Abstract
Grapes are among the widely cultivated fruit crops in the world. Grape berries like other nonclimacteric fruits undergo a complex set of dynamic, physical, physiological, and biochemical changes during ripening. Muscadine grapes are widely cultivated in the southern United States for fresh fruit and wine. To date, changes in the metabolites composition of muscadine grapes have been well documented; however, the molecular changes during berry development and ripening are not fully known. The aim of this study was to investigate changes in the berry proteome during ripening in muscadine grape cv. Noble. Isobaric tags for relative and absolute quantification (iTRAQ) MS/MS was used to detect statistically significant changes in the berry proteome. A total of 674 proteins were detected, and 76 were differentially expressed across four time points in muscadine berry. Proteins obtained were further analyzed to provide information about its potential functions during ripening. Several proteins involved in abiotic and biotic stimuli and sucrose and hexose metabolism were upregulated during berry ripening. Quantitative real-time PCR analysis validated the protein expression results for nine proteins. Identification of vicilin-like antimicrobial peptides indicates additional disease tolerance proteins are present in muscadines for berry protection during ripening. The results provide new information for characterization and understanding muscadine berry proteome and grape ripening.
Keywords: muscadine grape, berry, ripening, iTRAQ, Q-RT PCR
INTRODUCTION
North American native grape species Vitis rotundifolia Michx. Muscadine is a grape variety that is well adapted to the warm, humid climate of southeastern United States. This species is also an important commercial crop used in fresh consumption, food additives, dietary supplements, and the wine industry.1 Unlike other grape species, the inherent higher resistance of muscadine grape to pathogens enables a sustainable production in the region. They differ from other Vitis species in that muscadine grapes have an extra pair of chromosomes.2 Muscadine grapes have distinct morphology and significantly differ from V. vinifera in berry characteristics, composition, and quality traits. Despite the unique biochemical composition and novel traits found in muscadine grapes, little is known about its berry characteristics. One of the main tasks in grape research has been the investigation of berry development and ripening since grape producers are interested in improving cultural practices and increasing yield and quality of the final product. As grape berries develop, they change in size and composition from being small, firm, and acidic with little sugar and desirable flavors or aroma to becoming larger, softened, sweet, highly flavored, less acidic, and highly colored fruit.3 The development of these characteristics determines the quality of the final product.4
In this regard, proteomics has been successfully used to examine varietal and developmental differences as well as to analyze chemical and environmental effects on grapes.5–11 Currently, stable isotopic labeling followed by reverse phase high-performance liquid chromatography coupled with tandem mass spectrometry (HPLC–MS/MS) is successfully used to characterize and quantify changes in protein levels in V. vinifera.9,10,12
In the present study, isobaric tags for relative and absolute quantification (iTRAQ) was applied for the proteomic analysis of the muscadine berry to understand the unique genetic makeup and identify changes in protein expression of maturing and ripening muscadine berries. The aim of the study was to identify differentially expressed proteins between four time points during berry development and ripening of muscadine grape cv. Noble and compare protein expression patterns to reveal the developmental basis of the elite traits. Further selected proteins were used to correlate whether protein changes can be validated by transcript analysis. The results obtained provide a global insight into proteome changes of muscadine berry during development and ripening.
MATERIALS AND METHODS
Sample Collection
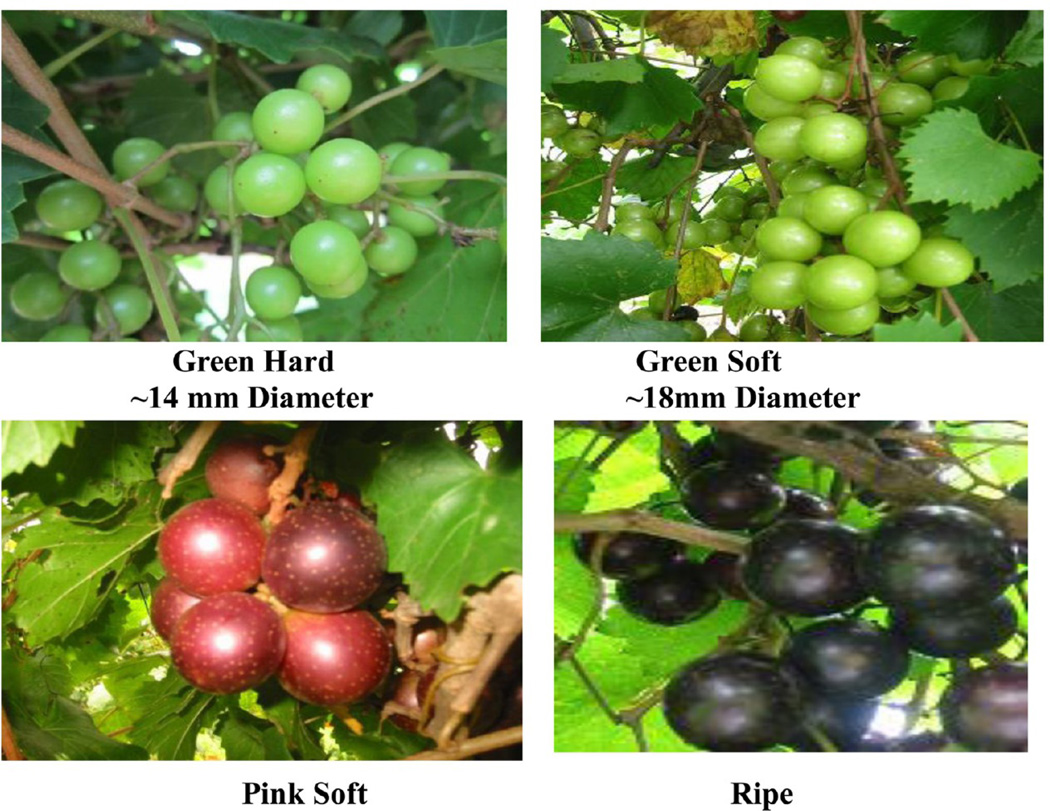
Grape berries were sampled from V. rotundifolia cv. Noble from the vineyard at the FAMU Center for Viticulture and Small fruit Research, Tallahassee, Florida, during the 2011 season. Sampling dates were based on the Brix content, titratable acidity, fresh weight, texture analysis, and anthocyanin content of the berry. Two hundred berries were collected from clusters of four different vines at each sampling date. Berry samples collected from different clusters of the same vine were considered as one biological replicate. The berry tissues were frozen immediately in liquid nitrogen and stored at −80 °C. At all stages, samples of whole fresh berries obtained as described above were used immediately to measure their total soluble solids, pH, texture analysis, and titratable acidity. Berries were sampled from four different time points: green hard (GH; EL-33), green soft (GS; EL-34), pink soft (PS; EL-35), and ripe (R; EL-38) maturity13 (EL refers to the modified Eichhorn and Lorenz developmental scale) categories.14 Berry development was characterized by monitoring berry diameter, total soluble solids, titratable acidity, texture analysis and anthocyanin content. Thirty berries from individual replicates were used for the measurements. Berry diameter was measured with a micrometer, and total soluble solids (°Brix) were assayed with a refractometer (Atago, USA) from juice crushed from harvested berries to estimate total sugar content. Titratable acidity (g/L) of the grape juice was measured by titration to an end point of pH 8.4 with 0.1 N NaOH. For firmness determinations, berries from green hard and green soft stages were subjected to a compression test with a texture analyzer (TA.XT plus, Texture Technologies Corp., Scarsdale, NY, U.S.A./Stable Micro Systems, Godalming, Surrey, U.K.). Settings were test speed of 10 mm s−1 and force at 10.0 g. Firmness was measured as the force needed to deform the berry by 20% at its equatorial diameter and expressed as loss of firmness with respect to the initial firmness.15
Protein Extraction, Digestion, and iTRAQ Labeling LC–MS/MS
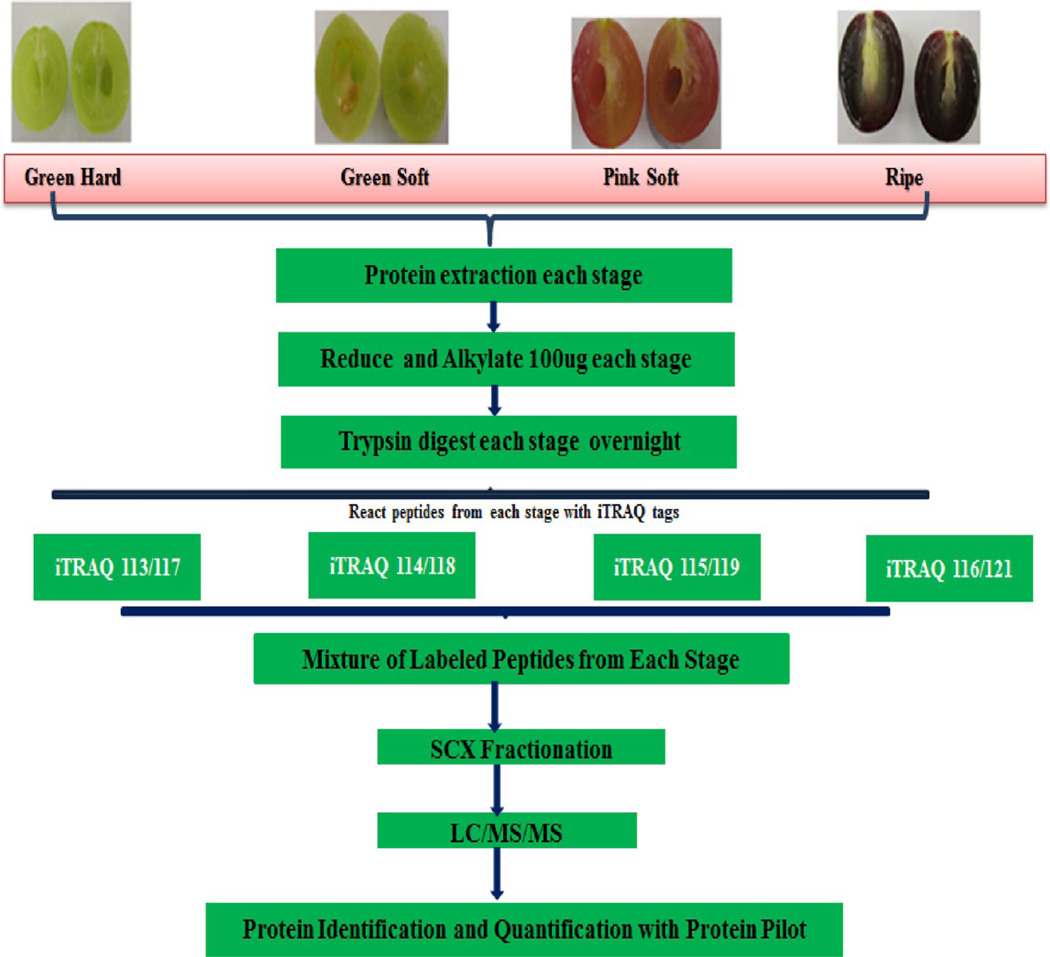
Protein was extracted from seed-free berry tissue (pericarp). Each replicate was subjected to independent protein extraction from frozen, finely ground berry samples (4 g powder) using two protocols: (1) TCA method: Berry samples were vortexed with 10 mL of 10% TCA/0.07% 2-mercaptoethanol(2-ME)/acetone w/v/v for 2 h @ −20 °C. The precipitate was then washed three times with 10 mL of 0.07% 2-ME/acetone v/v for 2 h at −20 °C and then vacuum-dried. The resulting pellet was resolubilized in a lysis buffer (9 M urea, 0.5% amidosulfobetaine 14 (ASB-14), 2% CHAPS, 1.2% hydroxyethyldisulfide (HED).16 (2) Phenol method: Berry samples were vortexed with 10 mL of extraction buffer (0.5 M Tris-HCl, 0.7 M sucrose, 0.05 M EDTA, 0.1 M KCL, 2 mM PMSF, 1% PVPP, and 2% 2ME, pH 7.5 with protease and phosphatase inhibitor cocktail, Roche Applied Science) and 10 mL of phenol saturated with Tris-HCl, pH 7.5. The extraction was performed under shaking at 4 °C for 2 h. Following centrifugation (5000g for 30 min@ 4 °C), the phenol phase containing proteins was collected and transferred to new tubes. The phenol phase was re-extracted with sucrose buffer. Proteins were precipitated with 5 vol of cold 0.1 M ammonium acetate in methanol overnight at −20 °C. The resulting protein pellet was washed thrice with 0.1 M ammonium acetate in 100% methanol, once with 100% acetone, and once with 100% acetone/ethanol (1:1). The final protein pellets were dissolved in buffer (7 M urea, 2 M thiourea, 4% CHAPS, 10 mM DTT). Protein yield was measured using the Bradford method.17 Aliquots of the protein samples were acetone precipitated, and the recovered pellets were resuspended in a 0.5 M Bicine buffer, pH 8.5 containing 0.1% w/v SDS and placed on a thermomixer (Eppendorf) overnight at 4 °C to facilitate the resuspension of the protein pellet.12 The resuspended protein pellet was quantified again using the Bradford method.17 Each sample of 100 µg protein was reduced using 2 µL of manufacturer supplied reducing agent and incubating at 60 °C for 1 h (iTRAQ 8-plex kit, AB Sciex). Cysteine residues were blocked using methyl methanethiosulfonate (MMTS) at room temperature for 10 min followed by trypsin digestion (modified trypsin, Promega) at 37 °C for 16 h. Before labeling, the reagents were dissolved in 50 µL of isopropanol, the contents of one vial were transferred to individual sample tubes, and it was ensured that the pH was 7.5. Samples were labeled with iTRAQ tags as follows: green hard (GH)-113, green soft (GS)-114, pink soft (PS)-115 and ripe-116 and (green hard-117, green soft-118, pink soft-119, and ripe-121) as illustrated in Figure 3 and incubated at room temperature for 2 h. This served as two replicates, and the same pattern was repeated for one more iTRAQ labeling experiment. After labeling, individual iTRAQ 8-plex samples were mixed and diluted into 0.1% trifluoricacetic acid, followed by loading on a MacroSpin Vydac C18 reverse phase mini-column. The washing and elution were conducted following instructions from the manufacturer (The Nestgroup Inc., USA). The eluates were dried down and dissolved in strong cation exchange (SCX) solvent A (25% v/v acetonitrile, 10 mM ammonium formate, pH 2.8). The peptides were fractionated on an Agilent HPLC system 1100 using a polysulfethyl A column (2.1 × 100 mm, 5 µm, 300 Å, PolyLC,Columbia, MD). Peptides were eluted at a flow rate of 200 µL/min with a linear gradient of 0–20% solvent B (25% v/v acetonitrile, 500 mM ammonium formate) over 50 min, followed by ramping up to 100% solvent B in 5 min and holding for 10 min. The absorbance at 280 nm was monitored, and a total of 32 fractions were collected and then combined into 12 fractions based on peak intensities.
Figure 3.
iTRAQ 8-plex workflow labeling of different stages of muscadine berry with iTRAQ 8-plex isobaric tags.
