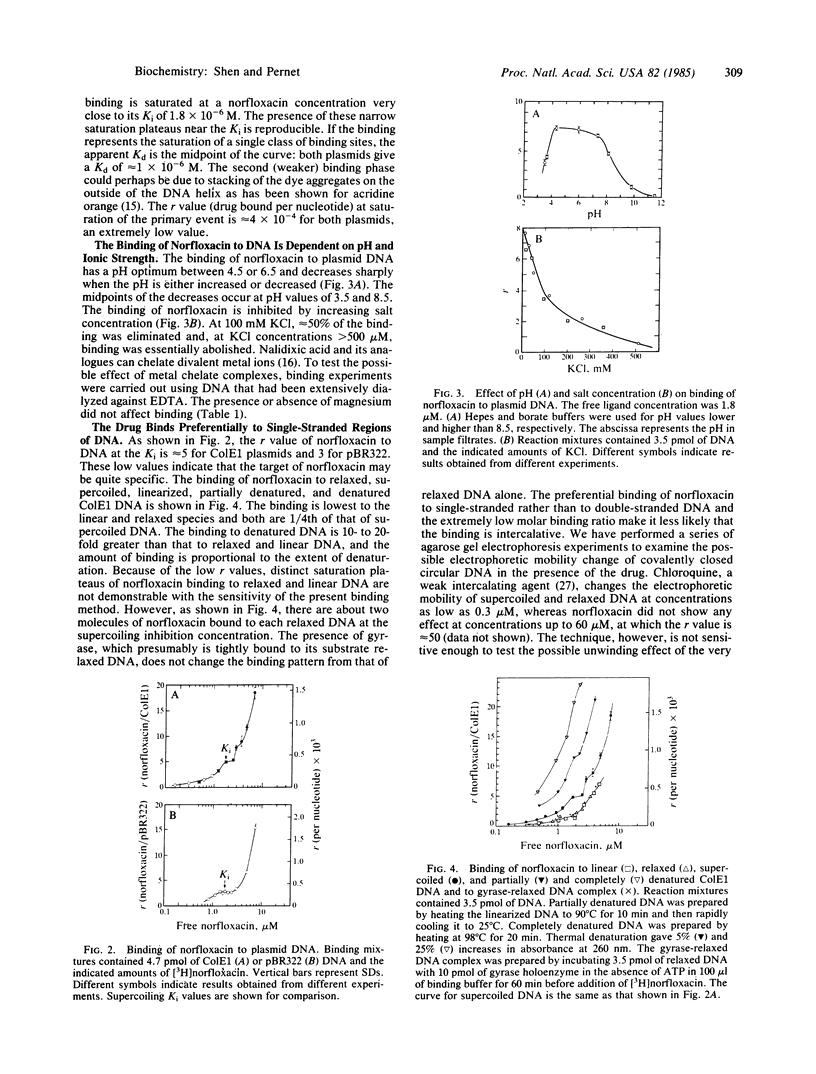
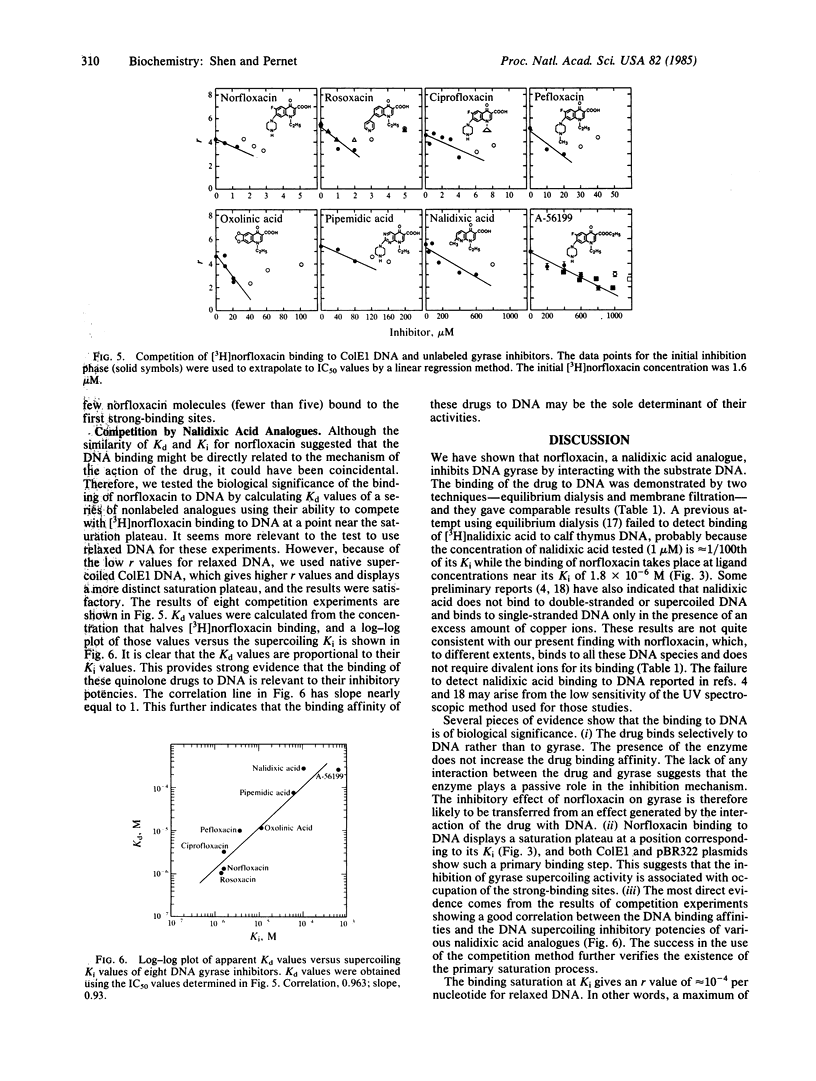
Abstract
Norfloxacin is a nalidixic acid analogue and one of the most potent DNA gyrase inhibitors. To study the mechanism of this important class of inhibitors, the binding of [3H]norfloxacin to gyrase and substrate DNA was measured. We found that, contrary to prior belief, norfloxacin does not bind to gyrase but instead binds to DNA. This was demonstrated by both equilibrium dialysis and membrane filtration techniques. Binding to ColE1 and pBR322 plasmids showed a primary process that is saturated at a norfloxacin concentration about equal to its supercoiling Ki (1.8 X 10(-6) M) and is followed by weaker secondary binding. The apparent Kd values are 1 X 10(-6) M for both plasmids. The molar binding ratio at this initial saturation point is extremely low: only 4 X 10(-4) norfloxacin per nucleotide for both plasmids. The binding of norfloxacin to DNA plasmids is nonintercalative, as shown by the fact that the drug binds preferentially to single-stranded DNA rather than to double-stranded DNA. The binding is reduced at high salt concentration, has a pH optimum between 4.5 and 6.5, and does not require divalent ions. The binding affinities of other nalidixic acid analogues were estimated by an indirect competition method. The calculated apparent Kd values of these analogues correlate well with their Ki values, providing strong evidence that the binding affinity of the drug to DNA determines biological potency.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bourguignon G. J., Levitt M., Sternglanz R. Studies on the mechanism of action of nalidixic acid. Antimicrob Agents Chemother. 1973 Oct;4(4):479–486. doi: 10.1128/aac.4.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Prusoff W. H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973 Dec 1;22(23):3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- Cozzarelli N. R. DNA gyrase and the supercoiling of DNA. Science. 1980 Feb 29;207(4434):953–960. doi: 10.1126/science.6243420. [DOI] [PubMed] [Google Scholar]
