Abstract
Early disruption of FGF signaling alters left-right (LR) asymmetry throughout the embryo. Here we uncover a role for FGF signaling that specifically disrupts brain asymmetry, independent of normal lateral plate mesoderm (LPM) asymmetry. When FGF signaling is inhibited during mid-somitogenesis, asymmetrically expressed LPM markers southpaw and lefty2 are not affected. However, asymmetrically expressed brain markers lefty1 and cyclops become bilateral. We show that FGF signaling controls expression of six3b and six7, two transcription factors required for repression of asymmetric lefty1 in the brain. We found that Z0-1, atypical PKC (aPKC) and β-catenin protein distribution revealed a midline structure in the forebrain that is dependent on a balance of FGF signaling. Ectopic activation of FGF signaling leads to overexpression of six3b, loss of organized midline adherins junctions and bilateral loss of lefty1 expression. Reducing FGF signaling leads to a reduction in six3b and six7 expression, an increase in cell boundary formation in the brain midline, and bilateral expression of lefty1. Together, these results suggest a novel role for FGF signaling in the brain to control LR asymmetry, six transcription factor expression, and a midline barrier structure.
Keywords: brain asymmetry, FGF signaling, sine occulis
Introduction
Alterations of left-right (LR) asymmetries in the human brain are correlated with neurological disorders, including dyslexia, schizophrenia, depression and autism (reviewed in [1, 2]). The most widely studied example of asymmetric brain development occurs in the zebrafish epithalamus. Within the epithalamus lies the asymmetrically placed pineal organ, which participates in sleep-wake regulation [3–5], and parapineal organ, with an unknown function. The pineal complex innervates the habenular nuclei, which project into the interpeduncular nucleus (IPN) in the ventral midbrain [6].
Prior to the establishment of brain asymmetry, asymmetric fluid flow is generated by motile cilia within Kupffer’s vesicle (KV) at the caudal end of the embryo. Asymmetric KV fluid flow is required for the asymmetric initiation of southpaw (spaw; a nodal homolog) expression in the left lateral plate mesoderm (LPM) [7, 8]. spaw expression then progresses from posterior to anterior LPM in a wavelike fashion eventually turning on the expression of other TGFβ family members including lefty1, lefty2, and cyclops [9–12]. Brain expression of lefty1 and cyclops localizes to the left dorsal diencephalon, a division of the forebrain. Altered expression of lefty1 and cyclops randomizes the placement of the parapineal gland and randomizes the expression of asymmetric markers in the habenulae [4, 13].
The effect of Nodal on brain asymmetry is dependent on the developmental timing of Nodal activity. Loss of Nodal during late gastrulation, as seen in oep or cyc mutants, results in bilaterally symmetric signals which in turn leads to randomized orientation of asymmetric brain structures [4, 13–15]. In contrast, loss of Nodal during midsomitogenesis, as seen in spaw morpholino knockdown, leads to absent expression of lefty1 and cyclops in the dorsal diencephalon [12, 13]. Current models hypothesize that the wave of spaw expression through the LPM activates downstream genes in both the heart field and the brain [12, 16].
The asymmetric expression of lefty1 in the forebrain relies on a the concerted functions of at least two members of the sine occulis family [17]. Specifically, knockdown of both six3b and six7, two six3 homologs, leads to bilateral expression of lefty1 in the dorsal diencephalon. Overexpression of six3b represses lefty1 expression in the brain but only when expression was upregulated before somite formation at 10 hpf (tailbud stage) [17]. When spaw and six7 were knocked down in clutches in which half the embryos were homozygous mutant for six3b, lefty1 expression was present in a half of the expected embryos (24% compared to 50% expected); the remainder of embryos had an absence of lefty1 expression (including some six3b homozygous mutants with six7 morpholino), a phenotype seen in spaw morphants alone [12, 17]. Inbal et al interpreted these results to indicate that Nodal activity from the LPM is required to relieve the repression by Six3 genes on lefty1 expression [17]. An alternative interpretation could be that Six3 activity is independent of spaw.
Wnt signaling also plays a role in the establishment of normal brain asymmetry. Activation of the Wnt pathway, either through the masterblind mutant (mutation in axin1 leading to activation of the Wnt pathway) or treatment of embryos with LiCl (chemical activation of Wnt signaling), converts the normally left-sided expression of lefty1 and pitx2c in the brain to bilateral expression. Activation of the canonical Wnt pathway during gastrulation alters asymmetric brain markers in a spaw independent manner, so that the patterns of asymmetrically expressed genes in the LPM are unaffected. However, later treatment with LiCl, during midsomitogenesis, causes asymmetric markers in the brain and in the LPM to be expressed bilaterally. Embryos injected with spaw MO and subsequently treated with LiCl, to activate Wnt signaling during midsomitogenesis, showed no expression (i.e. bilateral absence) of pitx2c or lefty1 [16]. This suggests that activation of brain Nodal during somitogenesis by Wnt signaling is dependent upon Nodal activity from the LPM [16].
Fibroblast growth factor 8 (FGF8) has been previously implicated in asymmetric positioning of the parapineal gland and asymmetric gene expression in the habenular nuclei, and cell fate decisions prior to asymmetric cell migration [18, 19]. Using an FGF8 null mutant, it was shown that loss of FGF8 reduced parapineal cell number, and cell fate analysis showed a corresponding increase in cone photoreceptor cells, which are also encompassed in the pineal organ [19, 20]. Epistasis experiments uncovered a cooperative role between Tbx2b and FGF8a, with Tbx2b specifying cells as pineal complex precursors and downstream FGF8a activity promoting differentiation to form parapineal cells [19]. After cell specification, FGF signaling is required for parapineal cell migration [18]. Down-regulation of FGF8 protein in hypomorphic fgf8 mutant embryos (acerebellar; ace) does not lead to early loss of asymmetrically expressed markers including lefty1, however migration of the parapineal gland fails to occur. Implantation of FGF8-soaked beads, regardless of implantation side, rescues this migration defect [18]. FGF8 presumably works through FGFR4, which is expressed in the parapineal cells [18]. Thus, FGF is required for the establishment of parapineal cell identity and also functions as a chemotactic signal for normal parapineal cell migration, after the establishment of LR gene expression in the brain.
Here we describe a role of FGF signaling that is distinct from the previously described roles, affecting earlier steps in brain asymmetry. We found that FGF signaling controls asymmetric gene expression in the brain by regulating expression of sine occulis homologs six3b and six7. Importantly, altered FGF signaling can perturb brain asymmetry in the context of normal LPM asymmetry. We also explore the possibility of a brain midline structure controlling brain asymmetry. To this end, we characterize a midline structure in the forebrain, marked by ZO-1, atypical PKC (aPKC) and β-catenin. We find that a fine-tuned level of FGF signaling is necessary for formation of this forebrain midline structure, suggesting that FGF signaling serves as a rheostat to control forebrain midline organization and LR asymmetric patterning.
Materials and Methods
Zebrafish stocks and embryo culture
Wild type zebrafish embryos were obtained from natural matings and cultured as previously described [21]. Tg(hsp70l:XlFgfr1, cryaa:DsRed)pd3 was a kind gift from Ken Poss [22].
Heat Shock
Hsp70:ca-FGFR embryos were obtained by mating Tg(hsp70l:XlFgfr1, cryaa:DsRed)pd3 transgenic fish to WT fish [22]. Embryos were incubated at 28°C until the desired developmental stage when they were placed in a 37°C water bath in two milliliter tubes filled with embryo media for 20 minutes for Hsp70:ca-FGFR embryos. To minimize embryo death, tubes were inverted two times during heat shock. Upon completion of heat shock, embryos were returned to culture dishes and incubated at 28°C until fixation for ISH or immunohistochemistry (IHC).
