Abstract
Until recently, mutations in histones had not been described in any human disease. However, genome-wide sequencing of pediatric high-grade gliomas revealed somatic heterozygous mutations in the genes encoding histones H3.1 and H3.3, as well as mutations in the chromatin modifiers ATRX and DAXX. The functional significance and mechanistic details of how these mutations affect the tumors is currently under intensive investigation. The information gained from these studies will shed new light on normal brain development as well as increase our understanding of the tumorigenic processes that drive pediatric high-grade gliomas.
The first histone mutations to be associated with human disease are mutations in histones H3.1 and H3.3. Their functional significance is currently under intensive investigation.
Histone proteins are among the most highly conserved proteins in eukaryotes, with the majority of the protein sequence being identical in organisms ranging from yeast to humans. Histones are involved in the basic packaging of DNA, allowing two meters of DNA to fit inside the nucleus of a single cell! There are four core histones: H2A, H2B, H3, and H4. DNA in the nucleus is wrapped around a histone octamer comprised of two of each of the core histones, forming a nucleosome. A string of nucleosomes is then further compacted to form chromatin. The amino-terminal tails of each of the core histones protrude from the nucleosome and receive a variety of posttranslational modifications (PTMs). Because essentially all cells in the body have the same genome but different sets of genes are expressed, it is the combination of PTMs of the histone tails—often referred to as the histone code—that largely determines the structure of the chromatin and whether genes will or will not be transcribed in each cell. This epigenetic regulation of gene expression is a key factor in cell determination and differentiation, and thus organismal development as a whole.
Until recently, there had not been any reports of histone mutations in any human disease. In January 2012, two studies simultaneously reported the first ever histone mutations in pediatric brain tumor patients. Both groups reported recurrent somatic heterozygous mutations in the gene encoding the histone variant H3.3 (i.e., H3F3A), in patients diagnosed with non-brainstem pediatric glioblastomas (non-BS-PGs) and diffuse intrinsic pontine gliomas (DIPGs; Schwartzentruber et al. 2012; Wu et al. 2012). One of the groups also reported heterozygous mutations in the gene encoding histone H3.1 (i.e., HIST1H3B) in a significant percentage of DIPGs (Wu et al. 2012). Strikingly, these mutually exclusive H3 mutations resulted in amino acid substitutions at two specific positions in the proteins: lysine-to-methionine substitutions at position 27 (K27M) in both H3.1 and H3.3 and either a glycine-to-arginine or glycine-to-valine substitution at position 34 (G34R, G34V) in H3.3. In fact, 78% of DIPG samples and 36% of non-BS-PG contained these histone mutations (Fig. 1B,C).
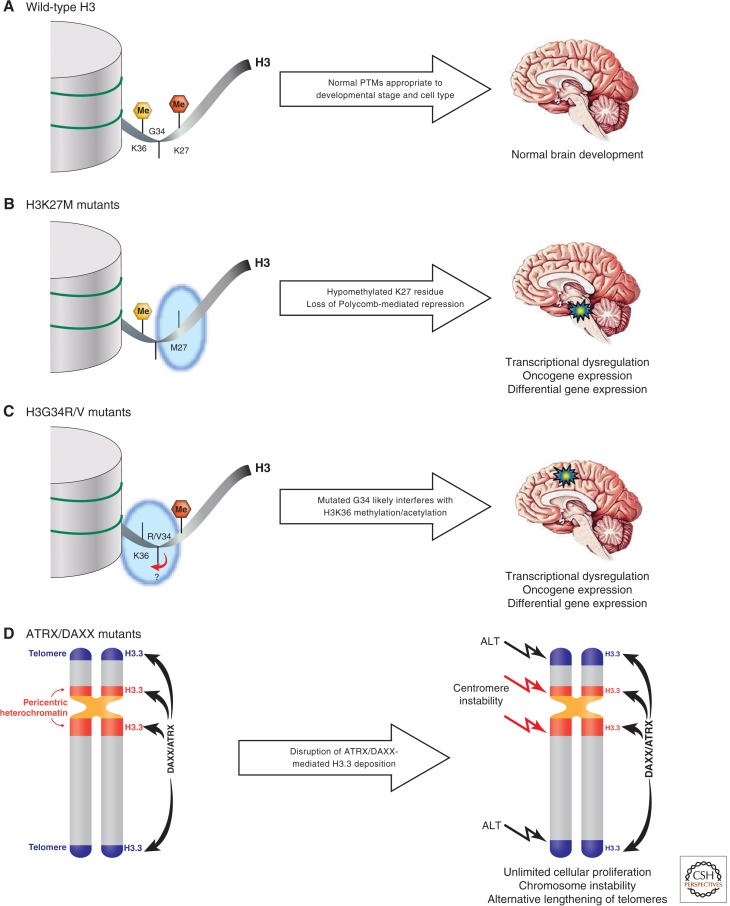
Figure 1.
