Abstract
Histone deacetylases (HDACs) are enzymes that catalyze the removal of acetyl functional groups from the lysine residues of both histone and nonhistone proteins. In humans, there are 18 HDAC enzymes that use either zinc- or NAD+-dependent mechanisms to deacetylate acetyl lysine substrates. Although removal of histone acetyl epigenetic modification by HDACs regulates chromatin structure and transcription, deacetylation of nonhistones controls diverse cellular processes. HDAC inhibitors are already known potential anticancer agents and show promise for the treatment of many diseases.
In humans, there are 18 histone deacetylase enzymes. Many, if not all, can also deacetylate nonhistone proteins.
OVERVIEW
The posttranslational modification (PTM) of histones can cause a functional change in gene expression or chromatin structure—many epigenetic phenomena have testified to this. A common form of histone modification and, indeed, one of the first discovered is acetylation, which occurs at the ε-amino group of lysines, largely in the amino-terminal tail of histones. Results from many early studies suggest that histone acetylation regulates gene transcription. The exact number and combination of acetylated lysine residues that occur in histones that are self-perpetuating and heritable in the cell is unknown. It is clear, nonetheless, that histone acetylation is an abundant source of potential epigenetic information.
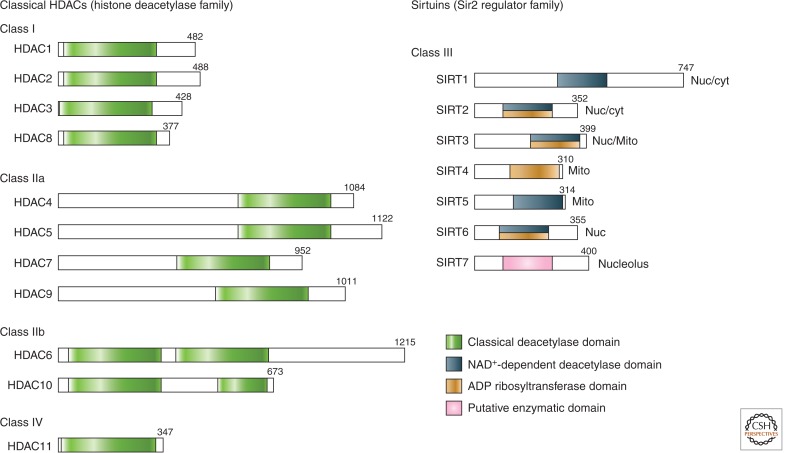
Histone lysine acetylation is highly reversible. A lysine residue becomes acetylated by the action of the histone/lysine acetyltransferase enzymes (HATs/KATs), and is removed by histone deacetylases (HDACs). In humans, there are 18 HDAC enzymes divided into four classes: the Class I Rpd3-like proteins (HDAC1, HDAC2, HDAC3, and HDAC8); the Class II Hda1-like proteins (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, and HDAC10); the Class III Sir2-like proteins (SIRT1, SIRT2, SIRT3, SIRT4, SIRT5, SIRT6, and SIRT7); and the Class IV protein (HDAC11). Like HATs, some HDACs possess substrate specificity. Accumulating evidence suggests that many, if not all, HDACs can also deacetylate nonhistone proteins. It is therefore important to take this fact into consideration when trying to ascertain an HDAC’s function.
Structural comparisons among different Class I and II HDACs, as well as HDAC homologs from different species that share significant homology with human classical HDACs, reveal a conserved group of active site residues, suggesting a common mechanism for the metal-dependent hydrolysis of acetylated substrates. The Class III HDACs use NAD+ as a reactant to deacetylate acetyl lysine residues of protein substrates forming nicotinamide, the deacetylated product, and the metabolite 2′-O-acetyl-ADP-ribose.
By removing acetyl groups from ε-amino lysines of proteins, HDACs not only alter transcription, but also promote either the establishment or erasure of alternative posttranslational lysine modifications such as methylation, ubiquitination, and sumoylation. Additionally, they may change the dynamics of histone modification “cross talk.” Like many important cellular enzymes, HDACs are subject to a variety of controlling mechanisms, including protein–protein interactions and posttranslational modifications. Abnormal HDACs play a key role in many human diseases. A thorough understanding of the functions and mechanisms of HDACs action is a prerequisite to further our understanding of how this family of enzymes impacts on human health and disease.
The availability of HDAC inhibitors has accelerated our understanding of HDAC functions and mechanism of actions. A number of compounds that inhibit HDAC activity have now been developed and characterized. They reportedly cause cell growth arrest, differentiation and/or apoptosis, and restrain tumor growth in animals. In parallel, research is increasingly showing that epigenetic abnormalities are tightly associated with a large number of human diseases, providing a rationale for the use of epigenetic-based therapies such as HDAC inhibitors.
1. INTRODUCTION
An enzyme activity that catalyzes the removal of acetyl functional groups from histones was first discovered in a calf thymus extract in 1969 (Inoue and Fujimoto 1969). The deacetylase activity has preference toward the ε-amino acetyl groups of lysine residues compared to the α-amino acetyl groups of amino-terminal amino acids in histones. Treatment of deacetylation reactions with proteinases destroyed most of the enzymatic deacetylation activity. A number of studies followed in the early 1970s further dissecting the biochemical characterization of the histone deacetylase activity in various tissues. Many different properties of histone deacetylase activity were learned from these early chromatography studies, including the finding that the activity contains multiple enzymes. However, initial attempts to purify HDACs to homogeneity using conventional chromatography were unsuccessful. It was not until 1996 that the histone deacetylation field exploded, when the first bona fide histone deacetylase, HDAC1, was isolated and cloned (Taunton et al. 1996). Since that time, more than 15,000 papers have been published on this topic (compared to less than 100 papers published up to that time). There is now overwhelming support showing that HDACs play crucial roles in gene transcription and most likely in all eukaryotic biological processes that involve chromatin. Notably, recent discussions on eukaryotic transcriptional repression refer to some aspects of histone deacetylation. This article will focus on important discoveries in the HDAC field in the last two decades since the cloning of the first HDAC. Particular emphasis will be placed on highlighting the structures, functions, mechanisms of action, and regulation of HDACs that will be beneficial to readers interested in histone modifications, chromatin, or epigenetics, as well as scientists not currently working in this area.
2. HDAC FAMILIES AND CLASSES: tWO FAMILIES AND FOUR CLASSES
Each HDAC belongs to either the histone deacetylase family or the Sir2 regulator family. In humans, HDACs are traditionally divided into separate categories called classes based on sequence similarities (Table 1; illustrated in Fig. 1). The Class I proteins (HDAC1, HDAC2, HDAC3, and HDAC8) have sequence similarity to the yeast Rpd3 protein. The Class II proteins (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, and HDAC10) have sequence similarity to the yeast Hda1 protein. Three proteins in Saccharomyces cerevisiae—Hos1, Hos2, and Hos3—however, have 35%–49% identity to Rpd3, and 21%–28% identity to Hda1. Thus, mammalian Class I and II HDACs are also related to the yeast Hos proteins. The Class III proteins (SIRT1, SIRT2, SIRT3, SIRT4, SIRT5, SIRT6, and SIRT7) have sequence similarity to the yeast Sir2 protein. The Class IV protein (HDAC11) shares sequence similarity to both Class I and II proteins. It should be noted that the different classes of HDACs are not to be confused with “Class” in taxonomy classification, or the Structural Classification of Proteins hierarchy classification, in which all HDACs belong to the α and β proteins class.
Table 1.
HDAC classification
| Superfamily | Family | Class | Protein (S. cerevisiae) | Subclass | Protein (human) |
|---|---|---|---|---|---|
| Arginase/deacetylase superfamily | Histone deacetylase family | Class I | Rpd3, Hos1, Hos2, Hos3 | HDAC1, HDAC2, HDAC3, HDAC8 | |
| Class II | Hda1 | Class IIa | HDAC4, HDAC5, HDAC7, HDAC9 | ||
| Class IIb | HDAC6, HDAC10 | ||||
| Class IV | HDAC11 | ||||
| Deoxyhypusine synthase like NAD/FAD-binding domain superfamily | Sir2 regulator family | Class III | Sir2, Hst1, Hst2, Hst3, Hst4 | I II III IV |
SIRT1, SIRT2, SIRT3 SIRT4 SIRT5 SIRT6, SIRT7 |
Figure 1.
Domain organization of human HDACs. The total number of amino acid residues in each HDAC is shown on the right of each protein. Many HDACs have multiple isoforms and, for simplicity, only the longest isoform is shown. Enzymatic domains (or putative enzymatic domains) are shown in colors. Sirtuin localizations: Nuc, nuclear; cyt, cytoplasmic; Mito, mitochrondial.
The Class I, II, and IV HDACs are numbered according to their chronological order of discovery. For example, HDAC1 was first reported several months before HDAC2, both in 1996 (Taunton et al. 1996; Yang et al. 1996). HDAC3 was discovered the following year (Yang et al. 1997). HDAC4, 5, and 6 were first described in 1999 (Grozinger et al. 1999), HDAC7 in early 2000 (Kao et al. 2000), and so on. It is important to clarify the term HDAC isoforms (or isoenzymes) that are frequently used in the literature. Many different forms of an HDAC may arise, such as by single-nucleotide polymorphisms or by alternative splicings. For example, the HDAC9 transcript is alternatively spliced to generate multiple protein isoforms with distinct biological activities. Thus, HDAC9 isoforms (several different forms of the same HDAC9 protein) clearly exist. However, HDAC4, HDAC5, HDAC7, and HDAC9 each originate from a distinct gene and, therefore, although functionally related, they are technically not isoforms of each other.
2.1. Arginase/Deacetylase Superfamily and the Histone Deacetylase Family
The Class I, II, and IV HDACs belong to the arginase/deacetylase superfamily of proteins (Table 1). This superfamily contains the arginase-like amidino hydrolases, as well as the histone deacetylases. It was proposed that eukaryotic HDACs originated from a prokaryotic enzyme similar to the acetylpolyamine amidohydrolases. The prokaryotic enzyme targeted the reversible acetylation and deacetylation of the aminoalkyl group of a DNA binding molecule to achieve a gene regulatory effect (Leipe and Landsman 1997).
The histone deacetylase family of HDACs (sometimes referred to as the classical HDAC family) is made up of three classes of proteins: Class I, II, and IV HDACs. Proteins within each class descend from a common ancestor and have similar three-dimensional structures, functions, and significant sequence homology. A phylogenetic analysis of all histone deacetylase family proteins and all proteins related to this family from all fully sequenced free-living organisms has been performed, together with the analysis of gene duplication events. Results indicate that the common ancestor of metazoan organisms contained two Class I, two Class II, and a single Class IV HDAC (Gregoretti et al. 2004). Functional prediction reveals that self-association is common among this family of HDACs. Interestingly, all Class I, II, and IV HDACs precede the evolution of histone proteins suggesting that the primary substrates for this HDAC family may be nonhistones.
2.1.1. Class I (HDAC1, HDAC2, HDAC3, HDAC8)
In S. cerevisiae, the histone deacetylase-A 1 (Hda1) protein, which shares sequence similarity to the transcription regulator reduced potassium dependency 3 (Rpd3), is a subunit of a large histone deacetylase complex, Hda. Hda1 also shares similarity to three yeast proteins designated Hos1, Hos2, and Hos3. Another yeast histone deacetylase complex, Hdb, contains Rpd3 as an associated factor. Using a trapoxin (an inhibitor of histone deacetylase) affinity matrix, Stuart Schreiber purified and cloned a human 55-kDa protein related to the yeast protein Rpd3 (Taunton et al. 1996). Immunoprecipitation of this 55-kDa protein, HDAC1 (initially called HD1), showed that it contains histone deacetylase activity. A second human histone deacetylase protein, HDAC2 (initially called mRPD3), with high homology with yeast Rpd3 was independently identified as a transcription factor (Yang et al. 1996). HDAC2 negatively regulates transcription by being recruited to DNA as a corepressor. The third human Rpd3-related protein, HDAC3, was discovered by searching the GenBank database for DNA and protein sequences with homology to HDAC1 and HDAC2 (e.g., Yang et al. 1997). Like HDAC1 and HDAC2, HDAC3 represses transcription, binds to and is recruited by transcription factors, and is expressed in many different cell types. Similar to the identification of HDAC3, a search of the GenBank database for protein sequences similar to HDAC1, 2, and 3, led to the discovery of HDAC8 (e.g., Hu et al. 2000). The highly conserved deacetylase domains of Class I HDACs share extensive homology with each other, with 45%–94% amino acid sequence identity.
It is widely stated in the literature that Class I HDACs are located in the nucleus and are ubiquitously expressed. Results from more thorough studies reveal that HDAC3 expression is restrictive to certain tissues, and HDAC1, HDAC2, HDAC3, and HDAC8 can be localized to the cytoplasm or specialized cellular organelles. Therefore, the generalization that Class I HDACs are nuclear and ubiquitously expressed may be oversimplified, and it is fair to predict that Class I HDACs might possess yet to be discovered extranuclear or tissue-specific functions.
2.1.2. Class II (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, HDAC10)
HDAC4, HDAC5, and HDAC6 were discovered together after GenBank databases search for human HDACs with sequence similarity to yeast Hda1 (e.g., Grozinger et al. 1999). These proteins possess certain features present in the conserved catalytic domains of Class I human HDACs, but also contain additional sequence domains that have no similarity to Class I enzymes. The divergence of the Class I and II HDACs appears to have occurred relatively early in evolution. Like Class I HDACs, immunopurified recombinant HDAC4, HDAC5, and HDAC6 possess in vitro HDAC activity, although at a much lower level. HDAC6, interestingly, contains an internal duplication of two deacetylase catalytic domains, which appear to function independently of each other.
HDAC7 was first isolated as a protein that interacts with the transcription corepressor silencing mediator for retinoid or thyroid-hormone receptors (SMRT) (Kao et al. 2000). HDAC7 possesses three repression domains, two of which contain autonomous repressor functions that are independent of the third deacetylase repressor activity. Shortly after the discovery of HDAC7, Paul Marks reported the identification of a protein, HDAC-related protein (HDRP), that shares 50% identity in deduced amino acid sequence to the noncatalytic amino-terminal domain of HDAC4 and HDAC5 (Zhou et al. 2000). Subsequently, HDAC9 was identified by a homology database search using the human HDAC4 amino acid sequence. HDAC9 has multiple alternatively spliced isoforms. One of these isoforms, which is an amino-terminal splice variant, is the HDRP or myocyte enhancer–binding factor 2–interacting transcriptional repressor (MITR). Like all Class I and II HDACs, HDAC9 possesses a conserved deacetylase domain, which represses gene activity when recruited to a promoter through deacetylation of histones. HDAC4, 5, 7, and 9 make up the Class IIa HDACs and share 48%–57% overall identity.
