Abstract
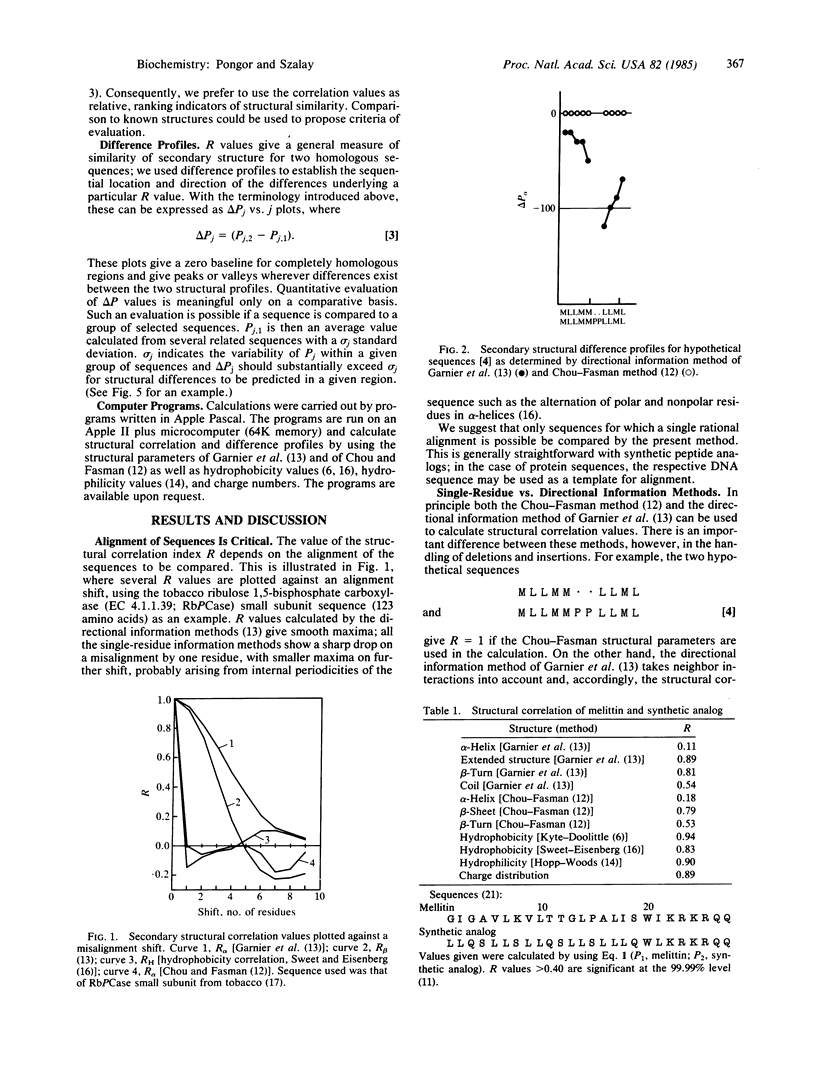
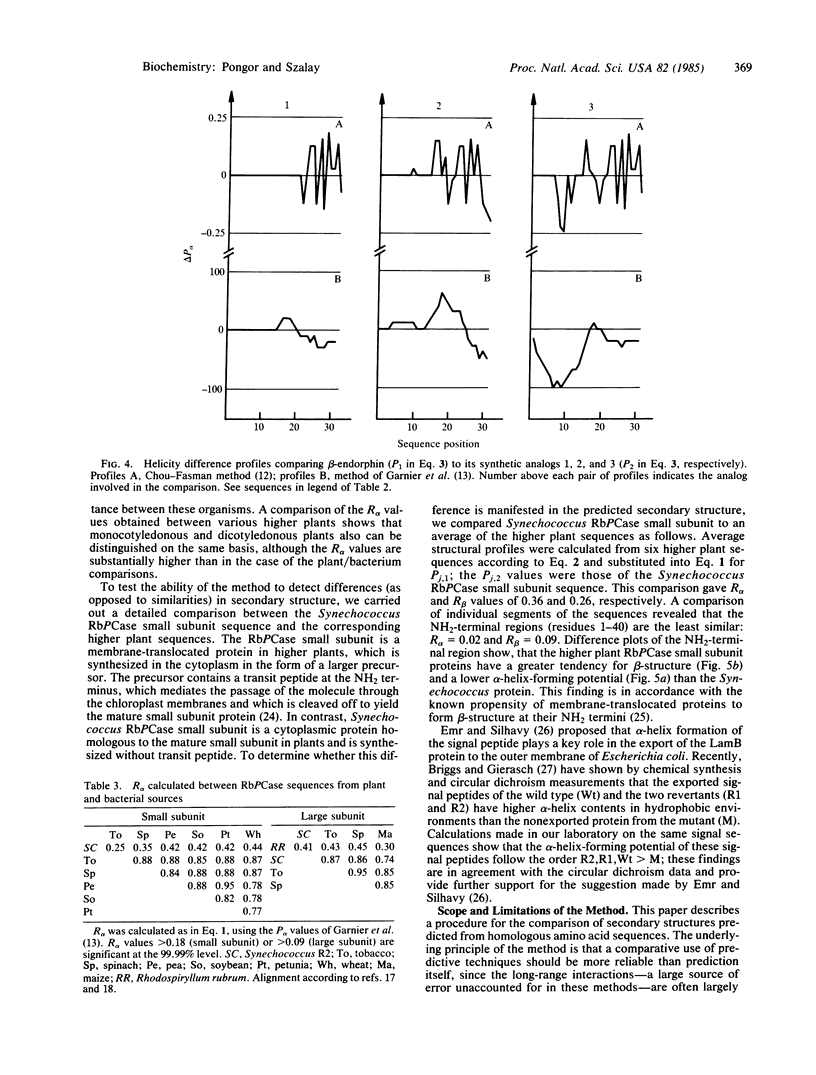
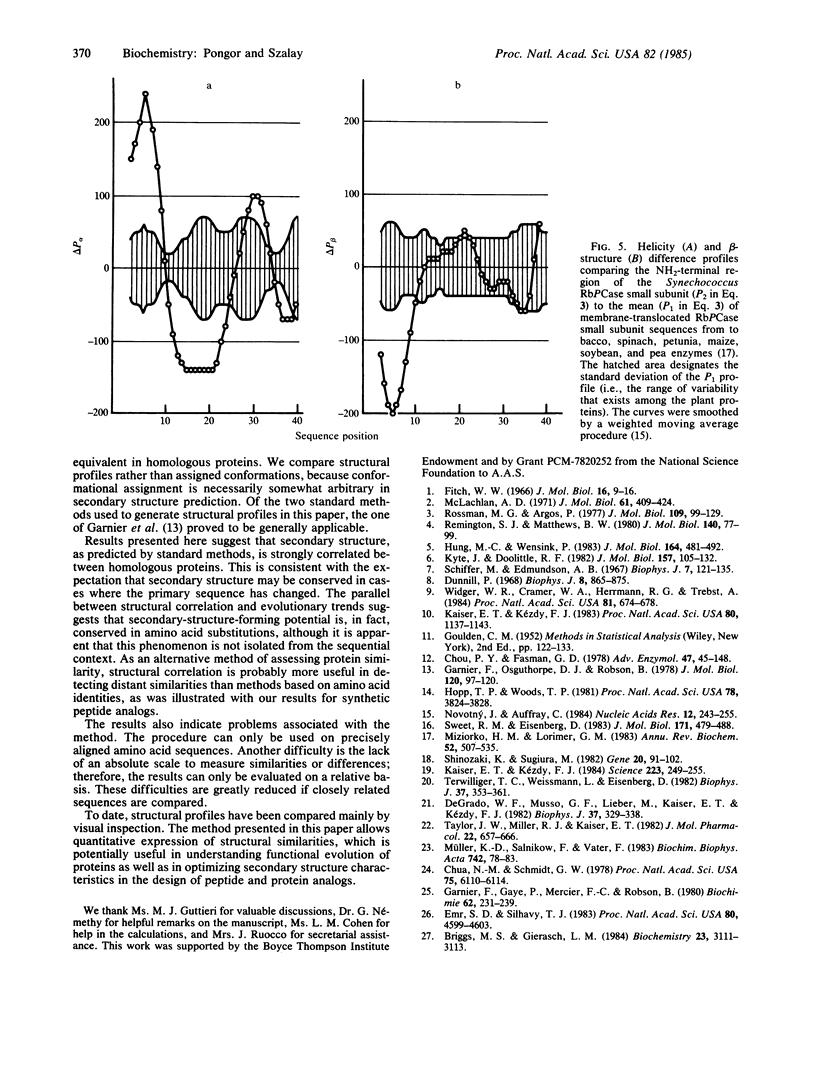
A quantitative procedure is described for the comparison of secondary structure of homologous proteins. Standard predictive methods are used to generate probability profiles from pairs of homologous amino acid sequences; correlation coefficients (R) are then computed between each pair of amino acids for alpha-helix (R alpha), extended structure (R beta), turn (R(t)), and coil (R(c)). R values are >0.2 for correctly aligned homologous sequences. Unrelated or incorrectly aligned sequences give R values near zero. Lack of correlation for a segment of otherwise well-correlated sequences is used to identify structural divergence, which is then evaluated graphically by using difference profiles. A combination of these techniques correctly predicts secondary structural differences between melittin or beta-endorphin and their respective synthetic analogs. The method is potentially useful to describe evolutionary changes in protein secondary structure as well as in the design of peptide analogs.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Briggs M. S., Gierasch L. M. Exploring the conformational roles of signal sequences: synthesis and conformational analysis of lambda receptor protein wild-type and mutant signal peptides. Biochemistry. 1984 Jul 3;23(14):3111–3114. doi: 10.1021/bi00309a001. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Chua N. H., Schmidt G. W. Post-translational transport into intact chloroplasts of a precursor to the small subunit of ribulose-1,5-bisphosphate carboxylase. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6110–6114. doi: 10.1073/pnas.75.12.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGrado W. F., Musso G. F., Lieber M., Kaiser E. T., Kézdy F. J. Kinetics and mechanism of hemolysis induced by melittin and by a synthetic melittin analogue. Biophys J. 1982 Jan;37(1):329–338. doi: 10.1016/S0006-3495(82)84681-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnill P. The use of helical net-diagrams to represent protein structures. Biophys J. 1968 Jul;8(7):865–875. doi: 10.1016/S0006-3495(68)86525-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emr S. D., Silhavy T. J. Importance of secondary structure in the signal sequence for protein secretion. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4599–4603. doi: 10.1073/pnas.80.15.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch W. M. An improved method of testing for evolutionary homology. J Mol Biol. 1966 Mar;16(1):9–16. doi: 10.1016/s0022-2836(66)80258-9. [DOI] [PubMed] [Google Scholar]
- Garnier J., Gaye P., Mercier J. C., Robson B. Structural properties of signal peptides and their membrane insertion. Biochimie. 1980;62(4):231–239. doi: 10.1016/s0300-9084(80)80397-x. [DOI] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Hopp T. P., Woods K. R. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung M. C., Wensink P. C. Sequence and structure conservation in yolk proteins and their genes. J Mol Biol. 1983 Mar 15;164(4):481–492. doi: 10.1016/0022-2836(83)90046-3. [DOI] [PubMed] [Google Scholar]
- Kaiser E. T., Kézdy F. J. Amphiphilic secondary structure: design of peptide hormones. Science. 1984 Jan 20;223(4633):249–255. doi: 10.1126/science.6322295. [DOI] [PubMed] [Google Scholar]
- Kaiser E. T., Kézdy F. J. Secondary structures of proteins and peptides in amphiphilic environments. (A review). Proc Natl Acad Sci U S A. 1983 Feb;80(4):1137–1143. doi: 10.1073/pnas.80.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- McLachlan A. D. Tests for comparing related amino-acid sequences. Cytochrome c and cytochrome c 551 . J Mol Biol. 1971 Oct 28;61(2):409–424. doi: 10.1016/0022-2836(71)90390-1. [DOI] [PubMed] [Google Scholar]
- Miziorko H. M., Lorimer G. H. Ribulose-1,5-bisphosphate carboxylase-oxygenase. Annu Rev Biochem. 1983;52:507–535. doi: 10.1146/annurev.bi.52.070183.002451. [DOI] [PubMed] [Google Scholar]