- Cozzarelli N. R. The mechanism of action of inhibitors of DNA synthesis. Annu Rev Biochem. 1977;46:641–668. doi: 10.1146/annurev.bi.46.070177.003233. [DOI] [PubMed] [Google Scholar]
- Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- Gellert M., Fisher L. M., Ohmori H., O'Dea M. H., Mizuuchi K. DNA gyrase: site-specific interactions and transient double-strand breakage of DNA. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):391–398. doi: 10.1101/sqb.1981.045.01.053. [DOI] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Itoh T., Tomizawa J. I. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins N. P., Cozzarelli N. R. The binding of gyrase to DNA: analysis by retention by nitrocellulose filters. Nucleic Acids Res. 1982 Nov 11;10(21):6833–6847. doi: 10.1093/nar/10.21.6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins N. P., Peebles C. L., Sugino A., Cozzarelli N. R. Purification of subunits of Escherichia coli DNA gyrase and reconstitution of enzymatic activity. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1773–1777. doi: 10.1073/pnas.75.4.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LESHER G. Y., FROELICH E. J., GRUETT M. D., BAILEY J. H., BRUNDAGE R. P. 1,8-NAPHTHYRIDINE DERIVATIVES. A NEW CLASS OF CHEMOTHERAPEUTIC AGENTS. J Med Pharm Chem. 1962 Sep;91:1063–1065. doi: 10.1021/jm01240a021. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Liu C. C., Alberts B. M. T4 DNA topoisomerase: a new ATP-dependent enzyme essential for initiation of T4 bacteriophage DNA replication. Nature. 1979 Oct 11;281(5731):456–461. doi: 10.1038/281456a0. [DOI] [PubMed] [Google Scholar]
- Lother H., Lurz R., Orr E. DNA binding and antigenic specifications of DNA gyrase. Nucleic Acids Res. 1984 Jan 25;12(2):901–914. doi: 10.1093/nar/12.2.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus G., Priore R. L., Wissler F. C. The binding of tranexamic acid to native (Glu) and modified (Lys) human plasminogen and its effect on conformation. J Biol Chem. 1979 Feb 25;254(4):1211–1216. [PubMed] [Google Scholar]