HDAC10 was discovered independently by four different groups (e.g., Kao et al. 2002). Homology comparison indicates that HDAC10 is most similar to HDAC6 (55% overall identity), and both contain a unique, putative second catalytic domain not found in other HDACs. Therefore, HDAC6 and HDAC10 are subclassified as Class IIb. An interesting feature of both Class IIa and IIb HDACs is their subcellular localization; each member of these classes shows at least some cytoplasmic localization, suggesting a major cytoplasmic functional role for Class II HDACs. The conserved deacetylase domains of Class II HDACs share 23%–81% amino acid sequence identity to each other.
2.1.3. Class IV (HDAC11)
HDAC11 is the sole member of the Class IV HDAC. It uniquely shares sequence homology with the catalytic domains of both Class I and II HDACs, and was first discovered by Basic Local Alignment Search Tool searches of GenBank databases using the yeast Hos3 protein as the query sequence (Gao et al. 2002). HDAC11 regulates the protein stability of DNA replication factor CDT1 (Glozak and Seto 2009) and the expression of interleukin 10. HDAC11, together with HDAC10, are probably the least studied and most poorly understood HDACs in the classical HDAC family.
2.2. Deoxyhypusine Synthase–Like NAD/FAD-Binding Domain Superfamily and the Sir2 Regulator Family
The deoxyhypusine synthase (DHS)-like NAD/FAD-binding domain superfamily of proteins includes silent information regulator 2 (Sir2) proteins, as well as deoxyhypusine synthase, carboxy-terminal domain of the electron transfer flavoprotein α subunit, pyruvate oxidase and decarboxylase middle domain, transhydrogenase domain III, and the ACDE2-like families. The founding member of the Sir2 regulator family of proteins, the S. cerevisiae Sir2, was originally identified in a genetic screen for genes involved in controlling expression of silent mating type loci. In yeast, Sir2 is required for transcription silencing (see Grunstein and Gasser 2013 for extensive description). The Sir2 regulator family has only one class (i.e., Class III) of nicotinamide adenine dinucleotide (NAD+)-dependent histone deacetylase, with seven Sir2-like proteins in humans (SIRT1, SIRT2, SIRT3, SIRT4, SIRT5, SIRT6, SIRT7). Sir2-like proteins (sirtuins) are phylogenetically conserved in eukaryotes, prokaryotes, and archaea, and based on phylogenetic relationships, they can be grouped into more than a dozen classes and subclasses. The first classification was organized into five major classes: I (SIRT1, SIRT2, SIRT3), II (SIRT4), III (SIRT5), IV (SIRT6, SIRT7), U (cobB in bacteria, no human homolog) (see Fig. 4 in Grunstein and Gasser 2013). All sirtuins contain a conserved core domain with several sequence motifs.
2.2.1. Class III (SIRT1, SIRT2, SIRT3, SIRT4, SIRT5, SIRT6, SIRT7)
S. cerevisiae homologs of Sir2 (Hsts) and the conservation of this protein family from bacteria to humans were first described by Lorraine Pillus and Jef Boeke (Brachmann et al. 1995). Subsequently, five human sirtuins (SIRT1, SIRT2, SIRT3, SIRT4, SIRT5) were identified in the GenBank database using S. cerevisiae Sir2 amino acid sequence as the probe (Frye 1999). Two additional human sirtuins (SIRT6 and SIRT7) were similarly identified using human SIRT4 as the probe. The seven sirtuins share 22%–50% overall amino acid sequence identity, and 27%–88% identity in the conserved catalytic domains. Of the seven human sirtuins, SIRT1 is most similar to the yeast Sir2 protein, possesses the most robust histone deacetylase activity, and has been most extensively studied.
A remarkable feature of sirtuins is that they have two enzymatic activities: mono-ADP-ribosyltransferase and histone deacetylase. SIRT5 possesses additional protein lysine desuccinylase and demalonylase activity in vitro (Du et al. 2011). Another interesting characteristic of sirtuins is their localizations (see Fig. 1), with SIRT1 and SIRT2 found in the nucleus and cytoplasm, SIRT3 in the nucleus and mitochondria, SIRT4 and SIRT5 exclusively in the mitochondria, SIRT6 only in the nucleus, and SIRT7 in the nucleolus. Like the Class I, II, and IV HDACs, sirtuins also have nonhistone substrates, at least in eukaryotes.
3. CATALYTIC MECHANISMS AND STRUCTURES
3.1. Catalytic Mechanisms and Structures of Classical HDACs (Class I and II)
The classical HDAC family of enzymes (Class I, II, IV) share a common catalytic mechanism that requires a zinc ion (Fig. 2). Insights into the catalytic mechanisms for the metal-dependent hydrolysis of the acetamide bond in acetylated lysine have been provided by structural, biochemical, and mutational analysis.
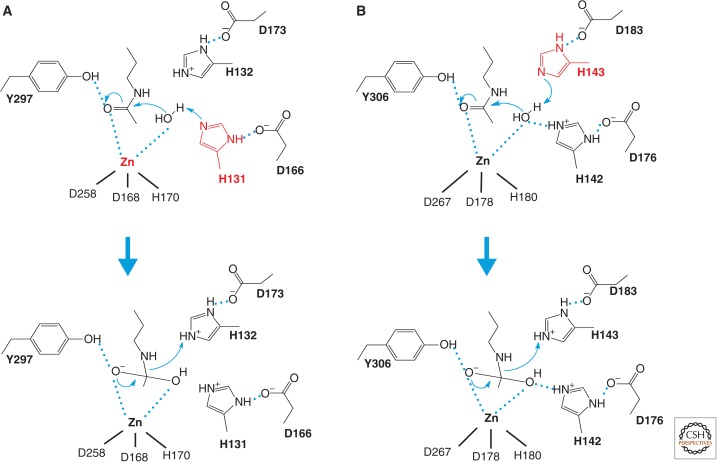
Figure 2.
Catalytic mechanism of HDACs. Two models for the catalytic mechanism of the Zn-dependent HDAC reaction have been proposed. (A) A model proposed from the HDLP structure. The HDLP catalytic core consists of a tubular pocket, a zinc-binding site, and active-site residues (in bold) of a tyrosine (Y297) and two histidines (H131 and H132) that make hydrogen bonds to two aspartic acids (D166 and D173). One of these catalytic histidines (red) facilitates nucleophilic attack at the substrate carbonyl by activating a water molecule coordinated with the zinc ion. Initially, two tandem histidine residues (H131 and H132) were proposed to function as Asp-His charge relay systems, typical of serine proteases such as chymotrypsin and chymotrypsinogen in the enzyme reaction. The active site zinc ion is coordinated by three residues (two aspartic acids and one histidine). (B) A model proposed from the HDAC8 structure, in which the other histidine residue (red) plays an essential role in the electron transfer. Hydrogen bond interactions are drawn in dotted lines.
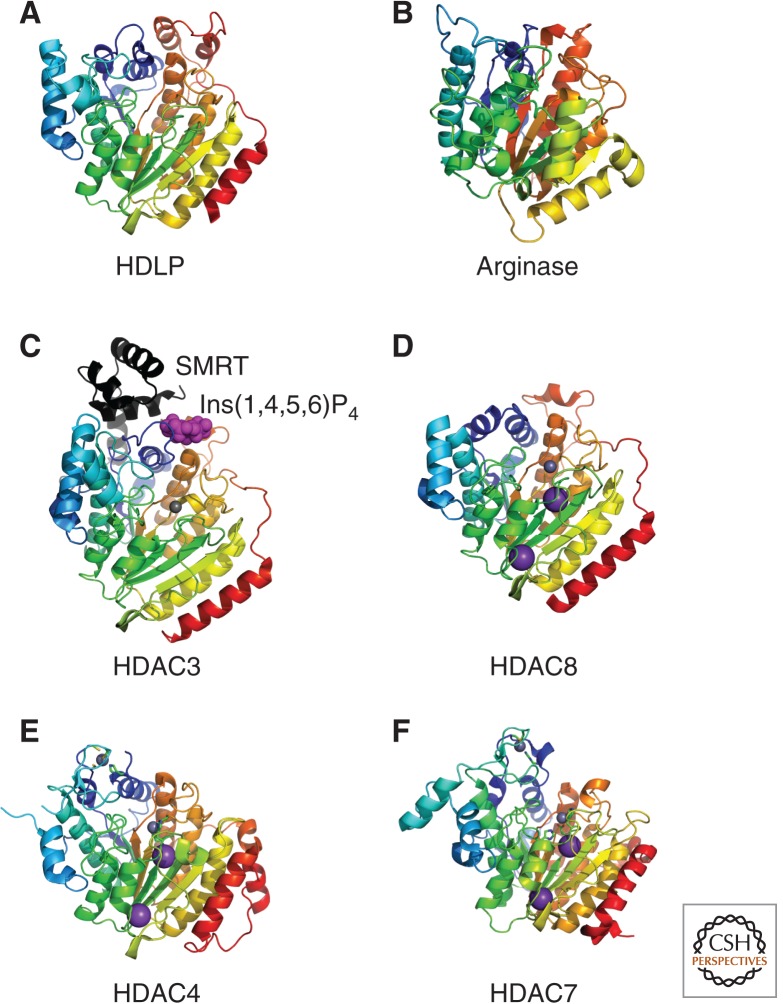
The first X-ray crystal structure of a classical HDAC family protein was determined for the histone deacetylase-like protein (HDLP) from a hyperthermophilic bacterium Aquifex aeolicus (Fig. 3A). HDLP has the same topology as arginase containing an α/β fold and an 8-stranded parallel β-sheet (Finnin et al. 1999). This similarity to arginase (Fig. 3B), a metalloenzyme that catalyzes the hydrolysis of arginine to ornithine, suggests that it evolved from a common metalloprotein ancestor.
Figure 3.
Structure of Class I and II HDACs. Crystal structures of the arginase/deacetylase superfamily of proteins are illustrated. Metal ions are represented as space filled spheres: red, gray, and violet spheres indicate Mn, Zn, and K atoms, respectively. (A) Aquifex aeolicus HDLP (1C3P), (B) rat arginase (PDB ID: 1RLA), (C) human Class I HDAC3 (4A69) in complex with Ins(1,4,5,6)P4 (highlighted by magenta space-filled spheres) and the deacetylase activation domain from the human SMRT corepressor (depicted with a ribbon model in black), (D) human Class I HDAC8 (3F07), (E) human Class IIa HDAC4 (2VQW), and (F) human Class IIa HDAC7 (3C0Y).
Structural studies of the HDLP catalytic core points to a catalytic reaction model illustrated in Figure 2A. Studies from X-ray crystallography of mammalian HDACs showed essentially the same catalytic domain structure as HDLP, in which the residues that make up the active site and contact inhibitors are conserved across the HDAC family. Structural analysis of HDAC8 (Fig. 3D) and its mutants, however, suggested a distinct model. This model proposes that one of the histidine residues acts as the general base (H143), whereas the other histidine (H142) serves as a general electrostatic catalyst (Fig. 2B). The HDAC8 H143A mutant has an almost complete loss of activity in contrast to the residual activity of an H142A mutant, in concordance with the proposed model of action (Gantt et al. 2010). Furthermore, quantum mechanical/molecular mechanical molecular dynamics simulations suggest that a neutral H143 first serves as the general base to accept a proton from the zinc-bound water molecule in the initial rate-determining nucleophilic attack step, and then shuttles it to the amide nitrogen atom to facilitate the cleavage of the amide bond (Wu et al. 2011). This model seems to be a more likely catalytic mechanism for HDAC-related enzymes, which is similar to the conventional metalloenzymes thermolysin and carboxypeptidase A.
All of the catalytic residues illustrated in Figure 2B are conserved in other HDACs except Class IIa enzymes, in which the tyrosine residue is replaced by a histidine residue. This tyrosine residue is positioned next to the zinc, opposite the histidine-aspartic acid residues, and is important for stabilizing the tetrahedral intermediate. Both the zinc and the tyrosine residue participate in polarizing the substrate carbonyl (C = O) for the nucleophilic attack. Thus the lack of the tyrosine residue in HDAC4 and other vertebrate Class IIa HDACs may be responsible for the low catalytic activity. Indeed, catalytic activity of HDAC4 was restored by a single His to Tyr substitution, reaching levels comparable to that of Class I enzymes (Lahm et al. 2007).
Most recently, X-ray crystallographic structure of HDAC3 complexed with the deacetylase activation domain of the human SMRT (NCoR) corepressor was determined (Fig. 3C). Surprisingly, inositol (1,4,5,6)-tetrakisphosphate was present in the interface between HDAC3 and SMRT acting as an “intermolecular glue.” This inositol phosphate molecule is essential for the interaction between the two proteins as well as the catalytic activity of HDAC3 (Watson et al. 2012).
3.2. Catalytic Mechanisms and Structures of Sirtuins (Class III)
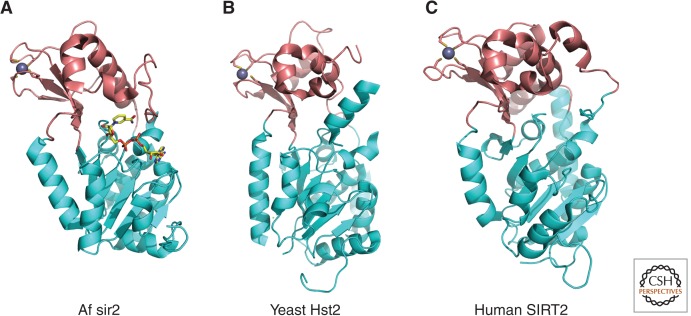
Class III HDACs require NAD+ as the cofactor for enzyme activity in contrast to the zinc-dependent catalysis by Class I, II, and IV enzymes (e.g., Imai et al. 2000). Structural studies of archaeal, yeast and human homologs of Sir2 have shown that the catalytic domain of sirtuins resides in a cleft formed between a large domain with a Rossmann-fold and a small zinc-binding domain (Fig. 4) (e.g., Finnin et al. 2001). The amino acid residues in the cleft are conserved in the sirtuin family and form a protein-tunnel in which the substrate interacts with NAD+. Recently, the crystal structure of SIRT5 was solved, revealing a larger substrate-binding site that may accept a larger acyl group on the lysine residue (Schuetz et al. 2007). This agrees with SIRT5 acting as a protein lysine desuccinylase and demalonylase rather than a deacetylase (Du et al. 2011).