Genotyping embryos
To identify presence of the Hsp70:caFGFR transgene in embryos after ISH or IHC, embryos were placed one to a tube and rinsed in 1X PBST (PBS+0.1% Tween-20) with one change of buffer over 2 hours. Embryos were incubated overnight at 55°C in DNA extraction buffer containing 20mM Tris pH 8.0, 50 mM KCl, 1.5 mM MgCl2 0.3% Tween-20, 0.3% NP-40 and 15μg/ml of ProteinaseK. After overnight incubation ProteinaseK was deactivated by 10 minutes at 95°C. A PCR reaction was set up to identify those embryos carrying the transgene using primers targeted against the DsRed marker in the construct and had the following sequence: ds-red1 5′-CATCCTGTCCCCCCAGTTCC- 3′ and ds-red2 5′-CCCAGCCCATAGTCTTCTTCTGC-3′ [23]. The following PCR conditions were used: 94°C for 5 minutes; 35 cycles of 94°C for 30 seconds, 65°C for 1 minute, 72°C for 30 seconds; 72°C for 5 minutes. Samples were then run on a 1% agarose gel and assessed for the presence of the transgene.
Embryo Injections
For gene knock-down, 1 nl of gene specific anti-sense morpholino (MO; GeneTools) was injected into one-cell to four-cell stages as previously described [24]. The following MO and injection amounts were used 8 ng spaw MO (5′-GCACGCTATGACTGGCTGCATTGCG-3′) [10, 12].
Pharmacological treatments
A reversible inhibitor of FGF signaling, SU5402 (Calbiochem and Tocris Biosciences), was applied to live zebrafish embryos still in their chorions during developmental stages of interest at a concentration of 40–60 μM (concentration dependent on experiment and drug lot) suspended in DMSO. As controls, sibling embryos were treated with the same concentration of DMSO without SU5402. To end FGF inhibition embryos were washed three times in embryo water and allowed to develop until fixed for in situ hybridization (ISH) or IHC.
In Situ Hybridization
ISH was performed as previously described [21] with digoxigenin RNA probes made using a Roche DIG RNA labeling kit. Templates include: spaw [12], erm [25], lefty1 [9], cyclops [26], lefty2 [9], sprouty2 [27], sprouty4 [28], six7 [29], and six3b [30], flh [31].
Immunohistochemistry
Embryos were fixed overnight in 4% PFA at 4°C and dehydrated stepwise into methanol for storage at −20°C. After stepwise re-hydration, embryos were blocked for 1 hour in PBS containing 5% sheep serum (Sigma), 1% bovine serum albumin (BSA), 1% DMSO and 0.1% Triton-X. Embryos were incubated with rabbit anti- β-catenin (1:100, Sigma C2206) or rabbit anti-atypical protein kinase C (1:100, Santa Cruz sc-216) and mouse anti-ZO1 (1:100 Zymed 33-9100) antibodies in blocking solution overnight. After washing embryos in 1% BSA, 1% DMSO, and 0.1% triton-X in PBS, embryos were incubated with fluorescent secondary antibodies (1:100, Molecular Probes) overnight. Embryos were then washed and mounted in glycerol. Embryos were dissected to show head region and were imaged using an Olympus Fluoview FV300 or FV1000 scanning laser confocal microscope with a 60X objective with images processed using ImageJ and Photoshop.
Results
FGF control of brain asymmetry is independent of LPM asymmetry
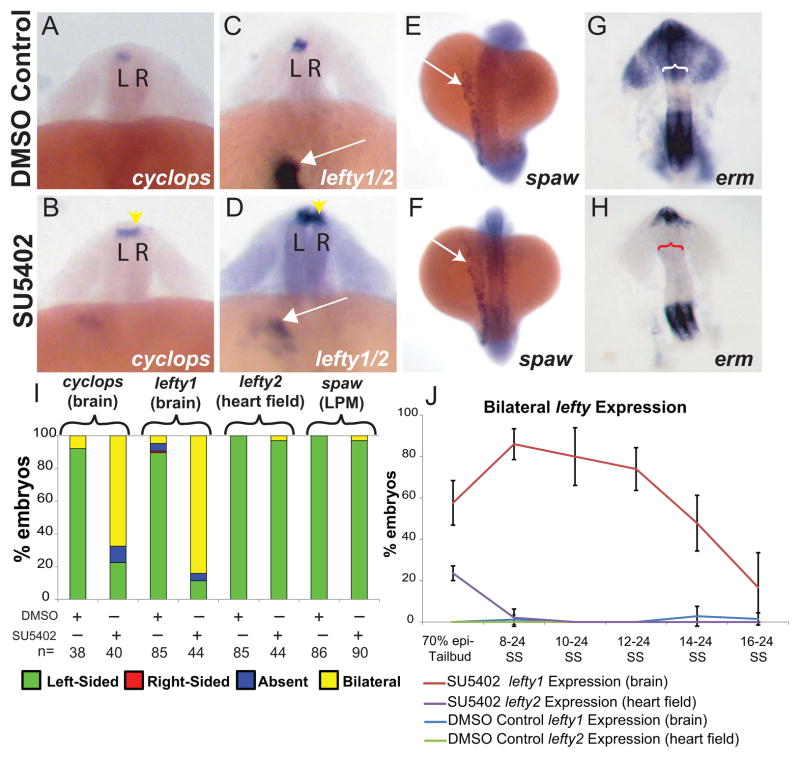
Previously we and others have shown that FGF signaling is required during early development for KV ciliogenesis. Disruption of cilia function alters downstream LR development, including asymmetric gene expression in LPM, cardiac and brain asymmetry [7, 21, 32, 33]. Here we ask whether FGF signaling is also important for brain asymmetry independent of its role in KV ciliogenesis. Using SU5402, an FGFR inhibitor [34], we found that inhibition of FGF signaling after KV formation alters brain asymmetry (Fig. 1). cyclops, a nodal homolog [35], is asymmetrically expressed in the left LPM and later in the left dorsal diencephalon in control embryos, providing an early marker of brain LR asymmetry (Fig. 1A) [9, 35, 36]. When FGF signaling was inhibited after the 8-somite stage (SS), the normal left-sided expression of cyclops was converted to bilateral expression in the brain (Fig. 1A–B, I). Similarly, lefty1 was also bilaterally expressed when FGF signaling was inhibited (Fig. 1C–D, I). Interestingly, inhibition of FGF signaling during the same developmental periods did not alter left-sided lefty2 expression in the heart field (Fig. 1C–D, I), nor did it perturb spaw expression in the lateral plate mesoderm (Fig. 1E–F, I), suggesting the effects were specific to brain asymmetry. DMSO control embryos had normal left-sided expression of cyclops, lefty1, and lefty2 in the brain and spaw in the LPM (Fig. 1).
Figure 1. FGF dependent control of brain asymmetry is uncoupled from earlier LR patterning.
