Abstract
Why does cancer risk increase as we age? Frequently attributed to the multi-hit hypothesis and the time required to accumulate genomic mutations, this question is a matter of ongoing debate. Here, we propose that the normal decline in oxidative metabolism during aging constitutes an early and important “hit” that drives tumorigenesis. Central to these metabolic changes are the sirtuins, a family of NAD+-dependent deacylases that have evolved as coordinators of physiological responses to nutrient intake and energetic demand. Thus, the modulation of sirtuins might be a fruitful approach to reversing the age-related metabolic changes that could underlie tumorigenesis.
Of all the factors that contribute to cancer, aging is the most potent (Frank, 2007). More than 60% of all cancers occur in those aged 65 and above. Why is this so? The most common explanation is the “multi-hit,” or Knudson, hypothesis, which states that cancer occurs more frequently as we age because time is necessary for cells to accumulate sufficient genetic mutations to push them over a certain mutagenic threshold and into full-blown carcinogenesis (Knudson, 1971). What this hypothesis fails to adequately explain is why cancer risk is greatly reduced by calorie restriction (CR) and physical exercise, and why calorie overload and a sedentary lifestyle has the opposite effect (Ligibel, 2012). Restriction of calories to a level 70% of ad libitum intake, for example, can completely block tumor growth even in situations where chemical carcinogens would normally evoke a 100% penetrance of cancer (Lagopoulos and Stalder, 1987; Wallace, 2005). The accumulation of genomic mutations from external causes such as sunlight and mutagenic compounds might be expected to occur regardless of diet or physical activity. Here, we propose that it is not simply the time taken to accumulate genomic hits that accounts for the increased rate of cancer with age, but the decline in metabolic homeostasis and gene regulation that occurs normally as we age. This hypothesis is consistent with the strong association between cancer prevalence and type 2 diabetes (Giovannucci et al., 2010), obesity (Renehan et al., 2008), exercise (Ligibel, 2012), and small molecules that modulate energy utilization, such as resveratrol (Baur et al., 2006; Oberdoerffer et al., 2008) and metformin (Lee et al., 2011).
Mitochondrial Homeostasis: A Unifying Link between Tumorigenesis and the Aging Process?
Recently, it has been established that dysregulated cellular energetic pathways are not just coincident with tumorigenesis but are a hallmark of it (Vander Heiden et al., 2009). Known as the Warburg effect, cancer cells can reprogram carbon metabolism by reducing energy production from oxidative phosphorylation and upregulating glycolysis. This change in mitochondrial metabolism appears to be advantageous to cancer cells: reduced oxidative phosphorylation diverts glycolytic and tricarboxylic acid (TCA) cycle intermediates into biosynthetic pathways including nucleotide biosynthesis and de novo lipogenesis, which allows the biosynthesis of macromolecules and organelles required for the rapid cell growth and division characteristic of cancer (Vander Heiden et al., 2009). Moreover, disruption of mitochondrial homeostasis is usually correlated with increased reactive oxygen species (ROS), which are not only powerful damaging agents that can induce mutagenesis but can also function as signaling molecules that contribute to cancer progression (Hamanaka and Chandel, 2010).
Though it is clear that metabolic reprogramming is necessary to support tumor growth, it is less clear what drives the cell to rewire its metabolism in the first place. Some clues come from rare genetic diseases caused by mutations in metabolic regulators. Peutz-Jegher’s syndrome, for example, is characterized by an increased risk of cancer in the gastrointestinal tract, pancreas, cervix, ovary, and breast. The disease is caused by mutations in Lkb1, a kinase that regulates modulators of the Warburg effect such as the energy sensor AMP-dependent kinase (AMPK) and the regulator of growth and proliferation mammalian target of rapamycin (mTOR; Faubert et al., 2013; Sedelnikova et al., 2004). Pharmacological agents have also provided clues to the importance of metabolic reprogramming in cancer. For example, metformin, a drug that activates AMPK, lowers cancer risk (Lee et al., 2011) and mTOR inactivation is viewed as an effective therapy against cancer, with several mTOR inhibitors in clinical trials (Guertin and Sabatini, 2007).
Another illustrative example is von Hippel Lindau disease, characterized by tumors predominately in the central nervous system, retina, kidney, and pancreas. This disease is caused by a mutation in Vhl, which encodes an E3 ubiquitin ligase that targets the oxygen-sensitive hypoxia-inducible factor α (HIF-α) transcriptional regulatory complex for degradation (Maxwell et al., 1999). Without functional VHL, the tissues of von Hippel Lindau patients accumulate HIF-1α, which suppresses oxidative metabolism and promotes aerobic glycolysis, a metabolic shift commonly observed in cancer cells (Semenza, 2011). How HIF-1α promotes these changes is largely understood. By binding to specific promoters and sequestering other transcriptional activators such as c-Myc away from promoters, HIF-1α alters metabolism to favor cell growth (Simon, 2006). HIF-1α was originally thought to be a survival adaptation by cells within the hypoxic core of solid tumors, however it is emerging that HIF-1α is frequently activated in cancers independently of oxygen availability (Zhong et al., 1999).
Other pathways have been implicated in mediating the metabolic shift during cancer, including the tumor suppressor p53, which maintains transcription of cytochrome c oxidase subunits and subsequent functional respiration through synthesis of cytochrome c oxidase 2 (Matoba et al., 2006), and the proto-oncogene B-Raf, which regulates mitochondrial biogenesis through its regulation of peroxisome proliferator-activated receptor γ co-activator 1-α (PGC-1α) and AMPK (Faubert et al., 2013; Haq et al., 2013). The M2 isoform of pyruvate kinase has also been implicated as a regulator of the metabolic shift in cancer (Christofk et al., 2008), although conflicting results have been reported (Bluemlein et al., 2011).