Potential effects of histone H3 and ATRX/DAXX mutations in the tumorigenic process of pediatric high-grade gliomas. (A) In the absence of mutations, the amino-terminal tail of histone H3 receives a myriad of cell-type- and developmental-stage-specific posttranslational modifications (PTMs) dictating the gene expression profiles required for normal brain development. (B) The H3K27M mutation results in a hypomethylated K27 residue (loss of red hexagon), preventing Polycomb-mediated repression of target genes. This likely results in oncogene expression and an altered gene expression profile suitable for the development of high-grade gliomas of the midline structures, most notably diffuse intrinsic pontine gliomas (blue/yellow star). (C) The H3G34R/V mutation diminishes the levels of H3K36 methylation (loss of yellow hexagon), which may impact transcriptional elongation in addition to generating a gene expression profile conducive to the formation of non-brainstem pediatric glioblastomas (blue/yellow star). The H3G34R/V may also impact on the acetylation of H3K36 (not shown for illustrative purposes). (D) Mutations in ATRX/DAXX alter the proper deposition of histone H3.3 at pericentric and telomeric heterochromatic loci, thus compromising chromatin structure and allowing for genomic instability and alternative lengthening of telomeres (ALT). H3K27me, red hexagon; H3K36me, yellow hexagon; high-grade glioma, blue/yellow star; telomeres, blue chromatin shading; pericentric heterochromatin, red chromatin shading.
The histone H3.1 and H3.3 variants are structurally similar proteins that differ at only five amino acid positions. H3.1 is termed a replication-dependent histone because it is expressed and incorporated into nucleosomes during S phase of the cell cycle. Conversely, H3.3 is replication-independent as it is expressed throughout the cell cycle and replaces existing nucleosomal histone H3 variants at a variety of loci along the genome (discussed in Henikoff and Smith 2014). In the hundreds of brain tumor samples sequenced between the two studies, only residues K27 and G34 of histone H3 were affected. This begs the question: Why is there such an extreme selective pressure for mutations affecting these residues?
Lysine 27 of histone H3 (H3K27) is a critical residue that, when trimethylated (me3), is involved in transcriptional repression via Polycomb repressive complexes 1 and 2. The H3K27me3 modification regulates the expression of genes associated with lineage commitment, cellular differentiation, and anterior–posterior patterning (Faria et al. 2011; Grossniklaus and Paro 2014). Thus, H3K27 has a role in normal brain development. Indeed, just a year after the discovery of these histone mutations, researchers are gaining some insight into the mechanistic details pertaining to the function of these mutations; namely, that the K27M mutation acts via a dominant-negative gain of function by competitively inhibiting the methyltransferase activity of EZH2 and thus abolishing Polycomb-mediated repression of numerous genes (Lewis et al. 2013).
The functional significance of the G34R/V mutation is less straightforward to interpret. Glycine 34 of histone H3 (H3G34) lies in close proximity to lysine 36 (H3K36), a residue that regulates transcriptional elongation. In fact, H3G34R/V mutant nucleosomes show reduced methylation of H3K36 by SETD2, the only human methyltransferase specific for H3K36 (Lewis et al. 2013). This suggests that the H3G34R/V mutation impacts the ability of histone-modifying complexes to methylate H3K36, thus altering the transcription of several target genes. Gene expression analyses revealed patterns of gene expression that were different in samples with the H3K27M mutation versus samples with the H3G34R/V mutation, both of which differ from the normal brain. These changes in gene expression could result in the transcription of oncogenes or microRNAs with oncogenic functions as well as prevent the expression of tumor-suppressor genes, promoting the growth of the respective tumors.
