Abstract
Epigenetics describes heritable alterations of gene expression and chromatin organization without changes in DNA sequence. Both hypermethylation and hypomethylation of DNA can affect gene expression and the multistep process of carcinogenesis. Epigenetic changes are reversible and may be targeted by dietary interventions. Bioactive compounds from green tea (GT) such as (–)-epigallocatechin gallate have been shown to alter DNA methyltransferase activity in studies of esophageal, oral, skin, Tregs, lung, breast and prostate cancer cells, which may contribute to the chemopreventive effect of GT. Three out of four mouse model studies have confirmed the inhibitory effect of (–)-epigallocatechin gallate on DNA methylation. A human study demonstrated that decreased methylation of CDX2 and BMP-2 in gastric carcinoma was associated with higher GT consumption. It is the goal of this review to summarize our current knowledge of the potential of GT to alter epigenetic processes, which may be useful in chemoprevention.
Keywords: cancer, COMT, DNA methylation, DNMT, (–)-epigallocatechin gallate, epigenomics, green tea, one-carbon metabolism, polyphenols
Cancer is a disease of genetic susceptibility as well as epigenetic abnormalities [1]. Epigenetics generally refers to changes in gene expression and chromatin organization that are independent of alterations in the DNA sequence [2]. Epigenetic phenomena are modifiable by dietary and environmental factors. Changes in DNA methylation can be passed on to the next generation [3]. Epigenetic changes have been identified as promising targets for the prevention and treatment of cancer [4].
DNA methylation is the most widely studied epigenetic modification in mammals. DNA methylation results in the addition of a methyl group to the carbon-5 position in the pyrimi-dine ring of cytosine in the CpG dinucleotide of genomic DNA. The distribution of CpG dinucleotides throughout the human genome is not uniform, and are frequently enriched in the promoter regions of genes, especially in regions of large repetitive sequences such as centromeric repeats, LINE-1 and ALU retrotransposon elements [5]. Short CpG-rich regions are also called ‘CpG islands’ and are present in more than 50% of human gene promoters [6]. Hypermethylation of CpG islands within gene promoters has been shown to lead to gene silencing, while promoters of transcriptionally active genes are typically hypomethylated [7].
In addition to DNA methylation, other epigenetic changes, such as histone modification and miRNAs, can affect gene expression. Histone modifications typically occur as post-translational alterations at the N-terminal of histones. These histone alterations include acetylation, methylation, phosphorylation, biotinylation and ubiquitination, and play a fundamental role in protein regulation throughout life [8–10]. miRNAs appear to have a fundamental role in the biology of the cell. They constitute a class of ncRNA molecules, which have now emerged as key players in regulating the activity of mRNA. miRNAs are small RNA molecules approximately 22 nucleotides in length, which affect the activity of specific mRNA by influencing their half-life through interference with the normal mRNA degradation process or mRNA translation into proteins [11]. This review will focus on the epigenetic effects of green tea (GT) mediated through changes in DNA methylation.
Nutrition can potentially affect epigenetic phenomena at multiple points in DNA methylation [12]. First, nutrients are the main source of methyl groups or act as coenzymes for the one-carbon metabolism that regulates methyl transfer and DNA synthesis. For example, B vitamins, such as folic acid, vitamin B2, B6 and B12, are involved as coenzymes with methionine, choline, betaine and serine as universal methyl donors [13]. Second, a number of phyto-chemicals found in plant foods and in dietary supplements alter the epigenetic processes by influencing enzyme activities such as 5-cytosine DNA methyltransferase (DNMT). Phyto-chemicals including polyphenols (green tea catechins, quercetin, myricetin), soy isoflavones (genistein), parthenolide, curcumin, resveratrol, isothiocyantes and butyrate, an intestinal product from fiber, affect the activities of methylation enzymes [9,12,14–16]. Third, dietary components, such as retinoic acid and vitamin D, bind to their receptors and modulate gene expression leading to competitive downregulation of methylating enzymes [17,18]. In addition, other phyto chemicals, such as garlic diallyl disulfide, sulfurophane and indol-3-carbinol, impact the epigenome through histone modulation and regulation of miRNAs [17].
DNA methylation is catalyzed by the enzyme 5-cytosine DNMT with S-adenosylmethionine (SAM) as the methyl donor. There are three main DNMT enzymes: DNMT1, DNMT3a and DNMT3b [19,20]. DNMT1 is a maintenance methyltransferase maintaining DNA methylation patterns in DNA replication during cell division [16], whereas both DNMT3a and DNMT3b are involved in de novo methyltransferase processes, providing an important function during development (differentiation) [21,22]. DNA methylation has evolved as an attractive target in cancer therapeutics. Altered DNMT gene expression and enzyme activity is seen in numerous diseases including cardiovascular diseases [23,24], Type 2 diabetes [25], obesity [26], possibly neurodegenerative diseases [27] and cancer [3,9,28]. In cancer, both DNA hypo- and hyper- methylation have been demonstrated to be associated with disease progression. Methy lation during cancer development includes hypermethylation of specific gene promoters, in addition to generalized hypomethylation. DNA hypermethylation in cancer often causes the silencing of tumor suppressors and other genes important for cellular growth, regulation and differentiation [29]. DNA hypomethylation has been shown to result in chromosomal instability and increased mutation events [30]. Changes in cellular DNA methylation in colorectal, pancreatic, prostate, bladder, breast and ovarian cancer have been reviewed by Heichman et al. [31]. For example, Yang et al. demonstrated a decrease in global cytosine hypomethylation comparing low-grade prostate epithelial neoplasia, high-grade prostate epithelial neoplasia and prostate cancer tissue, using immunohistochemistry in the prostate [32]. However, hypomethylation is not as commonly observed, with only a handful of specific genes being hypomethylated in prostate cancer. The majority of genes are characterized by site-specific hypermethylation [33]. The evaluation of a panel of methylation markers, such as APC, RARb2, TIG1 and GSTP1, demonstrated that utilizing the information derived from the methylation status of the gene panel, in combination with histological tissue evaluation, increased the percentage of detection of carcinoma from 64 to 97% compared with using histological tissue evaluation alone [34]. Analysis of the methylation status of 219 prostatectomy tissue samples using a panel of three genes (APC, HOXD3 and TGFβ) demonstrated that an increase in methylation was associated with prostate cancer progression [33]. Evaluation of DNA methylation of these three genes was superior for the prediction of biochemical recurrence compared with individual genes [33]. Importantly, many of these methylation events were also found in early high-grade prostatic intraepithelial neoplasia lesions [32], suggesting that aberrant DNA methylation changes occur early during carcinogenesis. The epigenetic changes in prostate cancer have been summarized in a review by Ho et al. [35].
The number of genes identified with altered methylation in breast cancer is rapidly growing. Breast cancer studies indicate that epigenetic alterations, such as promoter hypermethylation leading to gene silencing are involved in processes in carcinogenesis, including DNA repair (BRCA1), xenobiotic detoxification (GSTP1), apoptosis (HOXA5, RASSF1A and TWIST1), tissue invasion and metastatic processes (CDH1 and CDH13) [36,37]. These genes were not only hypermethylated in tumor cells, but also in normal epithelium surrounding the tumor site [37]. Methylated genes are utilized as cancer biomarkers in the clinical laboratory including not only breast but other major tumor tissues, such as colorectal, pancreatic, prostate, ovary, lung and bladder cancer [31].
Based on the reversible nature of epigenetic alterations, epigenetic therapy has strong potential for the prevention and treatment of chronic disease. Increasing interest in the potential of changing diet and lifestyle, or consuming dietary supplements to alter the epigenome, has led to a growing body of research focusing on the potential of dietary components and natural products as epigenetic agents in chemoprevention and cancer treatment. This review will summarize our current knowledge of the effect of green tea polyphenols on epigenetic processes in cell culture, animal and human epidemiological and clinical intervention studies.
Green tea polyphenol bioavailability & metabolism
The active compounds in green tea are polyphenols (GTPs), also known as flavan-3-ols, which include (–)-epigallocatechin gallate (EGCG), (–)-epicatechin gallate (ECG), (–)-epigallocatechin (EGC) and (–)-epicatechin (EC) (Figure 1) [38]. The most abundant polyphenol in green tea is EGCG and it is known to have anticancer effects through many different mechanisms [39–43]. In cell culture and animal models of lung, digestive tract, bladder, liver, prostate, breast and skin cancer, the most commonly observed anticancer mechanisms of EGCG include inhibition of proliferation, induction of apoptosis and cell-cycle arrest at G0/G1 [44]. EGCG induces apoptosis by multiple pathways including the inhibition of the PI3K/AKT/p-BAD cell survival pathway leading to downregulation of Bcl-2 and upregulation of Bax, as well as activation of the FASR/caspase-8 pathway [45]. EGCG also affects other factors implicated in proliferation and cell death, such as MAPK pathways (phosphoErk1/2) and growth factors (IGF1, IGF receptor and IGFBP-3) [46]. In addition, EGCG has been shown to affect cell cycle regulation through inhibition of class I histone deacetylase (HDAC) enzyme activity leading to increased accessibility of the promoter region and increased expression of p21/waf1 and Bax [47]. In protein-binding assays, it was demonstrated that EGCG binds several proteins (vimentin, IGF-1 receptor and GRP 78 kDa) with high specificity at very low concentrations. However, in cell culture experiments, due to nonspecific protein binding, higher concentrations of EGCG were required to inhibit the same targets [44]. Treatment with EGCG or GT extract also demonstrated inhibition of angiogenesis, invasion, VEGF, MMP2 and MMP9. Finally, EGCG has been demonstrated to exhibit anti-inflammatory activity via COX-2 and NFκB inhibition. The concentration of phosphorylated NFκB/p65 in HT-1080 cells was inhibited by EGCG treatment in a dose-dependent manner, while NFκB/p65 remained the same [48]. Most of these mechanisms still need to be confirmed in human intervention studies.
