of IL-2 and IFN-γ were measured in the day 3 culture supernatants by ELISA (one-way ANOVA/Tukey's posttest; n = 6-9 per group). Numbers in histograms represent percentages (mean ± standard error of the mean). *P < 0.05; **P < 0.01; ***P < 0.001.
Cellular Markers and Properties Associated With Liver-Primed CD8+ Treg Cells
Hallmarks of CD4+ Treg cells include FoxP3 and CD25 expression, lack of IL-2 production, and failure to undergo robust clonal expansion after TCR/antigen exposure. We analyzed a panel of markers and found that intrahepatic Ova Tet+CD8+ Treg cells had markedly diminished Il2 messenger RNA (mRNA) and lacked enhanced Foxp3 mRNA, but maintained elevated levels of Il10, Havcr2 (Tim-3), Pdc1 (PD-1), and Ikzf2 (Helios) mRNA, compared to Ova Tet−CD8+ T cells (Fig. 3A). Intracellular Helios and cell-surface glucocorticoid-induced TNF receptor-related protein (GITR) was expressed by Ova Tet+CD8+ Treg cells, but surface CD25 and surface and intracellular CTLA-4 was not detectable (Fig. 3B). It has been reported that expression of inhibitory Ly49 receptors, along with restriction of Qa-1 recognition, define a subset of CD8+ Treg cells.8 We could not detect surface expression of Ly49 receptors on these liver CD8+ T cells, but low-level expression of NKG2A and Qa-1 was detected (Fig. 3B; data not shown, respectively).
To explore the role of IL-10, we infected IL-10 transcriptional reporter enhanced green fluorescent protein (eGFP) expressing (Vert-X) mice with Ad-Ova and FACS-sorted eGFP+ and eGFP−CD8+ T cells from the day 7 liver. eGFP+CD8+ T cells maintained elevated Havcr2, Pdc1, and Ikzf2 mRNA, compared to eGFP−CD8+ T cells (Fig. 3C), and eGFP expression positively correlated with PD-1 and Tim-3 expression (Fig. 3D). Given these observations, we explored the IL-10 pathway and other regulatory mechanisms (e.g., PD-1/PD-L1 and Tim-3) to account for the suppressive activity of liver-primed CD8+ Treg cells.
CD8+ Treg Cells Suppress Teff Cells in a Tim-3-Dependent Manner
When we compared the suppressive activity of liver eGFP+ and eGFP−CD8+ Treg cells, we found that eGFP+CD8+ Treg cells were more effective at suppressing OT-I T cell division in vitro (Fig. 3E), presumably as a result of IL-10 secretion and possibly elevated expression of the regulatory molecules, PD-1/PD-L1 and Tim-3. However, anti-IL-10R Ab could not reverse suppression (Fig. 3F). These data indicate that IL-10 phenotypically marks the CD8+ Treg cell, but does not have a central role in suppression.
CD8+ Treg cells were then precoated with anti-PD-L1, anti-Tim-3, or both Abs. Anti-PD-L1 Ab had no effect, but anti-Tim-3 Ab partially rescued OT-I T cell division (Fig. 4A,B). Tim-3 blockade also resulted in enhanced release of IL-2 and IFN-γ into the supernatants collected from OT-I/IV CD8+ Treg cell cocultures (Fig. 4C). Thus, Tim-3 blockade appeared to inhibit CD8+ Treg cell ability to suppress CD8+ Teff cells.
Nascent CD8+ T-Cell Expansion During Liver and Lymph Node Antiviral Responses Is Suppressed by Liver-Primed CD8+ Treg Cells In Vivo
We next examined whether liver-primed CD8+ Treg cells could regulate the expansion of endogenous CD8+ Teff cells in vivo. To this end, naïve, Ad-Ova-, or Ad-LacZ-infected mice were reinfected with MCMV-Ova at day 7 post-Ad infection, and day 14 livers were analyzed. Seven days after MCMV-Ova infection, the absolute number of viable MNCs in livers of mice previously immunized with Ad-Ova was reduced, compared to Ad-LacZ-infected, MCMV-Ova-challenged animals (Fig. 5A). Furthermore, animals undergoing initial Ad-Ova infection had reduced frequencies of IFN-γ+ and TNF-α+IFN-γ+ antigen-specific CD8+ T cells (Fig. 5B). These results support and reinforce our in vitro data and implicate CD8+ Treg cells within the infected liver as the mediators of suppression operating in an antigen-specific manner.
Fig 5.