Reverse Phase Nanoflow HPLC and Tandem Mass Spectrometry
Each SCX fraction was lyophilized and dissolved in solvent A (3% acetonitrile v/v, 0.1% acetic acid v/v). The peptides were loaded onto a C18 capillary trap cartridge (LC Packings, USA) and then separated on a 15 cm nanoflow analytical C18 column (PepMap 75 µm id, 3 µm, 100 A) (LC Packings, USA) at a flow rate of 200 nL/min on a Tempo nanoflow multidimensional LC system (Applied Biosystems/MDS Analytical Technologies, USA). Peptides were eluted from the HPLC column by application of a linear gradient from 3% solvent B (96.9% acetonitrile v/v, 0.1% acetic acid v/v) to 40% solvent B for 2 h, followed by ramping up to 90% solvent B in 10 min. Peptides were sprayed into the orifice of a quadrupole time-of-flight mass spectrometer (QSTAR Elite MS/MS system, Applied Biosystems Inc., USA), which was operated in an information-dependent data acquisition mode where a TOF MS scan (m/z 300–1800, 0.25 s) followed by three MS/MS scans (m/z 50–2000, 30–2000 ms) of the three highest abundance peptide ions (with charge states 2–5) were acquired in each cycle. Former target ions were excluded for 60 s. Information-dependent acquisition features of Analyst QS software, such as automatic collision energy (smart CE), automatic MS/MS accumulation (smart exit), and dynamic exclusion were selected. The source nebulizing gas and curtain gas were set at 12 and 20, respectively. Ion spray voltage was 2200 V and the temperature was 80 °C.18
Protein Identification and Data Analysis
The MS/MS data from the two iTRAQ 8-plex experiments were processed through Protein Pilot v 4.5 (AB Sciex) and searched against the Uniprot Grape Data set (55,494 entries, March 2013). The analysis method used was the Paragon Algorithm, with search parameters as follows: “Thorough ID” mode with a 95% confidence level (Prot score 1.3), iTRAQ peptide labeling, Cys oxidized by methyl methanethiosulfonate (MMTS), and trypsin digestion. The final fold change was calculated as the average value obtained from all replicates. Accession numbers were compared between each iTRAQ data set and considered common if the accession number was identified in the two data sets. A change in expression was determined in comparison with the corresponding controls (green hard-113 and -117 iTRAQ tags). To increase the confidence level, we only focused on core proteins altered at least 1.5-fold. An identified protein was considered to be significantly increased or decreased in abundance at a given time point if the fold change met the threshold criteria of an iTRAQ ratio of 1.5.9,19,20 For further functional analysis, proteins with an increase/decrease of 1.5-fold and a minimum of two peptide match common in all the four replicates were used. Gene ontology analysis of identified proteins was carried out using Blast2GO software.21 Briefly, a file of FASTA format sequences of the identified and/or quantified protein set was batch retrieved from the Uniprot Web site. Blast2GO was performed using BLASTp search against NCBInr (e-value cutoff 1 × 10−50, 20 for the retrieved number of BLAST hits, 33 for the HSP (highest scoring pair) length cutoff), second to map GO, EC, and Interpro terms and then to annotate the sequences (E-Value Hit-Filter of 1 × 10−6, a Hsp-Hit Coverage Cutoff of 0, an Annotation Cutoff of 55, and a GO Weight of 5). Automatic annotation performed by Blast2GO was manually revised to guarantee accurate assignment. Fisher exact tests were performed for an enrichment analysis between two annotated protein lists to find the significant GO terms. A reference list containing all the quantified sequences in the experiment was compared with a selected subset of sequences based on the quantification ratio of 1.5-fold change proteins. Fisher’s exact test was performed using a term filter value of 0.05 and p-value as a term filter mode. Cluster Analysis was performed to demonstrate the distribution of expression pattern among four stages.22
Quantitative Real time PCR
To investigate whether gene expression is correlated between transcript and protein level and confirm two unique disease resistant proteins in the first iTRAQ data set, real time PCR was performed for 22 genes selected based on proteomic data. Primers were designed using Primer Express software (Applied Bio systems) and the sequences listed in Supporting Information, Supplementary Table 9. Total RNA from different stages of berries was extracted using the Spectrum Plant RNA kit (Sigma Aldrich, St. Louis, MO). Reverse transcription was performed on 500 ng of total RNA from each development stage and replicates using the Superscript II RT kit (Invitrogen, USA) according to the manufacturer’s instructions in a final volume of 20 µL. Quantitative real-time PCR was performed on a Bio-Rad iCycler using iQ SYBR Green Supermix (Bio-Rad), together with 1 µL of forward and reverse primers of the transcripts of interest and 1 µL of template cDNA. For each reaction, four biological replicates were included. Data were normalized against expression of the housekeeping gene ubiquitin. To determine relative fold differences for each gene in each experiment, the Ct value of the genes was normalized to the Ct value for ubiquitin (control gene), and relative expression was calculated relative to a calibrator using the formula 2−ΔΔCt. All the values shown are mean ± SE.
RESULTS AND DISCUSSION
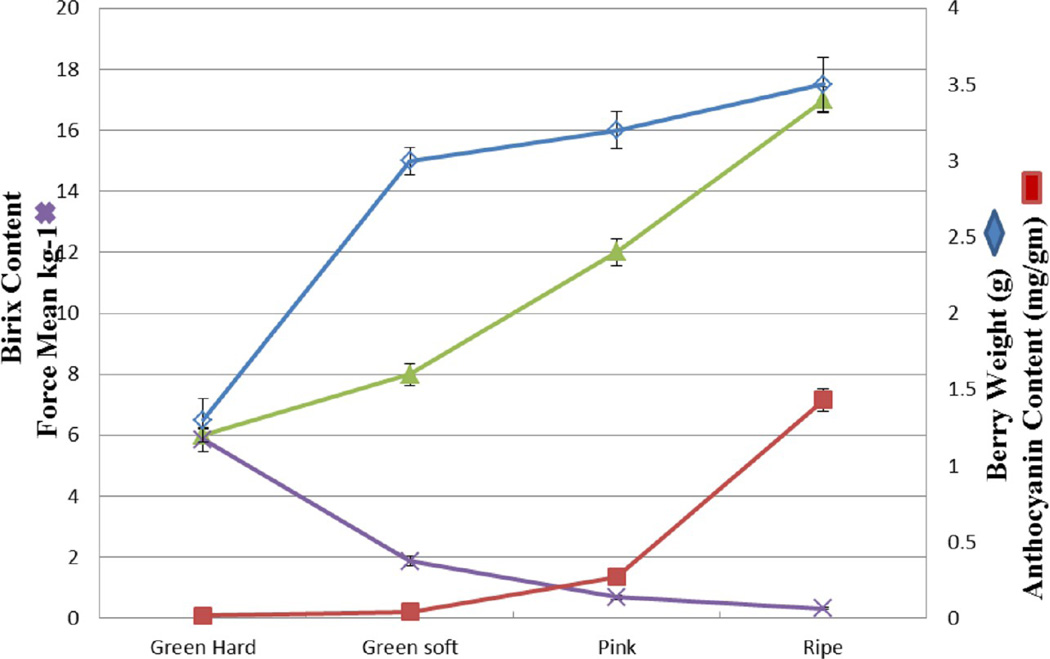
Berry samples were collected from green hard to commercially ripe stage. Developmental and ripening parameters were characterized. (Supplementary Table 1, Supporting Information, Figure 2). The sample for the earliest stage corresponded to the green hard stage was characterized by hard green berries of at least 15mm diameter with a maximum acid content (22 g/L) and minimum Brix of 6° (Supplementary Table 1, Supporting Information). The pink stage and fully ripe berries were selected based on berry color parallel to the anthocyanin content, Brix, and decrease in total acidity (Figure 2). Two protocols were opted for protein extraction from four developmental stages of muscadine berry. Applying the TCA/acetone based method, not more than 0.2 mg of protein/g of berry fresh weight was obtained in both the green stages and protein yield slightly increased up to 0.3 mg/g in ripe stages. In contrast, the phenol-based protocol yielded consistently higher yield up to 0.6 mg of protein/g of fresh berry in green stages and increased to 0.8 mg in pink stage and ripe berries (Supplementary Table 2, Supporting Information). Protein yield and concentration obtained in muscadine grape berries are equal to the published literature on V. vinifera.6,16
Figure 2.
Changes in fresh weight (blue diamonds), concentration of soluble solids (green triangles), anthocyanin content (red squares), and texture measurement (blue X’s) of Noble berries sampled at different developmental stages. GH-green hard; GS-green soft; PS-pink soft; and ripe.