- Novotný J., Auffray C. A program for prediction of protein secondary structure from nucleotide sequence data: application to histocompatibility antigens. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):243–255. doi: 10.1093/nar/12.1part1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington S. J., Matthews B. W. A systematic approach to the comparison of protein structures. J Mol Biol. 1980 Jun 15;140(1):77–99. doi: 10.1016/0022-2836(80)90357-5. [DOI] [PubMed] [Google Scholar]
- Rossmann M. G., Argos P. The taxonomy of protein structure. J Mol Biol. 1977 Jan 5;109(1):99–129. doi: 10.1016/s0022-2836(77)80048-x. [DOI] [PubMed] [Google Scholar]
- Schiffer M., Edmundson A. B. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys J. 1967 Mar;7(2):121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Sugiura M. The nucleotide sequence of the tobacco chloroplast gene for the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Gene. 1982 Nov;20(1):91–102. doi: 10.1016/0378-1119(82)90090-7. [DOI] [PubMed] [Google Scholar]
- Sweet R. M., Eisenberg D. Correlation of sequence hydrophobicities measures similarity in three-dimensional protein structure. J Mol Biol. 1983 Dec 25;171(4):479–488. doi: 10.1016/0022-2836(83)90041-4. [DOI] [PubMed] [Google Scholar]
- Taylor J. W., Miller R. J., Kaiser E. T. Structural characterization of beta-endorphin through the design, synthesis, and study of model peptides. Mol Pharmacol. 1982 Nov;22(3):657–666. [PubMed] [Google Scholar]
- Terwilliger T. C., Weissman L., Eisenberg D. The structure of melittin in the form I crystals and its implication for melittin's lytic and surface activities. Biophys J. 1982 Jan;37(1):353–361. doi: 10.1016/S0006-3495(82)84683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widger W. R., Cramer W. A., Herrmann R. G., Trebst A. Sequence homology and structural similarity between cytochrome b of mitochondrial complex III and the chloroplast b6-f complex: position of the cytochrome b hemes in the membrane. Proc Natl Acad Sci U S A. 1984 Feb;81(3):674–678. doi: 10.1073/pnas.81.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]