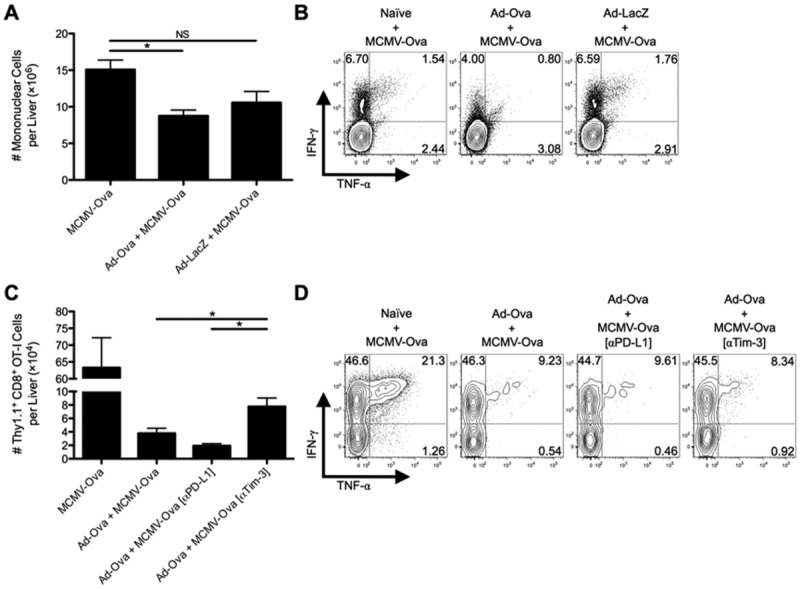
Tim-3 blockade improves antigen-specific hepatic secondary immune responses to viral infection. C57BL/6 mice were IV infected with 2.5 × 107 IU of Ad-Ova, 2.5 × 107 IU of Ad-LacZ, or left uninfected. At day 7, all three experimental groups were IV infected with 1 × 104 IU of MCMV-Ova. (A) The number of live MNCs isolated from livers of day 14 infected animals was enumerated. Trypan blue exclusion was used to assess the number of viable cells. (B) Endogenous CD8+ T cell TNF-α and IFN-γ were quantified at day 14 after a 5-hour restimulation with 2 μg/mL of SIINFEKL peptide (n = 3 per group). (C and D) In a parallel experiment, 5 × 105 naïve Thy1.1+CD8+ OT-I T cells were transferred at day 7. C57BL/6 mice were left untreated or were administered 300 μg of anti-PD-L1 Ab or anti-Tim-3 Ab IP at days 5 and 6 before a day 7 MCMV-Ova infection. The number of Thy1.1+CD8+ OT-I T cells and their TNF-α and IFN-γ production was assessed in livers of infected animals at day 14. Cytokine detection was achieved after a 5-hour restimulation with 2 μg/mL of SIINFEKL peptide (one-way ANOVA/Tukey's posttest; n = 3 per group). Numbers in the scatter plots represent percentages (mean ± standard error of the mean). *P < 0.05.
To further demonstrate the in vivo suppressive effect of CD8+ Treg cells, Thy1.1+CD8+ OT-I T cells were adoptively transferred into naïve and day 7 Ad-Ova-infected mice before MCMV-Ova infection. The numbers of Thy1.1+CD8+ OT-I T cells in livers of mice previously infected with Ad-Ova were reduced, but were partially recovered by IP delivery of anti-Tim-3 Ab at days 5 and 6 (Fig. 5C). No improvements in frequencies of IFN-γ+ and TNF-α+IFN-γ+ antigen-specific CD8+ T cells were observed (Fig. 5D). These observations support the concepts that the regulatory effect of CD8+ Treg cells is antigen-specific and that Tim-3 plays an important role in mediating suppression.
To more directly demonstrate the immunoregulatory effect of liver-primed CD8+ Treg cells on antigen-specific CD8+ Teff cell responses in vivo, day 7 Ad-Ova SC splenic CD8+ T cells, day 7 IV liver CD8+ Treg cells, and CFSE-labeled naïve Thy1.1+ CD8+ OT-I T cells were adoptively transferred into naïve Thy1.2+/+ C57BL/6 mice. IV CD8+ Treg cells were pretreated with anti-Tim-3 Ab (or control anti-PD-L1 Ab) before transfer. The recipients were then SC infected with Ad-Ova and the Ig LNs were analyzed at day 3. A notable reduction in size of the draining Ig LN was observed in mice that had received IV CD8+ Treg cells, and this was reversed with anti-Tim-3 Ab treatment (Fig. 6A). Within the draining Ig LN, IV CD8+ Treg cells suppressed OT-I T cell division. Pretreating IV CD8+ Treg cells with anti-Tim-3 Ab before adoptive transfer restored OT-I T cell proliferation kinetics in vivo comparable to animals that had received OT-I T cells alone (Fig. 6B,C). We also confirmed that both IV CD8+ Treg cells and OT-I T cells were trafficking to the T-cell zone within the Ig LN by day 2 (Fig. 6D). Collectively, these data imply that IV CD8+ Treg cells suppress CD8+ Teff cell proliferation by surface Tim-3 both in vitro and in vivo.
Fig 6.