Overall Proteome at Various Berry Developmental Stages
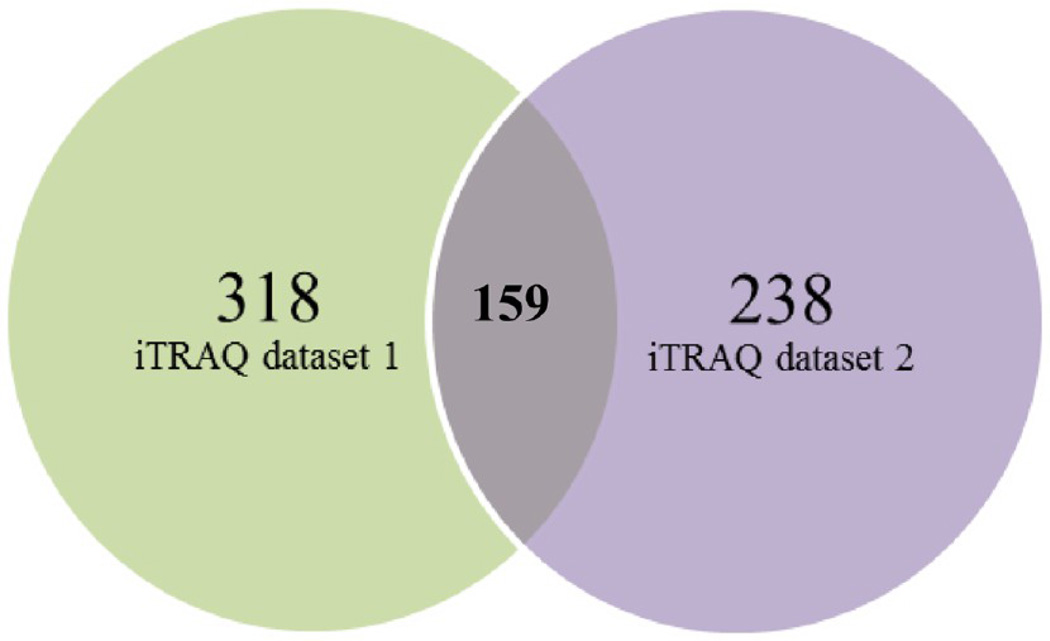
For each replicate, the validated peptides were assembled into proteins to produce a total number of 522 and 352 proteins respectively (Supporting Information, Supplementary Tables 3 and 4). The Prot scores, percent coverage, number of peptides matching individual proteins, and accession numbers assigned to each of the identified protein are given in Supplementary Table 5, Supporting Information. The raw files and results are deposited in the peptide atlas server with data identifier PASS00218. Protein description and annotation was automatically assigned using Blast2Go application. Of the 522 sequences, 465 were assigned a description and 440 were annotated in the first iTRAQ data set. In second iTRAQ data set, of the 352 sequences, 322 were assigned a description and 306 were annotated with an average number of 5 annotations per sequence and an average level depth of 5.0. The combination of the two protein lists resulted in a set of 674 unique proteins. The list of proteins obtained from individual iTRAQ data sets were exported to Excel, and proteins with single peptide match (204 proteins - 1 iTRAQ data set and 114 - 2 iTRAQ data set) were removed from both data sets to derive proteins only with two peptide match. A total of 318 and 238 proteins identified in the first and second iTRAQ 8-plex data set had proteins with a minimum of two peptide match. In contrast, the first iTRAQ 8-plex data set had proteins identified with a single peptide larger compared to the set of proteins with single peptide identified in the second iTRAQ 8-plex, indicating the proteins identified with single peptide in the first replicate were not repeatedly identified in the second replicate. To derive common proteins among both the iTRAQ data sets proteins with similar Uniprot IDs were sorted among all the four replicates. This resulted in identification of 159 proteins common to both the data sets with a minimum of two peptide match to proteins across four biological replications and two iTRAQ 8-plex experiments (Figure 4) (Supplementary Table 5C, Supporting Information). A fold change cutoff of 1.5-fold was used to select the protein subsets that resulted in 76 proteins. Four clusters were generated to determine the expression pattern of significantly up- and downregulated proteins during ripening. The number of proteins ascribed to each cluster is reported. Cluster 1 contained 10 proteins that increased in GS stage and then was downregulated during ripening. Cluster 2 included 28 proteins that increased during the GS stage and then was downregulated during the PS stage and again slightly upregulated in ripe berries. Twenty-five proteins were grouped into cluster 3 that were highly upregulated during the ripening stages, and cluster 4 refers to 13 proteins that were highly upregulated from GS to ripe stages. An additional file lists proteins by cluster number along with corresponding log2-transformed ratio metric data for each protein entry (Figure 5, Supplementary Table 7, Supporting Information).
Figure 4.
Venn diagram showing proteins identified with two peptide match among both the data sets and common proteins among both iTRAQ data sets.
Figure 5.
Cluster analyses of proteins significantly up- and downregulated among four replications. Four clusters were generated to classify proteins during four time points: GH (green hard), GS (green soft), PS (pink soft), and ripe.
Enrichment analysis using Fisher’s test was carried out by comparing the frequency of the annotation terms of each protein with that of the whole list of quantified proteins in individual clusters to identify GO terms enriched in each stage of berry development. Proteins involved in cellular process, cellular component organization, and nucleotide binding were overrepresented in the GS stage. GO terms over-represented in the GS stage and decreasing in PS and again increasing in ripe stages included proteins involved in plasmodesma, proteosomal protein catabolic process, cytoskeletal organization, translation, and gluconeogenesis proteins. GO terms overexpressed between the PS and ripening stage include response to salt, heat, water, and hydrogen peroxide, and proteins involved in the glucose catabolic process. GO terms overexpressed only in ripening include proteins responding to both abiotic and biotic stimulus. When classified into functional categories, the majority include the proteins involved in carbohydrate and organic acid metabolism (15%), stress (12%), and cellular organization and protein processing (9%) (Figure 6).
Figure 6.
GO term enriched broader functional classification of the 76 differentially expressed proteins during development and ripening of muscadine berry.
Profiles of the Protein’s Functional Clusters during Grape Berry Ripening
Muscadine berry tissue proteome relates to the final quality of the fruit and wine. It is responsible for traits such as color, astringency, flavor or resistance to pathogens, among others. As shown in Figure 7, the expression of various proteins at different ripening stages underlines their function and importance at each developmental stage. Below, the most important proteins according to their putative biological function are discussed.
Figure 7.
Cluster profiles of protein functional clusters during muscadine grape berry ripening. A heat map of the log 2 relative abundance of proteins throughout ripening in relation to the green hard stage was created using Genesis v1.7 with the iTRAQ-derived quantitative data. For each protein, an abbreviation, the Uniprot accession number, and the sequence description assigned with Blast2GO are provided. Proteins were grouped according to their known or putative role in metabolic pathways or cellular process.
Sucrose and Hexose Metabolism
The data show (Figure 7E) numerous changes in the expression of proteins involved in sucrose and hexose metabolism and glycolysis/gluconeogenesis. Several key enzymes involved in sucrose metabolism and glycolysis were differentially expressed in muscadine berry pericarp during ripening (Figure 7E). Six proteins involved in sucrose and hexose metabolism were identified to be up- and downregulated during ripening. Proteins belonging to phosphoglucomutase (PGM, tr|F6HFF7), pyruvate kinase (tr|F6HVY1), and glyceraldehyde 3-phosphate dehydrogenases (G3PDH, tr|F6GSG7) were downregulated during ripening, while enolase (ENO, E.C. 4.2.1.11) and glyceraldehyde 3-phosphate dehydrogenases (G3PDH; tr|F6HG44) were upregulated during ripening (Figure 7E). Enolase participates in the conversion of the cytosolic pool of 3-phosphoglycerate to phosphoenolpryruvate (PEP). PEP can be channeled to aromatic amino acid biosynthesis via the shikimic acid pathway in the choloroplast stroma through a PEP:Pi transporter.13 The upregulation of an enolase in PS and ripe stages may be due to the increase demand of PEP for phenylpropanoids and flavonoids biosynthesis.13 Glycolytic enzymes were reported to decrease after veraison in Trincadeira, Muscat Hamburg, Sangiovese, and Cabernet Sauvignon grape berry4,10,23,25 but were upregulated in ripening Barbera grapes.7 Differences in up- and downregulation of these enzymes might also be due to inhibition of plastid glycolysis at the onset and following véraison and cytoplasmic glycolysis activation.23 Upregulation of sucrose synthase was observed in Cabernet Sauvignon and Pinot Noir, whereas in Trincadeira this gene is downregulated, which is consistent with an increase in sucrose level.23 Sucrose synthase (SS) levels fluctuated during berry development in muscadines where it was upregulated 2-fold at the GS stage, then decreased 1-fold at PS stage, and then increased 3-fold in ripe berries (Figure 7E).
