Abstract
Protein sumoylation is a dynamic posttranslational modification involved in diverse biological processes during cellular homeostasis and development. Recently sumoylation has been shown to play a critical role in cancer, although to date there are few small molecule probes available to inhibit enzymes involved in the SUMO conjugation process. As part of a program to identify and study inhibitors of sumoylation we recently reported the discovery that 2’,3’,4’-trihydroxy flavone (2-D08) is a cell permeable, mechanistically unique inhibitor of protein sumoylation. The work reported herein describes an efficient synthesis of 2-D08 as well as a structurally related but inactive isomer. We also report an unanticipated Wessely-Moser rearrangement that occurs under vigorous methyl ether deprotection conditions. This rearrangement likely gave rise to 2-D08 during a deprotection step, resulting in 2-D08 appearing as a contaminant in a screening well from a commercial supplier.
Keywords: SUMO, synthesis, inhibitor, flavone
The posttranslational modification of protein substrates with the Small Ubiquitin-like Modifier (SUMO) has emerged as an important regulatory mechanism and a critical pathway in embryonic development and cancer.1 SUMO modification can result in a variety of consequences that vary broadly depending on the substrate. Documented effects of SUMO modification include altered subcellular localization,2 transcriptional regulation,3 and enzymatic activity.4 Protein sumoylation is also thought to be involved with the cellular stress response, playing important roles in recovery from heat shock,5 ischemia,6 and surviving the stress of tumorigenesis.7 Many of the known targets of sumoylation are transcription factors, and the modification of these targets is generally (but not always) seen as a repressive mark.1b In addition, a number of studies have demonstrated the role of sumoylation in cancer, as illustrated by the observation of high expression levels of the SUMO conjugating enzyme UBC9 in ovarian tumors.7 Additionally, high levels of SUMO E2 and E3 enzymes are correlated with decreased survival rates for multiple myeloma patients.8 While the genetic knockout mouse for UBC9 is embryonically lethal, a recent synthetic lethal screen identified sumoylation enzymes as required for the progression of Myc-driven cancers.9 New roles for sumoylation in cancer and developmental biology are still emerging.
A small molecule inhibitor of protein sumoylation would have significant value in studying of the role of sumoylation in basic and cancer biology, and would help define the therapeutic potential of sumoylation enzymes.10 However, to date little progress has been made in this regard.11 As part of a program to identify inhibitors of sumoylation, our laboratory recently reported the development of a novel medium throughput microfluidic electrophoretic mobility shift assay to monitor substrate sumoylation in vitro.12 This assay utilizes recombinant E1 (Aos1/Uba2) and E2 (UBC9) enzymes to effect sumoylation of a fluorescently labeled consensus sequence-containing peptide. Additionally, we discovered a synthetic oxygenated flavonoid that we named “2-D08”. This compound was found to block sumoylation of topoisomerase I in two different cancer cell lines dosed with camptothecin. Furthermore, our analysis indicated that 2-D08 inhibited sumoylation by preventing transfer of SUMO from the UBC9-SUMO thioester to the substrate, a mechanism of action that was unprecedented. Although 2 has been described in several papers,13 we were unable to find a synthetic procedure or commercial vendor for this compound in pure form.