The mechanisms underlying metabolic reprogramming in common cancers are still unclear. The prevailing view is that Warburg-like metabolic changes are genetic in origin, caused by mutations that accumulate in cells with high levels of genome instability (Vander Heiden et al., 2009). An alternative possibility is that the metabolic shift occurs early in a cancer cell lineage due to epigenetic changes during aging itself (Figure 1). In favor of this idea, aging in mammals is associated with a reduction in oxidative phosphorylation and a concomitant increase in aerobic glycolysis in many tissues including brain, liver, and muscle (Bowling et al., 1993; Hagen et al., 1997; Trounce et al., 1989). Old animals also have increased lactate levels both in tissues and in serum, a hallmark of increased glycolysis and reduced oxidative phosphorylation (Ross et al., 2010). Similarly, type 2 diabetes, a disease known to accelerate the rate of metabolic aging, is associated with a gene expression signature of glycolytic metabolism similar to that of hypoxia and HIF-1α accumulation (Ptitsyn et al., 2006) and a decline in oxidative phosphorylation (Petersen et al., 2004), paralleling the Warburg effect. Thus, in addition to oncogenic mutations, a shift toward Warburg metabolism during aging may be one of the “hits” required to push cells into carcinogenesis. This putative mechanism, which we refer to as “geroncogenesis,” may help explain why the greatest risk of carcinogenesis is age and why interventions that maintain metabolic health such as metformin and dietary restriction also prevent cancer.
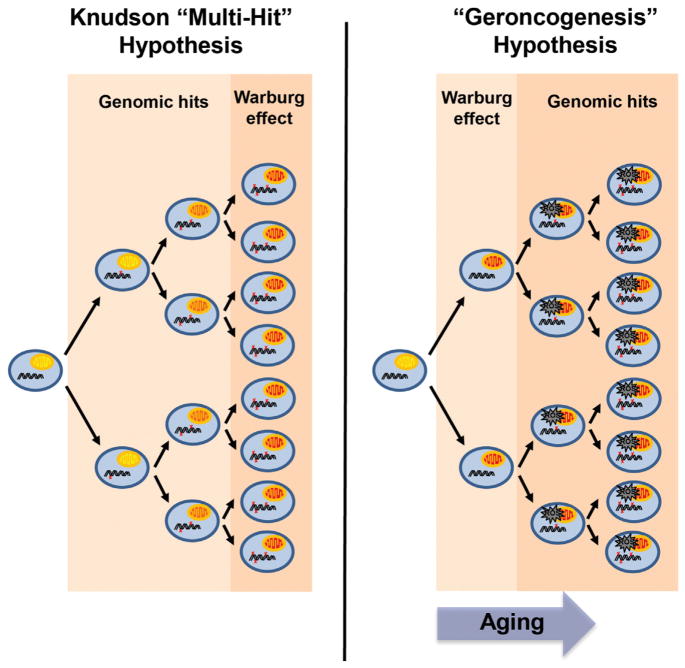
Figure 1. The Geroncogenesis Hypothesis: Aging-Induced Metabolic Decline as a Driver of Tumorigenesis.
According to the Knudson, or “multi-hit,” hypothesis, cancer occurs more frequently as we age because time is necessary for cells to acquire genetic mutations that drive carcinogenesis (indicated by red “x”). According to this model, the shift to oxidative glycolysis, known as the Warburg effect, occurs subsequent to these early hits. An alternative possibility is that the natural decline in oxidative metabolism as we age induces a Warburg-like metabolic state in normal tissues (indicated by red mitochondria). This increases the ROS production and sets the metabolic stage for later mutations to drive tumorigenesis. Low-calorie diets, exercise, and CR mimetic compounds delay this metabolic shift, thereby reducing the chance that oncogenic mutations will occur in a cell with optimal metabolism for tumorigenesis. The model predicts that CR mimetics could be used to reverse the metabolic reprogramming of tumors and even to prevent aging tissues from undergoing this switch in the first place.
If the natural decline in oxidative metabolism is a contributor to tumorigenesis, it is critical we understand why this decline occurs in the first place. As descendants of symbiotic α-proteobacteria, mitochondria maintain a separate genome that encodes tRNA and 13 subunits of the electron transport chain (Wallace, 2005). The mitochondrial genome is subjected to constant onslaught from ROS and is highly prone to mutation (Wallace, 2005). Accumulation of mitochondrial mutations is considered to be a major contributor to mitochondrial decline during aging (Trifunovic et al., 2004). Indeed, mice expressing error-prone mitochondrial DNA polymerase (Polgmut/mut) exhibit severe electron transport chain deficiencies along with a premature aging phenotype (Trifunovic et al., 2004). There is also some evidence that mitochondrial mutations can promote tumorigenesis: mitochondrial mutations are found in human cancers, and cybrid cell lines containing mitochondria with a point mutation in cytochrome oxidase I proliferate into tumors seven times the size of those in wild-type controls (Wallace, 2005).
Though mitochondrial mutations provide a satisfying explanation for aging, there are some confounding observations. Mice heterozygous for only one copy of the error-prone mitochondrial DNA polymerase (Polg+/mut) display a mitochondrial genome mutation rate over 500 times higher than that of normal aged mice but, unlike Polgmut/mut homozygous mice, display no change in lifespan (Vermulst et al., 2007). Also, the redundancy of having hundreds to thousands of mitochondria present in each cell, subject to continual fusion, fission, and mitophagy, allows for dysfunctional mitochondria to be quickly eliminated (Ono et al., 2001). A high mutation load therefore must be reached before major metabolic changes become apparent. Together, these findings suggest that mitochondrial mutations are but one part of a process that drives the metabolic shift during aging.