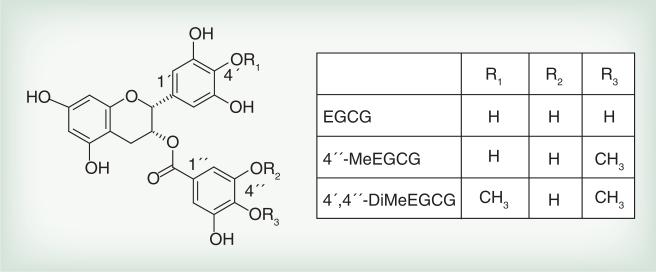
Figure 1. (–)-Epigallocatechin gallate and its methylated metabolites.
4″-MeEGCG: 4″-O-methyl-epigallocatechin gallate; 4′,4″-DiMeEGCG: 4′,4″-di-O-methyl-(–)-epigallocatechin gallate; EGCG: (–)-epigallocatechin gallate.
GTPs are mainly absorbed from the small intestine [49,50]. The absorption is regulated by several multidrug MRP transporters and monocarboxylated transporter [51]. According to their function, the transporters are either located at the basal membrane (MRP1) or at the apical membrane (MRP2). GTPs are taken up into epithelial cells and metabolized leading to enhanced excretion or reduced chemopreventive activity [52,53]. Nongallated GTPs such as EGC and EC undergo glucuronidation and sulfation, whereas gallated GTPs, EGCG and ECG, are mainly present in the free form [54]. All GTPs with at least one catechol group undergo methylation by catechol-O-methyl transferase (COMT). COMT catalyzes the transfer of methyl groups from SAM to one of the hydroxyl catechol groups, with the equimolar formation of S-adenosyl-l-homocysteine (SAH) [55]. EGCG and ECG contain two catechol ring structures. EGCG is readily methylated at 4′ and 4″ positions to form 4″-O-MeEGCG and 4′,4″-di-O-methyl-EGCG [56].
EGC and EC are the major catechins circulating in blood [49], with approximately 30% of EGC occurring in methylated form as 4′-O-MeEGC [57]. Although EGCG is not well absorbed in the small intestine due to its abundance in GT, it can be found in human tissue of men consuming six cups of GT daily [58,59]. In urine, EGC, 4′-O-MeEGC and EC are the predominant catechins, occurring in conjugated form [60]. In mouse and human prostate tissue, the major catechins present are EGCG and ECG [56,61,62]. Approximately 50% of EGCG occurs in the methylated form as 4″-O-MeEGCG or 4′,4″-O-di-O-methyl-EGCG in human prostate tissue obtained at prostatectomy after consumption of six cups (48 oz) of GT daily for 3–5 weeks [58,63]. A similar degree of methylation of EGCG was found after GT consumption in mouse tissues, including lung, kidney and xenograft prostate tumors [62]. However, there are strong differences in the methylation activity depending on the organ. For example, COMT activity is much higher in liver and kidney tissue compared with prostate and lung [64,65]. Methylation significantly decreases the biological activity of EGCG. Inhibition of proliferation and stimulation of apoptosis were significantly reduced in lymph node carcinoma of the prostate (LNCaP) cancer cells treated with 4″-O-MeEGCG compared with EGCG [58]. In human leukemia HL60 cells, the inhibition of proteasomal activity was significantly decreased using methylated EGCG compared with the nonmethylated form [66].
Enzyme kinetic analyses revealed that EGCG, in addition to being a substrate for COMT, also exhibits strong inhibition of COMT activity [67]. The most potent inhibitors among the catechins were those containing a galloyl-type D-ring (EGCG, 4″-O-methyl-EGCG, 4′,4″-di-O-methyl-EGCG and ECG), independent from their methylation status [68]. Protein aggregate formation, as well as computational molecular modeling studies, demonstrated a direct binding of EGCG to COMT [68,69].
Since GTPs are substrates for catechol methyl ation reactions, it has been suggested that COMT-mediated methylation might decrease the intracellular concentration of SAM and, at the same time, increase the concentration of SAH, a feedback inhibitor of various SAM-dependent methylation reactions. Therefore, excessive consumption of catechol-containing polyphenols may affect other methylation processes such as DNA methylation [68]. Potential mechanisms of DNMT inhibition by EGCG and other GTPs have been extensively investigated by Lee et al. [70]. Kinetic analyses using EC as a model inhibitor showed that DNA methylation was competitively inhibited in vitro mainly by increasing the formation of SAH. By comparison, the strong inhibitory effect of EGCG on DNMT-mediated methylation was largely due to its direct inhibition of the DNMT enzyme activity, independent of its own methylation [70].
GTPs & DNA methylation
Cell culture experiments
GTPs have been shown to inhibit DNA methylation leading to hypomethylation and activation of epigenetically silenced genes [71–77]. Extensive in vitro experiments have been performed in a variety of cancer cell lines to evaluate the effect of GTPs on DNA methylation (Table 1). Original studies on the investigation of the effect of EGCG on DNA methylation in cell culture were performed by the laboratory of CS Yang (NJ, USA) and are summarized in a review article by Fang et al. [55].
Table 1.
Cell culture studies to investigate the effect of green tea or epigallocatechin gallate on DNA methylation and gene expression.
| Study (year) | Tissue/cell line | Genes | GT/EGCG dose and duration of treatment (μmol/l, days) | Effect | Ref. |
|---|---|---|---|---|---|
| Fang et al. (2007) | Esophagus | p16, RARβ, hMLH1 and MGMT | 5–50 EGCG, 1-6 | Decreased DNMT1, gene reactivation | [55] |
| Wong et al. (2011) | Immune T cells, Jurkat | FOXP3 | 2–10 EGCG, 1–2 | Decreased DNMT1 | [71] |
| Kato et al. (2008) | Oral SCC9 and HSC3 | RECK tumor suppressor | 20–50 EGCG, 3–6 | Reversed hypermethylation, gene reactivation | [72] |
| Gao et al. (2009) | Lung H460 and A549 | WIF-1 | 10–50 EGCG, 3 | Demethylation | [83] |
| Li et al. (2010) | Breast MDAMB-231 | ER-α | 10 EGCG, 3 | Reversed hypermethylation, gene reactivation | [77] |
| Nandakumar et al. (2011) | Skin A431 | p16(INK4a) and p21/Cip1 | 10 EGCG, 6 | Decreased DNMT1, gene reactivation | [75] |
| Pandey et al. (2010) | Prostate | GSTP1 | 10 PolyE, 14 | Decreased DNMT1, gene reactivation | [84] |
| Chuang et al. (2005) | T24, HT29 and PC3 | p16, RARb, MAGE-A1, MAGE-B2 and Alu | 20–30 EGCG, 6 | No effect | [85] |
| Streseman et al. (2006) | TK6, Jurkat and KG-1 | TIMP3 | 2–50 EGCG, 3 | No effect | [86] |
EGCG: Epigallocatechin gallate; GT: Green tea; PolyE: Polyphenon E.
Esophageal cancer
Studies conducted by Fang et al. demonstrated that treatment of esophageal cancer cells with EGCG (5–50 μM, 1–6 days) exhibited a time- and dose-dependent inhibition of DNMT activity reversing hypermethylation in several tumor suppressor genes (p16, RARβ, hMLH1 and MGMT) [73].
Skin cancer
Treatment of A431 skin cancer cells with 10 μM of EGCG for 6 days decreased global DNA methylation as determined by 5-methylcytosine content using the QIAamp® DNA mini kit (Qiagen, Limburg, The Netherlands). In addition, EGCG treatment inhibited gene and protein expression of DNMT1, DNMT3a and DNMT3b leading to increased expression of p16(INK4a) and p21/Cip1 [75]. A comparison of the demethylation activity of the main four GTPs demonstrated that EGCG exhibited very similar activity compared with ECG, but the activity of both was higher than EGC and EC (EGCG = ECG > EGC > EC = GC) [75,78].
Tregs & Jurkat T cells
Wong et al. reported that treatment of Tregs and Jurkat T cells with 2–10 μM (1–2 days) of EGCG reduced global DNA methylation and DNMT gene expression leading to increased mRNA expression of IL-10 and Foxp3, a master switch that controls the development and function of Tregs and Jurkat T cells in vitro [71].