(A–D) Dorsal view of 22–24 somite embryos. L=left, R=right. (A–B) Cyclops is normally expressed in left diencephalon (A; DMSO controls) however when FGF signaling is inhibited, expression becomes bilateral (B; yellow arrowhead. (C–D) Normal left-sided expression of lefty2 within the prospective heart field in the LPM in both DMSO control (C) and SU5402 treated (D) embryos (white arrows). Note that SU5402 was suspended in DMSO in all experiments. Yellow arrowhead indicates bilateral expression of lefty1 in SU5402 treated embryos (D) compared with normal left-sided expression in DMSO treated embryos (C). (E, F) Dorsal view of 18–20 somite embryos showing spaw expression in the LPM. Normal left-sided expression of spaw in the left LPM of SU5402 (F) compared to the DMSO control embryo (E). (G, H) Images of the head dissected from 20 somite embryos showing erm expression (anterior is up). Erm expression is down-regulated in SU5402 treated embryos (H, n=28) in the dorsal diencephalon (red bracket) in comparison to a DMSO control embryo (G, white bracket, n=32). (I) Histogram showing the percentages of embryos displaying normal (left-sided), reversed (right-sided), absent and bilateral expression patterns of cyclops (brain), lefty1 (brain), lefty2 (heart field), spaw (LPM) in SU5402-treated embryos and DMSO-treated embryos. (J) Line graph showing a timeline of SU5402 and DMSO treatments affecting lefty expression in the brain (See Supplemental Figure 1 for all expression classes). 70% epiboly-Tailbud: DMSO n= 29, SU5402=42. 8–24 SS: DMSO n=53, SU5402 n=60. 10–24 SS: DMSO n=49, SU5402 n=57. 12–24 SS: DMSO n=45, SU5402 n=28. 14–24 SS: DMSO n=53, SU5402 n=48. 16–24 SS: DMSO n=64, SU5402 n=37.
To determine whether SU5402 treatment was effectively down-regulating FGF signaling, we examined markers for known downstream targets of FGF signaling. In embryos treated with SU5402 at 8 SS until fixation (at 24 SS) there was a global knockdown of FGF signaling downstream targets including erm (Fig. 1G –H, S1A–B), sprouty2, and sprouty4 (Fig. S1C–F). Despite prolonged treatment with FGFR inhibitor, some expression of FGF markers persisted. Although treatment with a higher concentration of drug further reduces expression of FGF response genes (data not shown), we selected a drug concentration that consistently knocked down FGF signaling in the dorsal diencephalon (Fig. 1G–H), and that has been shown to not affect PDGF signaling [34].
To determine when FGF signaling is needed during somitogenesis for control of asymmetric lefty1, we conducted developmental stage-specific SU5402 treatments. SU5402 treatments prior to KV formation (70% epiboly-Tailbud) altered both brain and LPM asymmetry (Fig. 1J, S1G–H). However, treatments initiated at 8 SS and continuing until embryos were collected for in situ hybridization (at 24 SS) affected brain asymmetry independently of LPM asymmetry, as seen by predominantly bilateral lefty1 in the brain and normal left-sided lefty2 in heart field (Fig. 1J, S1G–H). This effect persisted in treatments that were initiated at 10, 12 or 14 SS, but diminished when treatments were initiated at 16 SS. This suggests endogenous FGF signaling is necessary for control of asymmetric lefty1 specifically in brain structures until the 14–16 SS (Fig. 1J, S1G–H).
Hyper-activation of FGF signaling down-regulates lefty1 in the brain
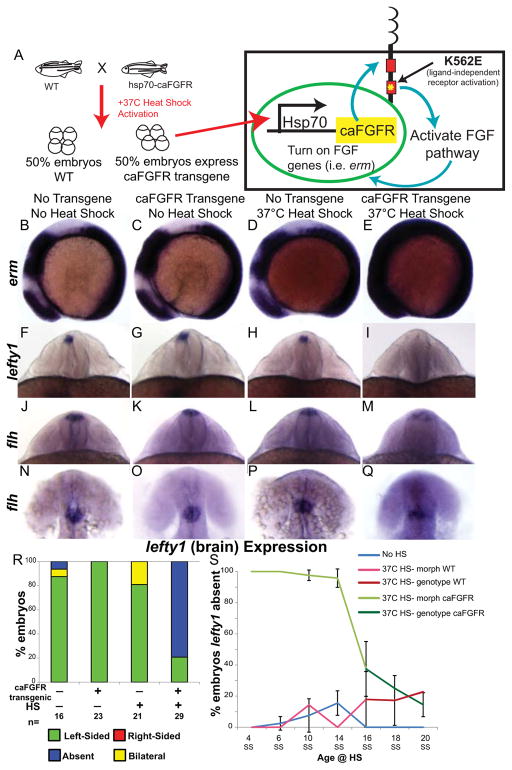
As shown above, loss of FGF signaling between the 8 and 16 SS leads to bilateral lefty1 expression in the brain. Conversely, to determine whether increased activation of FGF signaling leads to down-regulation of lefty1, we used a transgenic zebrafish line in which expression of a constitutively active FGFR (ca-FGFR) is regulated by a heat-shock promoter (hsp70:ca-FGFR; Fig. 2A) [22]. Heat shocking (HS) hsp70:ca-FGFR embryos allowed us to temporarily increase FGF signaling in a time-dependent manner. To test efficacy of FGF pathway up-regulation, embryos were collected one hour after the completion of the HS and erm expression was analyzed by whole mount in situ hybridization. Individual embryos were genotyped for the presence of the caFGFR transgene by PCR after in situ hybridization analysis. Compared to sibling embryos that did not undergo HS, HS’d caFGFR transgenic embryos had strong overexpression of erm throughout the embryo (Fig. 2B–C, E). As a control, non-transgenic siblings were also subjected to HS and found to have no up-regulation of erm expression compared to the non-HS siblings (Fig. 2B–D).
Figure 2. FGF pathway controls brain asymmetry.
(A) Diagram of heat-shock (HS) activation of the FGF pathway by expression of a constitutively active FGFR1 (caFGFR) protein with a K562E point mutation to activate downstream targets of FGF signaling. (B–E) Lateral view of erm expression in 8 somite embryos. Normal expression of erm in a non-HS’d non-transgenic embryo (B; WT expression 100%, n=24). No up-regulation of erm expression in either a caFGFR transgenic non-HS’d embryo (C; WT expression 100%, n=19) or a non-transgenic HS’d embryo (D; WT expression 93.3%, n=15). However, a dramatic increase in erm expression in a HS’d transgenic embryo (E; Increased expression 73.17%, n=41). (F–I) Dorsoposterior view of 22–24 somite embryos expressing lefty1 (brain). Normal left-sided expression of lefty1 in non-transgenic non-HS’d (F), transgenic non-HS’d (G), and non-transgenic HS’d (H) control embryos. Contrary to normal expression in control embryos lefty1 expression is absent in the brain of transgenic HS’d embryos (I). (J–M) Dorsoposterior and (N–Q) dorsal views of distinct 22–24 somite embryos expressing flh. Normal expression of flh in the dorsal diencephalons of non-transgenic non-HS’d (J, N; n=19 WT expression), transgenic non-HS’d (K, O; n=10 WT expression), and non-transgenic HS’d control embryos (L, P; n= 17 WT expression). Expression of flh is detected in the dorsal brain of caFGFR transgenic HS’d embryos (M, Q; n=23 present but dispersed expression), but appears to be more dispersed. (R) Histogram quantifying the expression of lefty1 in the brain of genotyped embryos. (S) Line graph illustrating the timeline over which FGF signaling controls lefty1 expression in the brain. Embryos that were HS’d caFGFR before 14SS can be clearly identified by changes in morphology, as confirmed by genotyping (panel 2R). Therefore, embryos HS from 4 SS to 14 SS were classed as follows: HS Morph WT (HS non-transgenic siblings), HS Morph caFGFR (HS transgenic siblings). Embryos HS from 16 SS to 20 SS cannot be distinguished by morphological phenotype, so they were individually genotyped, and were classed as follows: HS non-transgenic siblings (HS genotype WT), HS transgenic siblings (HS genotype caFGFR). 4 SS: No HS n=50, 37C HS- morph WT n=27, 37C HS- morph caFGFR n=30; 6 SS: No HS n=91, 37C HS- morph WT n=42, 37C HS- morph caFGFR n=35; 10 SS: No HS n=40, 37C HS- morph WT n=28, 37C HS- morph caFGFR n=29; 14 SS: No HS n=39, 37C HS- morph WT n=34, 37C HS- morph caFGFR n=26; 16 SS: No HS n=51, 37C HS- genotype WT n=27, 37C HS- genotype caFGFR n=30; 18 SS: No HS n=33, 37C HS- genotype WT n=24, 37C HS- genotype caFGFR n=28; 20 SS: No HS n=42, 37C HS- genotype WT n=23, 37C HS- genotype caFGFR n=16.
