Abstract
Purpose:
MicroRNA-21 (miRNA-21) has proto-oncogenic properties, though no miRNA-21 specific targets have been found in head and neck squamous cell carcinoma (HNSCC). Further study of miRNA-21 and its specific targets is essential to understanding HNSCC biology.
Experimental Design:
miRNA expression profiles of 10 HNSCC and 10 normal mucosa samples were investigated using a custom miRNA microarray. 13 HNSCC and 5 normal mucosa primary tissue specimens underwent mRNA expression microarray analysis. To identify miRNA-21 downstream targets, oral keratinocyte cells were subjected to microarray analysis after miRNA-21 transient transfection. miRNA and mRNA expression were validated by RT-qPCR in a separate cohort of 16 HNSCC and 15 normal mucosal samples. Microarray and bioinformatics analyses were integrated to identify potential gene targets. In vitro assays looked at the function and interaction of miRNA-21 and its specific gene targets.
Results:
miRNA-21 was upregulated in HNSCC and stimulated cell growth. Integrated analyses identified Clusterin (CLU) as a potential miRNA-21 gene target. CLU was downregulated after forced expression of miRNA-21 in normal and HNSCC cell lines. The activity of a luciferase construct containing the 3’UTR of CLU was repressed by the ectopic expression of miRNA-21. CLU was also downregulated in primary HNSCC and correlated with miRNA-21 over-expression. CLU variant 1 (CLU-1) was the predominant splice variant in HNSCC, and showed growth suppression function that was reversed by miRNA-21 over-expression.
Conclusions:
CLU is a specific, functional target of oncogenic miRNA-21 in HNSCC. CLU-1 isoform is the predominant growth suppressive variant targeted by miRNA-21.
Keywords: Clusterin, microRNA-21, gene target, tumor-suppressor gene, head and neck cancer
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common malignancy worldwide. Over 48,000 new cases of HNSCC are diagnosed in the United States annually. The 5-year overall survival remains around 50% partly due to advanced disease stage at diagnosis and the relatively high local and regional recurrence rates (1, 2). The development of reliable biomarkers and more effective therapeutic agents are necessary to improve patient outcomes.
A major advance in cancer biology in the last decade is the discovery of small regulatory noncoding RNAs including microRNA (miRNA). A single miRNA may regulate the expression of many genes, and it has been proposed that over one third of all protein-coding genes are under translational control by miRNA (3). These specific gene targets of miRNA are thought to be involved in cellular processes including differentiation, apoptosis, and proliferation, and seem to play an important role in a variety of human diseases including cancer (4). Expression profiles of miRNA can be established and seem to be unique for various types of cancers (5).
In this study, we verify that miRNA-21 is overexpressed in HNSCC and has proto-oncogenic properties through the regulation of Clusterin (CLU). Though no miRNA-21 specific targets have been found in HNSCC to date, several recent reports identified miRNA-21 alterations in primary HNSCC tissue samples (6-9). CLU only recently has been associated with cancer promotion and metastasis (10-12). CLU is considered as an enigmatic molecule because of the difficulties in the definition of its precise function. The role of CLU in carcinogenesis is unclear, with evidence that CLU has either a proapoptotic or a prosurvival role (11, 13-15). It is now clear that CLU encodes at least two major mRNA variants, but still very little is known about their biological relevance and regulation (16). Expression pattern of CLU and its transcript variants is unknown in HNSCC and normal tissues.
The aims of this study were: 1) to profile the miRNA expression in HNSCC, 2) to identify miRNA-21 targets that play a crucial role in HNSCC using an integrative microarray and bioinformatics techniques, and 3) to reveal the function of CLU and its regulation in HNSCC by a variant specific approach.
Materials and Methods
Human tissue samples
All human HNSCC tissue samples and normal mucosal tissues were obtained and used according to the policies of the JHMI institutional review board. Surgical specimens were obtained from surgical patients at John Hopkins Hospital. All specimens were quick-frozen in liquid nitrogen and stored at −80 °C. Microdissection of frozen tumor tissue was performed by a head an neck pathologist to assure HNSCC tumor yield of greater than 80%.
10 normal and 10 HNSCC tissues were obtained for the microRNA microarray analysis. Normal tissues were obtained from patients that underwent uvulopalatopharyngoplasty (UPPP) for sleep apnea, and the cancer tissues consisted of 9 stage IV and 1 stage III HNSCC (Supplementary Table 1). A separate cohort of primary tissue including 13 HNSCC and 5 normal mucosa UPPP specimens underwent mRNA expression microarray analysis. The tumors consisted of 7 oropharynx, 6 oral cavity, 2 larynx and a hypopharynx cancers (Supplementary Table 2). For miRNA-21 and CLU expression qRT-PCR validation study, another separate cohort of 16 HNSCC tumors and 15 normal mucosal UPPP tissues was used (Supplementary Table 3).
Cell lines and culturing conditions
Normal oral keratinocyte - spontaneously immortalized cell line (NOK-SI) was developed by National Institute of Dental and Craniofacial Research collaborators at the National Institutes of Health as previously described (17). Nok-SI cells were grown in keratinocyte serum-free medium supplemented with bovine pituitary extract, and epidermal growth factor, and 1% Penicillin-Streptomycin. The HNSCC cell lines FaDu and SCC9 were expanded and passaged in DMEM/F12 medium supplemented with 10% FBS, 1% penicillin-streptomycin and Hydrocortisone (0.4 ug/ml). JHU-O28 HNSCC cell line was expanded and passaged in RPMI-1640 medium supplemented with 10% FBS and 1% penicillin-streptomycin. All media components were obtained from Gibco Invitrogen Corporation (Carlsbad, CA). Cell growth conditions were maintained at 37 degrees Celsius in an atmosphere of 5% carbon dioxide and 95% relative humidity.