Figure 4.
Overall structure of sirtuins (Class III). Crystal structures of the sirtuin family of proteins are illustrated as cartoons highlighting the large Rossmann-fold domains (cyan) and small zinc-binding domains (brown). (A) Archaeoglobus fulgidus sir2 (PDB ID: 1ICI). NAD+ is drawn in stick model form in which yellow, blue, red, and orange represent C, N, O, and P atoms, respectively. (B) Yeast Hst2 (1Q14). (C) Human SIRT2 (1J8F). Zinc ions are represented as space-filled spheres in gray.
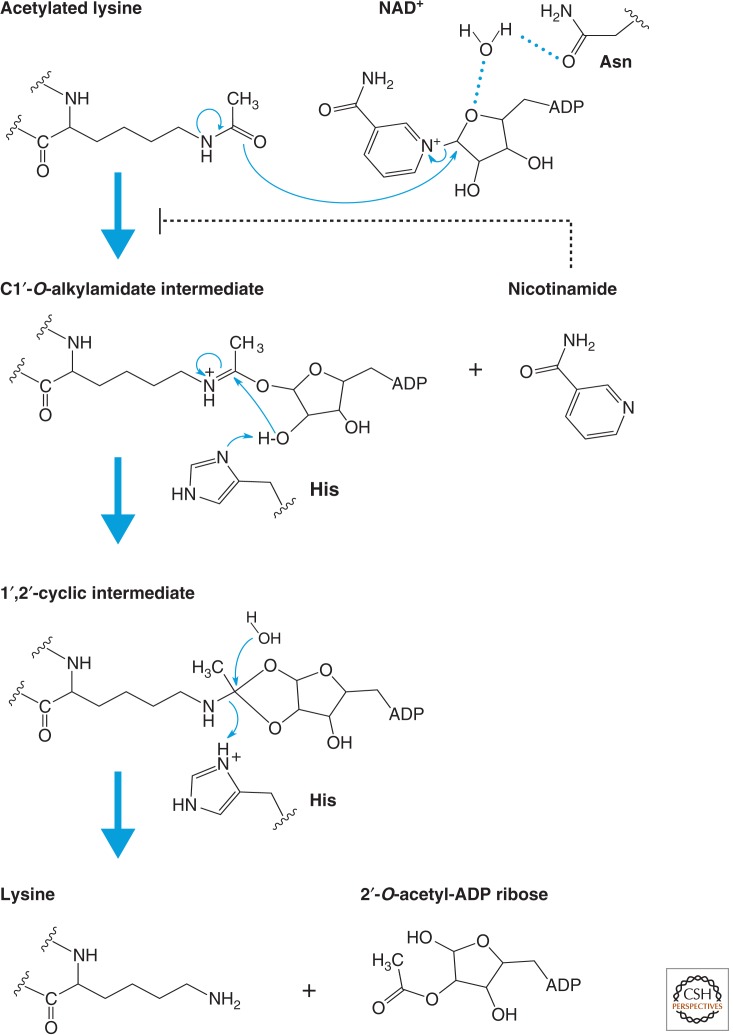
A proposed chemical mechanism for nicotinamide cleavage from NAD+ and ADP-ribose transfer to an acetylated lysine is illustrated in Figure 5. This is based on detailed structural analysis of substrates and/or cofactor analogs complexed with Sir2 homologs (e.g., Avalos et al. 2004). The first step of the reaction involves nucleophilic addition of the acetamide oxygen to the C1′ position of the nicotinamide ribose to form a C1′-O-alkylamidate intermediate and free nicotinamide. The C1′-O-alkylamidate intermediate is then converted to a 1′,2′-cyclic intermediate from which lysine and 2′-O-acetyl-ADP ribose are eventually released. Nicotinamide, one of the byproducts, acts as an inhibitor of sirtuins.
Figure 5.
Catalytic mechanism of sirtuins (Class III). Proposed mechanism of the NAD+-dependent deacetylase reaction. The first step of the reaction involves nucleophilic addition of the acetamide oxygen to the C1′ position of the nicotinamide ribose to form a C1′-O-alkylamidate intermediate and free nicotinamide. Next, the 2′-hydroxy group of the NAD+ ribose is activated by an active site histidine residue that, in turn, attacks the C1′-O-alkylamidate to form the 1′,2′-cyclic intermediate. The 1′,2′-cyclic intermediate is then attacked by an activated water molecule resulting in the formation of deacetylated lysine and 2′-O-acetyl-ADP ribose. 2′-O-acetyl-ADP ribose can be readily converted to 3′-O-acetyl-ADP ribose in aqueous solution by nonenzymatic intramolecular transesterification. Thus, nicotinamide, the deacetylated peptide, and a mixture of 2′- and 3′-O-acetyl-ADP ribose are the final reaction products.
Some sirtuin family members may also possess intrinsic mono-ADP-ribosyltransferase activity. The ADP-ribosyltransferase activity of sirtuins has been thought of as a low efficiency side-reaction caused by the partial uncoupling of intrinsic deacetylation and acetate transfer to ADP-ribose. Recently, however, mono-ADP-ribosyltransferase activity was found to be the main enzymatic activity of at least SIRT4, whereas SIRT2 and SIRT6 display both deacetylase and mono-ADP-ribosyltransferase activities (e.g., Frye 1999; Liszt et al. 2005). It is currently unclear whether NAD+-dependent deacetylation and mono-ADP-ribosylation of proteins can occur simultaneously.
4. HDAC SUBSTRATES
4.1. Histone Substrates
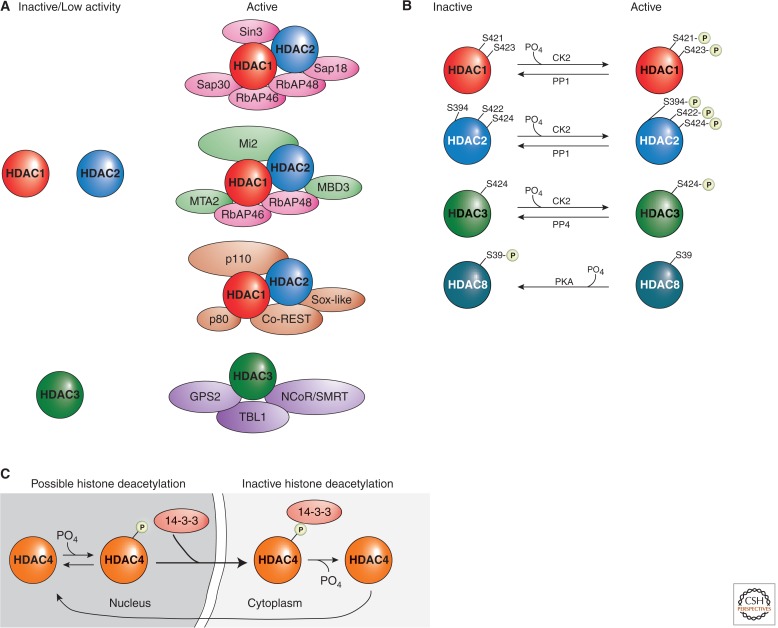
Soon after the discovery of HATs, it became apparent that each may have a particular histone substrate specificity (discussed in Marmorstein and Zhou 2014). The search for Class I, II, and IV HDAC histone substrate specificity has turned out to be much more difficult than for HATs. One obstacle to identifying HDAC substrate specificity is that most HDACs within this family possess very low measurable histone deacetylase activity when purified to homogeneity. Functional redundancy of many HDACs also contributes to the difficulty in deciphering substrate specificity. For example, knockdown of one classical HDAC can be compensated by the activity of another HDAC within the same class or even from a different class. Additionally, some Class I HDACs exist in several different complexes, with each complex potentially having different substrate preferences. For example, HDAC1 is present in at least three different stable protein complexes, with each complex possibly targeting different substrates (Fig. 6A). Finally, substrate specificity may differ depending on the source of the substrate, such as nucleosomal histones versus free histones.
Figure 6.
Examples of HDAC regulation. (A) Class I HDACs are commonly regulated by protein complex formations. HDAC1, HDAC2, and HDAC3 possess low enzymatic activities when in isolation, and their activities increase significantly when present in holoenzyme complexes. (B) The activities of Class I HDACs are modulated by phosphorylation and dephosphorylation. In general, phosphorylation activates HDAC1, HDAC2, and HDAC3, although it represses HDAC8 activities. (C) Phosphorylation of a Class II HDAC, HDAC4, promotes its interaction with the 14-3-3 protein and, subsequently, changes its localization. Multiple residues (S246, S467, and S632 on HDAC4, and corresponding conserved sites on HDAC5, HDAC7, and HDAC9) confer the HDAC–14-3-3 interactions.
Early studies indicated that purified HDAC1 alone or an HDAC1/2 complex deacetylated all four core histones. The same results for HDAC4, HDAC5, and HDAC6 suggested a lack of specificity (Grozinger et al. 1999). A later study, however, suggested that HDAC1 can deacetylate all four core histones at all lysines tested, but with varying efficiency (Johnson et al. 2002). Another study suggested that HDAC8 preferentially deacetylates histones H3 and H4 (Hu et al. 2000), whereas HDAC11 might specifically deacetylate H3K9 and H3K14.
The complexity in elucidating Class I, II, and IV HDAC histone substrate specificity can be illustrated by attempts to identify HDAC3 substrates. Using an immunoprecipitated HDAC3 complex and purified nucleosomes, it was shown that HDAC3 deacetylates histone H4 more efficiently than HDAC1. Using a similar approach, however, showed that although HDAC3 immunocomplexes completely deacetylated H2A, H4K5, and H4K12, it only partially deacetylated H3, H2B, H4K8, and H4K16 (Johnson et al. 2002). Interestingly, compared to HDAC1, HDAC3 preferentially deacetylates H4K5, H4K12, and H2AK5 in HeLa cells, based on chromatin immunoprecipitation (ChIP) experiments. Although these studies show that HDAC3 (and perhaps all Class I HDACs) has distinct substrate specificity, it is important to note that immunopurified HDAC3 complexes contain other proteins that themselves possess HDAC activity, thus complicating the interpretation of these results. In fact, in a study using homozygous HDAC3-deficient DT40 cells, the acetylation levels of H4K8 and H4K12 were similar to those detected in wild-type cells, suggesting that histone H4 might not be a major target of HDAC3 in vivo. Also, in an in vitro reconstituted chromatin system, an HDAC3-containing protein complex selectively deacetylated histone H3, compared to an HDAC1/2 complex that deacetylated both histones H3 and H4 (Vermeulen et al. 2004). In another study, there was no change in the overall acetylation status of histones H3 or H4 or the acetylation of H4K8 or H4K12 in HeLa cells depleted of HDAC3 (Zhang et al. 2004). Knockdown of HDAC3 by expression of HDAC3-specific siRNA, however, increased the acetylation of H3K9 and H3K18. This knockdown increased the acetylation of H3K9 to a greater extent than the HDAC1-specific siRNA did, whereas the reverse was true for H3K18. Although these incompatible results are quite puzzling, they do support the general conclusion that each Class I, II, IV HDAC might possess distinct histone substrate specificity. The possibility that each HDAC within the classical family targets specific lysines in certain histones, however, still requires more comprehensive investigations.
The case for sirtuins histone substrate specificity is less ambiguous (Table 2). Yeast Sir2 deacetylates H3K9, H3K14, and H4K16 (Imai et al. 2000). Biochemical studies revealed that SIRT1 deacetylates histones H4K16 and H3K9, and interacts with and deacetylates histone H1K26, mediating heterochromatin formation (Vaquero et al. 2004). Although there are reports that SIRT1 can deacetylate all four core histones in vitro, SIRT1’s chief target site on histones is H4K16 (Vaquero et al. 2004). Also, SIRT1 associates with lysine-specific histone demethylase 1 (LSD1) and together play a concerted role in deacetylating histone H4K16 and demethylating H3K4 to repress gene expression (Mulligan et al. 2011).
Table 2.
Sirtuin histone substrates
| Sirtuin | Histone substrate | Biological relevance |
|---|---|---|
| SIRT1 | H3K9 H3K14 H3K56 H4K16 H1K26 |
Chromatin organization, DNA repair/genome stability, cancer |
| SIRT2 | H4K16 H3K56 |
Chromatin condensation/mitosis, DNA repair, cancer |
| SIRT3 | H4K16 | Chromatin silencing, DNA repair, cellular stress |
| SIRT4 | None | |
| SIRT5 | None | |
| SIRT6 | H3K9 H3K56 |
Telomeric chromatin/senescence, DNA repair/genome stability |
| SIRT7 | H3K18 | Cellular transformation |
Like SIRT1, SIRT2 and its yeast ortholog, Hst2, have a strong preference for histone H4K16 (Vaquero et al. 2006). Although SIRT2 is located mainly in the cytoplasm, it localizes to chromatin during the mammalian G2/M transition phase of the cell cycle and deacetylates H4K16. Further, SIRT3 is a predominant mitochondrial protein, but like SIRT2, it also deacetylates H4K16 when transported to the nucleus under certain conditions.
Both SIRT4 and SIRT5 are located exclusively in the mitochondria and, therefore, do not deacetylate histones. Initially, SIRT6 was determined to be a mono-ADP-ribosyltransferase, and not a histone deacetylase enzyme (Liszt et al. 2005). Later studies, however, reveal that SIRT6 deacetylates H3K9 and H3K56 (e.g., Michishita et al. 2008), although biochemical and structural analyses argue that SIRT6 has very low deacetylase activity compared to other sirtuins. Deacetylation of H3K9 by SIRT6 modulates telomeric chromatin. SIRT7 is a highly selective H3K18 deacetylase that plays a key role in cellular transformation (Barber et al. 2012).