- Morrison A., Cozzarelli N. R. Contacts between DNA gyrase and its binding site on DNA: features of symmetry and asymmetry revealed by protection from nucleases. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1416–1420. doi: 10.1073/pnas.78.3.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A., Cozzarelli N. R. Site-specific cleavage of DNA by E. coli DNA gyrase. Cell. 1979 May;17(1):175–184. doi: 10.1016/0092-8674(79)90305-2. [DOI] [PubMed] [Google Scholar]
- Nelson E. M., Tewey K. M., Liu L. F. Mechanism of antitumor drug action: poisoning of mammalian DNA topoisomerase II on DNA by 4'-(9-acridinylamino)-methanesulfon-m-anisidide. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1361–1365. doi: 10.1073/pnas.81.5.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otter R., Cozzarelli N. R. Escherichia coli DNA gyrase. Methods Enzymol. 1983;100:171–180. doi: 10.1016/0076-6879(83)00053-1. [DOI] [PubMed] [Google Scholar]
- Pedrini A. M., Ciarrocchi G. Inhibition of Micrococcus luteus DNA topoisomerase I by UV photoproducts. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1787–1791. doi: 10.1073/pnas.80.7.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudenbauer W. L., Orr E. DNA gyrase: affinity chromatography on novobiocin-Sepharose and catalytic properties. Nucleic Acids Res. 1981 Aug 11;9(15):3589–3603. doi: 10.1093/nar/9.15.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers K., Sternglanz R. Ionization and divalent cation dissociation constants of nalidixic and oxolinic acids. Bioinorg Chem. 1978 Aug;9(2):145–155. doi: 10.1016/s0006-3061(00)80286-0. [DOI] [PubMed] [Google Scholar]
- Waring M. Variation of the supercoils in closed circular DNA by binding of antibiotics and drugs: evidence for molecular models involving intercalation. J Mol Biol. 1970 Dec 14;54(2):247–279. doi: 10.1016/0022-2836(70)90429-8. [DOI] [PubMed] [Google Scholar]