Transfection of miRNA-21, CLU transcript variant 1 and Luciferase constructs
The miRNA-21 over-expression vector pMIF-cGFP-Zeo-hsa-pre-miRNA-21 (MIFCZ307PA-1) and pMIF-cGFP-Zeo (MIFCZ300PA-1) empty vector control were purchased from System Biosciences (Mountain View, CA). CLU variant 1 (CLU-1) expression vector (Origene, Rockville, MD) was used to confirm the function of CLU-1.
Luciferase constructs containing the 3’ untranslated region (3’UTR) region of CLU and control luciferase vectors (GeneCopoeia, Rockville, MD) were used to conduct luciferase reporter assays. The miRNA-21 binding seed sequence was removed from the CLU 3’UTR region by cutting out a 120bp segment using double restriction enzyme digestion and reannealing the CLU luciferase vector per standard protocol and conditions. DNA sequencing was performed to verify deletion of the miRNA-21 seed sequence from the CLU 3’UTR sequence within the CLU luciferase vector. Transient transfections were performed in NOK-SI cells using FuGENE®6 reagent (Roche Applied Science, Indianapolis, IN) according to the manufacturer's protocols. Each experiment was performed independently in triplicate and at least three times.
Cell proliferation assay
NOK-SI, FaDu, JHU-O28 and SCC9 cells were plated at a confluency of 20% percent per well of a 96 well plate. Growth was assayed at 24, 48, and 72 hours by Cell Counting Kit-8 cell (CCK-8) proliferation assay (Dojindo Molecular Technologies, Inc., Rockville, MD). The standard manufacturer protocol was followe. All experimental conditions were performed in sextuplicate. Average fluorescence absorbance readings were taken with SpectraMax M2e Microplate reader (Molecular Devices, Sunnyvale, CA) at 450 nm and 650 nm wavelengths. Confirmatory cell proliferation studies were performed in 6 well plates in triplicate. Gross cell number at each time point was measured via direct cell counting using a hemacytometer (Fisher Scientific, Pittsburgh, PA). The average of three cell counts per well was reported.
miRNA and RNA isolation, quantification, and cDNA formation
Total RNA from primary human tissue and cell lines, was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA). For miRNA array experiments total RNA was extracted using the mirVana™ miRNA Isolation Kit (Ambion/Applied Biosystems, Austin, TX). Reverse transcription of RNA was performed using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). For miRNA specific cDNA formation, each sample was subjected to a reverse transcription reaction as part of the TaqMan® MicroRNA Assay Kit (Applied Biosystems, Foster City, CA). All RNA samples were stored at −80°C, while cDNA samples at −20C. All experiments were performed according to manufacturer’s protocols.
miRNA array
Northern blots were performed to assure that the quality of RNA was adequate. RNA integrity was evaluated using denaturing gels and detection of the 18S and 28S rRNA bands. This assured that the RNA lacked DNA contamination and that the RNA was not degraded, both of which could confound the array results. A custom-manufactured Affymetrix GeneChip® from Asuragen (DiscovArray) was designed to miRNA probes derived from Sanger mirBase v9.2 database and published reports (18, 19). Antigenomic probe sequences were provided by Affymetrix and derived from a larger set of controls used on the Affymetrix human exon array for estimating background signal, as described below. Other non-miRNA control probes were designed to have no sequence homology to the human genome and were used for spike-in external reference controls. The 3’ ends of the RNA molecules were labeled with biotin using the mirVana™ miRNA Labeling Kit (Life Technology, Austin, TX). The Kit’s dNTP mixture in the tailing reaction was replaced with a nucleotide mixture containing biotin-modified nucleotides (PerkinElmer, Waltham, MA). Labeled RNA was profiled by hybridization to the above described custom microchip. Hybridization, washing, staining, imaging, and signal extraction were performed according to Affymetrix protocols. Briefly, 100 ng of total RNA was labeled and hybridized to the microarray with ~13,000 probes including 600 known miRNA as well as putative miRNAs.
miRNA array data analysis
The signal processing implemented for the custom miRNA chip (DiscovArray) is a multi-step process involving probe-specific signal detection calls, background estimate and correction, constant variance stabilization (20) and either array scaling or global normalization. For each probe, an estimated background value was subtracted that is derived from the median signal of a set of GC content-matched anti-genomic controls. Arrays within a specific analysis experiment were normalized together according to the variance stabilization normalization (VSN) method described by Huber et al. Detection calls were based on a Wilcoxon rank-sum test of the miRNA probe signal compared to the distribution of signals from GC-content matched anti-genomic probes. Comparison between tumor and normal was tested with a two sample t-test with assumption of equal variance. One-way ANOVA was used for experimental designs with more than two experimental groupings (normal vs. tumor). These tests defined which probes were considered to be significantly differentially expressed, or "significant", based on a default p-value of less than 0.05 and at least 2-fold-change (log2 difference ≥1). Array-based differentially expressed miRNAs were further tested by qPCR as described below.