To mimic the timing of SU5402 treatments above, HS was initiated at 6 SS to allow time for up-regulation of the FGF signaling pathway. We included three classes of controls: non-transgenic sibling with HS, non-transgenic siblings without HS, and caFGFR transgenic siblings without HS. All of these controls displayed normal lefty1 expression (Fig. 2F–H, R, S3). In contrast, HS activation of the FGF pathway in caFGFR transgenic embryos resulted in an absence of lefty1 expression in the brain (Fig. 2I, R, S3).
We next asked whether the absence of lefty1 expression in HS’d caFGFR transgenic embryos was due to an absence of dorsal diencephalon cells. floating head (flh; a homeodomain transcription factor) is a reliable a marker for diencephalon cells [31]. flh was expressed in all four classes of embryos: with or without the transgene and with or without HS (Fig. 2J–Q, S2). Three distinct views of flh expression (dorsoposterior, Fig. 2M; dorsal, Fig. 2Q; lateral, Fig. S2D) show that dorsal diencephalon cells were present, albeit more dispersed, in embryos in which caFGFR has been activated (Fig. 2M, Q). Together these results suggest that hyper-activation of FGF signaling does not prevent the specification of flh-expressing dorsal diencephalon cells, and more specifically inhibits lefty1 expression in the brain.
To determine the developmental time period during which FGF signaling could control lefty1 expression, caFGFR transgenic and sibling non-transgenic embryos were HS’d at seven different stages between 4SS and 20SS. Transgenic embryos that were HS’d at 4 SS had a complete absence of lefty1 in the brain (Fig. 2S, S3). However, hyper-activation of FGF signaling at 4SS to 6SS leads to morphological defects such as shortened body axis, yolk extension defects, and midbrain-hindbrain defects (Fig. S2). Additionally, early HS leads to bilateral lefty2 expression in the heart field (Fig. S3). Thus, early hyperactivation of FGF signaling affects lefty1 expression in the brain, but it also alters gross morphology and other aspects of LR pattering. Therefore it is not known whether altered FGF signaling at these early stages has a direct role or indirect role in aberrant brain patterning. In contrast, later activation of FGF signaling resulted in absence of lefty1 in the brain but maintained a normal body axis, yolk extension and midbrain hindbrain morphology (Fig. 2S, S3; data not shown). Susceptibility of lefty1 to suppression by hyper-activated FGF signaling persisted through activation at 14 SS (Fig. 2S, S3), but at the same time having little effect on lefty2 expression in the LPM (Fig. S3). Later activation of FGF signaling at 18 and 20 SS had less effect on lefty1 expression, consistent with the results seen from the FGFR inhibitor experiments (Fig. 1J).
FGF signaling inhibits brain lefty1 independently of spaw/nodal
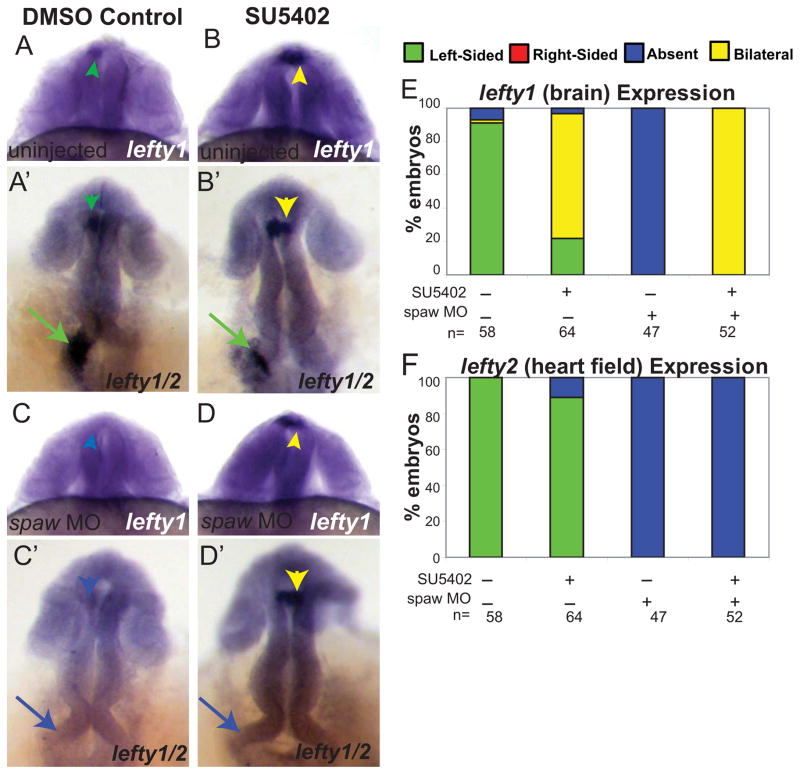
LR development in the zebrafish is dependent on asymmetric fluid flow in Kupffer’s vesicle, which activates asymmetric gene expression in the left LPM [7, 8]. The first of these asymmetrically expressed genes is spaw, a Nodal family member hypothesized to turn on lefty1 expression in the brain, and lefty2 expression in the heart field [12]. In the absence of spaw expression neither lefty1 nor lefty2 are expressed [12]. We wanted to determine whether the bilateral expression of lefty1 that is induced in the brain when FGF signaling is inhibited requires spaw function. To do so, we injected spaw MO into 1–4 cell embryos and inhibited FGF signaling in these embryos by SU5402 treatment from 8–24 SS, and then examined expression of lefty1. As reported above, uninjected embryos treated with SU5402 had normal left-sided lefty2 in the heart field and abnormal bilateral expression of lefty1 in the brain and (Fig. 3B, B′, E–F), in contrast to normal left-sided expression in uninjected DMSO control embryos (Fig. 3A, A′, E–F). In embryos injected with spaw MO and treated with DMSO as a control, both lefty1 in the brain and lefty2 in the heart field were absent (Fig. 3C, C′, E–F). However, embryos injected with spaw MO and then treated with SU5402 showed bilateral lefty1 expression in the brain but absent lefty2 expression in the LPM (Fig. 3D–F). Therefore, we conclude that FGF signaling is required for suppression of lefty1 expression in the brain independently of spaw expression in the LPM.
Figure 3. FGF signaling regulates lefty1 expression in the brain independently of spaw activity.