Over the past few years, a role for “epigenetic” alterations in age-related metabolic decline has become increasingly appreciated. There is evidence, for example, that metabolism becomes Warburg-like during old age because of a shift in balance between members of the lactate dehydrogenase complex to favor the production of lactate, resulting in diversion of pyruvate away from the TCA cycle and subsequent oxidative phosphorylation (Ross et al., 2010). The mechanism underlying these changes remains to be determined, and the importance of these changes to normal physiology has been the subject of debate (Quistorff and Grunnet, 2011). Another emerging idea is that the decline in metabolism during aging is due to a loss in activity of longevity regulators that are critical for the maintenance of cellular homeostasis. Central to this longevity regulation are the “sirtuins,” a seven-member family of nicotinamide adenine dinucleotide (NAD)+-dependent lysine deacylases.
Sirtuins: Relevance for Age-Induced Tumorigenesis and the Warburg Effect
The founding member of the sirtuin family was Sir2, a yeast transcriptional silencing protein that delays aging in response to low calorie intake (Lin et al., 2000). In mammals, the seven sirtuins (SIRT1–SIRT7) play key roles in the regulation of metabolism, inflammation, DNA repair, circadian rhythms, and aging. Sirtuins impart their effects largely via their catalytic activity, removing acyl-lysine moieties from proteins via a multi-step reaction that consumes NAD+ (Feldman et al., 2012). The number of lysine modifications that are known to be removed by sirtuins has grown in recent years to include acetyl-, succinyl-, malonyl-, and long-chain fatty acyl groups. Decreased activity of sirtuin family members during aging, especially SIRT1, SIRT3, and SIRT6, has been strongly implicated in the susceptibility of organs to aging and age-related diseases (Baur et al., 2006; Brown et al., 2013; Kanfi et al., 2012). A major cause of the decline in sirtuin activity is a decrease in NAD+ levels with age, a decline that is accelerated by obesity and counteracted by CR and physical activity (Koltai et al., 2010; Yang et al., 2007).
Over the past few years, it has become increasingly clear that the maintenance of sirtuin activity is likely to be critical for preventing tumorigenesis and slowing tumor growth in many tissues, but there are also studies indicating the opposite (Table 1). Mechanisms of tumor suppression by sirtuins initially focused on their ability to halt the cell cycle, inactivate oncogenic transcription factors, and promote DNA repair, but more recent studies have shown that their effects on energy metabolism may be equal, if not more important, for tumor suppression (Figure 2; Csibi et al., 2013; Finley et al., 2011; Firestein et al., 2008; Herranz et al., 2010; Jeong et al., 2013; Kim et al., 2010; Narayan et al., 2012; Oberdoerffer et al., 2008; Sebastián et al., 2012; Serrano et al., 2013). The sirtuin with the strongest known influence on tumorigenesis is SIRT3, a mitochondrial enzyme that regulates ROS production and enzymes that facilitate the TCA cycle, oxidative phosphorylation, and fatty acid metabolism (Finley et al., 2011; Hirschey et al., 2010). Deletion of Sirt3 results in chromosomal instability in vitro and causes spontaneous mammary tumorigenesis in mice, with metabolic changes that include increased glucose uptake, decreased ATP generation, and a metabolic reprogramming that parallels the Warburg effect (Kim et al., 2010). This metabolic switch is mediated by increased ROS that stabilize HIF-1α, thereby upregulating glycolysis and decreasing mitochondrial respiration (Bell et al., 2011; Finley et al., 2011). In one study, Sirt3 was deleted in 30% of breast cancer samples (Finley et al., 2011). Though these data are compelling, the tumor-suppressive role of SIRT3 is not clear cut: other studies indicate that Sirt3 is overexpressed in breast cancer compared to healthy mammary tissue (Alhazzazi et al., 2011b; Ashraf et al., 2006) and knockdown of this protein reduces tumor burden in an oral cancer model (Alhazzazi et al., 2011a).
Table 1.