Validation of miRNA array results by qRT-PCR
cDNA was synthesized from total RNA using gene-specific primers according to the TaqMan® MicroRNA Assay protocol (Applied Biosystems, Foster City, CA). Reverse transcription was performed in 10 ul reactions containing 15 ng of total RNA isolated from the same human tissue samples used for the miRNA array. Reactions were incubated in a 9700 Applied Biosystems (ABI) ThermaCycler in a 96-well plate for 30 min at 16°C, 30 min at 42°C, 5 min at 85°C. 2 ul of the RT reaction were used in the 15 ul real-time qPCR reaction performed on an Applied Biosystems 7900 Sequence Detection system using ABI’s Taqman primer and probes for PCR amplification. The reaction conditions were according to the TaqMan MicroRNA Assay protocol. Each sample was measured in duplicate RT reactions. We used the DCt values to identify candidate biomarkers for distinguishing HNSCC from normal tissues using the DDCt method (21).
Validation of miRNA-21 results by RT-qPCR in separate human tissue sample cohort
Extracted total RNA from HNSCC tumor and normal tissue was subjected to miRNA specific RT-qPCR as per the manufacturer protocol (Applied Biosystems, Foster City, CA). RT-qPCR for miRNA was performed as described previously (7). The expression was normalized to U47 expression. The expression level was again determined by the DDCt method (21). The average miRNA-21 expression level in normal versus tumor tissue was determined using the Mann Whitney U-test.
mRNA microarray
Total RNA extraction from HNSCC and normal mucosal human tissue samples and NOK-SI cells were performed using Trizol reagent, as above. RNA was further purified using the RNeasy Kit (Qiagen, Valencia, CA). Northern blots were performed to assure that the quality of RNA was adequate. RNA integrity was evaluated on a denaturing gel, and evaluating the presence of 18S and 28S rRNA. mRNA microarray analysis was carried out using the Affymatrix U133 Plus 2.0 array platform, both for the primary tissue and the NOK-SI cell line experiments samples. Significance analysis of microarrays (SAM) was performed to determine differential mRNA expression. A q-value or FDR was set at 5% to determine significant genes.
Quantification of CLU expression by RT-qPCR
Applied Biosystems gene expression kit Hs00971651_m1 specific for CLU was used for qPCR studies. The expression was normalized to 18s expression, using the Applied Biosystems gene expression kit Hs99999901_s1. The expression level was again determined by the DDCt method (21). The CLU expression level in normal versus tumor tissue was determined using the Mann Whitney U-test.
Absolute quantification of CLU differential transcript expression by RT-qPCR
In order to further elucidate the baseline specific expression pattern of CLU in NOK-SI cell line, CLU transcript variant 1 (CLU-1) and CLU transcript variant 2 (CLU-2) specific primers were used to quantify the absolute expression ratio of each variant. Variant specific primer sequences were as follows (22): CLU-1, 5’- ACAGGGTGCCGCTGAC -3’ (forward) and 5’- CCAGGACCTGCCCACTCT -3’ (reverse); CLU-2, 5’- ATGCAGATGGATTCGGTGT -3’ (forward) and 5’- AGTCTTTGCACGCCTCTGA -3’ (reverse). Purified PCR products were synthesized using CLU-1 and CLU-2 specific primers. Using standard curves based on the diluted purified PCR products, the input copy number of each transcript was determined by SYBR Green (Applied Biosystems) qRT-PCR.
Luciferase reporter assay
At 48 hours after transfection of miRNA-21 and the luciferase plasmids into NOK-SI cells, luciferase assays were performed using the dual-luciferase reporter assay system (Promega, Madison, WI) as per the manufacturer's instructions. Luminescent signal was quantified by the luminometer (Monolight 3020; BD Biosciences). Each Renilla luminescence value was first normalized to the control Firefly luciferase assay value contained in each luciferase construct. Each value is a mean of three separate transfections measured in triplicate.
Results
Identification of upregulated miRNAs in HNSCC
miRNA expression profiles of HNSCC were investigated using a miRNA microarray platform. Profiles from tumor samples (n=10) and non-tumor tissues (n=10) were compared (Supplementary Table 4). Thirty-two differentially expressed miRNAs were identified by miRNA array (Supplementary Table 5). To validate the microarray data, expression profiles of these miRNAs were analyzed by RT-qPCR in the same cohort of clinical samples that was used in miRNA array. Twenty-one significantly upregulated miRNAs were identified (Supplementary Table 6). miRNA-21, miRNA-155, and miRNA-375 have been previously reported as differentially expressed in HNSCC, but no specific gene targets of these miRNAs have been reported as of yet in HNSCC tumors (7, 8).
Among this group, miRNA-21 was one of the most notably upregulated in tumor tissues (P=0.0002). miRNA-21 plays a potential important role in carcinogenesis as there are increasing number of reports showing aberrant expression of this miRNA in many different tumor types. miRNA-21 has been implicated in tumor growth, carcinogenesis, and response to chemotherapy in different malignancies (7, 23-29). Given its potential active role across many different tumor types, including HNSCC, miRNA-21 was selected as the miRNA of choice for target identification experiments. For further validation miRNA-21 expression was examined in a separate validation cohort of 16 HNSCC tumors and 15 normal mucosal tissues. Upregulation of miRNA-21 in tumors was again confirmed (Supplementary Figure 1, P=0.0007).
Impact of miRNA-21 on cell growth
To confirm the well-known fact that miRNA-21 is an oncogenic miRNA (23, 30, 31), impact of miRNA-21cell growth was examined. The miRNA-21 over-expression vector pMIF-cGFP-Zeo-hsa-pre-miRNA-21and empty control vector pMIF-cGFP-Zeo were transfected in the NOK-SI cell line. Overexpression of miRNA-21 was confirmed by qRT-PCR. The over-expression vector produced over a five thousand-fold increase in pre-miRNA-21 and almost a four-fold increase in miRNA-21 expression when compared to the control vector (Supplementary Figure 2). In NOKSi, over-expression of miRNA-21 significantly stimulated proliferation compared with the negative empty vector control. At the 72 hour time point after transfection, the miRNA-21 overexpressed cells exhibited a 40.4% (p=0.0022) increase in cell growth compared to the empty vector (Supplementary Figure 3).