4.2. Nonhistone Substrates
ε-amino lysine acetylation and deacetylation of nonhistone proteins were first reported for high-mobility group proteins 1 and 2 (HMG-1 and HMG-2; Sterner et al. 1979). It was shown that an enzyme preparation that deacetylates histone H4 can also deacetylate HMG-1 and HMG-2. In the last 15 years, a large number of acetylated nonhistone proteins have been discovered and shown to be deacetylated by HDACs. Their functional consequences have been a subject of intense research. Perhaps the best characterized nonhistone HDAC substrate is the tumor suppressor protein p53. HDAC1 interacts with and deacetylates p53, promoting p53 ubiquitination and degradation (Luo et al. 2000). In addition, SIRT1 also binds to and deacetylates p53 (e.g., Vaziri et al. 2001). Deacetylation of p53 by SIRT1 decreases the ability of p53 to transcriptionally activate the cell cycle inhibitor p21, which causes cells to reenter into the cell cycle following DNA repair. Another well-characterized nonhistone HDAC substrate is the cytoskeletal protein α-tubulin. HDAC6 deacetylates lysine 40 of α-tubulin and regulates microtubule-dependent cell motility (Hubbert et al. 2002). These examples illustrate the ability of HDACs to regulate important biological processes without modifying histones.
More recently, using high-resolution mass spectrometry, more than 3600 acetylation sites on 1750 proteins were identified (Choudhary et al. 2009). The acetylation sites are present on nuclear, cytoplasmic, and mitochondrial proteins involved in many different cellular processes. The use of suberoylanilide hydroxamic acid (SAHA) and MS-275, two broad spectrum Class I, II, IV HDAC inhibitors, up-regulated ∼10% of all acetylation sites by at least a factor of 2, suggesting that many of these acetylations are regulated by classical HDACs. In a similar study, using a proteomics approach, 213 nuclear proteins were unambiguously identified to be hyperacetylated in the absence of Sirt1 (Peng et al. 2012). Again, these proteins have a range of diverse functions including DNA damage repair, apoptosis and survival, cell cycle, transcription, RNA processing, translation, metabolism, and chromatin structure. Also, high-throughput genetic interaction profiling revealed that HDACs functionally regulate nonhistone substrates that govern a wide array of biological processes (Lin et al. 2012). Profiling of protein acetylation in bacteria (which do not possess histones), likewise, has indicated that HDACs deacetylate a significant number of proteins in lower organisms, emphasizing the highly conserved nature of HDACs (Wang et al. 2010).
Using computational prediction tools, it is anticipated that many more ε-amino lysine acetylation sites on a wide variety of proteins are yet to be discovered. Thus, nonhistone lysine acetylation is prevalent and comparable with that of other major posttranslational modifications. Determining which of these acetylated proteins are functionally regulated by HDACs, therefore, is an important endeavor in future research. Considering the large number of potential nonhistone HDAC substrates, coupled with the fact that functional HDACs are present in cells devoid of histones, it is tempting to speculate that the chief function of HDACs is to regulate nonepigenetic phenomena. However, it is interesting that the activities of DNMT1, a key enzyme responsible for DNA methylation, and many histone modification enzymes (HATs, HDACs, HMTs) that are critical for heritable changes in gene expression, are regulated by HDACs (Choudhary et al. 2009; Peng et al. 2012). Therefore, although deacetylation of nonhistones in itself may not directly serve to erase epigenetic modifications or help establish an epigenetic off chromatin state, it could modify inheritance of chromatin states indirectly by deacetylation of other epigenetic-modifying enzymes.
5. REGULATION OF HDAC ACTIVITY
Like nearly all enzymes that are involved in critical cellular functions, the activities of HDACs are highly regulated. This regulation is achieved by a variety of different mechanisms at the transcription, posttranscription, translation, and posttranslational levels. The most well-defined mechanisms of HDAC regulation are protein–protein interactions and posttranslational modifications (PTMs). Less studied, but perhaps equally important, is the regulation of some HDACs by control of expression, alternative RNA splicing, availability of cofactors, subcellular localization, and proteolytic processing.
5.1. Protein Complexes
Many important molecular processes in the cell are performed by large multisubunit protein complexes. A protein may interact, sometimes transiently, with another protein to activate or repress the other protein’s activities and functions. This is indeed a common mechanism used to regulate many HDACs (Fig. 6A). Early studies indicated that the isolation of HDAC1 or HDAC2 individually, without associated proteins, generally yielded very low enzymatic activity. Subsequent biochemical analyses revealed that human HDAC1 and HDAC2 exist together in at least three distinct multiprotein complexes called the Sin3, the NuRD, and the CoREST complexes (reviewed by Ayer 1999). Sin3 and NuRD complexes share a common core, comprised of four proteins: HDAC1, HDAC2, RbAp46, and RbAp48. Additionally, each complex contains unique polypeptides (Sin3, SAP18, and SAP30 in the Sin3 complex; Mi2, MTA-2, and MBD3 in the NuRD complex). In a NuRD reconstitution experiment using purified subunits, it was shown that HDAC activity of the core complex was severely compromised compared to the native NuRD holo-complex. The addition of certain cofactors to the core complex, however, was sufficient to direct the formation of an enzymatically active complex. In a different study, it was shown that a protein in the Sin3 corepressor complex augments enzymatic activity of HDAC1 in vivo. Similarly, in the CoREST complex, the association of HDAC1/2 with CoREST is essential for HDAC enzymatic activity.
Another well-defined example of HDAC regulation by protein–protein interaction came from studies of HDAC3. Early studies suggested that SMRT and the nuclear receptor corepressor (NCoR) function as platforms for recruitment of HDACs. Subsequent studies surprisingly revealed that the interaction between HDAC3 and SMRT/NCoR resulted in the stimulation of HDAC3 enzymatic activity (e.g., Wen et al. 2000). In contrast, SMRT/NCoR mutants that did not bind HDAC3 could not activate HDAC3. Activation of this deacetylase enzymatic activity by SMRT/NCoR is specific to HDAC3.
For Class III HDACs, several proteins have been reported to interact and regulate SIRT1. Of these, the best described is the association of SIRT1 with deleted in breast cancer 1 (DBC1; e.g., Zhao et al. 2008). DBC1 negatively regulates SIRT1 deacetylase activity, which leads to an increase in p53 acetylation and up-regulation of p53-mediated function. Depletion or down-regulation of DBC1 conversely stimulates SIRT1-mediated deacetylation of p53 and inhibits p53-dependent apoptosis. SIRT1 also positively self-regulates via a small region in the carboxy-terminal of the protein, which competes with DBC1 binding (Kang et al. 2011). Another nuclear protein called active regulator of SIRT1, or AROS, when bound, enhances SIRT1-mediated deacetylation of p53, and inhibits p53-mediated transcriptional activity (Kim et al. 2007). Depletion of AROS enhances p21 expression and increases both the G0/G1 population and apoptosis in response to DNA damage, whereas AROS overexpression improves cell survival. The activation of SIRT1 by protein–protein interaction with AROS is reminiscent of the activation of Sir2 deacetylase activity in S. cerevisiae by interaction with Sir4 (discussed in Grunstein and Gasser 2013).
5.2. Posttranslational Modifications
HDACs can undergo a variety of posttranslational modifications including acetylation, glycosylation, S-nitrosylation, sumoylation, ubiquitination, and phosphorylation. The most extensively studied modification, phosphorylation, affects HDAC functionality (Fig. 6B,C). HDAC1 is phosphorylated by cAMP-dependent kinase PKA and protein kinase CK2 (Pflum et al. 2001). The two phospho-acceptor sites on the carboxyl-terminal of HDAC1, S421, and S423 are essential for enzymatic activity and when mutated to alanine result in a significant reduction in enzymatic activity. These mutations also disrupt protein complex formation of HDAC1 with RbAp48, MTA2, Sin3, and CoREST. HDAC2 is similarly phosphorylated at residues S394, S422, and S424 (corresponding to S393, S421, and S423 of HDAC1; Tsai and Seto 2002). Its phosphorylation also promotes enzymatic activity and affects protein complex formation with Sin3 and Mi2. Intriguingly, in cancer cells HDAC1 is phosphorylated at Y221 (which corresponds to Y222 of HDAC2; Rush et al. 2005). Because HDAC1-Y221 (HDAC2-Y222) is conserved in humans, mice, Xenopus laevis, and Caenorhabditis elegans, tyrosine phosphorylation may also be important in the regulation of HDAC1 and HDAC2 activities.
Phosphorylation of HDAC1 and HDAC2 is reversibly regulated by the protein phosphatase PP1 (e.g., Galasinski et al. 2002). Interestingly, mitotic arrest but not G1/S arrested cells results in hyperphosphorylation of HDAC2 (without any change in HDAC1), suggesting that spindle checkpoint activation provides a physiological stimulus that leads to HDAC2 hyperphosphorylation.
Phosphorylation of HDAC3 by CK2 and DNA-PKcs significantly enhances HDAC3 activity (e.g., Zhang et al. 2005). HDAC3 can also be phosphorylated by GSK-3β, and inhibition of GSK-3β protects against HDAC3-induced neurotoxicity (Bardai and D’Mello 2011). HDAC3’s phospho-acceptor site, S424, which is a nonconserved residue among the Class I HDACs, when mutated to alanine severely compromises enzymatic activity reminiscent of HDAC1 and HDAC2. Unlike HDAC1 and HDAC2, however, HDAC3 associates with the catalytic and regulatory subunits of the protein serine/threonine phosphatase 4 complex (PP4c/PP4R1), and dephosphorylation of HDAC3 by PP4 down-regulates HDAC3 enzymatic activity (Zhang et al. 2005).
HDAC8 regulation is quite different to other Class I enzymes. For one, phosphorylation inhibits, rather than increases its enzymatic activity (Lee et al. 2004). Mutation of S39 to alanine enhances HDAC8 activity, whereas an activator of HDAC8 phosphorylation causes a reduction in HDAC8 activity. Also, HDAC8 is refractory to phosphorylation by protein kinase CK2, but is instead phosphorylated by PKA. Crystal structures of HDAC8 revealed that S39 lies at the surface of HDAC8, roughly 20 Å from the opening to the HDAC8 active site (Somoza et al. 2004). Phosphorylation of S39 predictably leads to a major structural disruption of this region of the surface, which ultimately would negatively affect HDAC8’s activity.
The phosphorylation state and modifying enzymes specific to Class IIa HDACs have been extensively studied and found to affect enzyme activity, partly through cellular localization. The subcellular localization is regulated by the binding of the 14-3-3 proteins to a phosphorylated HDAC4 (Fig. 6C). A working model proposes that phosphorylation of HDAC4 at residues S245, S467, and S632 induces 14-3-3 binding to HDACs, preventing access of importin to the nuclear localization signal on HDAC4. This results in cytoplasmic sequestration of HDACs. No less than six groups of kinases have been shown to phosphorylate the 14-3-3 binding sites (reviewed by Seto and Yang 2010): Ca2+/calmodulin-dependent kinases, protein kinase D, microtubule affinity-regulating kinases, salt-inducible kinases, checkpoint kinase-1, and AMP-activated protein kinase (AMPK). In addition to nuclear export and cytoplasmic retention, phosphorylation of Class IIa HDACs may lead to their ubiquitination and proteasomal degradation.
Two phosphatases have been implicated in the regulation of Class IIa HDACs activities and functions: protein phosphatase 1β (PP1β), including myosin phosphatase targeting subunit 1 (MYPT1, a regulatory subunit of PP1), and protein phosphatase 2A (PP2A). In addition to 14-3-3 binding sites, PP2A also dephosphorylates S298 of HDAC4, which is required for nuclear import of HDAC4.
For Class IIb HDACs, global proteomic profiling of phosphopeptides, revealed that HDAC6 is phosphorylated at S22 and T30 (e.g., Beausoleil et al. 2004). However, the functional significance of phosphorylation on these two sites is unknown. Aurora A (AurA) phosphorylates HDAC6 to activate tubulin deacetylase activity (Pugacheva et al. 2007). At the basal body of cilia, phosphorylation and activation of HDAC6 promotes ciliary disassembly, and this pathway is both necessary and sufficient for ciliary resorption. In line with these observations, small molecule inhibitors of AurA and HDAC6 selectively stabilize cilia from regulated resorption cues. Other HDAC6 kinases include GSK3β, G protein–coupled receptor kinase 2, protein kinase CK2, and epidermal growth factor receptor.
For Class III HDACs, SIRT1 is a known phosphoprotein (Beausoleil et al. 2004) containing 13 residues that can be phosphorylated based on mass spectrometry data (Sasaki et al. 2008). Dephosphorylation of SIRT1 results in a decrease in SIRT1 deacetylase activity, arguing that SIRT1 activity is regulated by phosphorylation. Cell cycle–dependent kinase cyclin B/Cdk1 phosphorylates SIRT1, and mutations of two Cdk1 target sites, T530 and S540, disturbs normal cell cycle progression and fails to rescue proliferation defects in SIRT1-deficient cells. Mutation of the Cdk1 phosphorylation sites, however, did not lead to a reduction in deacetylase activity, suggesting that other phosphorylation sites might be more important for deacetylase activity. The protein levels of SIRT1 may be regulated by phosphorylation of SIRT1 at S27 (Ford et al. 2008). Cells depleted of c-JUN amino-terminal kinase 2 (JNK2), but not JNK1, in cells resulted in a SIRT1 protein with a reduced half-life and a lack of phosphorylation at S27. However, it is not clear in this study if the decrease in protein levels corresponds to a decrease in SIRT1 deacetylase activity in these cells. In contrast, a different study showed that JNK1 phosphorylates SIRT1 at S27, S47, and T530, and this phosphorylation of SIRT1 increased its nuclear localization and enzymatic activity (Nasrin et al. 2009). Like many Class I and II HDACs, SIRT1 is phosphorylated by CK2, and the target sites are S154, S649, S651, and S683 (Kang et al. 2009). In this case, phosphorylation of SIRT1 increases its deacetylation rate and its substrate-binding affinity. Also, CK2-mediated phosphorylation increases the ability of SIRT1 to deacetylate p53 and protects cells from apoptosis after DNA damage.