Organic Acid Metabolism
Organic acids such as malic and tartaric acids are well-known for their role in wine flavor and taste. Malate dehydrogenase (MDH) catalyzes a reversible reaction between oxaloacetate and malate where malate dehydrogenase may be involved in malate synthesis, which occur mainly during pre-veraison, while malate degradation occurs at post-veraison. Mitochondrial malate dehydrogenase, tr|E0CSB6 was upregulated during berry ripening (Figure 7D). In the cytoplasm, malate can be produced from phosphoenolpyruvate produced in glycolysis through the activities of phosphoenolpyruvate carboxylase (PEPC) and malate dehydrogenase.23 PEPC protein was downregulated in muscadine berries from GS to the ripe stage that is consistent with the decrease in acid content in berries (Supplementary Table 1, Supporting Information). Malic enzyme (ME) catalyzes the reversible conversion between malate and pyruvate. NADP-malic enzyme was found to be upregulated in the GH stage (Figure 7D). Contrasting reports exist for NADP-malic enzyme levels in ripening berry where it has been shown to decrease after veraison,25 remain at a steady level,26 and increase7 at veraison and may support the existence of several isoforms of this proteins. Environmental factors such as temperature may activate particular pathways of malate degradation, but it is also possible that different tissues behave differently.23
Protein Synthesis and Folding
Proteins involved in the synthesis and protein processing were upregulated in the green and ripe stages. Heat shock proteins functionally participate to minimize aggregation of newly synthesized proteins facilitating the folding process.27 Heat shock proteins 70 (tr|F6HLD8) and 80 were upregulated during the green soft stage. Endoplasmin, a molecular chaperone that optimizes the folding and assembly of newly synthesized secretory proteins and another HSP 70 protein (F6HNX5), was upregulated in ripe stages. Increase in protein synthesis and processing proteins during these stages has been related to the large changes in metabolism for the synthesis of new proteins.7,25 Leucine aminopeptidase (LAP) was upregulated from GH to ripe stage. Leucine aminopeptidases (LAPs) are metallopeptidases that cleave N-terminal residues from proteins and peptides. Aminopeptidases are involved in many different roles in plants, but few members have been characterized in relation to their function to date. LAPs are known to play a role in regulating wounding responses in tomato.28 Increases in LAPs during ripening also suggest higher protease activity might be required due to increased protein catabolism. The translationally controlled tumor protein (TCTP, 44) is highly regulated by a wide range of extracellular signals and has been implicated in important cellular processes, such as cell growth, cell cycle progression, and in the protection of cells against various stress conditions. TCTP expression was abundant when the grape berry was green and hard based on frequency analysis of ESTs and MPSS signatures.29 In muscadine, high expression of TCTP was observed in the PS stage and might be involved in various cellular processes during ripening that need further investigation.
Secondary Metabolism
Grape berries accumulate a wide range of secondary metabolites. Proteins involved in the synthesis of various polyphenolic compounds were significantly altered during pink soft and ripe berry stages (Figure 7A). Anthraniloyl-CoA:methanol anthraniloyl transferase (AMAT) was upregulated 2-fold from GH to PS. AMAT shows strong homology with other acyltransferases responsible for synthesis of flavor compounds, but it may be involved in the synthesis of methyl anthranilate, the volatile compound responsible for the distinctive “foxy” aroma and flavor of the Washington Concord grape (V. labrusca).30 Two enzymes participating in flavonoid biosynthesis were detected during ripening in muscadine grapes. Flavonoid synthase increased 2-fold during veraison and ripening, while anthocyanidin synthase decreased during ripening. The increase in flavonoid synthase is similar to transcript and protein abundance in the post veraison stage in V. aestivalis (Norton grape),65 and the decrease in anthocyanidin synthase during ripening is consistent with the previous observed results in V. viniera cv. Shiraz where anthocyanidin synthase gene expression decreased post veraison.31 Gluathione S-transferase (GST) (EC:2.5.1.18) is involved in the accumulation of anthocyanins in vacuole during berry ripening.10 GST (tr|Q56AY1-GST-4 type) was upregulated 3-fold during the PS stage and decreased slightly during ripening. An increase in the levels of glutathione was previously observed during ripening of Koshu and Cabernet Sauvignon grapes. GST increase during ripening is consistent with the increase in anthocyanin content and might play a similar role in muscadine grape. Terpenes, which are precursors for important aroma compounds, accumulate at veraison.9 Enzymes involved in the production of isoprenoids, isopentenyl-diphosphate isomerase (IPI) that catalyzes the conversion of the relatively unreactive isopentenyl pyrophosphate (IPP) to the more-reactive electrophile dimethylallyl pyrophosphate (DMAPP), increased 7-fold in PS and then decreased in ripe muscadine berries.
Fatty Acid and Phospholipid Metabolism
Lipoxygenase-derived hydroperoxy fatty acids are metabolized through major pathways involving enzymes such as the hydroperoxide lyase.32 A protein coding for hydroperoxide lyase (HPL 1) (EC 4.1.2) was upregulated almost 2-fold in GH and then downregulated in PS and slightly upregulated during ripening. VvHPL1 has the function of 13-HPL that metabolizes 13-HPOT/HPOD (13(S)-hydroperoxy-(9Z,11E,15Z)-octadecatrienoic acid (13-HPOT), (13(S)-hydroperoxy-(9Z,11E)- octadecadienoic acid (13-HPOD) to form C6 compounds. VvHPL1 expression increased and their corresponding hexanal and two hexenals 13-HPOT/HPOD during berry maturation and veraison in Cabernet Sauvignon.33 Hexenal produced by HPL can be converted to hexanol by alcohol dehydrogenases (ADH).23 In grapevine, ADH genes belong to a small multigene family which has been well characterized. Vv-ADH1 and Vv-ADH3 transcripts accumulate transiently in young developing berry, while Vv-ADH2 transcripts strongly increase at the onset of veraison.34 In muscadines, ADH 1 (EC:1.1.1.1) was upregulated 2-fold in GS and decreased during ripening. Alcohol dehydrogenase activity is suggested to contribute to the development of taste and aroma in fruits34 and may be involved in production of flavor and aroma compounds in muscadine grapes during ripening. Phospholiapase (PLD) (EC: 3.1.4.4) was downregulated from GS to ripening stages (Figure 7L). PLD-generated phosphatidic acid is known to act as a signaling molecule. Recent studies have indicated that PLDs play many regulatory roles in diverse plant processes such as ABA signaling and stress tolerance.3
PR Proteins
During berry development, defense-related proteins are highly expressed in muscadine grape berry pericarp as reported in V.vinifera4,10,23,35,36 and are likely to play important roles in protecting against fungal pathogens and possibly against other stresses. Up to five PR proteins were upregulated from green to ripe stage (Figure 7B). PR-4 and PR-10 protein increased between GH and ripe stages (Figure 7B). PR-4 and PR-10 (VvPR10.2) proteins are known to have antifungal activity and are expressed under pathogen attack.37,38 Up to 17 PR10 related sequences were identified in the whole V. vinifera genome, and all belonged to a single compact cluster on the chromosome 5.39 VvPR10.2 was expressed in root, stem, leaf, and flower and is known to be expressed under pathogen attack.38,39 Another group of PR proteins, major latex protein (MLP) 28, was upregulated during ripening in muscadine. Two MLP-28 proteins (tr|Q9M4G9) were upregulated 6-fold from the GH stage to PS and decreased slightly during ripening, and MLP 28 protein (tr|F6HFH2) maintained a 5-fold increase from green to ripe berries. The function of MLPs have been have been associated with fruit and flower development and in pathogen defense responses based in their modest sequence similarity with members of the Bet v 1 super family.40 MLPs are differentially expressed through plant development and seem to have similar functions to PR-10 proteins.41 MLP protein was reported to decrease during ripening in V.vinifera cv. Muscat Hamburg, while high expression of MLP-28 is unique to muscadine grape that might be developmentally regulated during ripening and have a role in defense response.