Breast cancer
Studies conducted by Li et al. found that treatment with EGCG at 10 μM for 3 days led to reactivation of estrogen receptor (ER)-α expression in ER-α-negative MDAMB-231 breast cancer cells [77]. A combination treatment of the same cells with EGCG and a HDAC inhibitor, trichostatin A, synergistically increased the activation of ER-α expression in ER-α-negative MDAMB-231 breast cancer cells [77]. More recent work demonstrated that sulforaphane (10 μM) enhanced the DNMT1-inhibitory effect of EGCG (20 μM) in paclitaxel-resistant ovarian cancer cells (KOV3TR-ip2) and, at the same time, enhanced the inhibition of hTERT and Bcl-2 [79].
Although overwhelming in vitro evidence supports the anticarcinogenic properties of GT treatment, the limited chemical stability of EGCG at alkaline pH under normal physiological conditions is of concern for the translation of our findings to clinical studies. To increase the chemical stability of EGCG, synthetic analogs have been generated, which show stronger anti-cancer activity with more stability and efficacy [80,81]. For example, studies comparing EGCG to an EGCG analog (peracetylated EGCG [pEGCG] and EGCG octa-acetate) demonstrated increased inhibition of hTERT expression by inducing DNA hypomethylation and promoter deacetylation, mediated by inhibition of DNMTs and histone acetylases, respectively [76,82]. Both EGCG (40 μM) and pEGCG (20 μM) inhibited the proliferation of human ER-positive (MCF-7) and ER-negative (MDAMB-231) breast cancer cells in a dose- and time-dependent way, but showed no effect on normal MCF10A cells.
Lung cancer
Treatment with 10–50 μM of EGCG for 3 days led to promoter demethylation and restoration of WIF-1, an antagonist of Wntproto-oncogene in non-small-cell lung cancer (H460 and A549) cells [83].
Oral carcinoma
EGCG (20–50 μM for 3 and 6 days) treatment of oral carcinoma cells (SCC9 and HSC3) partially reversed the hypermethylation status of the RECK tumor suppressor gene leading to a significant increase of the expression of RECK mRNA [72].
Prostate cancer
Treatment of LNCaP human prostate cancer cells with a GT extract (polyphenon E) was associated with a time- and dose-dependent activation of GSTP1. The hypermethylation and downregulation of GSTP1 has been associated with the development of several types of cancer, including cancer of the prostate [84]. The inhibition of DNMT1 protein expression in LNCaP prostate cancer cells treated with 10 μg/ml of polyphenon E was strongest at 14 days of treatment and was associated with decreased methylation of the promoter region of GSTP1 [84].
Despite the abovementioned studies, the ability of EGCG to inhibit DNA methylation remains controversial since there are also two published studies that did not find evidence that EGCG treatment will alter the methylation status and reverse gene silencing. Chuang et al. reported that purified EGCG did not inhibit DNA methylation at single copy loci or repetitive DNA elements in three different human cancer cell lines [85]. They examined the effect of EGCG compared with 5-aza-2′-deoxycytidine (5-Aza-Cd) on DNA methylation and RNA expression of six different genes/repetitive elements (p16, RARβ, LINE-1, MAGE-A1, MAGE-B2 and Alu) in three separate cell lines (T24, HT29 and PC3) [85]. Similarly, Stresemann et al. reported that EGCG treatment did not produce a significant effect on DNA methylation in HCT116 human colorectal cancer cells [86]. Treatment with 20–30 μM of EGCG for 6 days did not alter global methylation or the methylation status of TIMP3 in TK6, Jurkat and KG-1 cancer cell lines, including 2–50 μM for 3 days of EGCG [86]. The authors suggested potential reasons for the discrepancies between their studies and previously published studies, including different methods of analysis, possible gene specificity or cell line specificity of EGCG, or that the treatment method might have been ineffective to show efficacy. Stresemann et al. argued that in some in vitro cell culture conditions, cellular effects induced by EGCG could probably be attributed to the oxidative stress induced by this compound [86]. At neutral or alkaline pH, EGCG undergoes auto-oxidation, resulting in dimerization of EGCG and EGC to form homo- and hetero-dimers in an alkaline environment with concurrent formation of H2O2 [87,88]. This process is ubiquitous in in vitro experiments and during the intestinal digestion, but the degree of auto-oxidation depends on the cell culture medium. In cell culture medium, the indirect contribution of H2O2 formation can be avoided by the addition of superoxide dismutase or catalase prior to adding the GTPs [89]. However, the majority of cell culture experiments did not address the H2O2 formation. In summary, in vitro cell culture studies provide clear evidence that GTP treatment can alter DNA methylation, leading to re-expression of silenced genes. To achieve changes in DNA methylation, concentrations of 20–50 μmol/l of EGCG for 3–6 days is needed. These concentrations are much higher than physiologically achievable in mouse or human tissue. Therefore, GT interventions may not be suitable for therapeutic purposes, but may play a role in long-term treatment effects on DNA methylation in cancer prevention.
Therapeutic use of DNA methylation inhibitors
In general, two classes of DNA methylation inhibitors have been developed. One class includes the nucleoside analogs, such as 5-azacytidine (azacytidine) and 5-Aza-Cd (decitabine), and the second class includes non-nucleoside analogs, such as EGCG, genistein, hydralazine and procainamide, which are not incorporated into the DNA [90]. Two nucleoside analogs have been approved by the US FDA in 2004 and 2006 for the treatment of hematologic conditions, such as myelodysplastic syndrome, and are under development to treat acute myeloid leukemia; azacytidine has also been approved for chronic myelomonocytic leukemia [90]. However, the treatment of solid tumors with nucleoside analogs only led to limited response [91]. Myelosupression is the main toxicity when nucleoside analogs are used in higher doses [91]. The short plasma half-life of nucleoside analogs remains a challenge due to high levels of hepatic cytidine deaminase, an enzyme that deactivates the nucleoside analog. A second-generation prodrug with improved pharmacological profile, increased stability and decreased toxicity, is in Phase II clinical trials in the treatment of myelodysplastic syndrome and acute myeloid leukemia [90]. In the effort to minimize side effects, promising results have been shown with a combination of several cycles of treatment with very low doses of azacytidine and the HDAC inhibitor etinostat in patients with recurrent metastatic non-small-cell lung cancer [92].
Direct comparison of the inhibition of DNA methylation by nucleoside to non-nucleoside analogs in prostate (PC-3), colon (HT29) and bladder (T24) cell lines demonstrated that nucleoside analogs such as 5-Aza-Cd are far more effective in inhibiting DNA methylation leading to reactivation of silenced genes [85]. A concentration of 1 μmol/l of 5-Aza-Cd reactivated MAGE-A1, MAGE-B2 and p16 gene expression, while 30 μmol/l of EGCG, 20 μmol/l hydralazine and 200 μmol/l procainamide were inactive [85]. Another comparison of 5-Aza-Cd to GT showed that treatment of LNCaP prostate cancer cells with 10 μM 5-Aza-Cd for 3 days induced global hypomethylation of a CpG island within the LINE-1 promoter determined by methyl-specific PCR analysis, while exposure of cells to 10 μM of GTP did not result in global hypomethylation, but in re-expression of GSTp1 and promoted maintenance of genomic integrity [84]. Sustained and long-term exposure to very high doses of GTPs, comparable in activity with pharmacologic agents, could theoretically lead to chronic changes of DNA methylation [18]. However, these considerations do not apply to any of the current chemoprevention studies being carried out with GT.
On the other hand, potential interference of EGCG with the one-carbon metabolism has been observed. In a purified enzyme system of dihydrofolate reductase (DHFR) extracted from Stenotrophomonas maltophilia, EGCG was shown to inhibit DHFR enzyme activity in vitro at concentrations of 0.1–1 μM [93]. In Caco2 colon cancer cells, treatment with approximately 40 μM of EGCG interfered with the production of nucleotides, thus compromising DNA and RNA synthesis and acting as an antifolate agent [94]. DHFR catalyzes the reduction of 7,8-dihydrofolate to 5,6,7,8-tetrahydrofolate, which in turn is required as a coenzyme for many one-carbon group transfer reactions, including the nucleotide biosynthesis [95]. This interference with DNA biosynthesis may explain why neural tube defects, such as anencephaly and spina bifida, which are usually associated with folic acid deficiency, were linked to high levels of GT consumption of the mother during the periconceptional period [96].
Studies by Lemos et al. observed that EGCG also exhibited a dose-dependent inhibitory effect on 3H-folic acid uptake with an IC50 value of 7.7 μM (95% CI: 2.8–20.7) [97]. In addition, EGCG interfered with the uptake of methotrexate (IC50: 10.1 μM), which may result in reduced chemotherapeutic efficacy in patients taking methotrexate by the oral route [97].