(A–D) Dorsal views of 22–24 somite embryos showing lefty1 in the brain and (A′-D′) different focal plans showing lefty1 in the brain and lefty2 in heart field within the same embryo. Uninjected embryos showing normal left-sided expression of lefty2 in the heart field (green arrows) of both DMSO control embryos and SU5402 treated embryos (A, A′, B, B′). In contrast, bilateral lefty1 expression (yellow arrowhead) in the brain of SU5402 treated embryos in comparison to the normal left-sided expression (green arrowhead) of lefty1 in the DMSO control embryos (A, A′, B, B′). spaw MO injected embryos treated with either DMSO or SU5402 show absence of lefty2 expression in the heart field (blue arrows; C, C′, D, D′). However, SU5402 treated embryos still exhibit bilateral expression of lefty1 in the brain whereas DMSO control embryos no longer express lefty1 (blue arrowhead; C, C′, D, D′). (E) Histogram indicating percentages of embryos displaying normal (left-sided), reversed (right sided), absent and bilateral heart expression patterns of lefty1 for uninjected DMSO control embryos, uninjected SU5402 treated embryos, spaw MO injected DMSO embryos, and spaw MO injected embryos treated with SU5402. (F) Histogram indicating percentages of embryos displaying normal (left-sided), reversed (right-sided), absent and bilateral brain expression patterns of lefty2 for WT DMSO control embryos, WT SU5402 treated embryos, spaw MO injected DMSO embryos, and spaw MO injected embryos treated with SU5402.
FGF signaling controls six gene expression
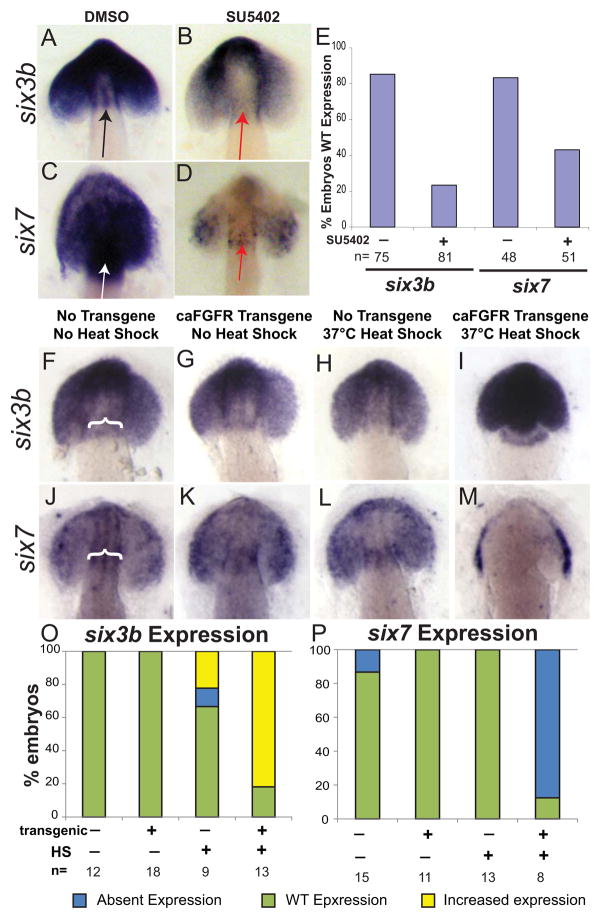
Two symmetrically expressed transcription factors, six3b and six7 [29, 30], both homologs of sine occulis, are required for brain asymmetry. Similar to inhibition of FGF signaling, double knockdown of both six3b and six7 causes bilateral lefty1 expression in the brain, even in the absence of spaw [17]. To determine whether these pathways intersect to control brain asymmetry, expression patterns of six3b and six7 were examined in FGF-signaling inhibited embryos (Fig. 4A–E). Expression of both six3b and six7 were decreased in the dorsal diencephalon of SU5402-treated embryos from 8–12 SS as compared to DMSO-treated control embryos (Fig. 4A–E).
Figure 4. FGF signaling controls brain asymmetry through regulation of six3 transcription factor expression.
(A, B) Dorsal view of a 12–14 somite embryo with head dissected away from yolk, showing expression of six3b. six3b is expressed broadly in the eye fields and brain, including the dorsal diencephalon (black arrow) of the developing embryo in DMSO control (A; n=75), but is diminished in SU5402 embryos (B; WT=81), including in the diencephalon (red arrow). (C, D) Dorsal view of a 12–14 somite embryo with head dissected away from yolk, showing expression of six7. six7 is expressed broadly in the eye fields and brain, including the dorsal diencephalon (black arrow), of the developing embryo in DMSO control (C; n=48), but is diminished in SU5402 embryos (D; n=51), including in the diencephalon (red arrow). (E) Histogram quantifying the % of embryos expressing WT levels of both six3b and six7 in DMSO Control and SU5402 treated embryos. (F–I) Dorsal view of a 12–14 somite control or caFGFR transgenic embryos, with head dissected away from yolk, showing expression of six3b Embryos were HS activated at 4–6 somites to have maximal activation of FGF signaling at 8 SS to parallel the SU5402 studies. Six3b expression is upregulated in caFGFR transgenic HS’d embryos (I) compared to non-transgenic non HS’d (F; white bracket showing dorsal diencephalon staining), caFGFR transgenic HS’d (G), and non-transgenic HS’d embryos (H). (J–M) Dorsal view of a 12–14 somite control or caFGFR transgenic embryos, with head dissected away from yolk, showing expression of six7. (M) Six7 expression is absent in the dorsal diencephalon of caFGFR transgenic HS’d embryos compared with non-transgenic non HS’d (J; white bracket showing dorsal diencephalon staining), caFGFR transgenic non HS’d (K), and non-transgenic HS’d embryos (L). (O) Histogram quantifying six3b expression in caFGFR genotyped embryos. (P) Histogram quantifying six7 expression in caFGFR genotyped embryos.
While both six3b and six7 expression were decreased when FGF signaling was inhibited, up-regulation of the FGF pathway in hsp70:ca-FGFR embryos revealed that the level of FGF signaling differentially controls the expression of six3 homologs. When caFGFR transgenic embryos were HS activated at 4–6 SS, there was a dramatic up-regulation of six3b expression compared to controls (Fig. 4F–I, O). In contrast, Six7 expression became restricted in much of the brain after HS-induced up-regulation of the FGF pathway (Fig. 4J–M, P).
To determine whether control over six genes persists in HS’d caFGFR embryos, we activated the FGF pathway at 14 SS and observed up-regulation of six3b expression in HS’d caFGFR transgenic embryos comparable to the earlier heat shocks (data not shown). Six7 is no longer expressed at 14 SS and a later up-regulation of FGF signaling does not cause ectopic expression (data not shown). Together these results demonstrate that FGF signaling levels differentially control six3 homolog expression; decreased FGF signaling decreases six3 homolog expression and conversely, increased FGF signaling increases six3b but decreases six7 expression.