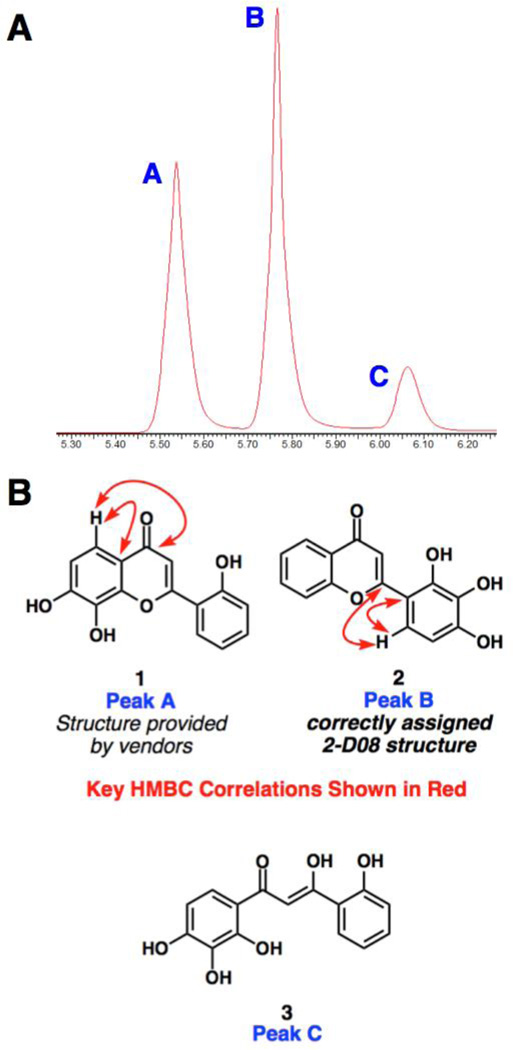
We identified 2 as a contaminant from a well in a commercially-supplied screening collection. Upon completion of our pilot screen, a well believed to contain compound 1 (Figure 1) was identified as active. We subsequently purchased samples of this compound from two other vendors, and found all three samples to be equally and reproducibly active in the microfluidic biochemical assay. LC/MS analysis using a short gradient indicated the presence of one broad peak, with two ions appearing at m/z 271 (presumed to be [M + H]+) and m/z 289 (Presumed to be [M + H + H2O]+). However, upon inspection of the 1H NMR spectrum in several different solvents, it was apparent that multiple species were present. Through extensive HPLC analysis and purification, it was established that three distinct species were present within all three commercial samples.
Figure 1.
A: Partial HPLC chromatogram of a commercial sample of 2-D08 containing three components. B: Structures of components identified from commercial samples.
In order to accurately characterize the components of the mixture, purification by preparative HPLC was pursued (Figure 1). Each of the three peaks was collected and analyzed by NMR and LC/MS. Peak C was identified as β–diketone 3 (present as a mixture of tautomers in multiple solvents). However, peaks A and B, were not easily assignable by inspection of one-dimensional 1H or 13C spectra. HMBC experiments enabled an assignment of peak A as flavone 1, the vendor-provided structure. Peak B, however, appeared to be a different flavonoid that was assigned the structure 2, on the basis of HMBC experiments. Each of the three commercial samples contained roughly the same ratio of these three components (A:B:C = 1.9:2.5:1).
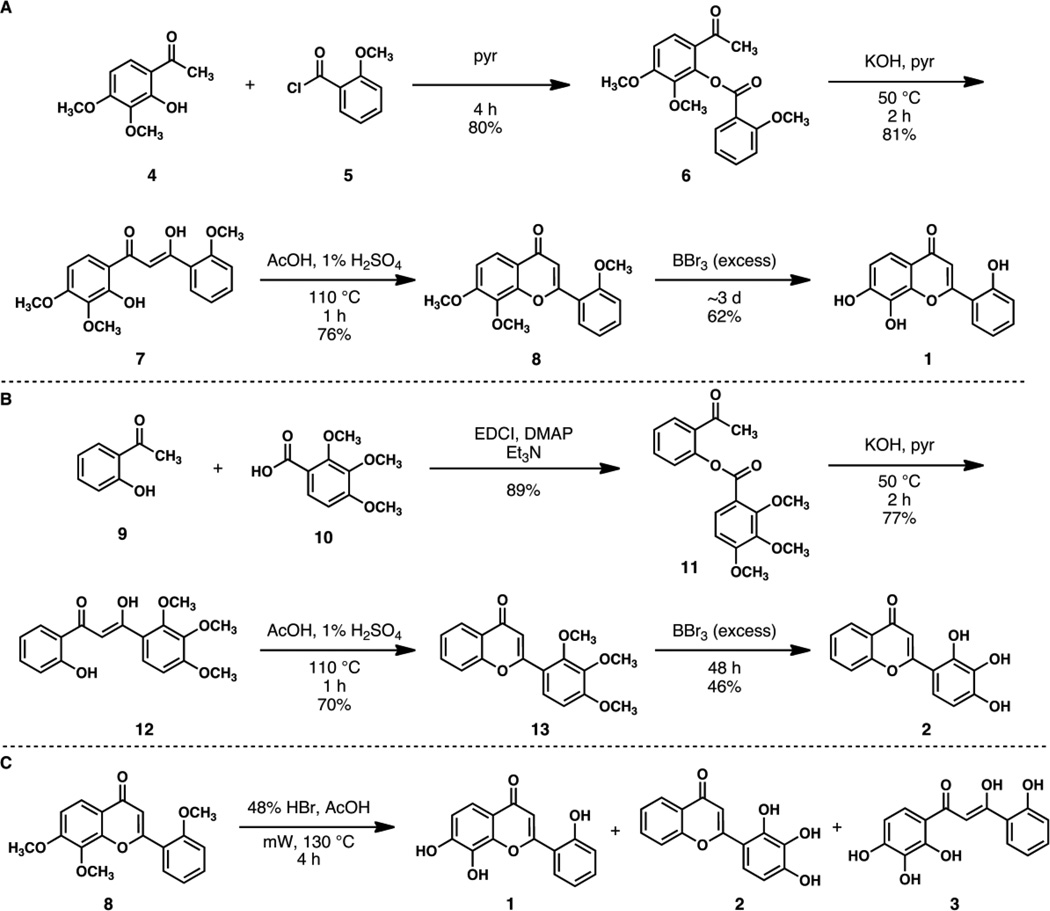