AMPK has been shown to increase intracellular NAD+ levels, which in turn enhances SIRT1 deacetylation activity (e.g., Canto et al. 2009). However, in a later study, it was shown that activation of the cAMP signaling pathway induces rapid deacetylation of SIRT1 substrates independent of changes in NAD+ levels (Gerhart-Hines et al. 2011). Activation of PKA phosphorylates S434, a residue located in the NAD+ binding pocket of the catalytic domain of SIRT1 and is phylogenetically conserved across all Sir2 orthologs. Other kinases downstream of PKA might also be responsible for the phosphorylation of SIRT1.
In summary, HDAC activities are modulated by multiple mechanisms. Both protein–protein interactions and posttranslational modifications (particularly phosphorylation) fine-tune HDAC activities. Multisubunit complexes dictate the activity of many HDAC deacetylases and their substrate specificity. Likewise, multiple kinases and phosphatases, through a variety of signaling pathways, can up- or down-regulate HDAC activities. A thorough understanding of HDAC regulation will not only provide tremendous insights into histone and protein deacetylation but also potential diagnostic and therapeutic approaches for the treatment of diseases that result from abnormal acetylation/deacetylation.
6. BIOLOGICAL IMPORTANCE OF HDACs
At the molecular level, the most obvious biological importance of HDACs is to oppose the functions of HATs. HDACs are critical in maintaining a dynamic equilibrium of protein acetylation. HDACs also exert profound effects on other protein posttranslational modifications. Deacetylation of histones and nonhistones may change chromatin conformation or modify the activities of transcription factors leading to a change in gene expression. Significantly, the molecular changes induced by HDACs impact on human health and disease. Abnormal HDACs have been documented to play a key role in many human diseases including (but not limited to) cancer, neurological diseases, metabolic disorders, inflammatory diseases, cardiac diseases, and pulmonary diseases.
6.1. HDACs Indirectly Regulate Many Posttranslational Modifications
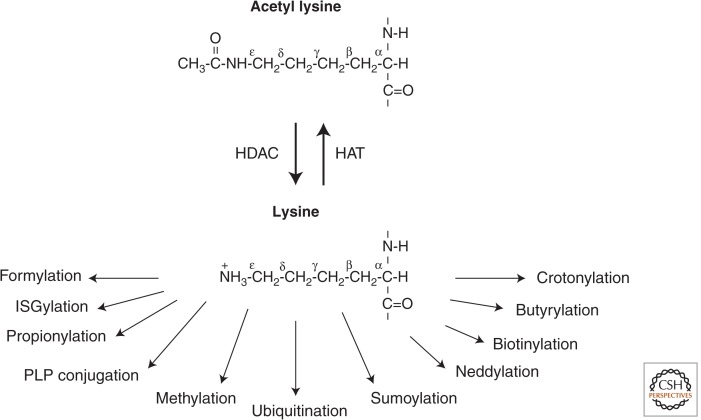
The lysine ε-amino group is prone to many different PTMs including acetylation, methylation, ubiquitination, sumoylation, neddylation, biotinylation, propionylation, butyrylation, and crotonylation (Tan et al. 2011). Introduction of an acetyl group, therefore, could exclude another modification on the same lysine residue within a protein (Fig. 7). An example of how lysine acetylation can interfere with other lysine modifications is the competition it often has with ubiquitination. Acetylation inhibits ubiquitination-dependent, proteasome-mediated protein degradation (Caron et al. 2005). Consequently, the stability of a number of proteins has been shown to increase after acetylation as a result of lysine site competition preventing ubiquitination. Conversely, removal of the acetyl group promotes lysine ubiquitination. Thus, one of the biological functions of HDACs is to accelerate protein degradation by exposing the ε-amino lysines for ubiquitination.
Figure 7.
Examples of the many different potential posttranslational modifications on an ε-amino lysine subsequent to HDAC deacetylation.
HDACs also play a key biological role in regulating histones (i.e., within a nucleosome) and chromatin (i.e., larger internucleosomal scale) cross talk. It has been fascinating to elucidate the many PTMs that communicate and interact with each other at multiple levels (e.g., see Fig. 10 in Allis et al. 2014). One of the best studied examples of histone modification cross talk occurs between acetylation and methylation (Fischle et al. 2003). Acetylation of histone H3K9 not only inhibits methylation of the same residue, but also promotes H3K4 methylation, which results in permissive chromatin and transcription activation. By logic, then, HDACs that deacetylate H3K9 will inhibit H3K4 methylation and ultimately repress transcription. Thus, HDACs not only provide access of previously acetylated lysines for another modification, but also promote histone and chromatin cross talk. Interestingly, several histone-modifying enzymes copurify with HDACs. For example, HDAC1, HDAC2, and G9a (a lysine methyltransferase) coordinate histone modifications from the same protein complex (Shi et al. 2003). Because thousands of combinations of histone modifications are possible, an abundance of regulatory potentials thus exist for HDACs. Cross-regulatory events between acetylation/deacetylation and other posttranslation modifications even extend to nonhistones (reviewed by Yang and Seto 2008). Furthermore, some HDAC proteins may possess other protein modification enzymatic activities in addition to deacetylase activity, suggesting that the effects of HDACs on protein modifications extend beyond the simple removal of acetyl groups from lysines.
6.2. HDACs Alter Gene Transcription
Early studies indicate that “relatively minor” histone modifications, particularly acetylation, could influence the rate of transcription (Allfrey et al. 1964). The thinking that emerged is that transcriptional activity is dictated, in part, by acetylation of nucleosomal histones. By conventional wisdom this would infer that the biological function of HDACs is to provide a reversible means of switching RNA synthesis off at different times and at different chromosomal loci. Mechanistically, it was believed that acetylation of core histones weakens its interaction with DNA, corroborated by the finding that acetylation markedly reduces the binding constant of the H4 tail to DNA in thermal denaturation studies (Hong et al. 1993). In contrast, deacetylation of histones by HDACs increases the positive charges on histones and, thereby, may strengthen histone–DNA interaction and repress transcription. Other, more recent evidence is accumulating in support of acetylation/deacetylation generating specific docking surfaces for proteins. This, in turn, regulates transcription without a significant change in the overall electrostatic charge of histones. In other words, although hyperacetylated histones interact with transcription activators, HDACs may furnish deacetylated interaction sites for transcriptional repressors.
One of the first evidences that directly links HDACs to transcription repression came from the findings that Rpd3 is required for both full repression and full activation of transcription of target genes in yeast (Vidal and Gaber 1991). Subsequently, it was shown that a Gal4 DNA binding domain-HDAC2 (mRPD3) fusion protein strongly represses transcription from a promoter containing Gal4 binding sites (Yang et al. 1996). Similar observations were obtained using Gal4-HDAC1 (Yang et al. 1997), Gal4-HDAC3 (Yang et al. 1997), Gal4-HDAC7 (Kao et al. 2000), Gal4-HDAC9 (Zhou et al. 2000), and Gal4-HDAC10 (Kao et al. 2002). Based on these promoter-targeting reporter assay studies, it was suggested that the recruitment of HDAC enzymes is a very common, although maybe not universal, mechanism by which repressors and corepressors alter transcription. However, whether the deacetylase activity of these HDACs per se is necessary for this repression is unclear. In one study, HDAC5 and HDAC7 were shown to possess autonomous repressor functions that were independent of their deacetylase activity (Kao et al. 2000). Therefore, a “one-size-fits-all” model in which HDACs mediate transcriptional repression via deacetylation of histones may not always be correct. Using yeast Rpd3 mutants that lack detectable histone deacetylase activity, Kevin Struhl confirmed that histone deacetylase activity of Rpd3 is important, but not absolutely required, for transcriptional repression in vivo (Kadosh and Struhl 1998).
In a yeast genome-wide mapping study, using ChIP followed by analysis of the precipitated DNA on microarrays (ChIP-chip), Richard Young showed that histone acetylation is associated with transcription activity, and the modification occurs predominantly at the beginning of genes (Pokholok et al. 2005). In a set of complementary experiments, it was shown that in histone H4, mutation of K16 alone changed specific transcription, whereas mutations of K5, K8, and K12 change transcription nonspecifically in a cumulative manner (Dion et al. 2005). Consistent with this finding, it was reported that a single histone modification, acetylation of H4K16, modulates both higher-order chromatin structure and functional interactions between a nonhistone protein and the chromatin fiber (Shogren-Knaak et al. 2006). These results suggest that acetylation, at least for histone H4, is operated by two distinct mechanisms: one that is specific for H4K16 and a nonspecific mechanism for H4K5, H4K8, H4K12. By inference, then, HDACs may exert their effects on transcriptional repression depending on which particular lysine it deacetylates. That is, deacetylation of H4K16 may result in a significant global transcription repression, whereas deacetylation of H4K5, H4K8, or H4K12 alone has little outcome but together generates a cumulative effect on transcription.
It is commonly believed that HDACs are bound to repressed genes and are cyclically replaced by HATs on gene activation. However, in another genome-wide mapping study, HDACs were found bound to chromatin at active genes but not silent genes (Wang et al. 2009). HDAC1 and HDAC3 were mainly detected in promoters, whereas HDAC2 and HDAC6 localized to both promoter and gene body regions of active genes. The majority of these HDACs in the human genome function to reset chromatin by removing acetylation at active genes. Related to this unexpected observation is that HDACs do not always repress transcription. For example, although HDAC3 represses transcription when targeted to promoters and serves as a corepressor (Yang et al. 1997), paradoxically HDAC3 also is required for the transcriptional activation of at least one class of retinoic acid response elements (Jepsen et al. 2000). In cells derived from Hdac3 knockout mice, both up-regulation and down-regulation of gene expression were detected (Bhaskara et al. 2008). Also, in gene expression profiling studies comparing cells treated and untreated with HDAC inhibitors, the number of genes down-regulated were comparable to up-regulated genes (e.g., LaBonte et al. 2009). One possibility is that HDACs may down-regulate transcription of transcriptional repressors, which leads to derepression of gene expression. Alternatively, HDACs may deacetylate and, consequently, activate transcription activators or inhibit the functions of transcription repressors independent of histone modifications. In summary, there is overwhelming evidence that a key biological function of HDACs is to modulate transcription, especially in repression. However, whether HDACs can directly activate transcription and the exact detailed mechanisms by which they regulate transcription still remain to be determined.
6.3. HDACs' Impact on Human Health and Diseases
One of the major motivations in studying HDACs is the expectation that an understanding of deacetylases will increase our understanding of histone modifications, chromatin biology, transcription regulation, and of course epigenetics. Equally important, however, is the need to understand the relevance of deacetylases in health and disease. Given the significant number of genes whose expression is regulated by HDACs, coupled with the finding that HDACs modulate the function of many proteins through nonhistone deacetylation, HDACs potentially play a role in nearly every aspect of health and disease. Numerous reports document the involvement of HDACs in cancer, neurodegenerative diseases, metabolic disorders, inflammatory diseases, immunological disorders, cardiac diseases, and pulmonary diseases. Because it is impractical to present a comprehensive discussion of HDACs in human diseases here, only a few examples are given.
The notion that HDACs are important in normal development, and that abnormal HDACs lead to disease is reinforced in HDAC knockout mice studies. For example, Hdac2-, Hdac5-, or Hdac9-null animals have cardiac defects (reviewed by Haberland et al. 2009). Conditional deletions of Hdac3 reveal that HDAC3 is important in liver homeostasis and heart functions. Hdac4 knockout mice expose the importance of HDAC4 in skeleton formation, and Sirt1-deficient mice have retinal, bone, and cardiac defects (e.g., McBurney et al. 2003).
Of the many roles HDACs play in human diseases, the most frequently discussed is cancer. Many correlative studies link cancer to epigenetic abnormalities. Somatic mutations in DNA and histone-modifying enzymes contribute to human malignancies (Dawson and Kouzarides 2012), and HDACs are no exception. For example, a frameshift mutation of HDAC2 has been found in sporadic carcinomas with microsatellite instability and in tumors arising in individuals with hereditary nonpolyposis colorectal cancer (Ropero et al. 2006). This mutation causes a loss of HDAC2 protein expression and enzymatic activity and renders these cells more resistant to the usual antiproliferative and proapoptotic effects of HDAC inhibitors. Loss of function HDAC2 mutations may derepress key genes from multiple cellular transformation pathways. HDACs can also be involved in cancer when recruited to specific loci in which repressive complexes are formed. This is the case for mutations in several oncogenic proteins generated by chromosome translocation such as PML-RARα and AML1-ETO fusion proteins that recruit HDACs (reviewed by Cress and Seto 2000).
Many studies have reported increased or decreased HDAC levels in various tumors compared to normal tissues. Although most of these studies focused on quantification of HDAC messenger RNA or proteins and not on HDAC enzymatic activities, conceivably any changes in HDACs can lead to changes in histone acetylation states, which in turn may lead to increased transcription of oncogenes or growth promoting factors, as well as a decrease in transcription of tumor suppressors or antiproliferative factors. It is widely known that HDACs can alter the expression of many cell cycle regulators. For example, early studies suggested that the cyclin-dependent kinase inhibitor p21 is an HDAC-response gene, and subsequent studies show that p21 expression is inversely correlated with HDAC2 in colorectal cancer cells (Huang et al. 2005). Some of the contributions that HDACs play in the development and progression of cancer may also be mediated through deacetylation of nonhistone proteins, including many oncogenes, tumor suppressors, and proteins that regulate tumor cell invasion and metastasis (reviewed by Glozak et al. 2005).
Like cancer, synaptic plasticity and cognition disorders can be manifestations of epigenetic dysregulation. HDAC inhibitors are therefore actively being pursued as potential therapeutics for neurodegenerative disorders. For example, SIRT2 deacetylates α-tubulin, and may promote α-synuclein toxicity and modified inclusion morphology in Parkinson’s disease (Outeiro et al. 2007). Other HDACs are likely to be involved in these disease etiologies, as administration of two HDAC inhibitors, sodium butyrate and SAHA (which notably do not affect SIRT2 activity), also protect against α-synuclein-dependent neurotoxicity in a Parkinson’s disease transgenic fly model. In the studies of Alzheimer’s disease, mice overexpressing HDAC2 result in decreased synaptic plasticity, synapse number, and memory formation, and SAHA rescues the synaptic number and learning impairments in HDAC2-overexpressing mice. Conversely, Hdac2 deficiency results in increased synapse number and memory facilitation similar to chronic treatment with HDAC inhibitors in mice (e.g., Graff et al. 2012). HDAC6 protein levels in Alzheimer’s disease brains is significantly increased, and it interacts with tau, a microtubule-associated protein that forms neurofibrillary tangles in Alzheimer’s disease (Ding et al. 2008). Furthermore, SIRT1 is significantly reduced in the parietal cortex of Alzheimer’s disease patients, and the accumulation of Aβ and tau in these patients may be associated with the loss of SIRT1 (Gao et al. 2010). HDACs may be linked to other neurological disorders, particularly psychiatric conditions including depression, anxiety disorders, and schizophrenia, as HDAC inhibitors show some potential promise in the treatments of these diseases.