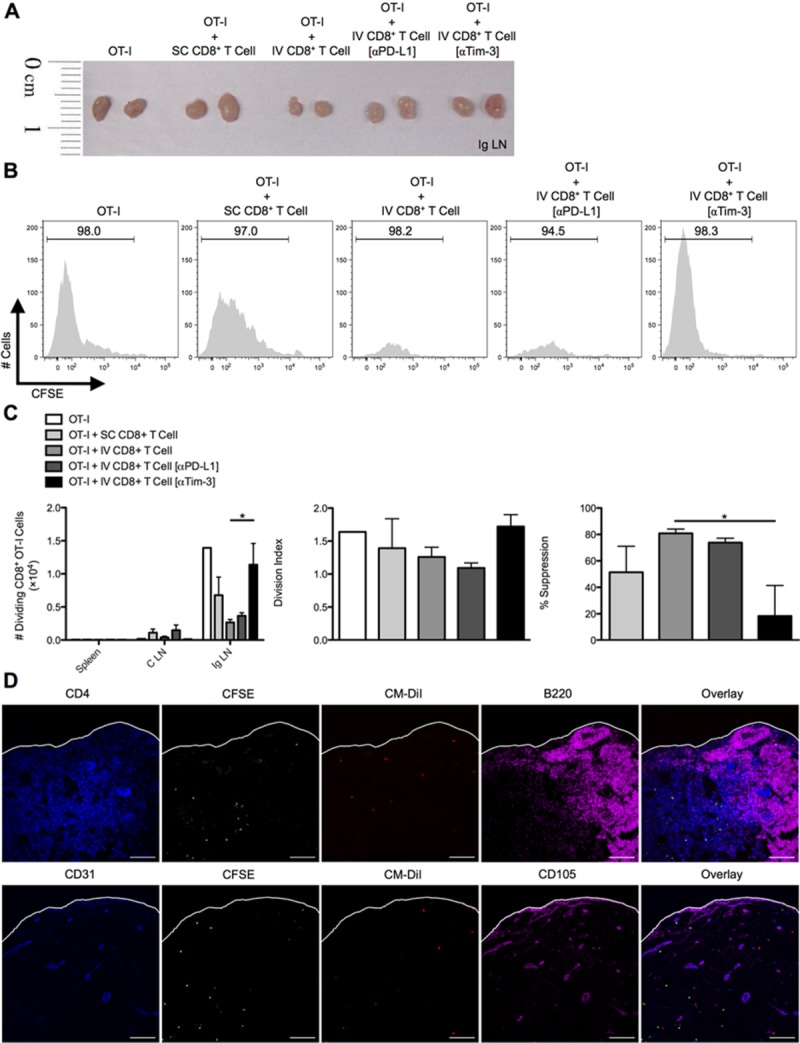
In vivo CD8+ Treg cell suppression of SC-primed OT-I cells entering draining lymph nodes is regulated by Tim-3. C57BL/6 mice were SC or IV infected with 2.5 × 107 IU of Ad-Ova, and bulk CD8+ T cells were isolated from day 7 SC spleens and IV livers. D7 CD8+ Treg cells from IV-infected livers were left alone or precoated with anti-PD-L1 Ab or anti-Tim-3 Ab. CFSE-labeled naïve Thy1.1+CD8+ OT-I T cells were adoptively transferred alone or in combination with CD8+ T cells from infected spleens and livers at a 1:2 ratio, respectively, into naïve C57BL/6 mice. Shortly thereafter, these recipient mice were SC infected. At day 3 postinfection, spleens, C LN, and (A and B) Ig LN were harvested, and (C) the number of dividing OT-I T cells was determined in each organ. The division index and percent suppression of OT-I T cells in the Ig LN is displayed (one-way ANOVA/Tukey's posttest; n = 3-6 per group). (D) CM-DiI-labeled CD8+ Treg cells from day 7 IV-infected liver (red) and CFSE-labeled naïve Thy1.1+CD8+ OT-I T cells (green) were transferred in a similar experiment, and Ig LNs were harvested at day 2 SC postinfection. PLP-fixed/OCT-frozen Ig LN cross-sections were stained with anti-CD4 (blue, upper panel), anti-CD31 (blue, lower panel), anti-B220 (magenta, upper panel), and CD105 (magenta, lower panel; n = 3 per group). Scale bar, 100 μm. Numbers in histograms represent percentages (mean ± standard error of the mean). *P < 0.05.