Evidence for Sirtuins as Tumor Suppressors or Promoters
| Sirtuin | Examples of Tumor Suppression | Examples of Tumor Promotion |
|---|---|---|
| SIRT1 | Sirt1 overexpression suppresses hepatocellular carcinoma in diethylnitrosamine-treated mice (Herranz et al., 2010) | small molecule inhibition of SIRT1 reduces growth of transplanted Bcr-Abl chronic myeloid leukemia cells (Li et al., 2012) |
| Sirt1 overexpression suppresses colon cancer in ApcMin/+ mice (Firestein et al., 2008) | ||
| Sirt1 overexpression and resveratrol suppress lymphoma in irradiated p53+/− mice (Oberdoerffer et al., 2008) | SIRT1 inhibits the tumor suppressor p53 (Luo et al., 2001) | |
| tumorigenesis in Sirt1 knockout mice (Wang et al., 2008) | Sirt1 overexpression promotes thyroid tumorigenesis in Pten+/− mice (Herranz et al., 2013) | |
| SIRT1 promotes degradation of c-Myc; overexpression represses colony formation in HO15 and Myc3 Rat1 cells (Yuan et al., 2009) | SIRT1 promotes stabilization of c-Myc; partial reduction in proliferation of Myc transformed U937 monoblasts in vitro (Menssen et al., 2012) | |
| SIRT1 allosteric activator resveratrol suppresses DMBA-induced skin cancer (Jang et al., 1997) | Sirt1 overexpression increases tumor growth in orthotopic xenografted hereditary colon cancer cell lines (Portmann et al., 2013) | |
| SIRT2 | Sirt2 deletion causes spontaneous tumorigenesis in mice (Narayan et al., 2012) | SIRT2 inhibition reduces proliferation of BE(2)-C and MiaPaca cell lines in vitro (Liu et al., 2013) |
| Sirt2 deletion causes spontaneous tumorigenesis in mice (Serrano et al., 2013) | ||
| SIRT3 | Sirt3 knockdown increases, and overexpression reduces tumor size in orthotopic xenografts (Bell et al., 2011) | Sirt3 knockdown increases tumor burden in orthotopic xenograft tumor model (Alhazzazi et al., 2011a) |
| Increased soft agar colony formation in knockout MEF cells, Sirt3 deletion in 30% of breast cancers (Finley et al., 2011) | increased Sirt3 expression in node positive breast cancer (Ashraf et al., 2006) | |
| tumor formation in Myc, Ras transformed immortalized MEF cells (Kim et al., 2010) | ||
| SIRT4 | decreased tumor growth in orthotopic immortalized Tsc2−/− MEF cells overexpressing Sirt4 (Csibi et al., 2013) | no strong evidence |
| increased tumor growth in immortalized Sirt4−/− MEF cells (Jeong et al., 2013) | ||
| SIRT6 | increased tumorigenesis in immortalized Sirt6−/− MEF cells (Sebastián et al., 2012) | no strong evidence |
| Sirt6 overexpression induces apoptosis in cancer cells but not normal cells in vitro (Van Meter et al., 2011) | ||
| SIRT7 | SIRT7 negatively regulates HIF-1α and HIF-2α (Hubbi et al., 2013), potentially underlying aspects of the Warburg effect | decreased growth of U251 xenografts with Sirt7 knockdown (Barber et al., 2012) |
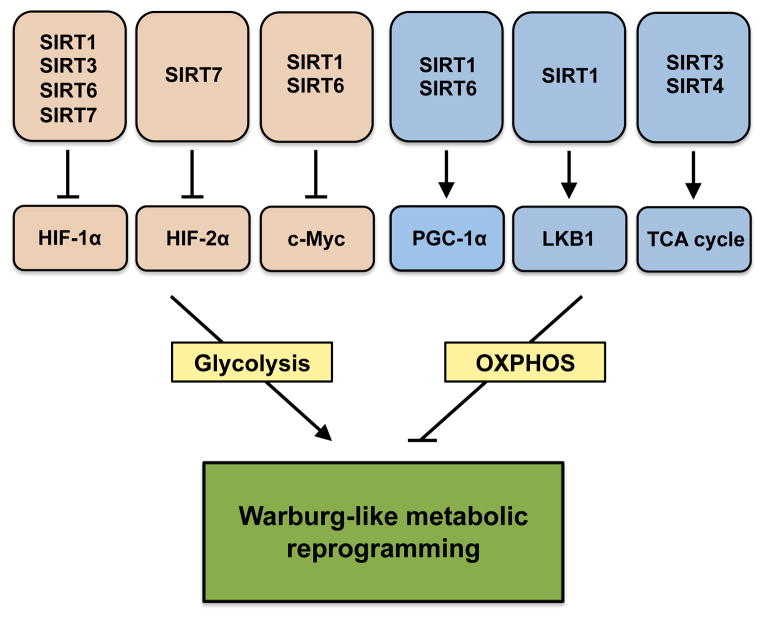
Figure 2. Sirtuins Are Central to Metabolic Reprogramming during Aging and Cancer.
Sirtuins control key nodes in the regulation of glycolysis (HIF-1α, HIF-2α, and c-Myc) and oxidative phosphorylation (PGC-1α, LKB1, and TCA enzymes). A decline in the activity of the sirtuin family of enzymes in old age is hypothesized to lead to a shift toward a predominantly glycolytic, Warburg-like metabolism that could contribute to the exponential increase in cancer susceptibility during aging.
Another sirtuin implicated in tumorigenesis is SIRT6, a chromatin-associated enzyme with deacetylase and long-chain deacylase activities. Sirt6 deletion increases HIF-1α and c-Myc transcriptional activity, with a corresponding upregulation of glycolysis (Sebastián et al., 2012; Zhong et al., 2010). Remarkably, knockdown of Sirt6 in otherwise normal mouse embryonic fibroblasts (MEFs) transforms them, independently of activation of known oncogenes (Sebastián et al., 2012). Although SIRT6 is required for genomic stability (Mostoslavsky et al., 2006), re-introduction of Sirt6 into knockout MEF cells in which genomic instability might already have been expected to take place represses tumor formation, effectively ruling out mutations as a cause (Sebastián et al., 2012). These findings further underscore the idea that metabolic alterations are required, if not sufficient, to induce tumor growth.
SIRT1, a nuclear sirtuin, was the first family member shown to act as a tumor suppressor. Pharmacological activation or genetic overexpression of Sirt1 increases genomic stability in cells treated with DNA-damaging agents, delays lymphoma, and improves the survival of irradiated p53+/− mice (Oberdoerffer et al., 2008), while Sirt1 deletion has the opposite effect (Wang et al., 2008). By localizing to sites of DNA damage and facilitating the recruitment of DNA repair factors such as histone deacetylase 1, Rad51, and Nbs1, SIRT1 plays a key role in promoting genome stability, a function that declines with age (Dobbin et al., 2013; Oberdoerffer et al., 2008). One of the strongest effects of SIRT1 in vivo is its ability to protect mice in the heterogeneous diethylnitrosamine-induced model of hepatocellular carcinoma (Herranz et al., 2010), potentially by suppressing inflammatory responses in this organ. SIRT1 can also suppress tumorigenesis by negatively regulating oncogenic transcription factors, including β-catenin (Firestein et al., 2008) and c-Myc (Yuan et al., 2009), though opposing findings for Myc have been reported (Menssen et al., 2012).