There is much evidence to support the fact that heart diseases are linked to abnormal HDAC expression or activity (reviewed by Haberland et al. 2009). For example, HDAC9 is highly expressed in cardiac muscle, and one of the targets of HDAC9 is the transcription factor MEF2, which has been implicated in cardiac hypertrophy. In a series of elegant studies, Eric Olson showed that activation of the cardiac myocyte fetal gene program by a range of potent hypertrophic inducers could be blocked by expressing mutated HDAC9/MITR. Furthermore, mutant mice lacking HDAC9 are sensitized to hypertrophic signals and show stress-dependent cardiomegaly. Mice lacking HDAC5 display a similar cardiac phenotype and develop profoundly enlarged hearts in response to pressure overload, suggesting redundant functions of HDAC5 and HDAC9 in the control of cardiac development (Chang et al. 2004). Additionally, results from two independent studies clearly suggested that HDAC2 has an important role in heart biology (e.g., Trivedi et al. 2007).
HDACs also play an important role in inflammation and lung diseases. Perhaps this is best illustrated by the observation that HDAC2 expression and activity are reduced in lung macrophages, biopsies, and blood cells from patients with chronic obstructive pulmonary disease (COPD) and asthma (Ito et al. 2005). This decrease in expression and activity correlates with disease severity and the intensity of the inflammatory response. In patients with very severe COPD, the reduction in HDAC activity is accompanied by an increase in H4 acetylation at the IL-8 promoter. These results, taken together, indicate that HDAC2 is a key regulator of IL-8 gene transcription in inflammatory lung diseases. Results from a more recent study show that HDAC3 may be responsible for activation of almost half of the inflammatory gene expression under lipopolysaccharide-stimulation in macrophages, again underscoring the importance of HDACs in inflammation (see Busslinger and Tarakhovsky 2014 for more discussion on the epigenetics of the inflammatory response; Chen et al. 2012).
7. INHIBITORS
7.1. Discovery of HDAC Inhibitors
During biochemical analysis of HDAC activity in nuclei in the 1970s, it was found that millimolar concentrations of n-butyrate induced accumulation of acetylated histones in cells (Riggs et al. 1977). Soon after this finding, it was reported that n-butyrate inhibits deacetylation (e.g., Candido et al. 1978). Unfortunately, however, a causal relationship between the phenotypic consequence, such as cell cycle inhibition, and histone hyperacetylation induced by n-butyrate was doubted because of its nonspecific action on other enzymes and membranes. In 1990, potent HDAC inhibition by the natural product trichostatin A (TSA) was discovered. TSA was isolated from a Streptomyces strain and was originally identified as an antifungal antibiotic and a powerful inducer of murine erythroleukemia cell differentiation (Yoshida et al. 1987). TSA inhibited the activity of partially purified HDACs with a low nanomolar inhibition constant. Importantly, TSA-resistant mutant cells possessed a TSA-resistant HDAC enzyme providing genetic evidence that HDACs are the primary TSA target responsible for the cell cycle inhibition (Yoshida et al. 1990). TSA has a hydroxamic acid group, which can chelate a metal ion. Trapoxin, a fungal cyclic peptide that had been identified as an inducer of morphological change in transformed cells, was also found to strongly inhibit HDAC (Kijima et al. 1993). Unlike TSA, trapoxin irreversibly inhibits HDAC activity depending on its epoxyketone moiety. The potent ability of trapoxin to bind to HDACs was used for isolating the first HDAC protein (HDAC1) by means of trapoxin-affinity matrix (Taunton et al. 1996).
In 1998, two clinically important HDAC inhibitors were reported: SAHA, which had been designed and synthesized as a hybrid polar compound that strongly induces erythroid differentiation (Richon et al. 1998); and FK228 (romidepsin), an antitumor cyclic depsipeptide isolated from Chromobacterium violaceum (Nakajima et al. 1998). Like TSA, the zinc-interacting group of SAHA is its hydroxamic acid. On the other hand, there was no apparent zinc-interacting group in FK228. FK228 has an intramolecular disulfide bond, which is readily reduced in cells by the cellular reducing activity yielding a thiol side chain that coordinates to the active site zinc (Furumai et al. 2002). Although in vitro inhibition by FK228 was reversible, fluorescent live imaging of histone acetylation revealed that FK228 action in cells was sustained for several hours after removal, suggesting that efflux of the reduced form of FK228 is not efficient (Ito et al. 2011). Phase I clinical trials conducted by Susan Bates at the National Cancer Institute revealed that FK228 is effective for the therapy of cutaneous and peripheral T-cell lymphoma. This finding consequently accelerated the development of HDAC inhibitors as anticancer drugs. SAHA (Vorinostat) was the first HDAC inhibitor approved for cancer chemotherapy in 2006 (Bolden et al. 2006). Following SAHA, FK228 (Istodax) was also approved in 2009. Another class of clinically important HDAC inhibitors is benzamides, which includes CI-994 and MS-275 (Entinostat). MS-275, a synthetic benzamide derivative with HDAC inhibitory activity (Saito et al. 1999), showed moderate in vitro inhibitory activity, but the activity in cells was relatively strong because of the slow binding and time-dependent inhibition by the benzamide class of inhibitors (Bressi et al. 2010).
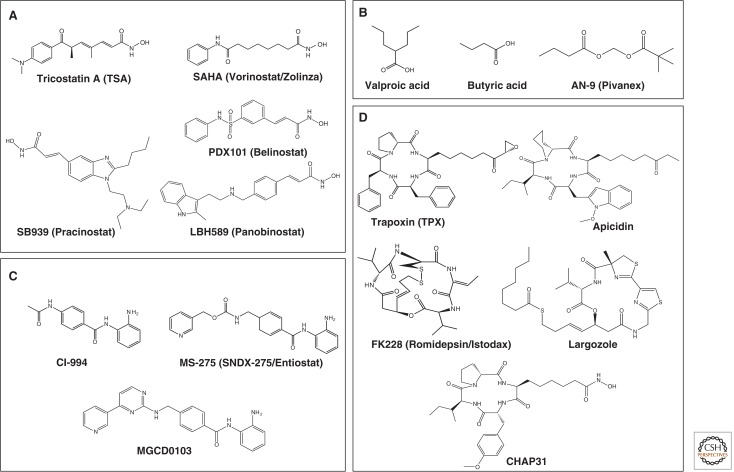
Figure 8 shows the large number of structurally diverse HDAC inhibitors. These have been discovered from natural sources or synthetically developed, and many are being studied clinically.
Figure 8.
HDAC inhibitors. Four classes of HDAC inhibitors categorized according to their chemical structures are shown. (A) Hydroxamic acid, (B) short-chain fatty acid, (C) benzamide, and (D) cyclic peptide.
7.2. Mechanisms of HDAC Inhibition
HDAC inhibitors can be divided into four classes based on their chemical structures: hydroxamate, short-chain fatty acid (carboxylate), benzamide, and cyclic peptide (Fig. 8). The most studied class is hydroxamate inhibitors. Structural studies of TSA or SAHA (hydroxamate class inhibitors) cocrystallized with the HDLP HDAC showed that the inhibitors bind by inserting their long aliphatic chain into the HDLP pocket with multiple contacts to the tubular pocket. Also, the hydroxamic acid coordinates the zinc ion in a bidentate fashion through its carbonyl and hydroxyl groups (Fig. 9A). The hydroxamic acid also makes hydrogen bonds with both histidines and tyrosines located around the zinc, thereby replacing the zinc-bound water molecule with its hydroxy group. On the other hand, the aromatic ring group contacts residues at the rim of the pocket, which allows inhibitors to lock in the pocket.
Figure 9.
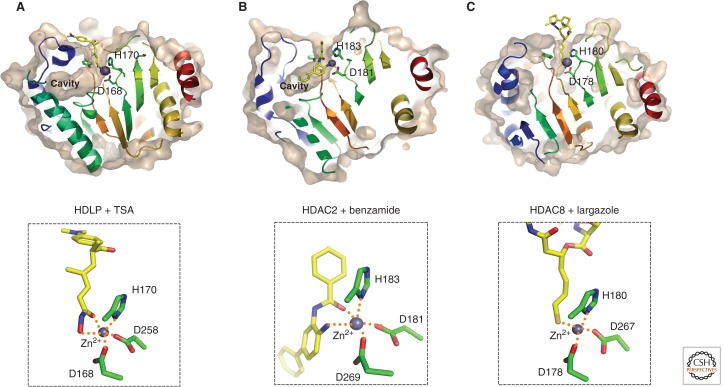
Crystal structure of HDAC proteins complexed with inhibitors. Molecular surfaces of the inhibitor-bound HDLP/HDACs cut at the level of the cavity are illustrated. (A) HDLP with TSA (PDB ID: 1C3R), (B) HDAC2 with benzamide (3MAX), and (C) HDAC8 with largazole (3RQD). The outer surfaces are depicted in light brown, and zinc atoms are shown in space-filled spheres in gray. Close-up views of the active sites are shown in the lower panels. Inhibitors are depicted in a stick model, in which yellow, blue, red, and gold represent C, N, O, and S atoms, respectively. Three Zn-coordinating residues (two aspartic acid and one histidine residue) are shown in a stick model. Hydrogen bond interactions are drawn in dotted lines.
The crystal structure of a benzamide inhibitor complexed with HDAC2 showed that the inhibitor coordinate (i.e., binds) to the catalytic zinc ion through both the carbonyl and amino groups to form a chelate complex (Fig. 9B). A simulation suggests that the transiently bound forms can be converted to the tightly bound, pseudoirreversible form over time, providing a basis for a time-dependent inhibition by this class of inhibitors (Bressi et al. 2010). Long-lasting effects on cellular histone acetylation levels after removal of MS-275 also suggest tight binding to the enzymes (Ito et al. 2011).
The cyclic peptide class contains different zinc-binding groups such as electrophilic ketones and thiols. Although crystal structure of the HDAC-FK228 (a cyclic peptide inhibitor) complex has not yet been solved, computer-modeling studies suggest that one of the thiol groups generated by reduction can coordinate to the active site zinc ion (Furumai et al. 2002). Recently, a similar inhibitory mechanism was proven by the crystal structure of HDAC8 with hydrolyzed largazole (Cole et al. 2011). Largazole has a thioester moiety, which can be hydrolyzed in cells to give an active thiol side chain (Fig. 8D). The HDAC8 structure complexed with the hydrolyzed largazole revealed that the thiol side chain coordinates to the catalytic zinc ion to inhibit the HDAC activity (Fig. 9C).
7.3. Target Enzyme Selectivity
TSA and SAHA, prototypical hydroxamate class inhibitors, are pan-HDAC inhibitors that block all Class I, II, and IV enzymes. The sensitivity to these inhibitors varies widely among different HDACs, with some HDAC activities inhibited only above pharmacologically relevant concentrations (Bradner et al. 2010). On the other hand, n-butyrate, trapoxin, FK228, and MS-275 cannot inhibit HDAC6 (a Class IIb enzyme), suggesting a difference in structure of the catalytic pockets between Class IIb and other enzymes (Matsuyama et al. 2002). Indeed, the different sensitivity of HDAC6 to TSA and trapoxin was used for identifying α-tubulin as an important HDAC6 substrate (Hubbert et al. 2002; Matsuyama et al. 2002). Benzamide class inhibitors such as MS-275 and MGCD0103 preferentially inhibit Class I enzymes except HDAC8. The class selective inhibition by nonhydroxamate inhibitors may be because of subtle differences in the structure of the catalytic pocket or the internal cavity among the HDAC classes. Importantly, benzamide inhibitors can interact with the cavity residues (Fig. 9B). Although Class IIa enzymes such as HDAC4 showed weak intrinsic deacetylase activity toward canonical HDAC substrates containing acetyl lysine, trifluoroacetyl lysine was found to be a Class IIa-specific substrate in vitro (Lahm et al. 2007). Based on this finding, 2-trifluoroacetylthiophene derivatives were designed as selective inhibitors for Class IIa HDACs.
Development of selective inhibitors is very challenging because of the high homology in the active site structure and catalytic mechanism of HDACs within each class. The first selective HDAC inhibitor was tubacin, which was obtained from a high-throughput screening campaign for an HDAC6 inhibitor that increases tubulin acetylation but not histone acetylation (Haggarty et al. 2003). Another strategy comparing HDAC8 complexed with different inhibitors gave insights into the design of a specific HDAC8 inhibitor (Somoza et al. 2004). Based on the flexible structure of the surface around the active site, an HDAC8-specific inhibitor, PCI-34051, was synthesized, which induced apoptosis in T-cell lymphoma without increasing histone and α-tubulin acetylation (Balasubramanian et al. 2008). In another approach, novel biaryl derivatives of benzamide inhibitors (SHI-1:2), which were designed based on homology modeling of HDAC1 and HDAC3, showed HDAC1/HDAC2-selective inhibitory activity. The SHI-1:2 inhibitors may access the internal cavity with different shapes between HDAC1 and HDAC3 (Methot et al. 2008).
The first generation of clinical HDAC inhibitors was largely nonselective. Considering the diverse functions of Class I, II, and IV HDACs, it is obviously important to develop class- and HDAC-selective inhibitors for the next-generation of therapeutic HDAC inhibitors.
7.4. Biological Activity of HDAC Inhibitors
Treatment of tumor cells with anticancer HDAC inhibitors changes the expression of ∼10% of the total genes, including many important regulatory genes that control the cell cycle and apoptosis. The most remarkable one is the p53-independent increase in p21, a CDK inhibitory protein, and GADD45, a regulator of DNA repair and senescence. The induction of p21 might be responsible for the HDAC inhibitor-induced G1 cell cycle arrest, with hypophosphorylation of pRb. In addition, repression of the cyclin D and cyclin A genes has been observed in a wide range of tumor cells on treatment, which is probably responsible in part for cell cycle inhibition. The therapeutic potential of HDAC inhibitors mainly comes from their capacity to selectively induce apoptosis in cancer cells. HDAC inhibitors activate expression of death receptors and their ligands in particular tumor cells. Furthermore, expression of proapoptotic BCL2 family proteins such as Bim or Bmf has been detected. Thus, both extrinsic and intrinsic apoptosis pathways, as well as reactive oxygen species production, may be involved in the HDAC-inhibitor-mediated apoptosis, although how important the activation of in vitro apoptosis pathways is for the in vivo therapeutic efficacy remains undefined (Bolden et al. 2006).