Disruption of FGF signaling alters forebrain midline development
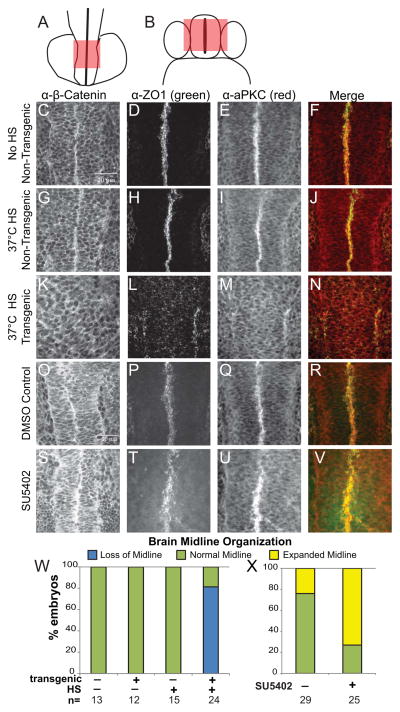
The notochord and neural floor plate have long been thought to serve as a midline barrier that separates asymmetric signals in LPM [37]. Previous studies of the brain have uncovered a midline in the forebrain which might be an analogous structure for brain laterality [38]. Considering the already characterized role of Wnt signaling in brain asymmetry [16] we used immunofluorescence with β-catenin antibodies in an attempt to uncover a link between Wnt and FGF signaling in the brain. While the overall levels of β-catenin appear to be only mildly affected in embryos that had either hyper-activated or downregulated FGF signaling (data not shown), we determined that the midline structure of the forebrain is affected by FGF signaling (Fig. 5). This midline structure is located in the forebrain at 14–16 hpf before ventricle lumen formation occurs (Fig. 5A, B). To obtain a better description of the midline structure labeled by anti-β-catenin antibody, the forebrain was scanned by confocal microscopy from dorsal to ventral; this structure continues down to the ventral floorplate (Fig. 5B). Focusing on the time period during which FGF signaling is required for brain asymmetry (12–16 somite), we found that HS’d caFGFR transgenic embryos showed decreased midline organization, as determined by β-catenin-labeling, compared to that of non-HS’d or non-transgenic siblings (Fig. 5C, G, K). To further explore the nature of this midline structure, we utilized other cell polarity markers including ZO1 to label tight junctions, and atypical protein kinase C (aPKC) as an apical marker (Fig. 5D–F, H–I, L–N). In non-HS’d transgenic embryos and HS’d non-transgenic control embryos all three of these markers were highly organized at the midline of the forebrain (Fig. 5C–J; Supplemental Movie 1; Table S1). In contrast, in HS’d caFGFR transgenic embryos there was a complete loss of midline organization, and the markers had either disrupted distribution or were absent (Fig. 5K–N, W; Supplemental Movie 2; Table S1).
Figure 5. Brain midline morphology is disrupted when the balance of FGF signaling is altered.
(A) Dorsal view of a 14 somite embryo, red box indicates area imaged by confocal microscopy. (B) To orient the reader, a cross sectional view through the forebrain, red box corresponds to the z-stacks imaged by confocal microscopy. (C–V) Dorsal view of the brain of 16 somite embryos, anterior down, taken at 60X magnification. Z-stacks were taken to encompass the organized midline of the brain, except in the cases where the midline organization was absent and then z-stacks were taken over the entire brain. (C, G, K, O, S) Representative images of α-β-Catenin. (D, H, L, P, T) Representative images of α-ZO1. (E, I, M, Q, U) Representative images of α-aPKC. (F, J, N, R, V) Merged images of both α-ZO1 (green) and α-aPKC (red). HS activated transgenic caFGFR embryos (K–N) appear to lose cellular organization in the midline of the brain compared to non-HS’d controls (C–F) and HS’d non-transgenic siblings (G–J). SU5402 embryos appear to have an increase of midline staining (S–V) compared to DMSO controls (O–R). (W–X) Histograms showing frequency of loss of midline (loss of midline), WT midline (normal) and disorganized midline organization of caFGFR transgenics and siblings (W) and SU5402 and DMSO embryos (X). See Supplementary Table 1 for quantification of midline organization and quantification of markers used.
To determine if decreasing FGF signaling lead to altered forebrain midline structure, we treated embryos with SU5402 from 8 SS to 14 SS (the time period during which FGF signaling affects brain asymmetry). Inhibiting FGF signaling altered the localization of all three forebrain midline markers (Fig. 5S–V), compared to normal midline organization in DMSO controls (Fig. 5O–R). In the SU5402 treated embryos, midline staining was disrupted and expanded, not organized into tightly opposing junctions seen in DMSO control embryos (Figure 5S–V, X, Table S1), suggesting that decreasing levels of FGF signaling also disrupts midline organization.
Discussion
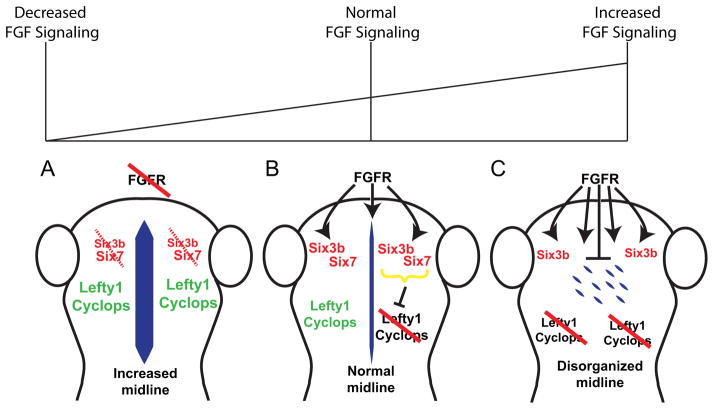
In this report we have uncovered an early role for FGF signaling in the establishment of brain asymmetry that can be altered without disrupting the highly conserved pathway establishing asymmetry in LPM and the heart. Inhibition of FGF signaling creates a bilaterally symmetric brain phenotype, with lefty1 expression in both the right and left dorsal diencephalon, whereas hyper-activation of FGF signaling leads to the converse phenotype, absent lefty1 expression. Together this suggests that too much FGF signaling negatively regulates asymmetric gene expression in the brain, an intermediate level of FGF signaling is required for asymmetric gene expression, and too little FGF signaling allows ectopic, bilateral expression of asymmetry genes (Fig. 6). Modulation of the FGF pathway affects brain expression of six3 transcription factors. Similarly, an appropriate balance of FGF signaling is required for normal forebrain midline organization, with excess FGF signaling resulting in loss of forebrain midline structures, and reduced signaling resulting in a disorganized forebrain midline.
Figure 6. A balance of FGF activity in the brain controls six3 gene expression, brain midline organization and Left-Right asymmetric lefty1 gene expression.
(A) Down-regulation of FGF signaling leads to decreased six3 expression on both sides of the brain and an expansion of brain midline structures. (B) When both FGF signaling and six3 activity are normal, a normal midline structure forms in the brain and allows normal left-sided expression of lefty1. (C) Hyper-activation of FGF signaling increases six3b expression, and other factors, while inhibiting six7 expression. Enhanced FGF-signaling also disrupts brain midline organization. Consequently, lefty1 expression is absent on both sides of the brain, leading to bilateral symmetry (absent expression) that is the opposite of the bilateral symmetry (bilateral expression) seen in the absence of FGF signaling and six3 gene function.
FGF signaling controls brain asymmetry distinct from LPM asymmetry
We and others have previously shown that FGF signaling has an early role in LR patterning through ciliogenesis and the downstream establishment of LPM asymmetry [21, 32, 33]. Here we present data showing a distinct and later role for FGF signaling in brain LR patterning, in the context of normal LPM asymmetry. Inhibition of FGF signaling creates bilateral expression of early markers of brain asymmetry. Although disruption of FGF signaling during gastrulation results in a bilateral brain phenotype, consistent with other studies of early brain development [16, 17], LPM asymmetry is affected. Here we show that later FGF signaling, during mid-somitogenesis, is required to control lefty1 expression in the brain, but not LPM asymmetry, resulting in embryos with normal LPM asymmetry but altered brain asymmetry.
Control of brain asymmetry by FGF signaling appears to be subsequent to spaw signaling from LPM. Knocking down spaw does not change the outcome of FGF inhibition: lefty1 expression in the brain was bilateral in the vast majority of embryos when FGF signaling was inhibited, either with or without spaw knockdown (Fig. 3). In contrast to this major effect, Inbal et al. [17] show that approximately half of six3b/six7/spaw knockdown embryos did not express lefty1. Similarly, lefty1 is bilateral in only 50% of masterblind mutants (axin1 mutant) when spaw is knocked down [16]. The high frequency of bilateral lefty1 we observe when FGF is inhibited, which is independent of spaw expression, indicates that FGF activity functions farther downstream in the control of lefty1 than Nodal signaling from the LPM. In addition, the effective window of transgenic Six3b overexpression ends [17] before the effective window of caFGFR or the FGFR inhibitor. The lack of a full epistatic relationship between LPM Nodal activity and Six3 genes, and differences in effective developmental windows for various manipulations, suggests that FGF works beyond the previously suggested Nodal-Six pathway to control asymmetric lefty1 expression.