Tim-3/HMGB-1 Binding and Internalization Mediates CD8+ Treg Cell Suppression Independently of Gal-9
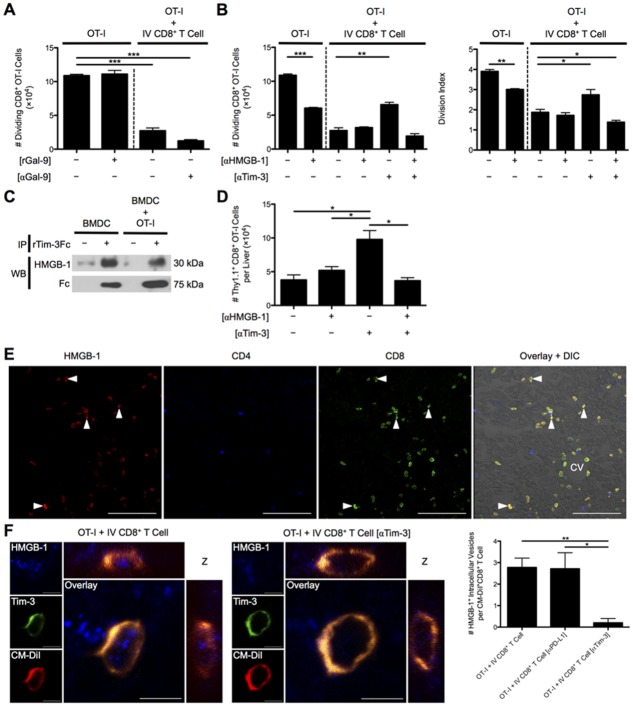
We next examined the mechanism for Tim-3-dependent suppression employed by liver-primed CD8+ Treg cells. Gal-9 appeared to be uninvolved because neither rGal-9 impeded OT-I cell division nor had the addition of anti-Gal-9 Ab to media of OT-I/IV CD8+ Treg cell cocultures reversed suppression (Fig. 7A). Tim-3 acted intrinsically on the IV CD8+ Treg cell highlighted by anti-Tim-3 Ab precoating of IV CD8+ Treg cells and lack of Tim-3 receptor expression on responder OT-I T cells (Supporting Fig. 5). HMGB-1 has been recently reported to bind Tim-3 within the early endosomes of DCs.17 These data raised the possibility that Tim-3 displayed by liver-primed CD8+ Treg cells may bind HMGB-1 to limit T-cell activation and subsequent proliferation. To test this possibility, we examined the effect of HMGB-1 blockade on OT-I T cell proliferation in culture with Ova257-264 pulsed BMDCs with or without CD8+ Treg cells. Consistent with published results, blockade of HMGB-1 diminished OT-I T cell proliferation in cultures devoid of CD8+ Treg cells. However, in OT-I/IV CD8+ T cell cocultures, the expected decrease in OT-I T cell proliferation was observed, which was unaffected by HMGB-1 blockade. Thus, CD8+ Treg cells might suppress independently of HMGB-1 or control the concentration of HMGB-1 through a Tim-3-dependent mechanism. When we examined the contribution of Tim-3 displayed by CD8+ Treg cells on OT-I proliferation by precoating CD8+ Treg cells with anti-Tim-3 Ab, Tim-3 blockade partially reversed CD8+ Treg cell-mediated suppression as before. Moreover, simultaneous blockade with anti-HMGB-1 and anti-Tim-3 Abs in coculture restored the suppressor function of CD8+ Treg cells (Fig. 7B). Tim-3 was indeed capable of binding HMGB-1, because rTim-3Fc chimeric protein coimmunoprecipitated with HMGB-1 present in supernatants collected from BMDC and BMDC/OT-I cultures (Fig. 7C).
Fig 7.
Tim-3 binding to HMGB-1 controls CD8+ Treg cell suppression independently of Gal-9. (A) The number of dividing OT-I T cells at D3 was determined when rGal-9 was included in media of CFSE-labeled naïve Thy1.1+CD8+ OT-I T cells cultured with SIINFEKL-pulsed BMDCs and when these cells were cocultured with day 7 Ad-Ova liver-primed CD8+ Treg cells in the presence of anti-Gal-9 Ab (n = 3 per group). (B) D7 Ad-Ova-primed CD8+ Treg cells were isolated from livers, left alone, or precoated with anti-Tim-3 Ab and cocultured with CFSE-labeled naïve Thy1.1+CD8+ OT-I T cells at a 1:1 ratio and SIINKFEKL-pulsed BMDCs. Select wells also contained anti-HMGB-1 Ab in media during culture. Day 3 CFSE dilution and number of dividing OT-I T cells and OT-I T cell division index were evaluated (one-way ANOVA/Tukey's posttest; n = 6 per group). (C) Supernatants from media obtained from BMDCs maturing in the presence of rIL-4 and rGM-CSF and from day 1 BMDC coculture with OT-I T cells were incubated with rTim-3Fc chimeric protein for 1 hour, followed by immunoprecipitation with protein A/G and immunoblotting analysis with anti-HMGB-1 Ab or anti-human Fc (n = 3 per group). (D) Naïve Thy1.1+CD8+ OT-I T cells (5 × 105) were transferred into day 7 Ad-Ova-infected mice, which were left untreated or received 300 μg of anti-HMGB-1 Ab or anti-Tim-3 Ab IP at days 5 and 6 before a day 7 MCMV-Ova infection. The number of Thy1.1+CD8+ OT-I T cells was assessed in livers of infected animals at day 14 (one-way ANOVA/Tukey's posttest; n = 3-6 per group). (E) PLP-fixed/OCT-frozen liver cross-sections from day 7 livers were stained with anti-HMGB-1 (red), anti-CD4 (blue), and anti-CD8 (green). Central vein (CV) is indicated (n = 6 per group). Scale bar, 100 μm. (F) Three-dimensional orthogonal view representative of PLP-fixed/OCT-frozen Ig LN cross-sections from C57BL/6 mice that received CM-DiI-labeled CD8+ Treg cells from day 7 infected liver (red) and CFSE-labeled naïve Thy1.1+CD8+ OT-I T cells stained with anti-HMGB-1 (blue) and anti-Tim-3 (green) is shown. Scale bar, 5 μm. Quantification of intracellular HMGB-1+ perinuclear vesicles within uncoated, anti-PD-L1 Ab precoated, and anti-Tim-3 Ab precoated CD8+ Treg cells was assessed per cell in images (one-way ANOVA/Tukey's posttest; n = 3 per group). Numbers in histograms represent percentages (mean ± standard error of the mean). *P < 0.05; **P < 0.01; ***P < 0.001.