Despite issues with bioavailability and stability, tea catechins have emerged as promising chemopreventive agents because many of the effects observed in vitro can be replicated in various animal models. However, to further increase the stability and bioavailability of GTPs, Saez-Ayala et al. synthesized two catechin-derived compounds, 3-O-(3,4,5-trimethoxybenzoyl)-(–)-catechin and 3-O-(3,4,5-trimethoxybenzoyl)-EC in an attempt to improve the chemopreventive activity of GTPs [98]. The antiproliferative and proapoptotic activities of both compounds were analyzed with various cancer cell lines. 3-O-(3,4,5-trimethoxybenzoyl)-(–)-catechin was superior compared with 3-O-(3,4,5-trimethoxybenzoyl)-EC in inhibiting proliferation and inducing apoptosis in both melanoma and non-melanoma cell lines [98].
Evidence from in vivo animal studies
The most convincing evidence to demonstrate that nutrition can modulate the epigenetic status of mammals was provided by studies with mice carrying the Avy gene [99,100]. In this mouse model, methylation affects the expression of the Avy gene. In the methylated form, the Avy gene is expressed only in the hair follicle (wild-type allele). However, in the unmethylated form, it is expressed ubiquitously resulting in the agouti (yellow) fur color and other characteristics of the agouti syndrome. Using the Avy model, Wolff and colleagues showed that feeding diets supplemented with high levels of folic acid (as methyl donor) to pregnant dams modified the expression of the agouti gene in the offspring [100].
Whether EGCG can reverse DNA hypermethylation and reactivate methylation-silenced genes in vivo still remains to be determined. Based on the evidence from in vitro cell culture studies, it is of interest to investigate the effect of GTPs on epigenetic processes in vivo (Table 2). Potential mechanisms are the inhibition of DNMT1 activity, directly or competitively, by depleting its substrate SAM or causing accumulation of the inhibitor SAH [18,70].
Table 2.
Animal studies to investigate the effect of green tea or epigallocatechin gallate on DNA methylation and gene expression.
| Study (year) | Tissue | Animal model | GT exposure | Effect | Ref. |
|---|---|---|---|---|---|
| Henning et al. (2012) | Prostate | C57BL/6J–LAPC4 xenograft | Brewed GT (0.075% GTP), 13 weeks | Decreased tumor volume, weight and decreased DNMT1 | [62] |
| Volate et al. (2009) | Intestine | Apc(Min/-) | GT 0.6% | Decreased CpG methylation of RXRα | [102] |
| Mittal et al. (2003) | Skin | UVB-exposed SKH-1 hairless mice | 1 mg/cm2 skin area EGCG cream prior to UV exposure | Decreased global DNA hypomethylation | [103] |
| Morey et al. (2009) | Prostate | TRAMP mice | GTP 0.1–0.6% in drinking water, 8–20 weeks | No decreased methylation and no decrease in tumor volume | [119] |
EGCG: Epigallocatechin gallate; GT: Green tea; GTP: Green tea polyphenol; Min: Multiple intestinal neoplasia; TRAMP: Transgenic adenocarcinoma of the mouse prostate.
The delay in the development of prostate cancer by administration of the DNA methyltransferase inhibitor 5-Aza-dC to transgenic adeno-carcinoma of the mouse prostate (TRAMP) mice has been demonstrated as a ‘proof of principle’ that cancer prevention may be achieved through epigenetic modifications [101]. Analysis of untreated TRAMP prostate lesions demonstrated elevated DNMT1 mRNA and protein levels in early stages of prostate cancer development (prostatic intraepithelial neoplasia), which continued through advanced prostate cancer and metastasis. In a 5-Aza-Cd intervention study, none of the 14 TRAMP mice receiving intraperitoneal injections twice weekly on consecutive days of 300 μl 5-Aza-Cd (0.25 mg/kg) developed prostate cancer at 24 weeks of age, whereas seven out of 13 (54%) control mice, injected with phosphate buffer solution, developed poorly differentiated prostate cancer [101].
In our investigation of the effect of drinking brewed GT instead of drinking water on tumor growth and DNMT activity in male severe-combined immunodeficiency mice, we determined an inhibition of DNMT1 protein and gene expression in prostate xenograft LAPC4 tumor tissue [62]. The GT contained a concentration of 0.07% of GTPs and was administered for 13 weeks. Tumor volume and weight were also decreased significantly in mice drinking the GT compared with the water control [62].
Intestinal cancer
Further evidence for the epigenetic activity of GT was provided by Volate et al. [102]. The administration of 0.6% (w/v) solution of GT as the only source of beverage from 8–16 weeks to Apc(Min/−) mice significantly increased the protein and mRNA levels of RXRa, which was downregulated in intestinal tumors of azoxy-methane-treated Apc(Min/+) control mice. Genomic bisulfite treatment of DNA extracted from the colon of these mice showed a significant decrease in CpG methylation with GT treatment in the promoter region of the RXRα gene, analyzed by pyrosequencing of 24 CpG sites [102]. GT treatment also reduced the number of newly formed intestinal tumors (28%; p < 0.05) compared with water control.
Skin cancer
In the studies by Mittal et al., global DNA hypomethylation was observed in chronically UVB-exposed SKH-1 hairless mice [103]. Topical treatment with EGCG in hydrophilic cream providing 1 mg/cm2 skin reversed DNA hypomethylation in chronic UVB-exposed skin determined by immunohistochemical detection of 5-methylcytosine [103]. The authors speculated that the epigenetic modulation contributed to the anticarcinogenic activity of EGCG. However, it is interesting that in this mouse model, the observation was contradictory to the concept that EGCG can prevent or reverse hypermethylation of certain specific genes [103].
Fang et al. addressed the question of whether the administration of EGCG in mice could lead to a decrease in SAM coinciding with the accumulation of SAH, which in turn could induce competitive inhibition of DNMT activity [55]. They examined this issue in their ongoing experiments on bioavailability, toxicity and cancer-preventive activities of EGCG. The results showed that only an acute intragastric treatment with high doses of EGCG (500–2000 mg/kg), significantly elevated plasma levels of homocysteine and at the same time decreased levels of plasma methionine and lowered the concentration of hepatic and intestinal SAM, as well as the SAM:SAH ratio [55]. The administration of EGCG (or polyphenon E) through drinking fluid (0.32% EGCG or 0.5% polyphenon E) decreased intestinal SAM concentrations moderately without increasing the level of SAH. No changes of hepatic SAM or SAH levels were observed with the administration of EGCG in the drinking water [55].
No effect on DNA methylation
Morey et al. recently tested whether oral consumption of GTPs could affect normal or cancer-specific DNA methylation in vivo, using the TRAMP mouse model [104]. TRAMP mice received 0.3% GTPs in drinking water beginning at 4 weeks of age. In these studies, GTP treatment did not inhibit tumor progression in TRAMP mice, whereas investigations by several other groups had demonstrated an inhibition of tumor size and number with GT treatment [105,106]. In addition, levels of 5-methyldeoxycytidine in the B1 repetitive element and methylation of the Mage-a8 gene were not affected. In these studies, no measurements of tissue GTP were performed to confirm the bioavailability of the GTPs.
Combinations of EGCG & other natural products
There is an increased interest in the use of combinations of natural products in order to overcome multidrug resistance, limited bioavailability or to target multiple mechanisms concurrently. For example, in our laboratory a combination of GT and quercetin increased the bioavailability and decreased EGCG methylation, leading to an increase in the anticarcinogenic activity in a prostate cancer xenograft mouse model [64]. In addition multiple cell culture studies demonstrated that the combination of GTPs with sulforaphane increased apoptosis and altered Nrf2- and AP-1-regulated gene expression in prostate and colon cancer cells [79,107,108]. Combining natural products that alter the epigenome will enhance the epigenetic effect, since some compounds may alter DNA methylation and other natural compounds may affect histone structure and miRNA regulation [109–111].
DNA methylation in human studies
There are only a limited number of clinical trials investigating the effects of dietary polyphenol sources on DNA methylation such as tea or diets limited in folate and one-carbon metabolites (Table 3). One study by Olthof et al. reported an increase in plasma homocysteine concentration associated with high polyphenol consumption from either black tea, chlorogenic acid or quercetin for 7 days [112].
Table 3.
Human studies to investigate the effect of green tea or epigallocatechin gallate on DNA methylation and gene expression.
| Disease | Participants (n) | GT exposure | Effect | Ref. |
|---|---|---|---|---|
| Healthy | – | Polyphenols, 7 days | Increased plasma SAH | [112] |
| Gastric carcinoma | Japanese (55) | More than seven cups compared with less than six cups GT/days | Methylation status of CDX2 was not significantly associated with low intake of tea | [113] |
| Gastric primary carcinoma | Japanese (106) | More than seven cups compared with <6 cups GT/days | Correlation between CDX2 and BMP-2 methylation and low tea intake | [114] |
EGCG: Epigallocatechin gallate; GT: Green tea; SAH: S-adenosyl-l-homocysteine.