Epistasis experiments to test the relationship of FGF signaling, six3b and six7 to lefty1 regulation were inconclusive due to severe, pleiotropic morphological defects. Activation of FGF signaling in either a double MO injection for six3b and six7 or six7 MO injections into six3b mutations had severe brain and axial defects at 22–24 SS, precluding the analysis of lefty1 expression. Further studies will be necessary to determine the presence of other downstream targets for brain midline formation and asymmetry.
Other reports indicate that FGF signaling also has roles in brain LR morphology at significantly later stages of development. Regan et al. show that the parapineal organ fails to migrate when FGF signaling is inhibited at 24–28 hpf, approximately 8–12 hours after the time window we describe here [18]. Clanton et al. show that inhibition of FGF signaling from 18–30 hpf results in a dramatic decrease in parapineal cell number through regulation of cell fates [19]. These studies demonstrate that the FGF ligand FGF8 regulates for parapineal migration and cell number. It is possible that FGF8 plays an additional role in early asymmetry establishment. However a single knock-down of this ligand does not affect lefty1 expression [18]. Combinatorial knockdown of multiple FGF ligands (FGF8, FGF3 and FGF24) produces a severe, compound phenotypes that make interpretation of brain development difficult [39, 40]. With the addition of our study, it appears that FGF signaling plays multiple and distinct roles in brain midline organization, asymmetric gene expression, cell division, and migration all of which are required for normal parapineal development to occur.
Unbalanced FGF signaling disrupts forebrain midline and LR asymmetry
Because of the importance of axial midline structures (notochord and neural floor plate) in patterning of asymmetric signals in the LPM [37, 41], we explored whether FGF signaling has an effect on brain midline structures. Altering the balance of FGF signaling reveals the importance of a midline structure in the developing forebrain. Remarkably, down-regulation of FGF signaling leads to mislocalization of cell polarity markers along the dorsal midline of the forebrain, and the conversion of brain asymmetry to symmetry as reflected in bilateral lefty1 expression. We propose that a decrease in FGF signaling leads to diminished six3 expression, abnormal expansion of midline and derepression of lefty1 expression (Fig. 6A). Conversely, hyperactivation of FGF signaling results in three correlated events: loss of forebrain midline organization, increased six3b expression, and loss of lefty1 expression (Fig. 6C). We also found flh expressing cells are present, indicating that cell fate has not changed, but that flh expression was abnormally dispersed, reflecting the disorganization of the cells in this region. Therefore a finely-tuned balance of FGF signaling is necessary for the establishment of normal brain LR asymmetry (Fig. 6B). Further experiments designed to directly disrupt the brain midline independent of FGF signaling will be necessary to determine the role of this midline structure in controlling brain asymmetry.
Conclusions
Control of early brain asymmetry requires a balance of FGF signaling, sine occulis homolog function, and formation of a forebrain midline structure demarked by ZO-1, aPKC and beta-catenin. Strikingly, FGF signaling functions at distinct stages in development, probably through different mechanisms, to control asymmetric gene expression throughout the embryo and also in the brain. FGF signaling controls expression of six3 homologs for normal brain asymmetry, but also utilizes novel downstream components.
Supplementary Material
Highlights.
FGF signaling controls brain laterality, independently of LPM asymmetry
FGF signaling controls expression of six3 transcription factors
Disruption of FGF signaling disrupts brain midline organization
Acknowledgments
We thank Megan Smith and Manju Karthikeyan for technical assistance; Jan Christian, Maureen Condic, Kara Cerveny and Rich Dorsky for discussion and comments on the manuscript; Lilianna Solnica-Krezel and Ken Poss for transgenic fish and reagents. This work was supported by grants to HJY from NHLBI (HL075472) and JMN (5T32DK007115).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Taylor RW, Hsieh YW, Gamse JT, Chuang CF. Making a difference together: reciprocal interactions in C. elegans and zebrafish asymmetric neural development. Development (Cambridge, England) 2010;137:681–691. doi: 10.1242/dev.038695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halpern ME, Gunturkun O, Hopkins WD, Rogers LJ. Lateralization of the vertebrate brain: taking the side of model systems. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:10351–10357. doi: 10.1523/JNEUROSCI.3439-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Concha ML, Signore IA, Colombo A. Mechanisms of directional asymmetry in the zebrafish epithalamus. Semin Cell Dev Biol. 2009;20:498–509. doi: 10.1016/j.semcdb.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Concha ML, Burdine RD, Russell C, Schier AF, Wilson SW. A nodal signaling pathway regulates the laterality of neuroanatomical asymmetries in the zebrafish forebrain. Neuron. 2000;28:399–409. doi: 10.1016/s0896-6273(00)00120-3. [DOI] [PubMed] [Google Scholar]
- 5.Liang JO, Etheridge A, Hantsoo L, Rubinstein AL, Nowak SJ, Izpisua Belmonte JC, Halpern ME. Asymmetric nodal signaling in the zebrafish diencephalon positions the pineal organ. Development (Cambridge, England) 2000;127:5101–5112. doi: 10.1242/dev.127.23.5101. [DOI] [PubMed] [Google Scholar]
- 6.Concha ML, Russell C, Regan JC, Tawk M, Sidi S, Gilmour DT, Kapsimali M, Sumoy L, Goldstone K, Amaya E, et al. Local tissue interactions across the dorsal midline of the forebrain establish CNS laterality. Neuron. 2003;39:423–438. doi: 10.1016/s0896-6273(03)00437-9. [DOI] [PubMed] [Google Scholar]
- 7.Essner JJ, Amack JD, Nyholm MK, Harris EB, Yost HJ. Kupffer’s vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development (Cambridge, England) 2005;132:1247–1260. doi: 10.1242/dev.01663. [DOI] [PubMed] [Google Scholar]
- 8.Kramer-Zucker AG, Olale F, Haycraft CJ, Yoder BK, Schier AF, Drummond IA. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer’s vesicle is required for normal organogenesis. Development (Cambridge, England) 2005;132:1907–1921. doi: 10.1242/dev.01772. [DOI] [PubMed] [Google Scholar]
- 9.Bisgrove BW, Essner JJ, Yost HJ. Regulation of midline development by antagonism of lefty and nodal signaling. Development (Cambridge, England) 1999;126:3253–3262. doi: 10.1242/dev.126.14.3253. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Yost HJ. Initiation and propagation of posterior to anterior (PA) waves in zebrafish left-right development. Dev Dyn. 2008;237:3640–3647. doi: 10.1002/dvdy.21771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rebagliati MR, Toyama R, Fricke C, Haffter P, Dawid IB. Zebrafish nodal-related genes are implicated in axial patterning and establishing left-right asymmetry. Developmental biology. 1998;199:261–272. doi: 10.1006/dbio.1998.8935. [DOI] [PubMed] [Google Scholar]