Tumor angiogenesis and cancer cell metastasis/invasion are also affected by HDAC inhibitors. Angiogenesis is an essential component of tumor growth and survival. The antiangiogenic activity of HDAC inhibitors has been associated with decreased expression of pro-angiogenic genes such as vascular endothelial growth factor, hypoxia-inducible factor-1α (HIFα), and chemokine receptor 4. Matrix metalloproteases (MMPs) also play a critical role in cancer metastasis/invasion. HDAC inhibitors can inhibit cancer cell invasion by up-regulating RECK, a tumor suppressor, to inhibit MMP-2 activation.
The increase in acetylation and the functional alteration of nonhistone substrates are likely to be involved in the biological activity of HDAC inhibitors. For example, the increase in p53 tumor suppressor protein acetylation increases the stability and DNA-binding capacity, leading to the enhanced expression of p53 target genes and apoptosis. Acetylation of HIFs represses the function of HIF by either reducing the protein level or decreasing the transactivation activity (Chen and Sang 2011). Cortactin is an F-actin binding protein controlling cell motility and invasion by remodeling actin filaments in the cell cortex. Acetylation occurs in the F-actin binding repeat domain, which attenuates actin binding and cell migration activity. HDAC6 is responsible for this deacetylation and thus influences actin-dependent motility in cancer cells (Zhang et al. 2007). Conceivably, HDAC6 inhibitors might alter cancer cell migration through this pathway.
Compared to conventional anticancer drugs, adverse effects of clinically available HDAC inhibitors are marginal. Dose-limiting toxicities include cardiac arrhythmia, thrombocytopenia, nausea, and fatigue, which are clinically manageable. As cardiac abnormalities were observed in Hdac5 null or Hdac9 null mice (Chang et al. 2004), it seems possible that cardiac arrhythmia in patients treated with HDAC inhibitors is related to cardiac function of these Class II HDACs.
Owing to the varied functions of HDAC proteins themselves, untangling the mechanisms and targets by which HDAC inhibitors bring about selective improvement of certain human disease is complex.
7.5. Sirtuin Inhibitors
Sirtuins, the Class III HDACs, are involved in a number of cellular processes including insulin secretion, the cell cycle, and apoptosis. A great deal of effort has been put into developing sirtuin inhibitors as potential therapeutics. Nicotinamide is widely used as a global inhibitor of sirtuins, although high concentrations are required for inhibition of activity in cell culture. The nicotinamide molecule is, in fact, a byproduct of the sirtuin enzyme reaction, and thus a physiological inhibitor that decreases gene silencing, increases rDNA recombination, and accelerates aging in yeast. In search of a specific sirtuin inhibitor, there has been much focus on SIRT1 as it importantly regulates mitochondrial biogenesis and metabolic pathways, cellular redox, angiogenesis, and notch signaling. Malfunction of SIRT1 may contribute to diabetes, obesity, abnormal cancer metabolism, cancer stemness, and neurological disorders. These diseases represent a huge burden to our societies’ health and healthcare systems, therefore sirtuin inhibitors represent hopeful therapeutics for many of these diseases.
The first synthetic sirtuin inhibitors, sirtinol and splitomicin, were identified through a screen based on telomere silencing in yeast (e.g., Bedalov et al. 2001). Following these, a number of compounds including cambinol, salermide, tenovin, EX-527, AGK2, etc. have been reported as sirtuin inhibitors (Fig. 10). The detailed mechanisms of how these compounds modulate sirtuin activity have mostly not yet been determined because structural information about their binding sites is currently limited. Sirtuin inhibitors may be roughly divided into two groups, those interacting with the NAD+ (nicotinamide and ADP-ribose) binding site and those interacting with the acetyl lysine binding site. Suramin, for example, was shown to inhibit the NAD+-dependent deacetylase activity of sirtuins (Fig. 10) (Schuetz et al. 2007). The crystal structure of suramin complexed with SIRT5 showed that the symmetrical suramin structure induces dimerization of SIRT5, mediated through interacting with both the NAD+ and acetylated peptide binding sites.
Figure 10.
Sirtuin inhibitors. Sirtinol and splitomicin are identified as the first small molecule sirtuin inhibitors that affect telomere silencing in yeast. Cambinol is a splitomicin-related β-naphthol that is more stable than splitomicin and increases p53 acetylation showing antitumor activity in BCL6-expressing Burkitt’s lymphoma. Salermide was designed based on the structure of sirtinol by molecular modeling and inhibits both SIRT1 and SIRT2 more effectively than sirtinol. Tenovin-1 and its water-soluble analog tenovin-6 induce p53 acetylation; their cellular targets were determined to be SIRT1 and SIRT2. A high-throughput screen revealed a number of indole compounds including EX-527, which selectively inhibits SIRT1 over SIRT2. Kinetic analysis suggests that EX-527 binds to the nicotinamide-binding site. AGK2, which was reported as a SIRT2 selective inhibitor, shows more than 10-fold selective inhibition relative to SIRT1 and SIRT3. Suramin, which was originally developed for treating trypanosomiasis and onchocerciasis, inhibits the NAD+-dependent deacetylase activity of sirtuins by inducing sirtuin dimerization.
Several acetyl lysine analogs have been shown to inhibit sirtuins as a mechanism-based inhibitor. For instance, thioacetyl peptide acts as a tight-binding inhibitor that stalls reaction at the ADP-ribose-peptide enzyme intermediate by forming 1′-S-alkylamidate (Smith and Denu 2007). Also, an ethyl malonyllysine-based small molecule was shown to inhibit SIRT1 and produce a covalent conjugate that may occupy the binding sites for NAD+ and acetyl lysine (Asaba et al. 2009). A variety of these acetyl lysine analogs have been useful for dissecting the catalytic mechanism of NAD+-dependent protein deacetylases as the mechanistic probes.
In agreement with the diverse function of sirtuins, these inhibitors show a variety of biological activities. Tenovin-6 induces p53 acetylation, inhibits tumor growth, and eliminates cancer stem cells in chronic myelogenous leukemia (CML) in combination with Imatinib, a BCR-ABL tyrosine kinase inhibitor (Li et al. 2012). Importantly, SIRT2 inhibition by AGK2 rescued toxicity caused by α-synuclein (α-Syn) insoluble fibril aggregation seen in synucleinopathies such as Parkinson’s disease. It did this by decreasing the number through increasing the size of aggregates in a cellular model of Parkinson’s disease. Thus, development of small molecules modulators of sirtuin activity has become one of the most active areas in drug discovery.
8. SUMMARY
Today, we know that there are at least 18 human HDAC proteins. They contribute to the erasure of epigenetic modifications, help establish epigenetic off chromatin states, and regulate heritable changes in gene expression. How HDACs achieve these ends is a question of immediate interest to many working in this field. The finding that HDACs function as transcriptional repressors and corepressors has ushered a surge of interest in this subject, and reports of genes that are regulated by HDACs are continuously expanding. Global expression profiling experiments estimate that the transcription of 10% of genes are regulated by HDACs. However, questions of how each HDAC uniquely regulates a specific set of genes remain largely unexplored. Similarly, although we know that there are potentially more than a thousand HDAC substrates, the biological consequences of deacetylation of these substrates require intense research. The mechanistic details of how histone deacetylation promotes modification cross talk, and the global consequences of deacetylation of other histone and epigenetic-modifying enzymes are ripe for further research.
HDAC inhibitors show promise in the treatment of cancer, inflammation, and neurological diseases. Many laboratories are actively working on developing more effective HDAC inhibitors with minimal side effects in patients. Who would have thought that a humble beginning in a biochemistry curiosity on an enzymatic activity purified from calf thymus would lead to a better understanding of epigenetics and have such a potential for impacting the improvement of human health?
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, and Danny Reinberg
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Allfrey VG, Faulkner R, Mirsky AE 1964. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci 51: 786–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Allis D, Jenuwein T, Reinberg D 2014. Overview and concepts. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018739 [DOI] [Google Scholar]
- Asaba T, Suzuki T, Ueda R, Tsumoto H, Nakagawa H, Miyata N 2009. Inhibition of human sirtuins by in situ generation of an acetylated lysine-ADP-ribose conjugate. J Am Chem Soc 131: 6989–6996 [DOI] [PubMed] [Google Scholar]
- Avalos JL, Boeke JD, Wolberger C 2004. Structural basis for the mechanism and regulation of Sir2 enzymes. Mol Cell 13: 639–648 [DOI] [PubMed] [Google Scholar]
- Ayer DE 1999. Histone deacetylases: Transcriptional repression with SINers and NuRDs. Trends Cell Biol 9: 193–198 [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Ramos J, Luo W, Sirisawad M, Verner E, Buggy JJ 2008. A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia 22: 1026–1034 [DOI] [PubMed] [Google Scholar]
- Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K, et al. 2012. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 487: 114–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardai FH, D’Mello SR 2011. Selective toxicity by HDAC3 in neurons: Regulation by Akt and GSK3β. J Neurosci 31: 1746–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP 2004. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci 101: 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedalov A, Gatbonton T, Irvine WP, Gottschling DE, Simon JA 2001. Identification of a small molecule inhibitor of Sir2p. Proc Natl Acad Sci 98: 15113–15118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara S, Chyla BJ, Amann JM, Knutson SK, Cortez D, Sun ZW, Hiebert SW 2008. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell 30: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolden JE, Peart MJ, Johnstone RW 2006. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov 5: 769–784 [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD 1995. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev 9: 2888–2902 [DOI] [PubMed] [Google Scholar]
- Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, Mazitschek R 2010. Chemical phylogenetics of histone deacetylases. Nat Chem Biol 6: 238–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressi JC, de Jong R, Wu Y, Jennings AJ, Brown JW, O’Connell S, Tari LW, Skene RJ, Vu P, Navre M, et al. 2010. Benzimidazole and imidazole inhibitors of histone deacetylases: Synthesis and biological activity. Bioorg Med Chem Lett 20: 3138–3141 [DOI] [PubMed] [Google Scholar]
- *.Busslinger M, Tarakhovsky S 2014. Epigenetic control of immunity. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candido EP, Reeves R, Davie JR 1978. Sodium butyrate inhibits histone deacetylation in cultured cells. Cell 14: 105–113 [DOI] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J 2009. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458: 1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron C, Boyault C, Khochbin S 2005. Regulatory cross-talk between lysine acetylation and ubiquitination: Role in the control of protein stability. Bioessays 27: 408–415 [DOI] [PubMed] [Google Scholar]
- Chang S, McKinsey TA, Zhang CL, Richardson JA, Hill JA, Olson EN 2004. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol Cell Biol 24: 8467–8476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Sang N 2011. Histone deacetylase inhibitors: The epigenetic therapeutics that repress hypoxia-inducible factors. J Biomed Biotechnol 2011: 197946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Barozzi I, Termanini A, Prosperini E, Recchiuti A, Dalli J, Mietton F, Matteoli G, Hiebert S, Natoli G 2012. Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc Natl Acad Sci 109: E2865–E2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325: 834–840 [DOI] [PubMed] [Google Scholar]
- Cole KE, Dowling DP, Boone MA, Phillips AJ, Christianson DW 2011. Structural basis of the antiproliferative activity of largazole, a depsipeptide inhibitor of the histone deacetylases. J Am Chem Soc 133: 12474–12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cress WD, Seto E 2000. Histone deacetylases, transcriptional control, and cancer. J Cell Physiol 184: 1–16 [DOI] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T 2012. Cancer epigenetics: From mechanism to therapy. Cell 150: 12–27 [DOI] [PubMed] [Google Scholar]
- Ding H, Dolan PJ, Johnson GV 2008. Histone deacetylase 6 interacts with the microtubule-associated protein τ. J Neurochem 106: 2119–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion MF, Altschuler SJ, Wu LF, Rando OJ 2005. Genomic characterization reveals a simple histone H4 acetylation code. Proc Natl Acad Sci 102: 5501–5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, et al. 2011. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334: 806–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP 1999. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401: 188–193 [DOI] [PubMed] [Google Scholar]
- Finnin MS, Donigian JR, Pavletich NP 2001. Structure of the histone deacetylase SIRT2. Nat Struct Biol 8: 621–625 [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD 2003. Histone and chromatin cross-talk. Curr Opin Cell Biol 15: 172–183 [DOI] [PubMed] [Google Scholar]
- Ford J, Ahmed S, Allison S, Jiang M, Milner J 2008. JNK2-dependent regulation of SIRT1 protein stability. Cell Cycle 7: 3091–3097 [DOI] [PubMed] [Google Scholar]
- Frye RA 1999. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun 260: 273–279 [DOI] [PubMed] [Google Scholar]
- Furumai R, Matsuyama A, Kobashi N, Lee KH, Nishiyama M, Nakajima H, Tanaka A, Komatsu Y, Nishino N, Yoshida M, et al. 2002. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res 62: 4916–4921 [PubMed] [Google Scholar]
- Galasinski SC, Resing KA, Goodrich JA, Ahn NG 2002. Phosphatase inhibition leads to histone deacetylases 1 and 2 phosphorylation and disruption of corepressor interactions. J Biol Chem 277: 19618–19626 [DOI] [PubMed] [Google Scholar]
- Gantt SL, Joseph CG, Fierke CA 2010. Activation and inhibition of histone deacetylase 8 by monovalent cations. J Biol Chem 285: 6036–6043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Cueto MA, Asselbergs F, Atadja P 2002. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol Chem 277: 25748–25755 [DOI] [PubMed] [Google Scholar]
- Gao J, Wang WY, Mao YW, Graff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH 2010. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 466: 1105–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Dominy JE Jr, Blattler SM, Jedrychowski MP, Banks AS, Lim JH, Chim H, Gygi SP, Puigserver P 2011. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+). Mol Cell 44: 851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glozak MA, Seto E 2009. Acetylation/deacetylation modulates the stability of DNA replication licensing factor Cdt1. J Biol Chem 284: 11446–11453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glozak MA, Sengupta N, Zhang X, Seto E 2005. Acetylation and deacetylation of non-histone proteins. Gene 363: 15–23 [DOI] [PubMed] [Google Scholar]