An independent synthetic route to both compounds 1 and 2 was developed so as to unambiguously confirm both structures (Figure 2).14 The preparation of 1 began with exposure of 3,4-dimethoxygalleacetophenone dimethyl ether 4 to 2-methoxybenzoyl chloride 5 in pyridine to afford ester 6 in 80% yield. Treatment of 6 with powdered KOH in pyridine under mild heating provided 7 in 81% yield. Flavone ring formation was accomplished by brief exposure to 1% H2SO4 in acetic acid. Finally, treatment with excess BBr3 for three days at room temperature afforded 1 in 62% yield (shorter reaction times afforded only partially deprotected products). In parallel, a similar route was employed to access 2 with comparable reaction conditions and yields (Figure 2B).
Figure 2.
A: Synthesis of compound 1. B: Synthesis of 2-D08 (compound 2). C: Wessely-Moser-type isomerization observed during deprotection of 8 with aqueous HBr. Abbreviations: pyr = pyridine, EDCI = 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide, DMAP = 4-dimethylaminopyridine.
We found that a sample of synthetic 1 and had identical 1H and 13C NMR spectra in comparison to material isolated from the commercial mixture (peak A). Similarly, synthetic 2 and peak B had identical 1H and 13C NMR spectra. These analyses unambiguously confirmed the structures of the two flavones. In an effort to understand the potential origin of the mixture of compounds, we exposed trimethyl ether 8 to more vigorous deprotection conditions (Figure 2C). Upon treatment with 48% aqueous HBr and acetic acid and heating to 130 °C in a microwave reactor, a mixture of products was observed containing 1, 2, and 3. The yields of this reaction were highly variable and appear to depend on a number of factors. This latter finding provides speculative evidence that the mixture of compounds identified from three commercial suppliers arose from a vigorous deprotection (e.g. exposure to hot HBr) during synthetic preparation by the vendors. The mechanism of this isomerization is complex, however 3 presumably arises by an acid-promoted ring opening reaction of 1, which may then undergo an acid-promoted ring closure to form either 1 or 2 (Figure S-5). This unanticipated reaction is a variant of the Wessely-Moser rearrangement.15