During the in vivo virus-induced hepatitis model, where Thy1.1+CD8+ OT-I T cells were adoptively transferred into day 7 Ad-Ova-infected mice before MCMV-Ova infection, IP delivery of anti-Tim-3 Ab at days 5 and 6 increased the absolute number of Thy1.1+CD8+ OT-I T cells in livers of mice previously infected with Ad-Ova as before. Consistent with in vitro results, animals that received both anti-HMGB-1 and anti-Tim-3 Abs IP failed to boost an OT-I T cell response to MCMV-Ova within the liver (Fig. 7D). These results suggest that blockade of HMGB-1 abrogates the enhancement in proliferation after Tim-3 blockade.
A histological analysis of day 7 Ad-Ova-infected livers revealed that CD8+ T cells were exclusively sequestering HMGB-1 in vivo on their surface and cytoplasm (Fig. 7E). Given that DCs are a potent source of HMGB-1 within the lymph node during T-cell activation, CD8+ Treg cells may also have sequestered HMGB-1 once transferred and trafficked to the Ig LN of SC-infected mice. Indeed, IV CD8+ Treg cells bound HMGB-1 within intracellular vesicles, which colocalized with CD8+ Treg cell Tim-3. Anti-Tim-3 Ab pre-coated IV CD8+ Treg cells were clearly defective in internalizing HMGB-1 (Fig. 7F). In human liver biopsy specimens from chronic HCV and HCC patients, HMGB-1 was also observed to bind the surface of a subpopulation of intrahepatic CD8+ T cells (Fig. 8). These results describe a novel mechanism where Tim-3 on the surface of IV CD8+ Treg cells binds HMGB-1 hampering CD8+ Teff cell proliferation, which have a potential for translation to human liver biology.
Fig 8.
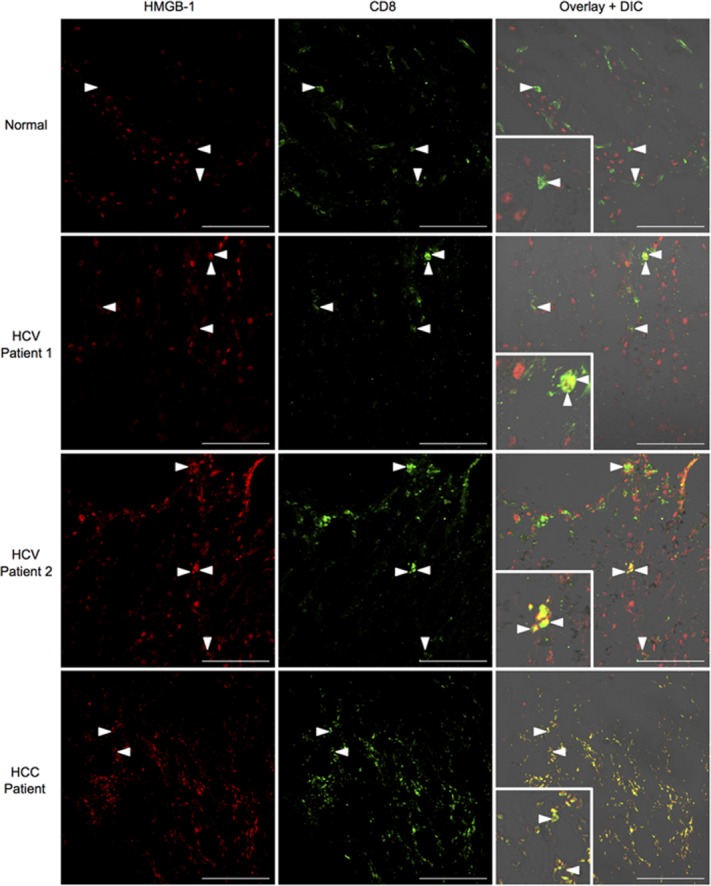
HMGB-1 binds to CD8+ T cells in livers of chronic HCV and HCC patients. Acetone-fixed liver cross-sections of human biopsies from normal donors, chronic HCV patients, and HCC patients were stained with anti-HMGB-1 (red) and anti-CD8 (green). Inlays depict areas of HMGB-1/CD8 colocalization in HCC/HCV patients (normal, n = 3 per group; HCV, n = 5 per group; HCC, n = 2 per group). Representative images are displayed. Scale bar, 100 μm.
Discussion
In this report, we demonstrate that hepatic viral infection results in the generation and expansion of a novel population of CD8+ Treg cells. Regulatory activity was independent of PD-L1 expression and IL-10 production by CD8+ Treg cells, but was dependent on Tim-3 expression. CD8+ Treg cell Tim-3 bound HMGB-1, preventing CD8+ Teff cell expansion. Given the role of HMGB-1 as a critical stimulus for expansion of CD8+ Teff cells after TCR-engagement, our results suggest a novel mechanism where intrahepatic CD8+ Treg cells limit CD8+ Teff cell expansion by Tim-3-mediated sequestration of HMGB-1.