Identification of miRNA-21specific target
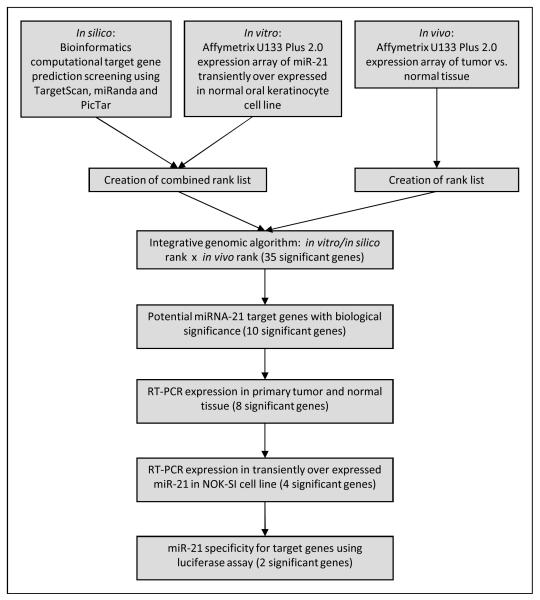
Bioinformatics tools (TargetScan, PicTar, miRanda) were used to predict the potential miRNA-21gene targets. Results revealed that miRNA-21 has over 1200 potential gene targets (Supplementary Table 7-9). This presented a logistical challenge to find a way to narrow this extensive list and identify gene specific targets of miRNA-21. The ultimate research strategy included in silico bioinformatics tools, as well as in vitro cell line and in vivo primary tissue studies (Figure 1). In vitro experiments using the normal oral keratinocyte (NOK-SI) cell line were used to over-express miRNA-21as described earlier. To capture and identify the potential expression changes that occur due to miRNA-21 over-expression and its downstream target genes, NOK-SI cells were subjected to mRNA microarray analysis after 72 hour transient transfection with miRNA-21 over-expression vector or its negative control. Affymetrix U133 Plus 2.0 array platform was used and 97 upregulated and 60 downregulated genes were identified (Supplementary Table 10).
Figure 1.
Study Flowchart.
In vivo studies included evaluation of the mRNA expression differences between normal mucosal and HNSCC tumor tissues. The Affymetrix U133 Plus 2.0 array platform was again used to evaluate differential mRNA expression in an independent tissue bank cohort that was separate from the primary tissue used in the miRNA discovery array. Analysis of the differential expression data revealed statistical significance in 368 upregulated and 6912 downregulated genes (Supplementary Table 11).
Results from all three arms of the experimental design were then combined in a novel integrative approach to narrow down possible miRNA-21gene targets in HNSCC. The 60 differentially downregulated mRNAs identified on the microarray analysis of in vitro miRNA-21 overexpressed NOK-SI cell line samples were only accepted as potential miRNA-21 targets if they could be verified by the in silico bioinformatics tools for gene target prediction. This preliminary list was then compared to the in vivo array results of downregulated mRNAs generated by comparing differential mRNA expression of HNSCC and normal mucosal tissues. An integrative approach was used to formulate a product rank order list based on potential miRNA-21 gene targets identified so far by combining the in vitro and in silico approaches and comparing them to downregulated mRNAs identified in the in vivo experimental arm (Figure 1).
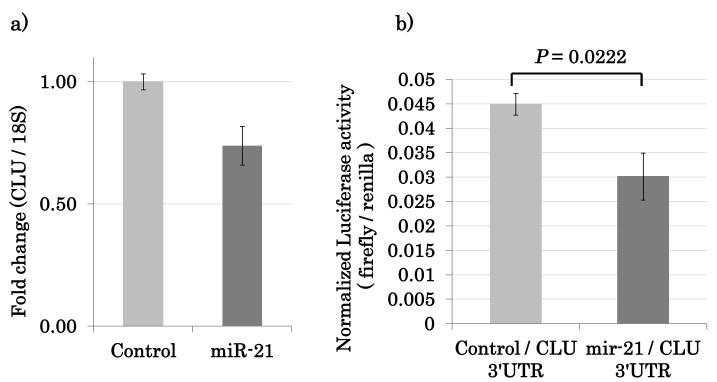
Combining these three approaches generated a candidate gene list (Table 1). Clusterin (CLU) was the most statistically significant downregulated gene target identified during this integrative analysis and CLU mRNA was downregulated by the forced expression of miRNA-21 in NOKSI cell line (Figure 2a).
Table 1.
Potential targets of miRNA-21
| Gene Name | Official Symbol |
|---|---|
| Clusterin | CLU |
| Basonuclin 2 | BNC2 |
| Desmin | FAM48a |
| Protein phosphatase 1, regulatory subunit 3E | PPP1R3E |
| Estrogen receptor 2 | ESR2 |
| Tumor necrosis factor 2 | TNF2 |
| Fibroblast growth factor 5 | FGF5 |
| B-cell CLL/lymphoma 2 | BCL2 |
| Programmed cell death 4 | PDCD4 |
| Pleiomorphic adenoma gene 1 | PLAG1 |
Figure 2.