A retrospective study published by Yuasa et al. examined the methylation status of six genes in primary gastric carcinomas from 106 patients in relation to past lifestyle, including dietary habits [113,114]. In this Japanese study of gastric carcinoma, high consumption of GT (seven cups or more per day compared with six cups or less) and cruciferous vegetables, as well as physical activity, was inversely related to the methylation of CDX2 and BMP-2, but not p16 (INK4A), CACNA2D3, GAT-5 and ER in gastric carcinoma tissue, which are frequently hypermethylated in gastric cancers [113]. It was pointed out by the authors that the epithelial surfaces of the esophagus and stomach may be particularly susceptible to beneficial DNMT1 inhibitory effects of GTPs, since the luminal intestinal side is exposed to high concentrations of GTPs before the polyphenols undergo metabolism.
Since it has been shown that the intestinal administration of large concentrations of GTPs will lead to a significant change in the SAM:SAH ratio [55], we also include human studies investigating the association of the one-carbon metabolism to colon cancer. A review by Lim and Song provided an excellent summary of these studies [115]. A landmark controlled feeding study to investigate the effect of a folate-deficient diet on global DNA methylation was performed in 1998. Global DNA methylation levels decreased in lymphocytes of postmenopausal women on a folate-deficient diet (56 μg/day for 5 weeks and 111 μg/day for 4 weeks) and increased on a folate-replete diet (286–516 μg/day for 3 weeks) [116]. Two previous human studies conducted in a metabolic unit demonstrated that marginal folate deficiency can change blood genomic DNA methylation [117]. Another study investigated the effect of the consumption of a sequence of a moderately deplete folate diet (118 μg/day for 7 weeks) followed by intake of a replete diet (415 μg/day) for 7 weeks in older women aged 60–85 years. During the moderate depletion phase, leukocyte global DNA methylation declined but did not significantly improve even after 7 weeks on the repletion diet [117]. Similarly, another clinical randomized and double-blinded intervention demonstrated that leukocyte global DNA methylation was increased in participants receiving folic acid supplements (400 μg/day for 10 weeks) [118]. These findings support the hypothesis that a reduction in total folate intake may be associated with reduced global DNA methylation and can be reversed with supplementation at levels of folic acid found in common multivitamin supplements (400 μg/day). However, older age may compromise or delay the recovery [117].
Taken together, the evidence from human studies, demonstrating significant dietary effects on DNA methylation in free-living human subjects, is very limited compared with in vitro and animal studies. Evidence for the role of GTPs in affecting DNA methylation in cancer development is mainly based on in vitro cell culture experiments. However, since dietary modifications induce relatively low impact changes on DNA methylation, with lower toxicity compared with epigenetic therapeutic drugs, dietary strategies may play an important role in the prevention of carcinogenesis. Moreover dietary exposures are long-term and potentially repeated several times daily in heavy tea drinkers. There is a critical need for future investigations in animal and human studies to reveal the potential of different bioactive and dietary components in the epigenetic regulation of chronic disease.
Conclusion
GTPs and other natural products have the potential to alter epigenetic processes through DNA methylation, histone modification and miRNA regulation. This area of research requires more information on the relative potency of these effects both for GTPs alone and for polyphenols in combination with other drugs and natural products or dietary supplements.
Future perspective
Owing to emerging technologies and decreasing cost of evaluating changes in DNA methylation, our understanding of the effects of nutrition and botanical dietary supplements on epigenetic processes will increase dramatically during the next decade. Although the effect of dietary exposure on epigenetics is subtle, due to the nature of dietary exposures being frequent and long-term, we expect that future evidence will support an emerging role of diet in regulating DNA methylation. In vitro cell culture studies provide clear evidence that extended GT treatment can change DNA methylation and reactivate gene expression. Future long-term in vivo GT studies are needed to ascertain whether permanent changes of DNA methylation are achievable. Results from these studies will contribute to cancer prevention and treatment.
Executive summary.
DNA methylation & cancer
■ Epigenetic phenomena are heritable and potentially modifiable by dietary, environmental and therapeutic factors.
■ Epigenetic processes are promising targets for interventions with nutrients and compounds derived from dietary supplements and natural products.
■ Nutrition affects epigenetic phenomena at multiple points: being a source of methyl groups, influencing enzyme activities (DNA methyltransferase and histone deacetylase), binding to receptors and modulating methylating enzymes and/or modulating histones and regulating miRNAs.
Green tea polyphenol bioavailability & metabolism
■ (–)-Epigallocatechin gallate (EGCG) is the main tea polyphenol under investigation.
■ Low bioavailability and extensive metabolic changes to less active methylated metabolites limits its potential.
■ Future research into combination treatments and using more stable EGCG preparations is needed.
Green tea polyphenols & DNA methylation
■ Green tea (GT) polyphenols have been shown to inhibit DNMT1 activity leading to reactivation of silenced genes in cultured cells from oral cavity, esophagus, lung, breast, prostate and immune cells.
■ Several animal studies confirmed the epigenetic effect of GT.
■ Additional animal and clinical studies are needed to confirm the epigenetic effect of GT.
■ Studies using combinations of different natural products such as EGCG with quercetin or sulforaphane may alter epigenetic processes more effectively.
Human studies
■ One human study demonstrated that the consumption of more than six cups of GT was inversely related to the methylation of CDX2 and BMP-2 in gastric carcinoma tissue.
■ Oral and gastric epithelial surfaces are particularly susceptible to beneficial DNMT1 inhibitory effects of tea polyphenols, since they are exposed to high concentrations of tea polyphenols before undergoing metabolism.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
■ of interest
■ of considerable interest
- 1.Baylin SB, Jones PA. A decade of exploring the cancer epigenome – biological and translational implications. Nat. Rev. Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 3.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 4.Ptak C, Petronis A. Epigenetics and complex disease: from etiology to new therapeutics. Annu. Rev. Pharmacol. Toxicol. 2008;48:257–276. doi: 10.1146/annurev.pharmtox.48.113006.094731. [DOI] [PubMed] [Google Scholar]
- 5.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Leung FC. An evaluation of new criteria for CpG islands in the human genome as gene markers. Bioinformatics. 2004;20:1170–1177. doi: 10.1093/bioinformatics/bth059. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 8.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Meeran SM, Ahmed A, Tollefsbol TO. Epigenetic targets of bioactive dietary components for cancer prevention and therapy. Clin. Epigenetics. 2010;1:101–116. doi: 10.1007/s13148-010-0011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chervona Y, Costa M. Histone modifications and cancer: biomarkers of prognosis? Am. J. Cancer Res. 2012;2:589–597. [PMC free article] [PubMed] [Google Scholar]
- 11.Kala R, Peek GW, Hardy TM, Tollefsbol TO. MicroRNAs: an emerging science in cancer epigenetics. J. Clin. Bioinforma. 2013;3:6. doi: 10.1186/2043-9113-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Link A, Balaguer F, Goel A. Cancer chemoprevention by dietary polyphenols: promising role for epigenetics. Biochem. Pharmacol. 2010;80:1771–1792. doi: 10.1016/j.bcp.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson OS, Sant KE, Dolinoy DC. Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J. Nutr. Biochem. 2012;23:853–859. doi: 10.1016/j.jnutbio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerhauser C. Cancer chemoprevention and nutriepigenetics: state of the art and future challenges. Top. Curr. Chem. 2013;329:73–132. doi: 10.1007/128_2012_360. [DOI] [PubMed] [Google Scholar]