Previous studies have shown that liver-infiltrating CD25+FoxP3+CD4+ Treg cells dampen immune responses to hepatic viral infection.21 FoxP3+CD4+ Treg cells are found in the naïve liver, but do not significantly expand after Ad infection (data not shown). In chronic HCV infection, Tim-3 is up-regulated on CD4+ iTreg cells.22 As a result, Tim-3 binding of HMGB-1 could play a similar role on CD25+FoxP3+CD4+ Treg cells. More research is needed to determine whether CD4+ and CD8+ Treg cells use Tim-3 to continuously sequester HMGB-1 released by necrotic hepatocytes and activated APCs during chronic HCV infection. This mechanism could contribute to the transition from acute to chronic infection and/or allow virus to persist during later stages of chronic disease.
The suppressive function of liver-primed CD8+ Treg cells was exerted in an antigen-specific manner, that is recognition of peptide/major histocompatibility complex (MHC) I complex on APCs by both CD8+ Treg and Teff cells was necessary and sufficient for Tim-3-mediated suppression. Rangachari et al. established that Tim-3 signaling is linked to TCR engagement by the Src-family kinase, Lck, through interaction with human leukocyte antigen/B-associated transcript 3 (Bat3). Gal-9 binding to Tim-3 prevented Bat-3 linkage to active Lck, thus modulating downstream TCR signaling.23 Our results support the notion that suppression of CD8+ Teff cells is dependent on both CD8+ Treg cell TCR recognition of antigen and Tim-3 binding to HMGB-1.
Although blockade of Tim-3 improved OT-I T cell proliferation in an HMGB-1-dependent manner, enhancement in IL-2 and IFN-γ production was only detected in culture supernatant and was not evident on a per-cell basis (data not shown). The elevated proinflammatory cytokine production likely reflected enhanced proliferation of responder OT-I cells. To date, HMGB-1 has been shown to improve proliferation of both CD4+ and CD8+ T cells, but enhanced levels of cytokines have only been associated with CD4+ Th1 cell responses.20 It is possible that HMGB-1 only affects proliferation of CD8+ Teff cells. Conversely, the TCR is one of the most complex signaling molecules in nature, having 10 immunoreceptor tyrosine-based activation motifs (ITAMs), whereas most receptors utilize two. Guy et al. revealed that different multiplicities of ITAM and TCR signal strength uncouples proliferation and cytokine production. This effect was dependent on TCR affinity for peptide, where superagonists and strong peptides induced proliferation whereas weak peptides sufficiently led to maximal IL-2 secretion.24 Ova was used as the model antigen in these experiments, where OT-I T cells receive a strong TCR signal. Because strength of signal dictates proliferation or cytokine production, we may have masked a role for Tim-3/HMGB-1 in regulating CD8+ Teff cell cytokine production. In the future, it is worth employing another Tg system where the responder T cells recognize a weaker peptide/MHC I complex. Alternatively, BMDCs could be pulsed with mutated Ova-derived peptides providing a suboptimal TCR signal.
Gal-9 and HMGB-1 are both present throughout the course of viral infection of the liver. During antiviral immune responses, including chronic HCV infection, Gal-9 is expressed by a wide variety of cells in response to elevated IFN-γ.25 Apart from activated APCs releasing HMGB-1, it can be passively released by virally lysed, cytolytically killed, hypoxic, and oxidatively stressed hepatocytes.26 Because the binding sites for Gal-9 and HMGB-1 are distally located on Tim-3 mucin and immunoglobulin variable regions, respectively, we cannot rule out the possibility of simultaneous recognition of both ligands. In this context, the concentrations of Gal-9 and HMGB-1, along with their different avidities for Tim-3 during viral infection and cancer, could balance the adaptive immune response in multiple ways. Previous research is limited because the RMT3-23 clone of anti-Tim-3 Ab is effective at blocking both Gal-9 and HMGB-1 binding to Tim-3.17,27
Approximately 600 million people worldwide are in danger of developing chronic liver disease resulting from HBV/HCV, and only a subset of these patients respond to IFN-α/ribavirin therapy.3 Simultaneous blockade of the IL-10, PD-1/PD-L1, and Tim-3 inhibitory pathways has potential clinical efficacy. However, elevated serum HMGB-1 during HCV infection directly correlates with liver disease progression.28 Therefore, HMGB-1 sequestration by Treg cell Tim-3 may not be sufficient to suppress Teff cells under conditions of extensive liver injury and HMGB-1 release. Herein, we report that HMGB-1 staining colocalizes to the surface of CD8+ T cell subpopulations found in chronic HCV and HCC liver biopsies; therefore, a role for HMGB-1 in advanced human diseases remains to be elucidated. Thus, disease progression, signaling pathway integration, and ligand concentration, accessibility, and avidity for receptors must all be considered in the design of immunotherapeutic strategies.
In summary, these data describe a novel mechanism where Tim-3 binds HMGB-1 on virus-specific CD8+ Treg cells suppressing proliferation of CD8+ Teff cells during acute Ad infection. This observation provides a framework for future studies of Tim-3, where consideration for ligand access of both Gal-9 and HMGB-1 is warranted. Translation to human liver disease is expected, given that a similar population of IL-10-producing, PD-1/PD-L1+Tim-3+CD8+ T cells exists in liver biopsies of chronic HCV patients.