CLU is a gene specific target of miRNA-21. (A) Overexpression of miRNA-21 downregulates CLU expression. (B) miRNA-21 specifically binds to CLU.
miRNA-21 targets the 3’ UTR of CLU
In order for CLU to be a specific gene target of miRNA-21 and undergo expression repression by this miRNA, the 3’UTR of the mRNA transcript should contain miRNA-21 specific binding sites. miRNA-21 must recognize these sites and directly bind to this 3’UTR of the mRNA transcript. To demonstrate this, the 3’UTR of CLU that contained all the predicted miRNA-21 binding sites was cloned into a luciferase reporter construct. Co-transfection of miRNA-21 over-expression vector along with the CLU 3’UTR sequence containing a luciferase reporter construct was performed to show specific binding and recognition of miRNA-21 for the CLU 3’UTR. Activity of a luciferase reporter containing the predicted microRNAs binding sequences of CLU 3’UTR was repressed by the ectopic expression of miRNA-21 (Figure 2b). miRNA-21 specificity for the CLU 3’UTR was lost when a short segment containing the miRNA-21 seed sequence was deleted from within the CLU 3’UTR region of the same luciferase reporter construct (Supplemental Figure 4).
Variant specific analysis of CLU expression
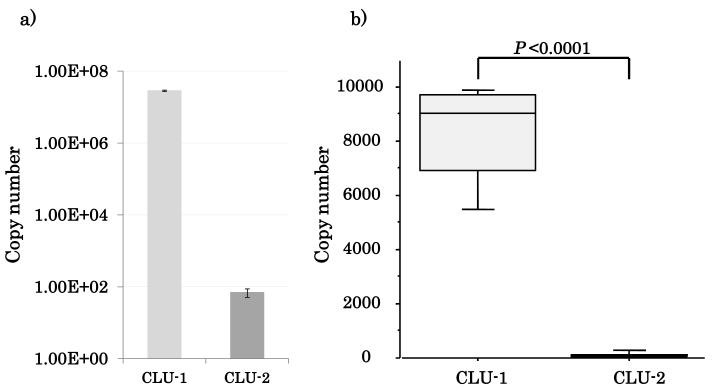
There are two main variants in CLU (16), and both of them have a common 3’ UTR sequence. Experimental evidence shows that these two different CLU transcript variants may have varying proapoptotic or prosurvival functions. Expression pattern of CLU and its transcript variants as well as the specific function of CLU is unknown in HNSCC and normal tissues to date. Therefore it was important to evaluate the variant specific expression pattern of the two major CLU mRNA transcript variants. Absolute quantification of CLU-1/CLU-2 was performed in NOKSI cell line and 16 HNSCC tumor samples (Supplementary Table 3). In both NOKSI and HNSCC tumors, CLU-1 was by far the dominant variant by RT-qPCT (copy numbers of CLU-1 and CLU-2 were 7717±2684 and 106±87 respectively, p<0.0001, Figure 3). CLU-1 expression was also verified in NOKSI and other HNSCC cell lines, with NOKSI showing the highest expression of CLU-1 (Supplementary Figure 5).
Figure 3.
CLU-1 is the dominant transcript variant in (A) NOKSI cell line and (B) HNSCC tumor samples.
Impact of CLU-1 on cell growth
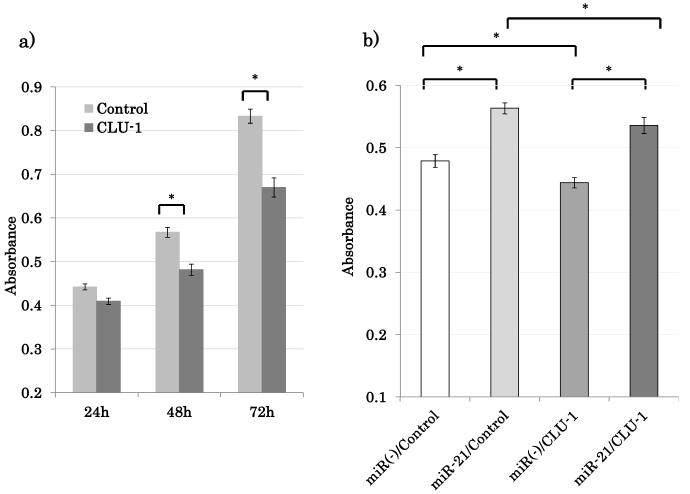
The goal of this study was to identify key biologically active gene targets of miRNA-21 in HNSCC. miRNA-21 is overexpressed in HNSCC primary tissue and has proto-oncogenic activity in vitro (7). Biologically significant targets should therefore in the least induce growth suppression when actively expressed. Growth effects of CLU-1 were then evaluated in vitro. CLU-1 was transiently overexpressed in NOKSI cell line to help clarify its potential effects on cell proliferation and results obtained using absorbance readings at regular 24 hour time point increments (Figure 4). Overexpression of CLU-1 was confirmed by RT-qPCR (Supplementary Figure 6).
Figure 4.
CLU-1 has growth inhibitory effects in NOKSI cell line. (A) Overexpression of CLU-1 inhibits cell proliferation. (B) miRNA-21 overexpression can reverse CLU-1 growth inhibitory effect. All measurements were obtained with the Cell Counting Kit -8 cell proliferation assay. *p<0.01.
In NOKSI, overexpression of CLU-1 inhibited proliferation compared to negative control. At the 72-hour time point after transfection, the CLU-1 overexpressed cells exhibited a 19.6% (p=0.0022) reduction in cell growth compared to the empty vector transfected cells (Figure 4a). Similar growth inhibitory results were obtained in HNSCC cell lines when CLU-1 was overexpressed in JHU-O28, FaDu and SCC9. Cell counts at various time points were obtained by direct real time cell counting as described previously (Supplementary Figure 7). All of the experimental analyses in this study clearly show that miRNA-21 specifically recognizes and downregulates a specific isoform of CLU, CLU-1, in head and neck mucosal tissue. miRNA-21 overexpression represses the growth inhibitory CLU-1 by specifically binding to the 3’UTR and effectively stimulates growth proliferation in cells.