- Graff J, Rei D, Guan JS, Wang WY, Seo J, Hennig KM, Nieland TJ, Fass DM, Kao PF, Kahn M, et al. 2012. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature 483: 222–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoretti IV, Lee YM, Goodson HV 2004. Molecular evolution of the histone deacetylase family: Functional implications of phylogenetic analysis. J Mol Biol 338: 17–31 [DOI] [PubMed] [Google Scholar]
- Grozinger CM, Hassig CA, Schreiber SL 1999. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci 96: 4868–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Grunstein M, Gasser SM 2013. Epigenetics in Saccharomyces cerevisiae. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a017491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN 2009. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat Rev Genet 10: 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL 2003. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci 100: 4389–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L, Schroth GP, Matthews HR, Yau P, Bradbury EM 1993. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 “tail” to DNA. J Biol Chem 268: 305–314 [PubMed] [Google Scholar]
- Hu E, Chen Z, Fredrickson T, Zhu Y, Kirkpatrick R, Zhang GF, Johanson K, Sung CM, Liu R, Winkler J 2000. Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. J Biol Chem 275: 15254–15264 [DOI] [PubMed] [Google Scholar]
- Huang BH, Laban M, Leung CH, Lee L, Lee CK, Salto-Tellez M, Raju GC, Hooi SC 2005. Inhibition of histone deacetylase 2 increases apoptosis and p21Cip1/WAF1 expression, independent of histone deacetylase 1. Cell Death Differ 12: 395–404 [DOI] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP 2002. HDAC6 is a microtubule-associated deacetylase. Nature 417: 455–458 [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800 [DOI] [PubMed] [Google Scholar]
- Inoue A, Fujimoto D 1969. Enzymatic deacetylation of histone. Biochem Biophys Res Commun 36: 146–150 [DOI] [PubMed] [Google Scholar]
- Ito K, Ito M, Elliott WM, Cosio B, Caramori G, Kon OM, Barczyk A, Hayash S, Adcock IM, Hogg JC, et al. 2005. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. N Engl J Med 352: 1967–1976 [DOI] [PubMed] [Google Scholar]
- Ito T, Umehara T, Sasaki K, Nakamura Y, Nishino N, Terada T, Shirouzu M, Padmanabhan B, Yokoyama S, Ito A, et al. 2011. Real-time imaging of histone H4K12-specific acetylation determines the modes of action of histone deacetylase and bromodomain inhibitors. Chem Biol 18: 495–507 [DOI] [PubMed] [Google Scholar]
- Jepsen K, Hermanson O, Onami TM, Gleiberman AS, Lunyak V, McEvilly RJ, Kurokawa R, Kumar V, Liu F, Seto E, et al. 2000. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell 102: 753–763 [DOI] [PubMed] [Google Scholar]
- Johnson CA, White DA, Lavender JS, O’Neill LP, Turner BM 2002. Human class I histone deacetylase complexes show enhanced catalytic activity in the presence of ATP and co-immunoprecipitate with the ATP-dependent chaperone protein Hsp70. J Biol Chem 277: 9590–9597 [DOI] [PubMed] [Google Scholar]
- Kadosh D, Struhl K 1998. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev 12: 797–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Jung JW, Kim MK, Chung JH 2009. CK2 is the regulator of SIRT1 substrate-binding affinity, deacetylase activity and cellular response to DNA-damage. PLoS One 4: e6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Suh JY, Jung YS, Jung JW, Kim MK, Chung JH 2011. Peptide switch is essential for Sirt1 deacetylase activity. Mol Cell 44: 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Downes M, Ordentlich P, Evans RM 2000. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev 14: 55–66 [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Lee CH, Komarov A, Han CC, Evans RM 2002. Isolation and characterization of mammalian HDAC10, a novel histone deacetylase. J Biol Chem 277: 187–193 [DOI] [PubMed] [Google Scholar]
- Kijima M, Yoshida M, Sugita K, Horinouchi S, Beppu T 1993. Trapoxin, an antitumor cyclic tetrapeptide, is an irreversible inhibitor of mammalian histone deacetylase. J Biol Chem 268: 22429–22435 [PubMed] [Google Scholar]
- Kim EJ, Kho JH, Kang MR, Um SJ 2007. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell 28: 277–290 [DOI] [PubMed] [Google Scholar]
- LaBonte MJ, Wilson PM, Fazzone W, Groshen S, Lenz HJ, Ladner RD 2009. DNA microarray profiling of genes differentially regulated by the histone deacetylase inhibitors vorinostat and LBH589 in colon cancer cell lines. BMC Med Genomics 2: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahm A, Paolini C, Pallaoro M, Nardi MC, Jones P, Neddermann P, Sambucini S, Bottomley MJ, Lo Surdo P, Carfi A, et al. 2007. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci 104: 17335–17340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Rezai-Zadeh N, Seto E 2004. Negative regulation of histone deacetylase 8 activity by cyclic AMP-dependent protein kinase A. Mol Cell Biol 24: 765–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, Landsman D 1997. Histone deacetylases, acetoin utilization proteins and acetylpolyamine amidohydrolases are members of an ancient protein superfamily. Nucleic Acids Res 25: 3693–3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang L, Li L, Wang Z, Ho Y, McDonald T, Holyoake TL, Chen W, Bhatia R 2012. Activation of p53 by SIRT1 inhibition enhances elimination of CML leukemia stem cells in combination with imatinib. Cancer Cell 21: 266–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YY, Kiihl S, Suhail Y, Liu SY, Chou YH, Kuang Z, Lu JY, Khor CN, Lin CL, Bader JS, et al. 2012. Functional dissection of lysine deacetylases reveals that HDAC1 and p300 regulate AMPK. Nature 482: 251–255 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liszt G, Ford E, Kurtev M, Guarente L 2005. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem 280: 21313–21320 [DOI] [PubMed] [Google Scholar]
- Luo J, Su F, Chen D, Shiloh A, Gu W 2000. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 408: 377–381 [DOI] [PubMed] [Google Scholar]
- *.Marmorstein R, Zhou M-M 2014. Writers and readers of histone acetylation: Structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama A, Shimazu T, Sumida Y, Saito A, Yoshimatsu Y, Seigneurin-Berny D, Osada H, Komatsu Y, Nishino N, Khochbin S, et al. 2002. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J 21: 6820–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M 2003. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol 23: 38–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methot JL, Chakravarty PK, Chenard M, Close J, Cruz JC, Dahlberg WK, Fleming J, Hamblett CL, Hamill JE, Harrington P, et al. 2008. Exploration of the internal cavity of histone deacetylase (HDAC) with selective HDAC1/HDAC2 inhibitors (SHI-1:2). Bioorg Med Chem Lett 18: 973–978 [DOI] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, et al. 2008. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452: 492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan P, Yang F, Di Stefano L, Ji JY, Ouyang J, Nishikawa JL, Toiber D, Kulkarni M, Wang Q, Najafi-Shoushtari SH, et al. 2011. A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol Cell 42: 689–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima H, Kim YB, Terano H, Yoshida M, Horinouchi S 1998. FR901228, a potent antitumor antibiotic, is a novel histone deacetylase inhibitor. Exp Cell Res 241: 126–133 [DOI] [PubMed] [Google Scholar]
- Nasrin N, Kaushik VK, Fortier E, Wall D, Pearson KJ, de Cabo R, Bordone L 2009. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS One 4: e8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, Volk CB, Maxwell MM, Rochet JC, McLean PJ, et al. 2007. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science 317: 516–519 [DOI] [PubMed] [Google Scholar]
- Peng L, Ling H, Yuan Z, Fang B, Bloom G, Fukasawa K, Koomen J, Chen J, Lane WS, Seto E 2012. SIRT1 negatively regulates the activities, functions, and protein levels of hMOF and TIP60. Mol Cell Biol 32: 2823–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflum MK, Tong JK, Lane WS, Schreiber SL 2001. Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. J Biol Chem 276: 47733–47741 [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527 [DOI] [PubMed] [Google Scholar]
- Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA 2007. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 129: 1351–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richon VM, Emiliani S, Verdin E, Webb Y, Breslow R, Rifkind RA, Marks PA 1998. A class of hybrid polar inducers of transformed cell differentiation inhibits histone deacetylases. Proc Natl Acad Sci 95: 3003–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs MG, Whittaker RG, Neumann JR, Ingram VM 1977. n-Butyrate causes histone modification in HeLa and Friend erythroleukaemia cells. Nature 268: 462–464 [DOI] [PubMed] [Google Scholar]
- Ropero S, Fraga MF, Ballestar E, Hamelin R, Yamamoto H, Boix-Chornet M, Caballero R, Alaminos M, Setien F, Paz MF, et al. 2006. A truncating mutation of HDAC2 in human cancers confers resistance to histone deacetylase inhibition. Nat Genet 38: 566–569 [DOI] [PubMed] [Google Scholar]
- Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ 2005. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol 23: 94–101 [DOI] [PubMed] [Google Scholar]
- Saito A, Yamashita T, Mariko Y, Nosaka Y, Tsuchiya K, Ando T, Suzuki T, Tsuruo T, Nakanishi O 1999. A synthetic inhibitor of histone deacetylase, MS-27–275, with marked in vivo antitumor activity against human tumors. Proc Natl Acad Sci 96: 4592–4597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Maier B, Koclega KD, Chruszcz M, Gluba W, Stukenberg PT, Minor W, Scrable H 2008. Phosphorylation regulates SIRT1 function. PLoS One 3: e4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz A, Min J, Antoshenko T, Wang CL, Allali-Hassani A, Dong A, Loppnau P, Vedadi M, Bochkarev A, Sternglanz R, et al. 2007. Structural basis of inhibition of the human NAD+-dependent deacetylase SIRT5 by suramin. Structure 15: 377–389 [DOI] [PubMed] [Google Scholar]
- Seto E, Yang XJ 2010. Regulation of histone deacetylase activities and functions by phosphorylation and dephosphorylation. In Handbook of cell signaling, 2nd ed (ed Bradshaw RA, Dennis EA), pp. 2379–2388 Elsevier Science, Amsterdam [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y 2003. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422: 735–738 [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847 [DOI] [PubMed] [Google Scholar]
- Smith BC, Denu JM 2007. Mechanism-based inhibition of Sir2 deacetylases by thioacetyl-lysine peptide. Biochemistry 46: 14478–14486 [DOI] [PubMed] [Google Scholar]
- Somoza JR, Skene RJ, Katz BA, Mol C, Ho JD, Jennings AJ, Luong C, Arvai A, Buggy JJ, Chi E, et al. 2004. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure 12: 1325–1334 [DOI] [PubMed] [Google Scholar]
- Sterner R, Vidali G, Allfrey VG 1979. Studies of acetylation and deacetylation in high mobility group proteins. Identification of the sites of acetylation in HMG-1. J Biol Chem 254: 11577–11583 [PubMed] [Google Scholar]
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. 2011. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146: 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taunton J, Hassig CA, Schreiber SL 1996. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272: 408–411 [DOI] [PubMed] [Google Scholar]
- Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, Floss T, Goettlicher M, Noppinger PR, Wurst W, et al. 2007. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 β activity. Nat Med 13: 324–331 [DOI] [PubMed] [Google Scholar]
- Tsai SC, Seto E 2002. Regulation of histone deacetylase 2 by protein kinase CK2. J Biol Chem 277: 31826–31833 [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D 2004. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell 16: 93–105 [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D 2006. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev 20: 1256–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107: 149–159 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Carrozza MJ, Lasonder E, Workman JL, Logie C, Stunnenberg HG 2004. In vitro targeting reveals intrinsic histone tail specificity of the Sin3/histone deacetylase and N-CoR/SMRT corepressor complexes. Mol Cell Biol 24: 2364–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Gaber RF 1991. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol Cell Biol 11: 6317–6327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K 2009. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138: 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. 2010. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327: 1004–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PJ, Fairall L, Santos GM, Schwabe JW 2012. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature 481: 335–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen YD, Perissi V, Staszewski LM, Yang WM, Krones A, Glass CK, Rosenfeld MG, Seto E 2000. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc Natl Acad Sci 97: 7202–7207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Lu Z, Cao Z, Zhang Y 2011. Zinc chelation with hydroxamate in histone deacetylases modulated by water access to the linker binding channel. J Am Chem Soc 133: 6110–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Seto E 2008. Lysine acetylation: Codified crosstalk with other posttranslational modifications. Mol Cell 31: 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WM, Inouye C, Zeng Y, Bearss D, Seto E 1996. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci 93: 12845–12850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WM, Yao YL, Sun JM, Davie JR, Seto E 1997. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem 272: 28001–28007 [DOI] [PubMed] [Google Scholar]
- Yoshida M, Nomura S, Beppu T 1987. Effects of trichostatins on differentiation of murine erythroleukemia cells. Cancer Res 47: 3688–3691 [PubMed] [Google Scholar]
- Yoshida M, Kijima M, Akita M, Beppu T 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem 265: 17174–17179 [PubMed] [Google Scholar]
- Zhang X, Wharton W, Yuan Z, Tsai SC, Olashaw N, Seto E 2004. Activation of the growth-differentiation factor 11 gene by the histone deacetylase (HDAC) inhibitor trichostatin A and repression by HDAC3. Mol Cell Biol 24: 5106–5118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ozawa Y, Lee H, Wen YD, Tan TH, Wadzinski BE, Seto E 2005. Histone deacetylase 3 (HDAC3) activity is regulated by interaction with protein serine/threonine phosphatase 4. Gene Dev 19: 827–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yuan Z, Zhang Y, Yong S, Salas-Burgos A, Koomen J, Olashaw N, Parsons JT, Yang XJ, Dent SR, et al. 2007. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol Cell 27: 197–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W 2008. Negative regulation of the deacetylase SIRT1 by DBC1. Nature 451: 587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Richon VM, Rifkind RA, Marks PA 2000. Identification of a transcriptional repressor related to the noncatalytic domain of histone deacetylases 4 and 5. Proc Natl Acad Sci 97: 1056–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]