Stress Response
Several proteins involved in general and oxidative stress response were upregulated during berry development (Figure 7C,F). In plants, a group of hydrophilic proteins, known as late embryogenesis abundant (LEA) proteins, accumulate to high levels during the last stage of seed maturation and during water deficit in vegetative organs, suggesting a protective role during water limitation.42,43 Dehydrins, also known as LEA D-11 or LEA II (late embryogenesis abundant), are proteins whose expression is induced by various environmental factors, which cause dehydration of the cells.44,45 Dehydrins are also found among the major mesocarp proteins of ripe grapevine berries.13 Stress proteins, late embryogenesis abundant (LEA), were upregulated 2-fold, and dehydrins were upregulated 6-fold during ripening in muscadine berry (Figure 7C). Rapid increase in LEAs and dehydrins during ripening may be induced in response to changes in ABA levels during berry ripening and seed maturation.35 High expression of dehydrin proteins in GH and PS stages might reflect variation in water status such as dehydration of seed and stress in berry flesh from hexose accumulation.
Enzymes participating in detoxification, such as ascorbate peroxidase, peroxidase, and peroxiredoxins, catalyze the direct reduction of hydrogen peroxide to water; the others (thioredoxins, glutaredoxins) regulate the balance of the oxidized and reduced forms of the antioxidants ascorbic acid and glutathione.46 Oxidative enzymes cationic peroxidase, ascorbate peroxidase, and thioredoxin-related protein, nucleoredoxin (NRX), that govern ROS-stimulated Wnt signaling31 was downregulated during ripening (Figure 7F), while only glutathione peroxidase (GPOX) was upregulated during ripening. Increase GPOX activity was observed in V. vinifera cv. Barbera skin.7 The profile of antioxidant enzyme expression in muscadines demonstrates that as ascorbate and cationic peroxidase activity decreased, GPOX activity had to take over to keep the redox homeostasis active.
Signaling Proteins
Proteins involved in signal transduction were identified and quantified during berry development and ripening. The 14-3-3 like proteins that interact with 0064iverse target proteins in a sequence-specific and phosphorylation-dependent manner47 were upregulated 2-fold in the GS stage (Figure 7J). 14-3-3 is also known to activate ATPase proton pumps.48 Ca2-binding proteins calreticulin (CRT) and calnexin (CNX) are the central players in the CRT/CNX cycle of glycoprotein folding quality control.49 CRT-1 family protein was upregulated during the course of ripening (Figure 7J).Calcium sensing proteins, calmodulin, involved in metabolism, ion transport, transcriptional regulation, protein phosphorylation, and other critical functions showed up- and downregulation during ripening (Figure 7J). Protein phosphatase 2A (PP 2A) has been shown to play a positive role in signaling, and recent publications50 indicate they are key components of stress signal transduction pathways, balancing the action of protein kinases. In muscadines PP 2A was highly expressed in GS stage and then downregulated during ripening. Small GTP-binding proteins play key roles in the signal pathway, vesicular trafficking, and targeting to the PM.51 GTP binding protein SAR-1 was upregulated during GS and decreased during ripening in muscadine grapes.
Protein Degradation
Protein degradation through the ubiquitin/26S proteasome system is the major pathway of protein degradation and plays a key role in cellular processes, removing misfolded or damaged proteins and controlling the level of some regulatory proteins in developing berries. The changes in expression among the ubiquitin/26S proteasome proteins reflect the complex regulatory mechanism for proteolytic machinery.52 26S proteasome regulatory subunit was downregulated during ripening (Figure 7I). Decrease in 26S proteasome protein indicates that as protein synthesis decreased during ripening the proteolytic activity of ubiquitin/26S proteasome pathway was less active.
Transport Proteins
Water permeation across biological membranes is facilitated by water channel proteins called aquaporins.53 Plasma membrane intrinsic protein PIP1 was up- and downregulated from GH to ripening. Expression analyses using cDNA macroarray indicated that aquaporin gene expression is strongly regulated along berry development and globally decreases during ripening in V. vinifera,54 suggesting aquaporins in muscadine grape might be differentially regulated. Cell expansion requires cell wall elongation and accumulation of solutes within the vacuole. Two distinct primary proton pumps, the vacuolar ATPase (V-ATPase) and vacuolar inorganic pyrophosphatase (V-PPase), generate a proton electromotive force, which, in turn, allows the secondary active transport of inorganic ions, sugars, and organic acids.55 V-ATPase expression is relatively high all over the growth of many fruits, such as peach, tomato, and cherry tomato suggesting a central role during fruit development.56 In the present study, V-PPase (EC 3.6.1.1) was upregulated in the PS stage and decreased in the ripening stage, while V-ATPase was upregulated in GS and ripe stages. During berry ripening, the V-ATPase activity is replaced by V-PPase activity and increases in parallel with the ripening in V. vinifera,57 and the switch between V-ATPase and V-PPAse during ripening is not observed in muscadine grape.
Amino Acid Metabolism
During ripening, proteins involved in amino acid metabolism increased mainly in the green and ripe stages (Figure 2H). Protein belonging to glutamate decarboxylase (GAD) was upregulated 3-fold during berry maturation and then gradually decreased during ripening (Figure 7H). Glutamate may be catabolized through glutamate decarboxylase, into γ-aminobutyric acid (GABA), a metabolite that increases during ripening. GAD was upregulated at veraison and ripening in Trincadeira grapes.23 Vitamin-B12 independent methionine 5-methyltetrahydropteroyltriglutamate- homocysteine participating in methionine metabolism was upregulated in GH berries and then downregulated during ripening. Up- and downregulation of proteins indicate that the metabolisms of glutamate, threonine, and methionine play a major role in muscadine grape ripening.
Cell Wall Remodeling and Cell Expansion Regulation
Proteins involved in cytoskeleton remodeling such as actin and tubulin were upregulated in green berries and then down-regulated during ripening (Figure 7K). High expression of cytoskeleton remodeling protein is expected during the first phase of berry development characterized by an active cell division that requires the synthesis of microtubules to drive chromosome segregation during mitosis.27 Protein belonging to the profilin family was upregulated during PS and then decreased during ripening (Figure 7K). Profilins are among the most highly expressed cytoplasmic proteins and are distributed throughout the cytoplasm. Profilins are best known for their ability to promote the exchange of nucleotide in actin monomers released from filaments.58 Another protein involved in cell division and growth, cell division cycle protein 48 homologue, was upregulated during ripening in muscadines (Figure 7K). CDC48 participates in the formation of spindle poles in proliferating tissue of plants.59 Protein involved in membrane-trafficking events associated with cell-plate expansion or maturation and involved in cell-plate biogenesis, patellin, was downregulated during ripening in muscadine grape berry (Figure 7K). Actin, tubulin, and cycle protein 48 homologue may interact with each other and participate in cell wall remodeling during muscadine berry development and ripening.
Photosynthesis and Energy
During the early stages of berry development, photosynthesis-related proteins may be involved in light-mediated CO2 assimilation and refixation of CO2 released by respiration or other metabolic processes,60 while after the onset of ripening, refixation of respiratory CO2 is likely to be the primary function.61,62,13 Proteins belonging to photosynthesis and energy are expressed prominently during the green soft stage and PS and declined toward ripening (Figure 7N). Expression of photosynthesis- related proteins in muscadine grape is consistent with previous studies on V. vinifera3,26 where gradual decline is observed which include photosystem ii proteins, light harvesting complex ii protein, NADH dehydrogenase and plastid lipid associated chloroplastic like.
Gene Expression and Protein Synthesis
In muscadine berry, three elongation factors (EF) (1, 2, and beta) and initiation factor 4a-8 like was highly expressed in green berries. The decrease in expression of elongation and initiation factor proteins after the GS stage suggests a gradual arrest of transcriptional activity post verasion in muscadine berries. The increase in protein expression of this group of proteins is reported due to an increase in protein synthesis and their proper folding to synthesize building blocks and provide energy to maintain cell and organ expansion,63 and this mechanism might be true in muscadine since a similar expression pattern is observed.
Other Proteins
Five proteins of unknown function were identified to be up- and downregulated during muscadine berry ripening that needs further investigation for their role in muscadine berry ripening.