Similar to SIRT3 and SIRT6, SIRT1 might influence tumorigenesis, not only through its ability to regulate genomic stability, but also by regulating cellular metabolism. SIRT1 regulates the transcriptional activity of HIF-1α (Lim et al., 2010), which is also an important regulator of the Warburg effect as well as angiogenesis and metastasis. SIRT1 also deacetylates and activates liver kinase B1 (LKB1; Lan et al., 2008), a known tumor suppressor that regulates mTOR and AMPK (Sedelnikova et al., 2004). Interestingly, the effects of SIRT1 on tumorigenesis are context dependent. For example, inhibition of SIRT1 improves the efficacy of a chemotherapeutic agent (Imatinib) against chronic myeloid leukemia (Li et al., 2012) and blocks the proliferation of hepatocellular carcinoma cell lines in vitro and in a xenograft model (Portmann et al., 2013). Conversely, Sirt1 overexpression can accelerate thyroid cancers in vivo (Herranz et al., 2013). These latter findings likely reflect the ability of SIRT1 to inhibit the tumor suppressor p53, which promotes survival under situations of cell stress (Luo et al., 2001).
There is evidence that SIRT2, the cytosolic sirtuin, is also a tumor suppressor. Deletion of Sirt2 results in spontaneous tumorigenesis in the liver and accelerates the 7,12-dimethyl-benz(a)anthracene (DMBA)/12-O-tetradecanoylphorbol-13-acetate model of skin cancer (Narayan et al., 2012; Serrano et al., 2013). One mechanism is likely to be cell cycle control, as SIRT2 deacetylates and regulates CDH1 and CDC20, members of the anaphase-promoting complex (Narayan et al., 2012). Moreover, SIRT2 transiently migrates to the nucleus during mitosis (North and Verdin, 2007), where it modulates the activity of the methyltransferase PR-Set7, resulting in H4K20 methylation (Serrano et al., 2013), a chromatin mark involved in genomic stability (Oda et al., 2009). Although primarily studied in the context of its cytosolic regulation of cell cycle, one interesting possibility is that SIRT2 influences mitochondrial function and the Warburg effect by deacetylating CDH1, a protein that limits glycolysis and proliferation of cancer cell lines through ubiquitination and degradation of the glycolysis-promoting enzyme 6-phosphofructo-2-kinase (Almeida et al., 2010). Again, the data are not clear cut; an in vitro study found that SIRT2 knockdown or small molecule inhibition reduced neuroblastoma cell growth through stabilization of Myc oncoproteins (Liu et al., 2013).
Recently, two other sirtuins, SIRT4 and SIRT7, have also been implicated in the regulation of tumorigenesis. The mitochondrial sirtuin SIRT4 promotes metabolic reprogramming by facilitating the cataplerotic diversion of carbons from the TCA cycle to aerobic glycolysis and lactate generation, forcing cancer cells to rely on glutamine for replenishment of the TCA cycle (Csibi et al., 2013; Jeong et al., 2013). Upon DNA damage, Sirt4 expression is upregulated, leading to a repression of glutamine metabolism through its inhibition of glutamate dehydrogenase, which converts glutamate into α-ketoglutarate (Csibi et al., 2013; Jeong et al., 2013). This shift prevents the cell from upregulating nonessential biosynthetic pathways and undergoing premature cellular division prior to genomic repair (Csibi et al., 2013; Jeong et al., 2013). The nucleolar sirtuin, SIRT7, may also regulate cellular metabolism by negatively regulating HIF-1α and HIF-2α (Hubbi et al., 2013), potentially underlying the Warburg effect. The role of SIRT7 in tumorigenesis, however, also seems context dependent: SIRT7 may help maintain a pro-oncogenic phenotype by interacting with the transcription factor ELK4 and deacetylating H3-K18, a modification that promotes tumor growth (Barber et al., 2012).
Therapeutic Strategies to Combat Age-Induced Tumorigenesis
Mutations that give rise to cancer are essentially irreversible. However, if age-related metabolic changes are an early driver of tumorigenesis, molecules that prevent and reverse metabolic aging may be useful as cancer therapies. Indeed, molecules such as metformin and HIF-1α inhibitors in development show promise as anticancer agents (Onnis et al., 2009). Given the key role of sirtuins in tumorigenesis, it is feasible that lifestyle interventions and/or small molecules that activate sirtuins could induce a youthful metabolic state and serve to prevent and treat cancer. Direct SIRT1 activators that work by allosteric mechanisms (Hubbard et al., 2013) have been developed. These molecules are in human clinical trials to treat metabolic and inflammatory diseases, but they are not yet under investigation as chemotherapy adjuncts.