- 15.Hardy TM, Tollefsbol TO. Epigenetic diet: impact on the epigenome and cancer. Epigenomics. 2011;3:503–518. doi: 10.2217/epi.11.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szarc vel Szic K, Ndlovu MN, Haegeman G, Vanden BW. Nature or nurture: let food be your epigenetic medicine in chronic inflammatory disorders. Biochem. Pharmacol. 2010;80:1816–1832. doi: 10.1016/j.bcp.2010.07.029. [DOI] [PubMed] [Google Scholar]
- 17.Stefanska B, Karlic H, Varga F, Fabianowska-Majewska K, Haslberger A. Epigenetic mechanisms in anti-cancer actions of bioactive food components – the implications in cancer prevention. Br. J. Pharmacol. 2012;167:279–297. doi: 10.1111/j.1476-5381.2012.02002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park LK, Friso S, Choi SW. Nutritional influences on epigenetics and age-related disease. Proc. Nutr. Soc. 2012;71:75–83. doi: 10.1017/S0029665111003302. [DOI] [PubMed] [Google Scholar]
- 19.Siedlecki P, Zielenkiewicz P. Mammalian DNA methyltransferases. Acta Biochim. Pol. 2006;53:245–256. [PubMed] [Google Scholar]
- 20.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 21.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baccarelli A, Ghosh S. Environmental exposures, epigenetics and cardiovascular disease. Curr. Opin. Clin. Nutr. Metab. Care. 2012;15:323–329. doi: 10.1097/MCO.0b013e328354bf5c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wierda RJ, Geutskens SB, Jukema JW, Quax PH, van den Elsen PJ. Epigenetics in atherosclerosis and inflammation. J. Cell. Mol. Med. 2010;14:1225–1240. doi: 10.1111/j.1582-4934.2010.01022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bramswig NC, Kaestner KH. Epigenetics and diabetes treatment: an unrealized promise? Trends Endocrinol. Metab. 2012;23:286–291. doi: 10.1016/j.tem.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simmons R. Epigenetics and maternal nutrition: nature v. nurture. Proc. Nutr. Soc. 2011;70:73–81. doi: 10.1017/S0029665110003988. [DOI] [PubMed] [Google Scholar]
- 27.Babenko O, Kovalchuk I, Metz GA. Epigenetic programming of neurodegenerative diseases by an adverse environment. Brain Res. 2012;1444:96–111. doi: 10.1016/j.brainres.2012.01.038. [DOI] [PubMed] [Google Scholar]
- 28.Das PM, Singal R. DNA methylation and cancer. J. Clin. Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 29.Khavan-Niaki H, Samadani AA. DNA methylation and cancer development: molecular mechanism. Cell Biochem. Biophys. 2013 doi: 10.1007/s12013-013-9555-2. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 30.Watanabe Y, Maekawa M. Methylation of DNA in cancer. Adv. Clin. Chem. 2010;52:145–167. doi: 10.1016/s0065-2423(10)52006-7. [DOI] [PubMed] [Google Scholar]
- 31.Heichman KA, Warren JD. DNA methylation biomarkers and their utility for solid cancer diagnostics. Clin. Chem. Lab. Med. 2012;50:1707–1721. doi: 10.1515/cclm-2011-0935. [DOI] [PubMed] [Google Scholar]
- 32.Yang B, Sun H, Lin W, et al. Evaluation of global DNA hypomethylation in human prostate cancer and prostatic intraepithelial neoplasm tissues by immunohistochemistry. Urol. Oncol. 2013;31(5):628–634. doi: 10.1016/j.urolonc.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Liu L, Kron KJ, Pethe VV, et al. Association of tissue promoter methylation levels of APC, TGFbeta2, HOXD3 and RASSF1A with prostate cancer progression. Int. J. Cancer. 2011;129:2454–2462. doi: 10.1002/ijc.25908. [DOI] [PubMed] [Google Scholar]
- 34.Tokumaru Y, Harden SV, Sun DI, Yamashita K, Epstein JI, Sidransky D. Optimal use of a panel of methylation markers with GSTP1 hypermethylation in the diagnosis of prostate adenocarcinoma. Clin. Cancer Res. 2004;10:5518–5522. doi: 10.1158/1078-0432.CCR-04-0108. [DOI] [PubMed] [Google Scholar]
- 35.Ho E, Beaver LM, Williams DE, Dashwood RH. Dietary factors and epigenetic regulation for prostate cancer prevention. Adv. Nutr. 2011;2:497–510. doi: 10.3945/an.111.001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nowsheen S, Aziz K, Tran PT, Gorgoulis VG, Yang ES, Georgakilas AG. Epigenetic inactivation of DNA repair in breast cancer. Cancer Lett. doi: 10.1016/j.canlet.2012.05.015. doi:10.1016/j.canlet.2012.05.015 (2012) (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 37.Khan SI, Aumsuwan P, Khan IA, Walker LA, Dasmahapatra AK. Epigenetic events associated with breast cancer and their prevention by dietary components targeting the epigenome. Chem. Res. Toxicol. 2012;25:61–73. doi: 10.1021/tx200378c. [DOI] [PubMed] [Google Scholar]
- 38■.Sharma V, Rao LJ. A thought on the biological activities of black tea. Crit. Rev. Food Sci. Nutr. 2009;49:379–404. doi: 10.1080/10408390802068066. [Provides details on tea chemistry.] [DOI] [PubMed] [Google Scholar]
- 39.Yang CS, Wang H, Li GX, Yang Z, Guan F, Jin H. Cancer prevention by tea: evidence from laboratory studies. Pharmacol. Res. 2011;64:113–122. doi: 10.1016/j.phrs.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 40.Henning SM, Wang P, Heber D. Chemopreventive effects of tea in prostate cancer: green tea versus black tea. Mol. Nutr. Food Res. 2011;55:1–16. doi: 10.1002/mnfr.201000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pandey M, Gupta S. Elite, editor. Green tea and prostate cancer: from bench to clinic. Front. Biosci. 2009;1:13–25. doi: 10.2741/s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Higdon JV, Frei B. Tea catechins and polyphenols: health effects, metabolism, and antioxidant functions. Crit. Rev. Food Sci. Nutr. 2003;43:89–143. doi: 10.1080/10408690390826464. [DOI] [PubMed] [Google Scholar]
- 43.Shirakami Y, Shimizu M, Moriwaki H. Cancer chemoprevention with green tea catechins: from bench to bed. Curr. Drug Targets. 2012;13:1842–1857. doi: 10.2174/138945012804545506. [DOI] [PubMed] [Google Scholar]
- 44■.Yang CS, Wang H. Mechanistic issues concerning cancer prevention by tea catechins. Mol. Nutr. Food Res. 2011;55:819–831. doi: 10.1002/mnfr.201100036. [Provides an excellent overview on mechanisms of chemoprevention by tea polyphenols.] [DOI] [PubMed] [Google Scholar]
- 45.Gupta K, Thakur VS, Bhaskaran N, et al. Green tea polyphenols induce p53-dependent and p53-independent apoptosis in prostate cancer cells through two distinct mechanisms. PLoS ONE. 2012;7:e52572. doi: 10.1371/journal.pone.0052572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khan N, Afaq F, Saleem M, Ahmad N, Mukhtar H. Targeting multiple signaling pathways by green tea polyphenol (–)-epigallocatechin-3-gallate. Cancer Res. 2006;66:2500–2505. doi: 10.1158/0008-5472.CAN-05-3636. [DOI] [PubMed] [Google Scholar]
- 47.Thakur VS, Gupta K, Gupta S. Green tea polyphenols causes cell cycle arrest and apoptosis in prostate cancer cells by suppressing class I histone deacetylases. Carcinogenesis. 2012;33:377–384. doi: 10.1093/carcin/bgr277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee MH, Han DW, Hyon SH, Park JC. Apoptosis of human fibrosarcoma HT-1080 cells by epigallocatechin-3-O-gallate via induction of p53 and caspases as well as suppression of Bcl-2 and phosphorylated nuclear factor-κB. Apoptosis. 2011;16:75–85. doi: 10.1007/s10495-010-0548-y. [DOI] [PubMed] [Google Scholar]
- 49.Renouf M, Marmet C, Guy PA, et al. Dose-response plasma appearance of green tea catechins in adults. Mol. Nutr. Food Res. 2013;57:833–839. doi: 10.1002/mnfr.201200512. [DOI] [PubMed] [Google Scholar]
- 50.Borges G, Lean ME, Roberts SA, Crozier A. Bioavailability of dietary (poly)phenols: a study with ileostomists to discriminate between absorption in small and large intestine. Food Funct. 2013;4:754–762. doi: 10.1039/c3fo60024f. [DOI] [PubMed] [Google Scholar]
- 51.Lambert JD, Sang S, Lu AY, Yang CS. Metabolism of dietary polyphenols and possible interactions with drugs. Curr. Drug Metab. 2007;8:499–507. doi: 10.2174/138920007780866870. [DOI] [PubMed] [Google Scholar]
- 52.Lambert JD, Sang S, Yang CS. Biotransformation of green tea polyphenols and the biological activities of those metabolites. Mol. Pharm. 2007;4:819–825. doi: 10.1021/mp700075m. [DOI] [PubMed] [Google Scholar]
- 53.Sang S, Lambert JD, Ho CT, Yang CS. The chemistry and biotransformation of tea constituents. Pharmacol. Res. 2011;64:87–99. doi: 10.1016/j.phrs.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 54.Henning SM, Choo JJ, Heber D. Nongallated compared with gallated flavan-3-ols in green and black tea are more bioavailable. J. Nutr. 2008;138:S1529–S1534. doi: 10.1093/jn/138.8.1529S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fang M, Chen D, Yang CS. Dietary polyphenols may affect DNA methylation. J. Nutr. 2007;137:S223–S228. doi: 10.1093/jn/137.1.223S. [DOI] [PubMed] [Google Scholar]