Correlation of Protein Fold Changes with Transcripts
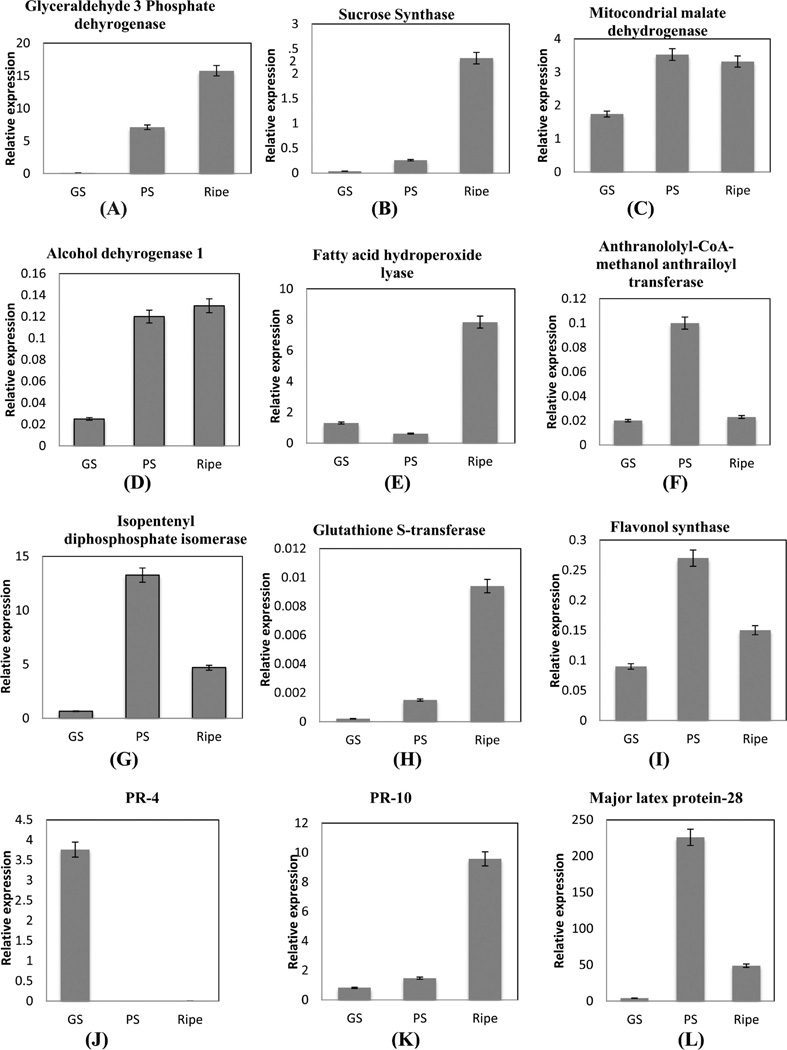
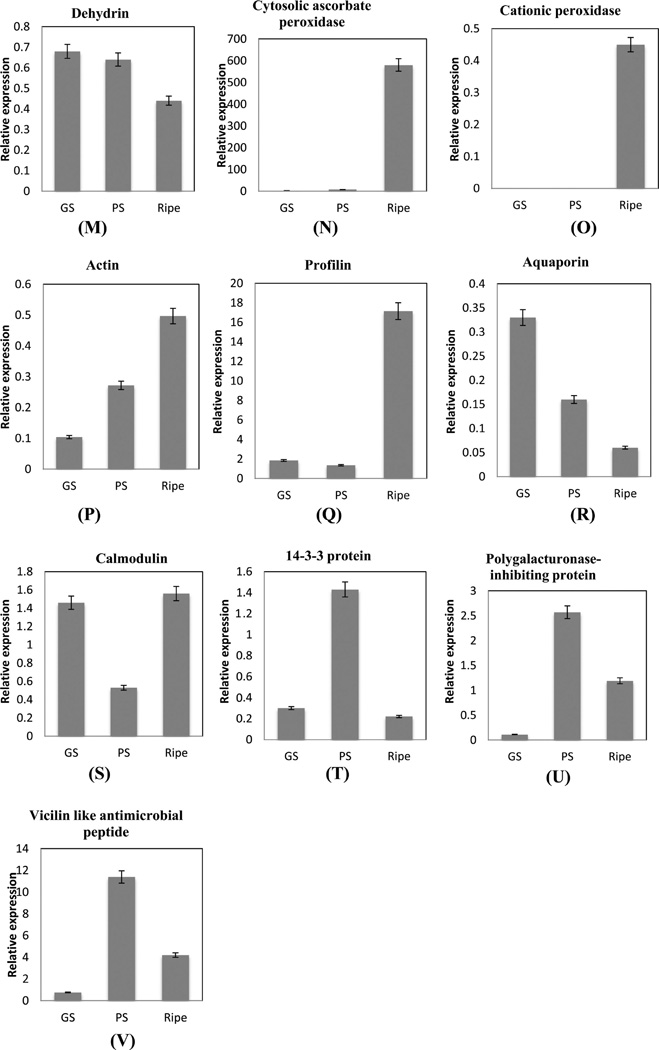
Q-RT PCR was performed to examine the transcript levels for proteins that were identified common to all four replicates and two unique proteins involved in disease resistance identified only in the first iTRAQ 8-plex data set (Supplementary Tables 5 and 8, Supporting Information). Proteins for real time PCR were chosen based on their involvement in various protein functional groups during ripening. Transcript analysis during berry ripening revealed mixed results with two sets of information. One set includes seven transcripts with a similar trend in expression during berry ripening matching with the proteome data. They include glyceraldehyde 3-phosphate dehydrogenase (tr|F6HG44), sucrose synthase (tr|A5C6H7), fatty acid hydroperoxide lyase (tr|F6HID6), isopentenyl diphosphate isomerase (tr|F6GXI9), PR-10 (tr|Q9FS43), flavonol synthase (tr|E0CR99), and major latex protein-28 (tr|Q9M4G9), (Figure 8A,B,E,G,I,K,L). The second set included proteins identified in the iTRAQ study that do not correlate with the transcript profile obtained in real-time PCR. These proteins are mitochondrial malate dehydrogenase (tr|E0CSB6), alcohol dehydrogenase (tr|F6I0F6), anthraniloyl-CoA:methanol anthraniloyl transferase (tr|Q3ZPN4), glutathione transferase (tr|Q56AY1), PR-4 (tr|G9JVT0), dehydrin (tr|F6H0C4), cytosolic ascorbate peroxidase (tr|A9UFX7), cationic peroxidase (tr|F6H0Z2), actin (tr|F6I0I5), profilin (tr|A5AQ89), aquaporin pip1 2 (tr|Q9LLL9), calmodulin (tr|D7T1F3), and 14-3-3 protein (tr|A5AEH1) (Figure 8C,D,F,H,J,M–T).
Figure 8.
Relative expression levels of transcripts. Data were normalized against expression of the housekeeping gene ubiquitin. To determine relative fold differences for each gene in each experiment, the Ct value of the genes was normalized to the Ct value for ubiquitin (control gene), and relative expression was calculated relative to a calibrator using the formula 2−ΔΔCt. All the values shown are mean ± SE.