An alternative approach to activating sirtuins, one that raises the activity of the entire family of enzymes, is to exploit their common requirement for NAD+. Increasing NAD+ levels has been shown to protect mice from metabolic decline in mouse models of obesity and aging (Cantó et al., 2012; Escande et al., 2013; Yoshino et al., 2011), but it is not yet considered a viable strategy for cancer, in part because raising overall NAD+ levels is not without risks. As described above, there is evidence that SIRT1 and SIRT7 may promote the growth of cancers. It is also important to consider the role of NAD+ as a redox carrier that is essential to glycolysis, which cancer cells heavily rely upon. Another consideration is the fact that raising NAD+ may not be as simple as it sounds. NAD+ is compartmentalized into cytosolic, nuclear, and mitochondrial pools (Nikiforov et al., 2011), and it is unclear what degree of flux exists between these pools and whether a particular pool of NAD+ influences the Warburg effect and tumorigenesis.
Some researchers have taken the opposite approach, seizing upon the fact that NAD+ is an essential redox carrier for glycolysis and cancer cell viability. FK866, a small molecule inhibitor of the NAD+ recycling enzyme Nampt, has strong antiproliferative effects in cancer cells and has entered clinical trials (Hasmann and Schemainda, 2003). Of course, NAD+ depletion can also be toxic to normal cells. If the approach does prove effective as a chemotherapeutic strategy, it will be important to optimize dosing to avoid toxicity to noncancerous cell types. Clearly, more work is required to determine whether raising or decreasing NAD+ levels will be beneficial for the treatment of cancer in humans and which specific pools of NAD+ influence tumorigenesis.
Conclusions and Future Perspectives
While the “multi-hit” concept of oncogenic DNA mutations has dominated cancer biology for the past few decades, aberrant tumor metabolism and disruption of mitochondrial homeostasis are emerging as key drivers of both initiation and progression of cancer. Metabolic reprogramming is currently viewed as a late stage of tumorigenesis, the result of mutations that accumulate over time. Here, we propose that one of the early drivers of tumorigenesis is aging-induced dysregulation of mitochondrial metabolism. This may be driven, in part, by an age-related decline in NAD+ and sirtuin activity. These metabolic changes likely occur independently of, and may even precede, genomic lesions, acting as an early “hit” that pushes cells toward complete cellular transformation. If seen though this lens, aging induces a gradual reprogramming of metabolism toward a “cancer-like” state, a transformation that is accelerated by increased calorie intake and a sedentary lifestyle and that is counteracted by low-calorie diets and exercise. This may explain why diet, exercise, and CR mimetics, which alter the pace of metabolic aging, strongly influence cancer susceptibility (Baur et al., 2006; Lee et al., 2011; Ligibel, 2012; Oberdoerffer et al., 2008; Renehan et al., 2008). If this hypothesis holds true, there may come a day when lifestyle interventions along with CR mimetics are used to reverse the metabolic reprogramming of tumors and even to prevent aging tissues from undergoing the metabolic switch in the first place.
Acknowledgments
D.A.S. is a consultant to Segterra and Horizon Science and a consultant to and inventor on patents licensed to GlaxoSmithKline, Ovascience, and Metrobiotech. L.E.W. is supported by an Early Career Fellowship from Cancer Institute New South Wales (CINSW). We are grateful for support from the Glenn Foundation for Medical Research, the Juvenile Diabetes Research Foundation, the United Mitochondrial Disease Foundation, Edward Schulak, the National Health and Medical Research Council of Australia, and the National Institutes of Health.
References
- Alhazzazi TY, Kamarajan P, Joo N, Huang JY, Verdin E, D’Silva NJ, Kapila YL. Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer. 2011a;117:1670–1678. doi: 10.1002/cncr.25676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhazzazi TY, Kamarajan P, Verdin E, Kapila YL. SIRT3 and cancer: tumor promoter or suppressor? Biochim Biophys Acta. 2011b;1816:80–88. doi: 10.1016/j.bbcan.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida A, Bolaños JP, Moncada S. E3 ubiquitin ligase APC/C-Cdh1 accounts for the Warburg effect by linking glycolysis to cell proliferation. Proc Natl Acad Sci USA. 2010;107:738–741. doi: 10.1073/pnas.0913668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf N, Zino S, Macintyre A, Kingsmore D, Payne AP, George WD, Shiels PG. Altered sirtuin expression is associated with node-positive breast cancer. Br J Cancer. 2006;95:1056–1061. doi: 10.1038/sj.bjc.6603384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K, et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487:114–118. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30:2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluemlein K, Grüning NM, Feichtinger RG, Lehrach H, Kofler B, Ralser M. No evidence for a shift in pyruvate kinase PKM1 to PKM2 expression during tumorigenesis. Oncotarget. 2011;2:393–400. doi: 10.18632/oncotarget.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling AC, Mutisya EM, Walker LC, Price DL, Cork LC, Beal MF. Age-dependent impairment of mitochondrial function in primate brain. J Neurochem. 1993;60:1964–1967. doi: 10.1111/j.1471-4159.1993.tb13430.x. [DOI] [PubMed] [Google Scholar]