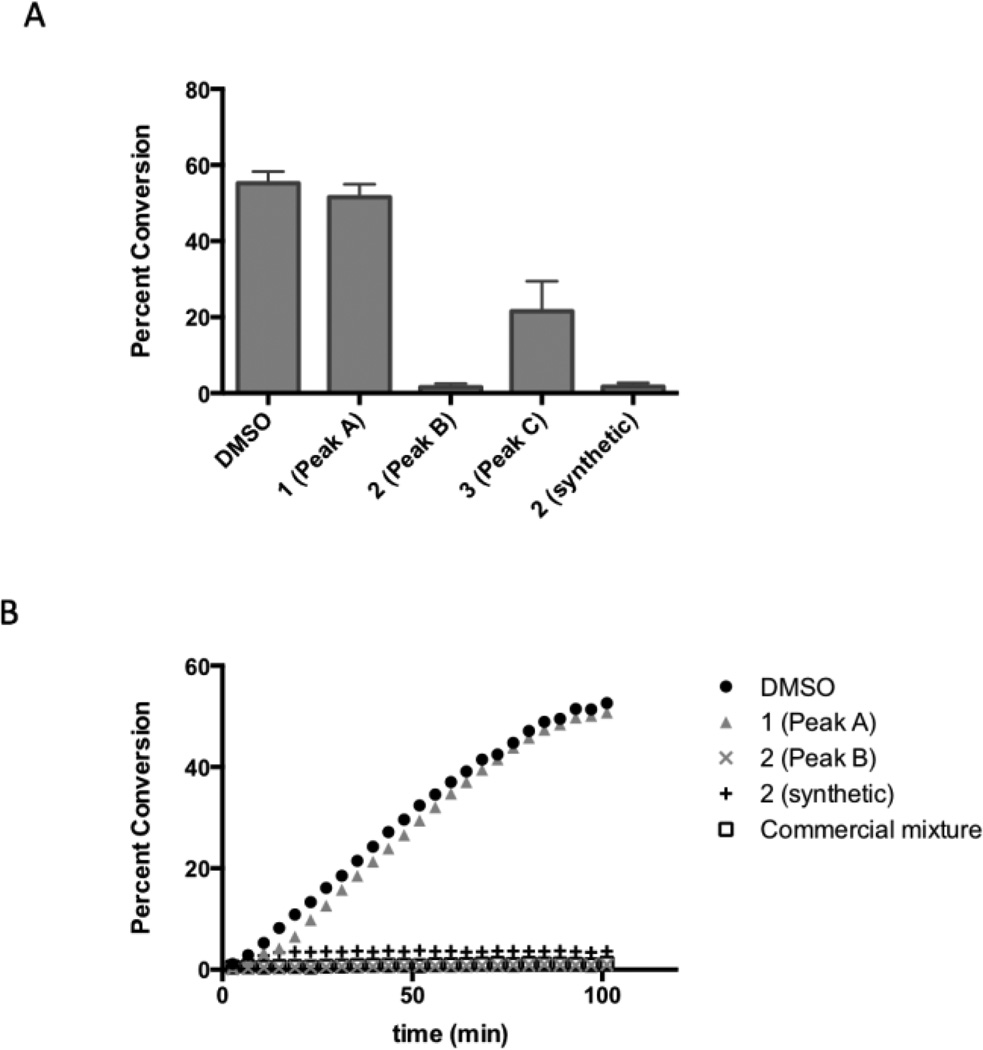
Each of the three compounds isolated from the mixture was evaluated in a biochemical sumoylation assay. In this assay, a microfluidic electrophoretic mobility shift protocol was used to monitor the conjugation of SUMO-1 to a fluorescent peptide substrate.12 Compounds were evaluated in the assay at a concentration of 30 µM, and inhibition relative to a DMSO control was measured at ~30% conversion after quenching with EDTA (Figure 3A). Only compound 2 showed complete inhibition at this concentration, with synthetic and purified commercial samples having roughly equal inhibitory potency. Compound 3 was comparatively weak, showing modest inhibitory activity at this concentration. Furthermore, we evaluated compounds 1 and 2 (synthetic and purified commercial) in a kinetic assay (Figure 3B). Again, only samples of compound 2 (both synthetic and purified commercial) showed substantial inhibitory activity, while compound 1 was inactive. Although compound 2 was the major product in all three commercial mixtures, neither its structure nor the structure of contaminant 3 was reported by any of the vendors.
Figure 3.
A: Inhibitory activity of selected compounds in an endpoint biochemical assay. Compounds were incubated with SUMO1, E1, E2 enzymes, a fluorescent substrate, and ATP in an appropriate buffer and quenched with EDTA after 90 min. Conversion was quantified by ratiometric peak height in a microfluidic electrophoretic mobility shift assay using a Perkin Elmer EZ Reader II. Values represent the mean of three replicates. B: Evaluation of selected compounds in kinetic biochemical sumoylation assays. Compounds were incubated with SUMO1, E1, E2 enzymes, a fluorescent substrate, and ATP in an appropriate buffer and monitored over the course of 110 minutes. Conversion was quantified by ratiometric peak height in a microfluidic electrophoretic mobility shift assay using a Perkin Elmer EZ Reader II.