To further validate miRNA-21 specificity and regulation of CLU-1, over-expression of CLU-1 and the resulting growth suppression should be inhibited if not reversed by miRNA-21 over-expression. To confirm the regulation of CLU by miRNA-21, cotransfection was performed using miRNA-21 expression vector and CLU-1 expression vector in the NOKSI cell line. Cell growth inhibitory effect of CLU-1 was reversed by cotransfection of miRNA-21 expression vector as measured with the CCK-8 cell growth kit (Figure 4b). These results were repeated and verified by quantifying cell growth with direct cell number counts at each time point (data not published).
Relation between the expression of miRNA-21 and CLU in HNSCC
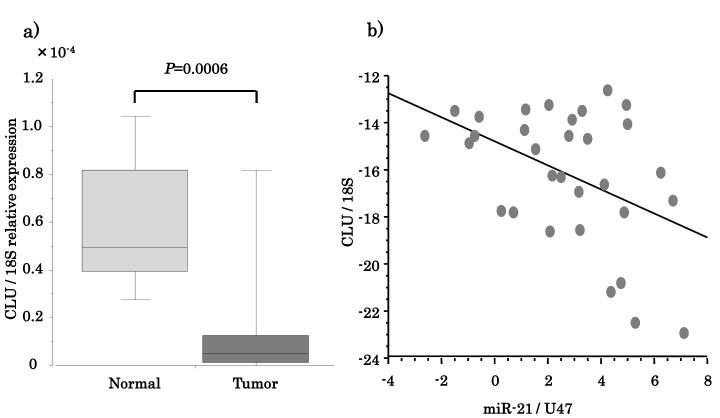
Given the specificity of miRNA-21 for CLU and the CLU-1 transcript variant, expression of both genes was analyzed in primary tissue samples. If CLU and CLU-1 play an important potential role in HNSCC carcinogenesis, miRNA-21 expression in tumors should correlate with repression of CLU. Analysis was performed in a separate cohort of 16 HNSCC tumors and 15 normal UPP mucosal samples (Supplementary Table 3). CLU was significantly downregulated in primary HNSCC compared with normal mucosa tissue (P=0.0006, Figure 5a). On the other hand, as shown in Supplementary Figure 1, miRNA-21 expression was significantly upregulated in tumor (P=0.0007). There was a significant inverse correlation between miRNA-21 expression and CLU expression. The Spearman correlation coefficient between miRNA-21 and CLU was −0.449 (P=0.0114, Figure 5b).
Figure 5.
miRNA-21 expression is inversely proportional to CLU expression in HNSCC. (A) HNSCC tissue has lower expression levels of CLU compared to normal. (B) Spearman correlation between CLU and miRNA-21 expression in HNSCC tissue.
Discussion
A large-scale survey to determine the miRNA signature of 540 tumor samples including lung, breast, stomach, prostate, colon, and pancreatic tumors revealed that miRNA-21 was the only miRNA upregulated in all these tumors (32). Recently emerging evidence indicates that miRNA-21 has oncogenic properties (23, 30, 31), and inhibition of miRNA-21 could provide a new therapeutic strategy in cancer through the Ras/MEK/ERK pathway and apoptosis (33). However, regarding HNSCC, little is known about the role of miRNA-21 and all of its potential targets.
Each miRNA targets numerous genes simultaneously (34, 35), so it is quite difficult to identify targets of miRNAs that have crucial role in regulation of cancer biology. In this study, we have successfully narrowed down the candidates of specific target of miRNA-21 by the combination of microarray analysis with bioinformatics target prediction tools. Our results indicated that miRNA-21 expression modulates cell growth via regulation of CLU and more specifically CLU-1, a CLU mRNA transcript variant with growth suppressive function. Furthermore, in clinical tissue samples, significant inverse correlation between miRNA-21 expression and CLU expression was observed. It is usually difficult to determine the significant correlation between miRNAs expression and their predicted targets in clinical samples (27, 36). So these results suggest that CLU is an important target of oncogenic miRNA-21 in HNSCC, and miRNA-21 might be a key regulator of CLU in HNSCC. The accumulated evidence indicates that CLU has many biological functions and plays an important role in cancer biology (37, 38). Now it is generally accepted that there are two alternative isoforms of CLU protein; the secreted CLU (sCLU) and the nuclear form of CLU (nCLU). The existence of sCLU and nCLU might explain the contradictory role of CLU. sCLU is usually reported as a survival factor, on the other hand, nCLU is considered as proapoptotic associated with cell death (13, 39-42). During the last decade, many studies have discussed the complexity of CLU, but there is still a lack of precise information about the complex regulation of its expression, so CLU has been defined as an enigmatic protein. Several reports have indicated that CLU-1 may account for the existence of the nuclear form of CLU (nCLU) (16, 43), and CLU-2 encodes for a secreted CLU (sCLU) (16, 44, 45). We identified CLU-1 as the dominant variant expressed in HNSCC that is regulated by miRNA-21. Our results also suggest that nCLU might originate at least partially from CLU-1 because forced expression of CLU-1 resulted in growth inhibition. It is now clear that CLU encodes more than one mRNA, but it still remains unclear how the protein isoforms are produced from the CLU gene and how each transcript relates to the diverse CLU isforms (46). Furthermore, it is important to note that none of studies have isolated and sequenced the intracellular CLU (46), so it is difficult to make a definitive conclusion regarding the origin of nCLU.