Acknowledgments
After pilot experiments established a CD8+ Treg cell phenotype, Jie Sun at Indiana University kindly aided in optimizing CD8+ T cell coculture experiments. The authors also thank Taeg Kim and Hai-Chon Lee of the University of Virginia for their contributions in experimental design. Sun-Sang Sung and the University of Virginia Advanced Microscopy Facility assisted in confocal imaging. Michael Solga and the University of Virginia Flow Cytometry Core Facility contributed to performing and outlining FACS experiments.
Glossary
- Ab
antibody
- Ad
adenovirus
- ANOVA
analysis of variance
- APC
antigen presenting cell
- Bat3
B-associated transcript 3
- β-Gal
beta-galactosidase
- BMDC
bone marrow-derived dendritic cell
- CFSE
carboxyfluorescein succinimidyl ester
- C LN
celiac lymph node
- CTLA-4
cytotoxic T-lymphocyte antigen 4
- DC
dendritic cell
- eGFP
enhanced green fluorescent protein
- ELISA
enzyme-linked immunosorbent assay
- FACS
fluorescence-activated cell sorting
- FMO
fluorescence minus one
- FoxP3
forkhead box protein 3
- Gal-9
galectin-9
- GITR
glucocorticoid-induced TNF receptor-related protein
- GrB
granzyme B
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HMGB-1
high-mobility group box 1
- HRP
horseradish peroxidase
- IFN-γ
interferon-gamma
- IgG
immunoglobulin G
- Ig LN
inguinal lymph node
- IL
interleukin
- IP
intraperitoneal
- ITAM
immunoreceptor tyrosine-based activation motif
- IV
intravenous
- MCMV
mouse cytomegalovirus
- MHC
major histocompatibility complex
- MNCs
mononuclear cells
- mRNA
messenger RNA
- NK
natural killer
- OCT
optimal cutting temperature
- Ova
ovalbumin
- PAMP
pathogen associated molecular pattern
- PD-1
programmed death 1
- PD-L1
PD-1 ligand
- PLP
periodate-lysine-paraformaldehyde fixative
- PMA
phorbol myristate acetate
- qPCR
quantitative polymerase chain reaction
- r
recombinant
- SC
subcutaneous
- TCR
T-cell receptor
- Teff
T effector
- Tg
transgenic
- TGF-β,; Tim-3
T-cell immunoglobulin and mucin 3
- TNF-α
tumor necrosis factor alpha
- Treg
T regulatory
Supporting Information
Additional Supporting Information may be found in the online version of this article.
References
- 1.Crispe IN. The liver as a lymphoid organ. Annu Rev Immunol. 2009;27:147–163. doi: 10.1146/annurev.immunol.021908.132629. [DOI] [PubMed] [Google Scholar]
- 2.Lukens JR, Dolina JS, Kim TS, Tacke RS, Hahn YS. Liver is able to activate naïve CD8+ T cells with dysfunctional anti-viral activity in the murine system. PLoS One. 2009;4:e7619. doi: 10.1371/journal.pone.0007619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callendret B, Walker C. A siege of hepatitis: immune boost for viral hepatitis. Nat Med. 2011;17:252–253. doi: 10.1038/nm0311-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tiegs G, Lohse AW. Immune tolerance: what is unique about the liver. J Autoimmun. 2010;34:1–6. doi: 10.1016/j.jaut.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 5.Mills KHG. Regulatory T cells: friend or foe in immunity to infection? Nat Rev Immunol. 2004;4:841–855. doi: 10.1038/nri1485. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi T, Tagami T, Yamazaki S, Uede T, Shimizu J, Sakaguchi N, et al. Immunologic self-tolerance maintained by CD25(+)CD4(+) regulatory T cells constitutively expressing cytotoxic T lymphocyte-associated antigen 4. J Exp Med. 2000;192:303–310. doi: 10.1084/jem.192.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strauss L, Bergmann C, Whiteside TL. Human circulating CD4+CD 25highFoxp3+ regulatory T cells kill autologous CD8+ but not CD4+ responder cells by Fas-mediated apoptosis. J Immunol. 2009;182:1469–1480. doi: 10.4049/jimmunol.182.3.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim H-J, Wang X, Radfar S, Sproule TJ, Roopenian DC, Cantor H. CD8+ T regulatory cells express the Ly49 class I MHC receptor and are defective in autoimmune prone B6-Yaa mice. Proc Natl Acad Sci U S A. 2011;108:2010–2015. doi: 10.1073/pnas.1018974108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salti SM, Hammelev EM, Grewal JL, Reddy ST, Zemple SJ, Grossman WJ, et al. Granzyme B regulates antiviral CD8+ T cell responses. J Immunol. 2011;187:6301–6309. doi: 10.4049/jimmunol.1100891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitazawa Y, Fujino M, Wang Q, Kimura H, Azuma M, Kubo M, et al. Involvement of the programmed death-1/programmed death-1 ligand pathway in CD4+CD25+ regulatory T-cell activity to suppress alloimmune responses. Transplantation. 2007;83:774–782. doi: 10.1097/01.tp.0000256293.90270.e8. [DOI] [PubMed] [Google Scholar]