- 56.Meng X, Sang S, Zhu N, et al. Identification and characterization of methylated and ring-fission metabolites of tea catechins formed in humans, mice, and rats. Chem. Res. Toxicol. 2002;15:1042–1050. doi: 10.1021/tx010184a. [DOI] [PubMed] [Google Scholar]
- 57.Renouf M, Redeuil K, Longet K, et al. Plasma pharmacokinetics of catechin metabolite 4′-O-Me-EGC in healthy humans. Eur. J. Nutr. 2011;50:575–580. doi: 10.1007/s00394-010-0164-1. [DOI] [PubMed] [Google Scholar]
- 58■.Wang P, Aronson WJ, Huang M, et al. Green tea polyphenols and metabolites in prostatectomy tissue: implications for cancer prevention. Cancer Prev. Res. (Phila.) 2010;3:985–993. doi: 10.1158/1940-6207.CAPR-09-0210. [First time (–)-epigallocatechin gallate was found in human prostate tissue in men consuming six cups of green tea (GT).] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Henning SM, Niu Y, Lee NH, et al. Bioavailability and antioxidant activity of tea flavanols after consumption of green tea, black tea, or a green tea extract supplement. Am. J. Clin. Nutr. 2004;80:1558–1564. doi: 10.1093/ajcn/80.6.1558. [DOI] [PubMed] [Google Scholar]
- 60.Stalmach A, Troufflard S, Serafini M, Crozier A. Absorption, metabolism and excretion of Choladi green tea flavan-3-ols by humans. Mol. Nutr. Food Res. 2008;53(Suppl. 1):S44–S53. doi: 10.1002/mnfr.200800169. [DOI] [PubMed] [Google Scholar]
- 61.Lambert JD, Lee MJ, Lu H, et al. Epigallocatechin-3-gallate is absorbed but extensively glucuronidated following oral administration to mice. J. Nutr. 2003;133:4172–4177. doi: 10.1093/jn/133.12.4172. [DOI] [PubMed] [Google Scholar]
- 62■.Henning SM, Wang P, Said J, et al. Polyphenols in brewed green tea inhibit prostate tumor xenograft growth by localizing to the tumor and decreasing oxidative stress and angiogenesis. J. Nutr. Biochem. 2012;23:1537–1542. doi: 10.1016/j.jnutbio.2011.10.007. [First evidence of in vivo inhibition of protein and gene expression of DNMT1 by GT administration in C57BL/6 mice.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Henning SM, Wang P, Abgaryan N, et al. Phenolic acid concentrations in plasma and urine from men consuming green and black and potential chemopreventive properties in colon cancer. Mol. Nutr. Food Res. 2013;57:483–493. doi: 10.1002/mnfr.201200646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang P, Heber D, Henning SM. Quercetin increased bioavailability and decreased methylation of green tea polyphenols in vitro and in vivo. Food Funct. 2012;3:635–642. doi: 10.1039/c2fo10254d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karhunen T, Tilgmann C, Ulmanen I, Julkunen I, Panula P. Distribution of catechol-O-methyltransferase enzyme in rat tissues. J. Histochem. Cytochem. 1994;42:1079–1090. doi: 10.1177/42.8.8027527. [DOI] [PubMed] [Google Scholar]
- 66.Landis-Piwowar KR, Wan SB, Wiegand RA, Kuhn DJ, Chan TH, Dou QP. Methylation suppresses the proteasome-inhibitory function of green tea polyphenols. J. Cell. Physiol. 2007;213:252–260. doi: 10.1002/jcp.21124. [DOI] [PubMed] [Google Scholar]
- 67.Zhu BT, Shim JY, Nagai M, Bai HW. Molecular modelling study of the mechanism of high-potency inhibition of human catechol-O-methyltransferase by (–)-epigallocatechin-3-O-gallate. Xenobiotica. 2008;38:130–146. doi: 10.1080/00498250701744641. [DOI] [PubMed] [Google Scholar]
- 68.Chen D, Wang CY, Lambert JD, Ai N, Welsh WJ, Yang CS. Inhibition of human liver catechol-O-methyltransferase by tea catechins and their metabolites: structure-activity relationship and molecular-modeling studies. Biochem. Pharmacol. 2005;69:1523–1531. doi: 10.1016/j.bcp.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 69.Weng Z, Greenhaw J, Salminen WF, Shi Q. Mechanisms for epigallocatechin gallate induced inhibition of drug metabolizing enzymes in rat liver microsomes. Toxicol. Lett. 2012;214:328–338. doi: 10.1016/j.toxlet.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 70.Lee WJ, Shim JY, Zhu BT. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol. Pharmacol. 2005;68:1018–1030. doi: 10.1124/mol.104.008367. [DOI] [PubMed] [Google Scholar]
- 71.Wong CP, Nguyen LP, Noh SK, Bray TM, Bruno RS, Ho E. Induction of regulatory T cells by green tea polyphenol EGCG. Immunol. Lett. 2011;139:7–13. doi: 10.1016/j.imlet.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kato K, Long NK, Makita H, et al. Effects of green tea polyphenol on methylation status of RECK gene and cancer cell invasion in oral squamous cell carcinoma cells. Br. J. Cancer. 2008;99:647–654. doi: 10.1038/sj.bjc.6604521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73■.Fang MZ, Wang Y, Ai N, et al. Tea polyphenol (–)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563–7570. [Original study of the effect of GT polyphenols on DNA methylation in cell culture.] [PubMed] [Google Scholar]
- 74.Montenegro MF, Saez-Ayala M, Pinero-Madrona A, Cabezas-Herrera J, Rodriguez-Lopez JN. Reactivation of the tumour suppressor RASSF1A in breast cancer by simultaneous targeting of DNA and E2F1 methylation. PLoS ONE. 2012;7:e52231. doi: 10.1371/journal.pone.0052231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nandakumar V, Vaid M, Katiyar SK. (–)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis. 2011;32:537–544. doi: 10.1093/carcin/bgq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meeran SM, Patel SN, Li Y, Shukla S, Tollefsbol TO. Bioactive dietary supplements reactivate ER expression in ER-negative breast cancer cells by active chromatin modifications. PLoS ONE. 2012;7:e37748. doi: 10.1371/journal.pone.0037748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Y, Yuan YY, Meeran SM, Tollefsbol TO. Synergistic epigenetic reactivation of estrogen receptor-a (ER-a) by combined green tea polyphenol and histone deacetylase inhibitor in ER-a-negative breast cancer cells. Mol. Cancer. 2010;9:274. doi: 10.1186/1476-4598-9-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Katiyar SK, Singh T, Prasad R, Sun Q, Vaid M. Epigenetic alterations in ultraviolet radiation-induced skin carcinogenesis: interaction of bioactive dietary components on epigenetic targets. Photochem. Photobiol. 2012;88:1066–1074. doi: 10.1111/j.1751-1097.2011.01020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen H, Landen CN, Li Y, Alvarez RD, Tollefsbol TO. Epigallocatechin gallate and sulforaphane combination treatment induce apoptosis in paclitaxel-resistant ovarian cancer cells through hTERT and Bcl-2 down-regulation. Exp. Cell Res. 2013;319:697–706. doi: 10.1016/j.yexcr.2012.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lambert JD, Sang S, Hong J, et al. Peracetylation as a means of enhancing in vitro bioactivity and bioavailability of epigallocatechin-3-gallate. Drug Metab. Dispos. 2006;34:2111–2116. doi: 10.1124/dmd.106.011460. [DOI] [PubMed] [Google Scholar]
- 81.Landis-Piwowar KR, Huo C, Chen D, et al. A novel prodrug of the green tea polyphenol (–)-epigallocatechin-3-gallate as a potential anticancer agent. Cancer Res. 2007;67:4303–4310. doi: 10.1158/0008-5472.CAN-06-4699. [DOI] [PubMed] [Google Scholar]
- 82.Meeran SM, Patel SN, Chan TH, Tollefsbol TO. A novel prodrug of epigallocatechin-3-gallate: differential epigenetic hTERT repression in human breast cancer cells. Cancer Prev. Res. (Phila.) 2011;4:1243–1254. doi: 10.1158/1940-6207.CAPR-11-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao Z, Xu Z, Hung MS, et al. Promoter demethylation of WIF-1 by epigallocatechin-3-gallate in lung cancer cells. Anticancer Res. 2009;29:2025–2030. [PubMed] [Google Scholar]
- 84.Pandey M, Shukla S, Gupta S. Promoter demethylation and chromatin remodeling by green tea polyphenols leads to re-expression of GSTP1 in human prostate cancer cells. Int. J. Cancer. 2010;126:2520–2533. doi: 10.1002/ijc.24988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85■.Chuang IC, Yoo CB, Kwan JM, et al. Comparison of biological effects of nonnucleoside DNA methylation inhibitors versus 5-aza-2′-deoxycytidine. Mol. Cancer Ther. 2005;4:1515–1520. doi: 10.1158/1535-7163.MCT-05-0172. [Demonstrates the difference in inhibition of DNA methylation between (–)-epigallocatechin gallate and nucleoside analog drugs.] [DOI] [PubMed] [Google Scholar]