Several oncogenes including H-Ras, cMyc, and N-Myc - generally downregulate CLU (47-49), so it is hypothesized that suppression of CLU by oncogenes is required for oncogene-dependent transformation (46). Our finding supports this hypothesis because miRNA-21 is an oncogenic microRNA. It is still hard to reveal the complexity of CLU regulation, but variant specific analysis of CLU at the transcriptional level may provide clues to understanding the complexity of CLU. However, further studies are necessary to reveal the mechanism for tissue-specific regulation of CLU.
Novel miRNA targets can be identified and validated utilizing a combined microarray analysis and bioinformatics target prediction methods. miRNA-21 is one of the crucial regulators for CLU, so there is a significant inverse correlation between miRNA-21 and CLU in clinical tissue samples. Further study is necessary to evaluate the potential diagnostic and prognostic significance and even possibly therapeutic importance of CLU in HNSCC.
Supplementary Material
Translational Relevance.
Understanding the molecular mechanisms underlying head and neck cancer is essential to successfully developing novel avenues for targeted therapy. MicroRNAs play a powerful role in cancer biology, but identifying their gene specific targets is challenging given they can target numerous genes and pathways. MicroRNA specific roles and targets in head and neck cancer are largely unknown and to date no specific gene targets have been identified. By utilizing a novel integrative discovery approach, Clusterin and more specifically the unique transcript variant CLU-1, was identified as a specific miRNA-21 gene target with functional significance. By understanding the function and molecular mechanism of miRNA-21 and its specific gene targets, targeted therapy may have therapeutic potential.
Acknowledgments
Financial Support: Drs. Califano and Sidransky are supported by a NCI/NIDCR SPORE (5P50CA096784-05) and Dr. Califano is supported by NIDCR Challenge Grant (RC1DE020324). Drs. Mydlarz and Hennessey are funded by the NIH T32 Research Training Grant (T32DC000027). Funders had no role in study design, data collection, analysis, decision to publish, or preparation of the manuscript. Study was also funded by Asuragen, Inc. The researchers at Asuragen, Inc. who participated in this study were not influenced by management or parties outside of their research group and had exclusive control over study design, data collection, analysis, decision to publish, and preparation of the manuscript.
REFERENCES
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Carvalho AL, Nishimoto IN, Califano JA, Kowalski LP. Trends in incidence and prognosis for head and neck cancer in the United States: a site-specific analysis of the SEER database. Int J Cancer. 2005;114:806–16. doi: 10.1002/ijc.20740. [DOI] [PubMed] [Google Scholar]
- 3.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 4.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Avissar M, Christensen BC, Kelsey KT, Marsit CJ. MicroRNA expression ratio is predictive of head and neck squamous cell carcinoma. Clin Cancer Res. 2009;15:2850–5. doi: 10.1158/1078-0432.CCR-08-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang SS, Jiang WW, Smith I, Poeta LM, Begum S, Glazer C, et al. MicroRNA alterations in head and neck squamous cell carcinoma. Int J Cancer. 2008;123:2791–7. doi: 10.1002/ijc.23831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hui AB, Lenarduzzi M, Krushel T, Waldron L, Pintilie M, Shi W, et al. Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res. 2010;16:1129–39. doi: 10.1158/1078-0432.CCR-09-2166. [DOI] [PubMed] [Google Scholar]
- 9.Tran N, McLean T, Zhang X, Zhao CJ, Thomson JM, O'Brien C, et al. MicroRNA expression profiles in head and neck cancer cell lines. Biochem Biophys Res Commun. 2007;358:12–7. doi: 10.1016/j.bbrc.2007.03.201. [DOI] [PubMed] [Google Scholar]
- 10.Mazzarelli P, Pucci S, Spagnoli LG. CLU and colon cancer. The dual face of CLU: from normal to malignant phenotype. Adv Cancer Res. 2009;105:45–61. doi: 10.1016/S0065-230X(09)05003-9. [DOI] [PubMed] [Google Scholar]
- 11.Shannan B, Seifert M, Leskov K, Willis J, Boothman D, Tilgen W, et al. Challenge and promise: roles for clusterin in pathogenesis, progression and therapy of cancer. Cell Death Differ. 2006;13:12–9. doi: 10.1038/sj.cdd.4401779. [DOI] [PubMed] [Google Scholar]
- 12.Pucci S, Bonanno E, Pichiorri F, Angeloni C, Spagnoli LG. Modulation of different clusterin isoforms in human colon tumorigenesis. Oncogene. 2004;23:2298–304. doi: 10.1038/sj.onc.1207404. [DOI] [PubMed] [Google Scholar]
- 13.Bettuzzi S, Davalli P, Davoli S, Chayka O, Rizzi F, Belloni L, et al. Genetic inactivation of ApoJ/clusterin: effects on prostate tumourigenesis and metastatic spread. Oncogene. 2009;28:4344–52. doi: 10.1038/onc.2009.286. [DOI] [PubMed] [Google Scholar]
- 14.Caccamo AE, Desenzani S, Belloni L, Borghetti AF, Bettuzzi S. Nuclear clusterin accumulation during heat shock response: implications for cell survival and thermo-tolerance induction in immortalized and prostate cancer cells. J Cell Physiol. 2006;207:208–19. doi: 10.1002/jcp.20561. [DOI] [PubMed] [Google Scholar]
- 15.Scaltriti M, Santamaria A, Paciucci R, Bettuzzi S. Intracellular clusterin induces G2-M phase arrest and cell death in PC-3 prostate cancer cells1. Cancer Res. 2004;64:6174–82. doi: 10.1158/0008-5472.CAN-04-0920. [DOI] [PubMed] [Google Scholar]