- 12.Long S, Ahmad N, Rebagliati M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development (Cambridge, England) 2003;130:2303–2316. doi: 10.1242/dev.00436. [DOI] [PubMed] [Google Scholar]
- 13.Gamse JT, Thisse C, Thisse B, Halpern ME. The parapineal mediates left-right asymmetry in the zebrafish diencephalon. Development (Cambridge, England) 2003;130:1059–1068. doi: 10.1242/dev.00270. [DOI] [PubMed] [Google Scholar]
- 14.Gamse JT, Kuan YS, Macurak M, Brosamle C, Thisse B, Thisse C, Halpern ME. Directional asymmetry of the zebrafish epithalamus guides dorsoventral innervation of the midbrain target. Development (Cambridge, England) 2005;132:4869–4881. doi: 10.1242/dev.02046. [DOI] [PubMed] [Google Scholar]
- 15.Aizawa H, Bianco IH, Hamaoka T, Miyashita T, Uemura O, Concha ML, Russell C, Wilson SW, Okamoto H. Laterotopic representation of left-right information onto the dorso-ventral axis of a zebrafish midbrain target nucleus. Curr Biol. 2005;15:238–243. doi: 10.1016/j.cub.2005.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carl M, Bianco IH, Bajoghli B, Aghaallaei N, Czerny T, Wilson SW. Wnt/Axin1/beta-catenin signaling regulates asymmetric nodal activation, elaboration, and concordance of CNS asymmetries. Neuron. 2007;55:393–405. doi: 10.1016/j.neuron.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inbal A, Kim SH, Shin J, Solnica-Krezel L. Six3 represses nodal activity to establish early brain asymmetry in zebrafish. Neuron. 2007;55:407–415. doi: 10.1016/j.neuron.2007.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Regan JC, Concha ML, Roussigne M, Russell C, Wilson SW. An Fgf8-dependent bistable cell migratory event establishes CNS asymmetry. Neuron. 2009;61:27–34. doi: 10.1016/j.neuron.2008.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clanton JA, Hope KD, Gamse JT. Fgf signaling governs cell fate in the zebrafish pineal complex. Development (Cambridge, England) 2013;140:323–332. doi: 10.1242/dev.083709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masai I, Heisenberg CP, Barth KA, Macdonald R, Adamek S, Wilson SW. floating head and masterblind regulate neuronal patterning in the roof of the forebrain. Neuron. 1997;18:43–57. doi: 10.1016/s0896-6273(01)80045-3. [DOI] [PubMed] [Google Scholar]
- 21.Neugebauer JM, Amack JD, Peterson AG, Bisgrove BW, Yost HJ. FGF signalling during embryo development regulates cilia length in diverse epithelia. Nature. 2009;458:651–654. doi: 10.1038/nature07753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marques SR, Lee Y, Poss KD, Yelon D. Reiterative roles for FGF signaling in the establishment of size and proportion of the zebrafish heart. Developmental biology. 2008;321:397–406. doi: 10.1016/j.ydbio.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez-Quevedo R, Lee Y, Poss KD, Wilkinson DG. Neuronal regulation of the spatial patterning of neurogenesis. Dev Cell. 2010;18:136–147. doi: 10.1016/j.devcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nature genetics. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 25.Munchberg SR, Ober EA, Steinbeisser H. Expression of the Ets transcription factors erm and pea3 in early zebrafish development. Mech Dev. 1999;88:233–236. doi: 10.1016/s0925-4773(99)00179-3. [DOI] [PubMed] [Google Scholar]
- 26.Rebagliati MR, Toyama R, Haffter P, Dawid IB. cyclops encodes a nodal-related factor involved in midline signaling. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:9932–9937. doi: 10.1073/pnas.95.17.9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furthauer M, Lin W, Ang SL, Thisse B, Thisse C. Sef is a feedback-induced antagonist of Ras/MAPK-mediated FGF signalling. Nature cell biology. 2002;4:170–174. doi: 10.1038/ncb750. [DOI] [PubMed] [Google Scholar]
- 28.Furthauer M, Reifers F, Brand M, Thisse B, Thisse C. sprouty4 acts in vivo as a feedback-induced antagonist of FGF signaling in zebrafish. Development (Cambridge, England) 2001;128:2175–2186. doi: 10.1242/dev.128.12.2175. [DOI] [PubMed] [Google Scholar]
- 29.Seo HC, Drivenes O, Ellingsen S, Fjose A. Transient expression of a novel Six3-related zebrafish gene during gastrulation and eye formation. Gene. 1998;216:39–46. doi: 10.1016/s0378-1119(98)00328-x. [DOI] [PubMed] [Google Scholar]
- 30.Seo HC, Drivenes, Ellingsen S, Fjose A. Expression of two zebrafish homologues of the murine Six3 gene demarcates the initial eye primordia. Mech Dev. 1998;73:45–57. doi: 10.1016/s0925-4773(98)00028-8. [DOI] [PubMed] [Google Scholar]
- 31.Talbot WS, Trevarrow B, Halpern ME, Melby AE, Farr G, Postlethwait JH, Jowett T, Kimmel CB, Kimelman D. A homeobox gene essential for zebrafish notochord development. Nature. 1995;378:150–157. doi: 10.1038/378150a0. [DOI] [PubMed] [Google Scholar]
- 32.Hong SK, Dawid IB. FGF-dependent left-right asymmetry patterning in zebrafish is mediated by Ier2 and Fibp1. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2230–2235. doi: 10.1073/pnas.0812880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu DW, Hsu CH, Tsai SM, Hsiao CD, Wang WP. A variant of fibroblast growth factor receptor 2 (Fgfr2) regulates left-right asymmetry in zebrafish. PloS one. 2011;6:e21793. doi: 10.1371/journal.pone.0021793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohammadi M, McMahon G, Sun L, Tang C, Hirth P, Yeh BK, Hubbard SR, Schlessinger J. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science (New York, N Y. 1997;276:955–960. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- 35.Sampath K, Rubinstein AL, Cheng AM, Liang JO, Fekany K, Solnica-Krezel L, Korzh V, Halpern ME, Wright CV. Induction of the zebrafish ventral brain and floorplate requires cyclops/nodal signalling. Nature. 1998;395:185–189. doi: 10.1038/26020. [DOI] [PubMed] [Google Scholar]
- 36.Feldman B, Dougan ST, Schier AF, Talbot WS. Nodal-related signals establish mesendodermal fate and trunk neural identity in zebrafish. Curr Biol. 2000;10:531–534. doi: 10.1016/s0960-9822(00)00469-3. [DOI] [PubMed] [Google Scholar]
- 37.Danos MC, Yost HJ. Role of notochord in specification of cardiac left-right orientation in zebrafish and Xenopus. Developmental biology. 1996;177:96–103. doi: 10.1006/dbio.1996.0148. [DOI] [PubMed] [Google Scholar]
- 38.Lowery LA, Sive H. Initial formation of zebrafish brain ventricles occurs independently of circulation and requires the nagie oko and snakehead/atp1a1a.1 gene products. Development (Cambridge, England) 2005;132:2057–2067. doi: 10.1242/dev.01791. [DOI] [PubMed] [Google Scholar]
- 39.Leger S, Brand M. Fgf8 and Fgf3 are required for zebrafish ear placode induction, maintenance and inner ear patterning. Mech Dev. 2002;119:91–108. doi: 10.1016/s0925-4773(02)00343-x. [DOI] [PubMed] [Google Scholar]
- 40.Draper BW, Stock DW, Kimmel CB. Zebrafish fgf24 functions with fgf8 to promote posterior mesodermal development. Development (Cambridge, England) 2003;130:4639–4654. doi: 10.1242/dev.00671. [DOI] [PubMed] [Google Scholar]
- 41.Bisgrove BW, Essner JJ, Yost HJ. Multiple pathways in the midline regulate concordant brain, heart and gut left-right asymmetry. Development (Cambridge, England) 2000;127:3567–3579. doi: 10.1242/dev.127.16.3567. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.