- 86.Stresemann C, Brueckner B, Musch T, Stopper H, Lyko F. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 2006;66:2794–2800. doi: 10.1158/0008-5472.CAN-05-2821. [DOI] [PubMed] [Google Scholar]
- 87.Neilson AP, Hopf AS, Cooper BR, Pereira MA, Bomser JA, Ferruzzi MG. Catechin degradation with concurrent formation of homo- and heterocatechin dimers during in vitro digestion. J. Agric. Food Chem. 2007;55:8941–8949. doi: 10.1021/jf071645m. [DOI] [PubMed] [Google Scholar]
- 88.Neilson AP, Song BJ, Sapper TN, Bomser JA, Ferruzzi MG. Tea catechin auto-oxidation dimers are accumulated and retained by Caco-2 human intestinal cells. Nutr. Res. 2010;30:327–340. doi: 10.1016/j.nutres.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang GY, Liao J, Li C, et al. Effect of black and green tea polyphenols on c-jun phosphorylation and H(2)O(2) production in transformed and non-transformed human bronchial cell lines: possible mechanisms of cell growth inhibition and apoptosis induction. Carcinogenesis. 2000;21:2035–2039. doi: 10.1093/carcin/21.11.2035. [DOI] [PubMed] [Google Scholar]
- 90.Gros C, Fahy J, Halby L, et al. DNA methylation inhibitors in cancer: recent and future approaches. Biochimie. 2012;94:2280–2296. doi: 10.1016/j.biochi.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 91.Karahoca M, Momparler RL. Pharmacokinetic and pharmacodynamic analysis of 5-aza-2′-deoxycytidine (decitabine) in the design of its dose-schedule for cancer therapy. Clin. Epigenetics. 2013;5:3. doi: 10.1186/1868-7083-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Juergens RA, Wrangle J, Vendetti FP, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011;1:598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Navarro-Peran E, Cabezas-Herrera J, Garcia-Canovas F, Durrant MC, Thorneley RN, Rodriguez-Lopez JN. The antifolate activity of tea catechins. Cancer Res. 2005;65:2059–2064. doi: 10.1158/0008-5472.CAN-04-3469. [DOI] [PubMed] [Google Scholar]
- 94.Navarro-Peran E, Cabezas-Herrera J, Campo LS, Rodriguez-Lopez JN. Effects of folate cycle disruption by the green tea polyphenol epigallocatechin-3-gallate. Int. J. Biochem. Cell Biol. 2007;39:2215–2225. doi: 10.1016/j.biocel.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 95.Abali EE, Skacel NE, Celikkaya H, Hsieh YC. Regulation of human dihydrofolate reductase activity and expression. Vitam. Horm. 2008;79:267–292. doi: 10.1016/S0083-6729(08)00409-3. [DOI] [PubMed] [Google Scholar]
- 96.Correa A, Stolley A, Liu Y. Prenatal tea consumption and risks of anencephaly and spina bifida. Ann. Epidemiol. 2000;10:476–477. doi: 10.1016/s1047-2797(00)00144-7. [DOI] [PubMed] [Google Scholar]
- 97.Lemos C, Peters GJ, Jansen G, Martel F, Calhau C. Modulation of folate uptake in cultured human colon adenocarcinoma Caco-2 cells by dietary compounds. Eur. J. Nutr. 2007;46:329–336. doi: 10.1007/s00394-007-0670-y. [DOI] [PubMed] [Google Scholar]
- 98.Saez-Ayala M, Sanchez-del-Campo L, Montenegro MF, et al. Comparison of a pair of synthetic tea-catechin-derived epimers: synthesis, antifolate activity, and tyrosinase-mediated activation in melanoma. Chem. Med. Chem. 2011;6:440–449. doi: 10.1002/cmdc.201000482. [DOI] [PubMed] [Google Scholar]
- 99.Yen TT, Gill AM, Frigeri LG, Barsh GS, Wolff GL. Obesity, diabetes, and neoplasia in yellow A(vy)/- mice: ectopic expression of the agouti gene. FASEB J. 1994;8:479–488. doi: 10.1096/fasebj.8.8.8181666. [DOI] [PubMed] [Google Scholar]
- 100.Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12:949–957. [PubMed] [Google Scholar]
- 101.McCabe MT, Low JA, Daignault S, Imperiale MJ, Wojno KJ, Day ML. Inhibition of DNA methyltransferase activity prevents tumorigenesis in a mouse model of prostate cancer. Cancer Res. 2006;66:385–392. doi: 10.1158/0008-5472.CAN-05-2020. [DOI] [PubMed] [Google Scholar]
- 102.Volate SR, Muga SJ, Issa AY, Nitcheva D, Smith T, Wargovich MJ. Epigenetic modulation of the retinoid X receptor a by green tea in the azoxymethane-Apc Min/+ mouse model of intestinal cancer. Mol. Carcinog. 2009;48:920–933. doi: 10.1002/mc.20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mittal A, Piyathilake C, Hara Y, Katiyar SK. Exceptionally high protection of photocarcinogenesis by topical application of (–)-epigallocatechin-3-gallate in hydrophilic cream in SKH-1 hairless mouse model: relationship to inhibition of UVB-induced global DNA hypomethylation. Neoplasia. 2003;5:555–565. doi: 10.1016/s1476-5586(03)80039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Morey K, Zhang W, Pascual M, et al. Lack of evidence for green tea polyphenols as DNA methylation inhibitors in murine prostate. Cancer Prev. Res. (Phila.) 2009;2:1065–1075. doi: 10.1158/1940-6207.CAPR-09-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Adhami VM, Siddiqui IA, Sarfaraz S, et al. Effective prostate cancer chemopreventive intervention with green tea polyphenols in the TRAMP model depends on the stage of the disease. Clin. Cancer Res. 2009;15:1947–1953. doi: 10.1158/1078-0432.CCR-08-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Harper CE, Patel BB, Wang J, Eltoum IA, Lamartiniere CA. Epigallocatechin-3-gallate suppresses early stage, but not late stage prostate cancer in TRAMP mice: mechanisms of action. Prostate. 2007;67:1576–1589. doi: 10.1002/pros.20643. [DOI] [PubMed] [Google Scholar]
- 107.Nair S, Hebbar V, Shen G, et al. Synergistic effects of a combination of dietary factors sulforaphane and (–)-epigallocatechin-3- gallate in HT-29 AP-1 human colon carcinoma cells. Pharm. Res. 2008;25:387–399. doi: 10.1007/s11095-007-9364-7. [DOI] [PubMed] [Google Scholar]
- 108.Nair S, Barve A, Khor TO, et al. Regulation of Nrf2- and AP-1-mediated gene expression by epigallocatechin-3-gallate and sulforaphane in prostate of Nrf2-knockout or C57BL/6J mice and PC-3 AP-1 human prostate cancer cells. Acta Pharmacol. Sin. 2010;31:1223–1240. doi: 10.1038/aps.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Choi KC, Jung MG, Lee YH, et al. Epigallocatechin-3-gallate, a histone acetyltransferase inhibitor, inhibits EBV-induced B lymphocyte transformation via suppression of RelA acetylation. Cancer Res. 2009;69:583–592. doi: 10.1158/0008-5472.CAN-08-2442. [DOI] [PubMed] [Google Scholar]
- 110.Lovat F, Valeri N, Croce CM. MicroRNAs in the pathogenesis of cancer. Semin. Oncol. 2011;38:724–733. doi: 10.1053/j.seminoncol.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 111.Tsang WP, Kwok TT. Epigallocatechin gallate up-regulation of miR-16 and induction of apoptosis in human cancer cells. J. Nutr. Biochem. 2010;21:140–146. doi: 10.1016/j.jnutbio.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 112.Olthof MR, Hollman PC, Zock PL, Katan MB. Consumption of high doses of chlorogenic acid, present in coffee, or of black tea increases plasma total homocysteine concentrations in humans. Am. J. Clin. Nutr. 2001;73:532–538. doi: 10.1093/ajcn/73.3.532. [DOI] [PubMed] [Google Scholar]
- 113.Yuasa Y, Nagasaki H, Akiyama Y, et al. Relationship between CDX2 gene methylation and dietary factors in gastric cancer patients. Carcinogenesis. 2005;26:193–200. doi: 10.1093/carcin/bgh304. [DOI] [PubMed] [Google Scholar]
- 114■.Yuasa Y, Nagasaki H, Akiyama Y, et al. DNA methylation status is inversely correlated with green tea intake and physical activity in gastric cancer patients. Int. J. Cancer. 2009;124:2677–2682. doi: 10.1002/ijc.24231. [Demonstrates a correlation between high GT intake and DNA methylation status.] [DOI] [PubMed] [Google Scholar]
- 115.Lim U, Song MA. Dietary and lifestyle factors of DNA methylation. Methods Mol. Biol. 2012;863:359–376. doi: 10.1007/978-1-61779-612-8_23. [DOI] [PubMed] [Google Scholar]
- 116.Jacob RA, Gretz DM, Taylor PC, et al. Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. J. Nutr. 1998;128:1204–1212. doi: 10.1093/jn/128.7.1204. [DOI] [PubMed] [Google Scholar]
- 117.Rampersaud GC, Kauwell GP, Hutson AD, Cerda JJ, Bailey LB. Genomic DNA methylation decreases in response to moderate folate depletion in elderly women. Am. J. Clin. Nutr. 2000;72:998–1003. doi: 10.1093/ajcn/72.4.998. [DOI] [PubMed] [Google Scholar]
- 118.Pufulete M, Al-Ghnaniem R, Khushal A, et al. Effect of folic acid supplementation on genomic DNA methylation in patients with colorectal adenoma. Gut. 2005;54:648–653. doi: 10.1136/gut.2004.054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Morey K, Zhang W, Pascual M, et al. Lack of evidence for green tea polyphenols as DNA methylation inhibitors in murine prostate. Cancer Prev. Res. (Phila.) 2009;2:1065–1075. doi: 10.1158/1940-6207.CAPR-09-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]