- 16.Trougakos IP, Djeu JY, Gonos ES, Boothman DA. Advances and challenges in basic and translational research on clusterin. Cancer Res. 2009;69:403–6. doi: 10.1158/0008-5472.CAN-08-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castilho RM, Squarize CH, Leelahavanichkul K, Zheng Y, Bugge T, Gutkind JS. Rac1 is required for epithelial stem cell function during dermal and oral mucosal wound healing but not for tissue homeostasis in mice. PLoS One. 2010;5:e10503. doi: 10.1371/journal.pone.0010503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–70. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 19.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, et al. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature. 2005;434:338–45. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 21.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic acids research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersen CL, Schepeler T, Thorsen K, Birkenkamp-Demtroder K, Mansilla F, Aaltonen LA, et al. Clusterin expression in normal mucosa and colorectal cancer. Mol Cell Proteomics. 2007;6:1039–48. doi: 10.1074/mcp.M600261-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 24.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 25.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer research. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 26.Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–29. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 27.Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–84. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 28.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 29.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nature reviews Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 30.Wang P, Zou F, Zhang X, Li H, Dulak A, Tomko RJ, Jr., et al. microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res. 2009;69:8157–65. doi: 10.1158/0008-5472.CAN-09-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–9. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 32.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–93. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 36.Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10:1813–9. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones SE, Jomary C. Clusterin. Int J Biochem Cell Biol. 2002;34:427–31. doi: 10.1016/s1357-2725(01)00155-8. [DOI] [PubMed] [Google Scholar]
- 38.Rosenberg ME, Silkensen J. Clusterin: physiologic and pathophysiologic considerations. Int J Biochem Cell Biol. 1995;27:633–45. doi: 10.1016/1357-2725(95)00027-m. [DOI] [PubMed] [Google Scholar]
- 39.Hara I, Miyake H, Gleave ME, Kamidono S. Introduction of clusterin gene into human renal cell carcinoma cells enhances their resistance to cytotoxic chemotherapy through inhibition of apoptosis both in vitro and in vivo. Jpn J Cancer Res. 2001;92:1220–4. doi: 10.1111/j.1349-7006.2001.tb02143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reddy KB, Jin G, Karode MC, Harmony JA, Howe PH. Transforming growth factor beta (TGF beta)-induced nuclear localization of apolipoprotein J/clusterin in epithelial cells. Biochemistry. 1996;35:6157–63. doi: 10.1021/bi952981b. [DOI] [PubMed] [Google Scholar]
- 41.Scaltriti M, Bettuzzi S, Sharrard RM, Caporali A, Caccamo AE, Maitland NJ. Clusterin overexpression in both malignant and nonmalignant prostate epithelial cells induces cell cycle arrest and apoptosis. Br J Cancer. 2004;91:1842–50. doi: 10.1038/sj.bjc.6602193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shannan B, Seifert M, Boothman DA, Tilgen W, Reichrath J. Clusterin and DNA repair: a new function in cancer for a key player in apoptosis and cell cycle control. J Mol Histol. 2006;37:183–8. doi: 10.1007/s10735-006-9052-7. [DOI] [PubMed] [Google Scholar]
- 43.Rauhala HE, Porkka KP, Saramaki OR, Tammela TL, Visakorpi T. Clusterin is epigenetically regulated in prostate cancer. Int J Cancer. 2008;123:1601–9. doi: 10.1002/ijc.23658. [DOI] [PubMed] [Google Scholar]
- 44.Wong P, Taillefer D, Lakins J, Pineault J, Chader G, Tenniswood M. Molecular characterization of human TRPM-2/clusterin, a gene associated with sperm maturation, apoptosis and neurodegeneration. Eur J Biochem. 1994;221:917–25. doi: 10.1111/j.1432-1033.1994.tb18807.x. [DOI] [PubMed] [Google Scholar]
- 45.Zoubeidi A, Ettinger S, Beraldi E, Hadaschik B, Zardan A, Klomp LW, et al. Clusterin facilitates COMMD1 and I-kappaB degradation to enhance NF-kappaB activity in prostate cancer cells. Mol Cancer Res. 2010;8:119–30. doi: 10.1158/1541-7786.MCR-09-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rizzi F, Bettuzzi S. The clusterin paradigm in prostate and breast carcinogenesis. Endocr Relat Cancer. 2010;17:R1–17. doi: 10.1677/ERC-09-0140. [DOI] [PubMed] [Google Scholar]
- 47.Chayka O, Corvetta D, Dews M, Caccamo AE, Piotrowska I, Santilli G, et al. Clusterin, a haploinsufficient tumor suppressor gene in neuroblastomas. J Natl Cancer Inst. 2009;101:663–77. doi: 10.1093/jnci/djp063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klock G, Storch S, Rickert J, Gutacker C, Koch-Brandt C. Differential regulation of the clusterin gene by Ha-ras and c-myc oncogenes and during apoptosis. J Cell Physiol. 1998;177:593–605. doi: 10.1002/(SICI)1097-4652(199812)177:4<593::AID-JCP10>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 49.Thomas-Tikhonenko A, Viard-Leveugle I, Dews M, Wehrli P, Sevignani C, Yu D, et al. Myc-transformed epithelial cells down-regulate clusterin, which inhibits their growth in vitro and carcinogenesis in vivo. Cancer Res. 2004;64:3126–36. doi: 10.1158/0008-5472.can-03-1953. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.