In conclusion, herein we report an efficient synthetic route to 2-D08, an inhibitor of protein sumoylation. Furthermore, the structural identification of the active component from a mixture of three compounds (provided by multiple separate commercial vendors) is described. Historically, the purity of commercial screening libraries has been problematic16 and the there have been many instances of unanticipated structures being identified from screening collections.17 In this particular case, we were able to show that an impurity in a screening collection likely arose from a vigorous methyl ether deprotection protocol, resulting in an unanticipated ring opening/ring closing Wessely-Moser-type rearrangement. This rearrangement ultimately gave rise to 2 (an isomer of the desired product 1), and 2 was identified as the active component (2-D08). The Wessely-Moser reaction was also shown to occur on synthetic material such as 8 upon heating with aqueous HBr to provide several products in variable yield. We found that more mild deprotection conditions with BBr3 effectively accomplished deprotection and completely suppressed isomerization, albeit over a longer reaction time. Flavone 2 and the biologically inactive isomer 1 are distinguishable, but not easily assignable, by one-dimensional 1H and 13C NMR experiments. However, HMBC analysis clearly establishes the two structures, both of which were also confirmed by synthesis. Once structurally confirmed, the flavonoids were evaluated in a biochemical assay. Compound 2 inhibited sumoylation in both kinetic and endpoint assays, while 1 was completely inactive under identical conditions. Another contaminant, 3, showed only modest activity. The combination of structural, synthetic, and biochemical studies allowed us to assign the structure 2 as 2-D08. Finally, the synthetic procedure reported herein will be useful to provide quantities of pure 2-D08 for further studies, and efforts to evaluate 2-D08 in a variety of other biological contexts are ongoing.
Supplementary Material
Acknowledgment
This work was supported by the Intramural Research Program of the NIH, Center for Cancer Research, and the National Cancer Institute, National Institutes of Health. Vectors encoding for SAE 1/2 and UBC9 were kindly provided by Dr. Christopher Lima, Memorial Sloan Kettering Cancer Institute. We thank Drs. Christopher Lai and James Kelley for obtaining high-resolution mass spectra for this work. We thank Drs. Martin Schnermann and Terrence Burke for helpful comments during the preparation of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.(a) Ulrich HD. Methods Mol. Biol. 2009;497:3. doi: 10.1007/978-1-59745-566-4_1. [DOI] [PubMed] [Google Scholar]; (b) Geiss-Friedlander R, Melchior F. Nat Rev Mol Cell Bio. 2007;8:947. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]; (c) Gareau JR, Lima CD. Nat. Rev. Mol. Cell Biol. 2010;11:861. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Majumdar A, Petrescu AD, Xiong Y, Noy N. J. Biol. Chem. 2011;286:42749. doi: 10.1074/jbc.M111.293464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong Y, Xing X, Li S, Bi H, Yang C, Zhao F, Liu Y, Ao X, Chang AK, Wu H. Plos One. 2011;6:e23046. doi: 10.1371/journal.pone.0023046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Truong K, Lee TD, Chen Y. J. Biol. Chem. 2012;287:15154. doi: 10.1074/jbc.M112.353789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. Science Signalling. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- 6.Lee YJ, Mou Y, Maric D, Klimanis D, Auh S, Hallenbeck JM. Plos One. 2011;6:e25852. doi: 10.1371/journal.pone.0025852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mo YY, Yu YN, Theodosiou E, Ee PLR, Beck WT. Oncogene. 2005;24:2677. doi: 10.1038/sj.onc.1208210. [DOI] [PubMed] [Google Scholar]