- 11.Blackburn SD, Wherry EJ. IL-10, T cell exhaustion and viral persistence. Trends Microbiol. 2007;15:143–146. doi: 10.1016/j.tim.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Jin H-T, Anderson AC, Tan WG, West EE, Ha S-J, Araki K, et al. Cooperation of Tim-3 and PD-1 in CD8 T-cell exhaustion during chronic viral infection. Proc Natl Acad Sci U S A. 2010;107:14733–14738. doi: 10.1073/pnas.1009731107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Couper KN, Blount DG, Riley EM. IL-10: the master regulator of immunity to infection. J Immunol. 2008;180:5771–5777. doi: 10.4049/jimmunol.180.9.5771. [DOI] [PubMed] [Google Scholar]
- 14.Fife BT, Pauken KE, Eagar TN, Obu T, Wu J, Tang Q, et al. Interactions between PD-1 and PD-L1 promote tolerance by blocking the TCR-induced stop signal. Nat Immunol. 2009;10:1185–1192. doi: 10.1038/ni.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golden-Mason L, Palmer BE, Kassam N, Townshend-Bulson L, Livingston S, McMahon BJ, et al. Negative immune regulator Tim-3 is overexpressed on T cells in hepatitis C virus infection and its blockade rescues dysfunctional CD4+ and CD8+ T cells. J Virol. 2009;83:9122–9130. doi: 10.1128/JVI.00639-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao E, Zang X, Ramagopal UA, Mukhopadhaya A, Fedorov A, Fedorov E, et al. T cell immunoglobulin mucin-3 crystal structure reveals a galectin-9-independent ligand-binding surface. Immunity. 2007;26:311–321. doi: 10.1016/j.immuni.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 17.Chiba S, Baghdadi M, Akiba H, Yoshiyama H, Kinoshita I, Dosaka-Akita H, et al. Tumor-infiltrating DCs suppress nucleic acid–mediated innate immune responses through interactions between the receptor TIM-3 and the alarmin HMGB1. Nat Immunol. 2012;13:832–842. doi: 10.1038/ni.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5:331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 19.Dumitriu IE, Baruah P, Valentinis B, Voll RE, Herrmann M, Nawroth PP, et al. Release of high mobility group box 1 by dendritic cells controls T cell activation via the receptor for advanced glycation end products. J Immunol. 2005;174:7506–7515. doi: 10.4049/jimmunol.174.12.7506. [DOI] [PubMed] [Google Scholar]
- 20.Sundberg E, Fasth AER, Palmblad K, Harris HE, Andersson U. High mobility group box chromosomal protein 1 acts as a proliferation signal for activated T lymphocytes. Immunobiology. 2009;214:303–309. doi: 10.1016/j.imbio.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Xu D, Fu J, Jin L, Zhang H, Zhou C, Zou Z, et al. Circulating and liver resident CD4+CD25+ regulatory T cells actively influence the antiviral immune response and disease progression in patients with hepatitis B. J Immunol. 2006;177:739–747. doi: 10.4049/jimmunol.177.1.739. [DOI] [PubMed] [Google Scholar]
- 22.Moorman JP, Wang JM, Zhang Y, Ji XJ, Ma CJ, Wu XY, et al. Tim-3 pathway controls regulatory and effector T cell balance during hepatitis C virus infection. J Immunol. 2012;189:755–766. doi: 10.4049/jimmunol.1200162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rangachari M, Zhu C, Sakuishi K, Xiao S, Karman J, Chen A, et al. Bat3 promotes T cell responses and autoimmunity by repressing Tim-3–mediated cell death and exhaustion. Nat Med. 2012;18:1394–1400. doi: 10.1038/nm.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guy CS, Vignali KM, Temirov J, Bettini ML, Overacre AE, Smeltzer M, et al. Distinct TCR signaling pathways drive proliferation and cytokine production in T cells. Nat Immunol. 2013;14:262–270. doi: 10.1038/ni.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez-Manzanet R, DeKruyff R, Kuchroo VK, Umetsu DT. The costimulatory role of TIM molecules. Immunol Rev. 2009;229:259–270. doi: 10.1111/j.1600-065X.2009.00772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klune J, Dhupar R. HMGB1: endogenous danger signaling. Mol Med. 2008;14:1. doi: 10.2119/2008-00034.Klune. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanzaki M, Wada J, Sugiyama K, Nakatsuka A, Teshigawara S, Murakami K, et al. Galectin-9 and T cell immunoglobulin mucin-3 pathway is a therapeutic target for type 1 diabetes. Endocrinology. 2012;153:612–620. doi: 10.1210/en.2011-1579. [DOI] [PubMed] [Google Scholar]
- 28.Jung JH, Park JH, Jee MH, Keum SJ, Cho MS, Yoon SK, et al. Hepatitis C virus infection is blocked by HMGB1 released from virus-infected cells. J Virol. 2011;85:9359–9368. doi: 10.1128/JVI.00682-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.