- Brown K, Xie S, Qiu X, Mohrin M, Shin J, Liu Y, Zhang D, Scadden DT, Chen D. SIRT3 reverses aging-associated degeneration. Cell Rep. 2013;3:319–327. doi: 10.1016/j.celrep.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, Fernandez-Marcos PJ, Yamamoto H, Andreux PA, Cettour-Rose P, et al. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Csibi A, Fendt SM, Li C, Poulogiannis G, Choo AY, Chapski DJ, Jeong SM, Dempsey JM, Parkhitko A, Morrison T, et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. 2013;153:840–854. doi: 10.1016/j.cell.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbin MM, Madabhushi R, Pan L, Chen Y, Kim D, Gao J, Ahanonu B, Pao PC, Qiu Y, Zhao Y, Tsai LH. SIRT1 collaborates with ATM and HDAC1 to maintain genomic stability in neurons. Nat Neurosci. 2013;16:1008–1015. doi: 10.1038/nn.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escande C, Nin V, Price NL, Capellini V, Gomes AP, Barbosa MT, O’Neil L, White TA, Sinclair DA, Chini EN. Flavonoid apigenin is an inhibitor of the NAD+ ase CD38: implications for cellular NAD+ metabolism, protein acetylation, and treatment of metabolic syndrome. Diabetes. 2013;62:1084–1093. doi: 10.2337/db12-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubert B, Boily G, Izreig S, Griss T, Samborska B, Dong Z, Dupuy F, Chambers C, Fuerth BJ, Viollet B, et al. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab. 2013;17:113–124. doi: 10.1016/j.cmet.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman JL, Dittenhafer-Reed KE, Denu JM. Sirtuin catalysis and regulation. J Biol Chem. 2012;287:42419–42427. doi: 10.1074/jbc.R112.378877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S, de Cabo R, Fuchs C, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS ONE. 2008;3:e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SA. In Dynamics of Cancer: Incidence, Inheritance, and Evolution. Princeton, NJ: Princeton University Press; 2007. Age of cancer incidence; pp. 17–35. [PubMed] [Google Scholar]
- Giovannucci E, Harlan DM, Archer MC, Bergenstal RM, Gapstur SM, Habel LA, Pollak M, Regensteiner JG, Yee D. Diabetes and cancer: a consensus report. Diabetes Care. 2010;33:1674–1685. doi: 10.2337/dc10-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Hagen TM, Yowe DL, Bartholomew JC, Wehr CM, Do KL, Park JY, Ames BN. Mitochondrial decay in hepatocytes from old rats: membrane potential declines, heterogeneity and oxidants increase. Proc Natl Acad Sci USA. 1997;94:3064–3069. doi: 10.1073/pnas.94.7.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka RB, Chandel NS. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem Sci. 2010;35:505–513. doi: 10.1016/j.tibs.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, Frederick DT, Hurley AD, Nellore A, Kung AL, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell. 2013;23:302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasmann M, Schemainda I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 2003;63:7436–7442. [PubMed] [Google Scholar]
- Herranz D, Muñoz-Martin M, Cañamero M, Mulero F, Martinez-Pastor B, Fernandez-Capetillo O, Serrano M. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun. 2010;1:3. doi: 10.1038/ncomms1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz D, Maraver A, Cañamero M, Gómez-López G, Inglada-Pérez L, Robledo M, Castelblanco E, Matias-Guiu X, Serrano M. SIRT1 promotes thyroid carcinogenesis driven by PTEN deficiency. Oncogene. 2013;32:4052–4056. doi: 10.1038/onc.2012.407. [DOI] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard BP, Gomes AP, Dai H, Li J, Case AW, Considine T, Riera TV, Lee JE, ESY, Lamming DW, et al. Evidence for a common mechanism of SIRT1 regulation by allosteric activators. Science. 2013;339:1216–1219. doi: 10.1126/science.1231097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbi ME, Hu H, Kshitiz Gilkes DM, Semenza GL. Sirtuin-7 inhibits the activity of hypoxia-inducible factors. J Biol Chem. 2013;288:20768–20775. doi: 10.1074/jbc.M113.476903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang M, Cai L, Udeani GO, Slowing KV, Thomas CF, Beecher CW, Fong HH, Farnsworth NR, Kinghorn AD, Mehta RG, et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275:218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- Jeong SM, Xiao C, Finley LW, Lahusen T, Souza AL, Pierce K, Li YH, Wang X, Laurent G, German NJ, et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell. 2013;23:450–463. doi: 10.1016/j.ccr.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Bar-Joseph Z, Cohen HY. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltai E, Szabo Z, Atalay M, Boldogh I, Naito H, Goto S, Nyakas C, Radak Z. Exercise alters SIRT1, SIRT6, NAD and NAMPT levels in skeletal muscle of aged rats. Mech Ageing Dev. 2010;131:21–28. doi: 10.1016/j.mad.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagopoulos L, Stalder R. The influence of food intake on the development of diethylnitrosamine-induced liver tumours in mice. Carcinogenesis. 1987;8:33–37. doi: 10.1093/carcin/8.1.33. [DOI] [PubMed] [Google Scholar]
- Lan F, Cacicedo JM, Ruderman N, Ido Y. SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1. Possible role in AMP-activated protein kinase activation. J Biol Chem. 2008;283:27628–27635. doi: 10.1074/jbc.M805711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Hsu CC, Wahlqvist ML, Tsai HN, Chang YH, Huang YC. Type 2 diabetes increases and metformin reduces total, colorectal, liver and pancreatic cancer incidences in Taiwanese: a representative population prospective cohort study of 800,000 individuals. BMC Cancer. 2011;11:20. doi: 10.1186/1471-2407-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang L, Li L, Wang Z, Ho Y, McDonald T, Holyoake TL, Chen W, Bhatia R. Activation of p53 by SIRT1 inhibition enhances elimination of CML leukemia stem cells in combination with imatinib. Cancer Cell. 2012;21:266–281. doi: 10.1016/j.ccr.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligibel J. Lifestyle factors in cancer survivorship. J Clin Oncol. 2012;30:3697–3704. doi: 10.1200/JCO.2012.42.0638. [DOI] [PubMed] [Google Scholar]