In addition to mutations in histone H3 genes, it was determined that there were frequent inactivating mutations in ATRX and DAXX in non-BS-PGs (Schwartzentruber et al. 2012). ATRX and DAXX encode chromatin-remodeling proteins responsible for the replication-independent incorporation of H3.3 at pericentric and telomeric heterochromatic loci. Inactivating mutations in these genes have indeed been previously found in pediatric and adult glioblastomas, neuroblastomas, and pancreatic neuroendocrine tumors and are among the many reported epigenetic regulators to be mutated in different cancers. Mutations in ATRX/DAXX may interfere with H3.3 incorporation at these heterochromatic loci, thus compromising the structural integrity of the chromosome (Fig. 1D). Interestingly, alternative lengthening of telomeres (ALT), a telomerase-independent telomere maintenance mechanism, was also frequently observed in tumors with ATRX mutations (Lovejoy et al. 2012; Schwartzentruber et al. 2012). Although it is not entirely clear how cancer-associated ALT operates, the genomic instability associated with mutations in ATRX/DAXX somehow results in telomeric dysfunction, allowing for ALT. This aberrant telomere lengthening mechanism provides tumor cells with the capacity for unlimited cellular proliferation, one of the hallmarks of cancer cells.
As there was a staggering frequency of histone H3 mutations, why were these mutations not found in earlier DNA sequencing studies? This is due in part to the new sequencing techniques now available. Rather than selecting and sequencing genes thought to be important, these two groups used different unbiased genome-wide sequencing methods to cover all protein-coding genes in samples from diseased and normal tissue. Furthermore, the clinical samples used in these two studies were from pediatric patients, not adults. These histone mutations are almost exclusively found in pediatric patients. This is of major significance as the human brain continues to develop postnatally. This then prompts the question: What is unique about the developing brain that enables these mutations to be tumorigenic in children and not adults? Surprisingly, the majority of tumors with H3K27M mutations were found in the thalamus or brainstem, structures in the midline of the brain. In contrast, H3G34R/V tumors arose in the cortex. The development of the normal brain is a very dynamic and complex process, involving numerous extracellular factors that are present at precise times in specific brain locations. It is possible that the different microenvironmental factors in these particular regions of the developing brain work in concert with the altered transcriptional profiles induced by the H3K27M and H3G34R/V mutations. Together, this may contribute to an increased transformation potential, resulting in the formation of these specific types of pediatric high-grade gliomas.
Often, the study of disease conditions leads to a deeper understanding of normal biological function. It is clear that more in-depth studies are needed to discern the functional roles of these mutations pertaining to tumor biology and normal brain development. The information and insight that will be gained from such studies will potentially provide clinically relevant diagnostic, prognostic, and/or therapeutic benefits for pediatric patients with this disease, as well as increasing our overall knowledge of epigenetics.
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, and Danny Reinberg
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Faria CMC, Rutka JT, Smith C, Kongkham P 2011. Epigenetic mechanisms regulating neural development and pediatric brain tumor formation. J Neurosurg Pediatr 8: 119–132 [DOI] [PubMed] [Google Scholar]
- *.Grossniklaus U, Paro R 2014. Transcriptional silencing by Polycomb group proteins. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Henikoff S, Smith M 2014. Histone variants and epigenetics. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Müller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD 2013. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340: 857–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, De Lange T, De S, Petrini JHJ, Sung PA, Jasin M, et al. 2012. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet 8: e1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu X-Y, Jones DTW, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang D-AK, Tönjes M, et al. 2012. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482: 226–231 [DOI] [PubMed] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, et al. 2012. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 44: 251–253 [DOI] [PMC free article] [PubMed] [Google Scholar]