- 8.Driscoll JJ, Pelluru D, Lefkimmiatis K, Fulciniti M, Prabhala RH, Greipp PR, Barlogie B, Tai YT, Anderson KC, Shaughnessy JD, Jr, Annunziata CM, Munshi NC. Blood. 2010;115:2827. doi: 10.1182/blood-2009-03-211045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, Rao M, Yu P, Dominguez-Vidana R, Liang AC, Solimini NL, Bernardi RJ, Yu B, Hsu T, Golding I, Luo J, Osborne CK, Creighton CJ, Hilsenbeck SG, Schiff R, Shaw CA, Elledge SJ, Westbrook TF. Science. 2012;335:348. doi: 10.1126/science.1212728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bedford L, Lowe J, Dick LR, Mayer RJ, Brownell JE. Nat. Rev. Drug Disc. 2011;10:29. doi: 10.1038/nrd3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Fukuda I, Ito A, Hirai G, Nishimura S, Kawasaki H, Saitoh H, Kimura K, Sodeoka M, Yoshida M. Chem Biol. 2009;16:133. doi: 10.1016/j.chembiol.2009.01.009. [DOI] [PubMed] [Google Scholar]; (b) Fukuda I, Ito A, Uramoto M, Saitoh H, Kawasaki H, Osada H, Yoshida M. J Antibiot (Tokyo) 2009;62:221. doi: 10.1038/ja.2009.10. [DOI] [PubMed] [Google Scholar]
- 12.Kim YS, Nagy K, Keyser S, Schneekloth JS. Chem. Biol. 2013;20:604. doi: 10.1016/j.chembiol.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Seyoum A, Asres K, El-Fiky FK. Phytochemistry. 2006;67:2058. doi: 10.1016/j.phytochem.2006.07.002. [DOI] [PubMed] [Google Scholar]; (b) Cotelle N, Bernier JL, Catteau JP, Pommery J, Wallet JC, Gaydou EM. Free Radical Biol. Med. 1996;20:35. doi: 10.1016/0891-5849(95)02014-4. [DOI] [PubMed] [Google Scholar]; (c) Cotelle N, Bernier JL, Henichart JP, Catteau JP, Gaydou E, Wallet JC. Free Radical Bio Med. 1992;13:211. doi: 10.1016/0891-5849(92)90017-b. [DOI] [PubMed] [Google Scholar]; (d) Laget M, DeMeo M, Wallet JC, Gaydou EM, Guiraud H, Dumenil G. Arch. Pharmacol. Res. 1995;18:415. [Google Scholar]
- 14.Liu X, Chan CB, Jang SW, Pradoldej S, Huang J, He K, Phun LH, France S, Xiao G, Jia Y, Luo HR, Ye K. J. Med. Chem. 2010;53:8274. doi: 10.1021/jm101206p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.(a) Larget R, Lockhart B, Renard P, Largeron M. Bioorg. Med. Chem. Lett. 2000;10:835. doi: 10.1016/s0960-894x(00)00110-4. [DOI] [PubMed] [Google Scholar]; (b) Shaw SC, Gupta AK, Kumar R. J. Indian Chem. Soc. 1991;68:615. [Google Scholar]; (c) Shaw SC, Azad R, Mandal SP, Gandhi RS. J. Indian Chem. Soc. 1988;65:107. [Google Scholar]; (d) Donnelly DMX, Green PB, Philbin EM, Smyth FTB, Wheeler TS. Chem. Ind. (london) 1958;28:892. [Google Scholar]
- 16.(a) Nat. Chem. Biol. 2009;5:127. doi: 10.1038/nchembio0309-127. [DOI] [PubMed] [Google Scholar]; (b) Thorne N, Auld DS, Inglese J. Curr. Opin. Chem. Biol. 2010;14:315. doi: 10.1016/j.cbpa.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Inglese J, Shamu CE, Guy RK. Nat. Chem. Biol. 2007;3:438. doi: 10.1038/nchembio0807-438. [DOI] [PubMed] [Google Scholar]
- 17.(a) Dounay AB, Anderson M, Bechle BM, Campbell BM, Claffey MM, Evdokimov A, Evrard E, Fonseca KR, Gan XM, Ghosh S, Hayward MM, Horner W, Kim JY, McAllister LA, Pandit J, Paradis V, Parikh VD, Reese MR, Rong S, Salafia MA, Schuyten K, Strick CA, Tuttle JB, Valentine J, Wang H, Zawadzke LE, Verhoest PR. ACS Med. Chem. Lett. 2012;3:187. doi: 10.1021/ml200204m. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kang MI, Henrich CJ, Bokesch HR, Gustafson KR, McMahon JB, Baker AR, Young MR, Colburn NH. Mol. Cancer Ther. 2009;8:571. doi: 10.1158/1535-7163.MCT-08-0811. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Peterson S, Wang L, Robertson K, Torchon G, Ouerfelli O, Gautier J. Nat. Chem. Biol. 2009;5:130. [Google Scholar]; (d) Dupre A, Boyer-Chatenet L, Sattler RM, Modi AP, Lee JH, Nicolette ML, Kopelovich L, Jasin M, Baer R, Paull TT, Gautier J. Nat. Chem. Biol. 2009;5:191. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Dupre A, Boyer-Chatenet L, Sattler RM, Modi AP, Lee JH, Nicolette ML, Kopelovich L, Jasin M, Baer R, Paull TT, Gautier J. Nat. Chem. Biol. 2008;4:119. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.