- Lim JH, Lee YM, Chun YS, Chen J, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol Cell. 2010;38:864–878. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Liu PY, Xu N, Malyukova A, Scarlett CJ, Sun YT, Zhang XD, Ling D, Su SP, Nelson C, Chang DK, et al. The histone deacetylase SIRT2 stabilizes Myc oncoproteins. Cell Death Differ. 2013;20:503–514. doi: 10.1038/cdd.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Menssen A, Hydbring P, Kapelle K, Vervoorts J, Diebold J, Lüscher B, Larsson LG, Hermeking H. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc Natl Acad Sci USA. 2012;109:E187–E196. doi: 10.1073/pnas.1105304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Narayan N, Lee IH, Borenstein R, Sun J, Wong R, Tong G, Fergusson MM, Liu J, Rovira II, Cheng HL, et al. The NAD-dependent de-acetylase SIRT2 is required for programmed necrosis. Nature. 2012;492:199–204. doi: 10.1038/nature11700. [DOI] [PubMed] [Google Scholar]
- Nikiforov A, Dölle C, Niere M, Ziegler M. Pathways and sub-cellular compartmentation of NAD biosynthesis in human cells: from entry of extracellular precursors to mitochondrial NAD generation. J Biol Chem. 2011;286:21767–21778. doi: 10.1074/jbc.M110.213298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Verdin E. Interphase nucleocytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS ONE. 2007;2:e784. doi: 10.1371/journal.pone.0000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, Torres-Padilla ME, Heard E, Reinberg D. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol. 2009;29:2278–2295. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onnis B, Rapisarda A, Melillo G. Development of HIF-1 inhibitors for cancer therapy. J Cell Mol Med. 2009;13:2780–2786. doi: 10.1111/j.1582-4934.2009.00876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Isobe K, Nakada K, Hayashi JI. Human cells are protected from mitochondrial dysfunction by complementation of DNA products in fused mitochondria. Nat Genet. 2001;28:272–275. doi: 10.1038/90116. [DOI] [PubMed] [Google Scholar]
- Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med. 2004;350:664–671. doi: 10.1056/NEJMoa031314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portmann S, Fahrner R, Lechleiter A, Keogh A, Overney S, Laemmle A, Mikami K, Montani M, Tschan MP, Candinas D, Stroka D. Antitumor effect of SIRT1 inhibition in human HCC tumor models in vitro and in vivo. Mol Cancer Ther. 2013;12:499–508. doi: 10.1158/1535-7163.MCT-12-0700. [DOI] [PubMed] [Google Scholar]
- Ptitsyn A, Hulver M, Cefalu W, York D, Smith SR. Unsupervised clustering of gene expression data points at hypoxia as possible trigger for metabolic syndrome. BMC Genomics. 2006;7:318. doi: 10.1186/1471-2164-7-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quistorff B, Grunnet N. The isoenzyme pattern of LDH does not play a physiological role; except perhaps during fast transitions in energy metabolism. Aging (Albany NY) 2011;3:457–460. doi: 10.18632/aging.100329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renehan AG, Tyson M, Egger M, Heller RF, Zwahlen M. Body-mass index and incidence of cancer: a systematic review and meta-analysis of prospective observational studies. Lancet. 2008;371:569–578. doi: 10.1016/S0140-6736(08)60269-X. [DOI] [PubMed] [Google Scholar]
- Ross JM, Öberg J, Brené S, Coppotelli G, Terzioglu M, Pernold K, Goiny M, Sitnikov R, Kehr J, Trifunovic A, et al. High brain lactate is a hallmark of aging and caused by a shift in the lactate dehydrogenase A/B ratio. Proc Natl Acad Sci USA. 2010;107:20087–20092. doi: 10.1073/pnas.1008189107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastián C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151:1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedelnikova OA, Horikawa I, Zimonjic DB, Popescu NC, Bonner WM, Barrett JC. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat Cell Biol. 2004;6:168–170. doi: 10.1038/ncb1095. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Regulation of metabolism by hypoxia-inducible factor 1. Cold Spring Harb Symp Quant Biol. 2011;76:347–353. doi: 10.1101/sqb.2011.76.010678. [DOI] [PubMed] [Google Scholar]
- Serrano L, Martínez-Redondo P, Marazuela-Duque A, Vazquez BN, Dooley SJ, Voigt P, Beck DB, Kane-Goldsmith N, Tong Q, Rabanal RM, et al. The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev. 2013;27:639–653. doi: 10.1101/gad.211342.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MC. Coming up for air: HIF-1 and mitochondrial oxygen consumption. Cell Metab. 2006;3:150–151. doi: 10.1016/j.cmet.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly-Y M, Gidlöf S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Trounce I, Byrne E, Marzuki S. Decline in skeletal muscle mitochondrial respiratory chain function: possible factor in ageing. Lancet. 1989;1:637–639. doi: 10.1016/s0140-6736(89)92143-0. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meter M, Mao Z, Gorbunova V, Seluanov A. SIRT6 overexpression induces massive apoptosis in cancer cells but not in normal cells. Cell Cycle. 2011;10:3153–3158. doi: 10.4161/cc.10.18.17435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermulst M, Bielas JH, Kujoth GC, Ladiges WC, Rabinovitch PS, Prolla TA, Loeb LA. Mitochondrial point mutations do not limit the natural lifespan of mice. Nat Genet. 2007;39:540–543. doi: 10.1038/ng1988. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino J, Mills KF, Yoon MJ, Imai S. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Minter-Dykhouse K, Lou Z. A c-Myc-SIRT1 feedback loop regulates cell growth and transformation. J Cell Biol. 2009;185:203–211. doi: 10.1083/jcb.200809167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, Simons JW. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59:5830–5835. [PubMed] [Google Scholar]
- Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]