qRT-PCR of the first set of genes with similar protein and mRNA profiles showed they are involved in sucrose and hexose metabolism, fatty acid cleavage, secondary metabolism, and defense. qRT-PCR determination of G3PDH and sucrose synthase (SS) mRNA levels displayed a 15-fold and 2.3-fold increase respectively in ripe berries over green hard berries (Figure 8A,B), and these results compared favorably to the 3-fold increase in protein expression of G3PDH (tr|F6HG44) and SS. Expression of G3PDH demonstrates that glycolysis is active in muscadine ripe berries and may occur in the cytoplasm as observed in V. vinifera.7 Sucrose synthase (SS) expression during green and ripe stages indicates that SS might be involved in conversion of sucrose to malate during green developmental stages3 and into glucose and fructose in ripe stages. mRNA and protein profile of mMDH was upregulated from GS to PS stage in muscadine berries (Figure 8C) and may participate in malate degradation, as previously reported.64 Similar expression patterns of isopentenyl diphosphate isomerase protein and mRNA (Figure 8G) demonstrate the active conversion of IPP to DMAPP in muscadine berries to produce volatile aroma compounds. The isoprenoid pathway produces monoterpenes (C10), isopentenyl-pyrophophate (IPP), and its isomer dimethylallyl pyrophosphate (DMAPP) are responsible for the formation of many volatile aroma precursors arising mainly from berry exocarp tissues.65 The increase in flavonoid synthase gene and protein expression was observed indicating active biosynthesis of flavonoids during ripening in muscadine berries (Figures 7A and 8I). Similarly, an increase in flavonol synthase was also observed in Norton grape skin after veraison.66 Transcript and protein levels of GST showed an increase during ripening (Figures 7A and 8H) and thus it may participate in anthocyanin sequestration in muscadine grapes. GSTs are known to be involved in anthocyanin sequestration in vacuoles.23 Transcript abundance of AMAT increased up to the PS stage and then decreased in the ripe stage (Figure 8F), while protein was upregulated 2-fold in ripe berries (Figure 7A). Developmental studies with V. labrusca berries demonstrated that AMAT gene expression, enzyme activity, and methyl anthranilate accumulation increased during ripening.30 Dehydrin transcript level was downregulated during ripening, while protein expression increased in the PS stage (Figures 7C and 8M). Dehydrins are found among the major proteins of ripe grapevine berries.67 The increase of dehydrins during ripening initiation in muscadine cv. Noble berries may reflect coincident changes in water status and osmotic stress that results from hexose accumulation.13 Poor correlations between mRNA and protein expression profiles of AMAT and dehydrins reflect the biological reality of protein regulation, as proteins with long half-lives are expected to maintain stable expression for some time after transcript abundance decreases.68
Cytosolic ascorbate peroxidase, cationic peroxidase, actin, and profilin transcripts were upregulated with concurrent downregulation of corresponding proteins in the ripe stage of muscadine berries (Figures 8N–Qand 7F,K). The lack of changes in protein abundance in these genes might be due to post-translational (down) regulation of the protein activity after the stress is relieved.69 Hydroperoxide lyase (HPL) protein and mRNA levels increased during ripening (Figures 8E and 7L), which might be due to the increase in ADH mRNA levels in ripening grape berry (Figure 8F). Increase in ADH and HPL transcripts were also reported in Cabernet Sauvignon.33 The increase in PR 10 and MLP-28 mRNA is consistent with the protein expression level (Figures 8K,L and 7B). PR-4 protein displayed contrasting mRNA and protein profiles. The increase in PR-4 mRNA level to 3.5-fold in the green soft stage (Figure 8 J) could lead to a sustained increase of PR-4 proteins during ripening in muscadine grapes. This can also be explained by the longer half-lives of proteins than mRNAs.68,69
Protein profile of calcium sensing proteins, calmodulin, and 14-3-3 decreased during ripening while transcript increased and then decreased in the ripe stage (Figure 8S,T). Higher expression of calmodulin was observed during heat stress, suggesting a role of Ca2+-mediated signals in the heat stress response in Cabernet Sauvignon.70 Regulation of transcript abundance can control the gene function, thus setting the increase or decrease in protein abundance. A combination of posttranscriptional, translational, and degradative regulation, acting through miRNAs or other mechanisms, then fine-tunes protein abundances to their preferred levels, rather than causing large expression changes.71
Two unique proteins of interest identified in first iTRAQ data set viz. vicilin-like antimicrobial peptides 2-1-like (tr|F6HI56 with two peptide match, Supplementary Tables 5A and 8, Supporting Information) and polygalacturonase-inhibiting protein (PGIP) (tr|G4XMZ9 with three peptide match, Supplementary Tables 5A and 8, Supporting Information) were analyzed by real-time PCR to visualize their presence and expression pattern. The two transcripts followed the trend of proteins identified in the first iTRAQ data set indicating high expression during PS and downregulation in ripe berries. Polygalacturonase-inhibiting protein (PGIP) was upregulated 2-fold and then decreased during ripening (Figure 8B), while transcript level was upregulated 5-fold in the PS stage and then decreased (Figure 8U) confirming that the presence and increase in expression of PGIP protein in the first iTRAQ data set is valid. PGIPs are plant cell-wall proteins that specifically inhibit fungal endopolygalacturonases (PGs) that contribute to the aggressive decomposition of susceptible plant tissues.72 Induction of PGIP gene has been reported in Norton grape,66 and reduction in PGIP activity was observed in Cabernet Sauvignon.9 Vicilin-7S-like antimicrobial peptides that belong to the cupin-2 superfamily73 protein was upregulated 6-fold (Figure 7B), while the transcript level increased 11-fold between the GH and PS stages (Figure 8V) confirming an increase in protein expression pattern. The difference in fold increase between the two transcripts can be explained by the efficiency and post-translational processing of transcripts that might affect the final protein level. Up- and downregulation of PGIP proteins is observed in V. vinifera66,9 during berry ripening, but the identification of vicilin-like antimicrobial peptides indicates that muscadine grape may possess additional disease tolerance proteins for berry protection during ripening.
CONCLUSION
The iTRAQ technique has allowed the detection of proteome changes in muscadine grape for the first time. Stringent filtering criteria were applied to improve confidence in the identification and quantification of proteins. Analysis of proteome changes has allowed the detection of new proteins and contributed to the elucidation of many aspects of primary and secondary metabolism in ripening muscadine berry. This study demonstrates changes in quantitative protein profiles of developing muscadine berry and shows an increase in PR group of proteins and GSTs, which is consistent with the published literature on V. vinifera.10 Flavor and aroma-related proteins (ADH1, HPL1, IPI, and AMAT) identified in this study could be used as candidate marker proteins linked to muscadine flavor and aroma.
High expression of cell expansion and cytoskeleton remodeling proteins viz. actin and tubulin observed in this study were consistent with berry softening. In addition, comparison between transcripts and protein expression showed similarity between the protein and transcript profiles for 40% of the transcripts tested. This study is the first attempt toward understanding the complexity of muscadine berry proteome.
Supplementary Material
Figure 1.
Sampling dates were based the Brix content, size, fresh weight, anthocyanin content, and titratable acidity of the berry. Clusters were tagged from four different vines. Individual berries representing each stage from same vine served as one biological replicate.
ACKNOWLEDGMENTS
We thank Dr. Neil James and Ms. Conchita Newman for providing assistance in texture analysis of grape berries at Food Science Laboratory, College of Agriculture and Food Science, Florida A&M University. We thank Dr. Sixue Chen and acknowledge the Proteomics Division, University of Florida’s Interdisciplinary Center for Biotechnology Research (ICBR) for LC−MS/MS analysis. This work was supported by Grant from USDA-NIFA-CBG-FLAX-SHEIKH. K.R. is thankful for grant support from NIH-NIMHD-RCMI 8 G12 MD007582-28.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Table 1: Physiological berry measurements of muscadine berry for two green and two ripening stages. Table 2: protein yield among the two protocols tested. Tables 3 and 4: Summary of proteins detected among two iTRAQ data sets. Table 5: iTRAQ -two replications data set - 95% Confidence 1.3 Protscore.xls. Table 6: Cluster Analysis of Muscadine Grape Berry Proteome.xls. Table 7: Fishers enrichment analysis was performed to find the significant GO terms in each cluster. Table 8: Functional classification of differential expressed proteins based on Blast2GO and available literature are listed in the table. Table 9: List of primers used for real time PCR. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Marshall DA, Stringer SJ, Spiers JD. Stilbene, ellagic acid, flavonol, and phenolic content of Muscadine Grape (V. rotundifolia Michx.) cultivars. Pharma. Crops. 2012;3:69–77. [Google Scholar]
- 2.Patel GI, Olmo HP. Cytogenetics of Vitis. I. The hybrid V. vinifera × V. rotundifolia. Amer. J. Bot. 1955;42(2):141–155. [Google Scholar]
- 3.Martínez-Esteso MJ, Sellés-Marchart S, Lijavetzky D, Pedreño MA, Bru-Martínez R. A DIGE-based quantitative proteomic analysis of grape berry flesh development and ripening reveals key events in sugar and organic acid metabolism. J. Exp. Bot. 2011;62(8):2521–2269. doi: 10.1093/jxb/erq434. [DOI] [PubMed] [Google Scholar]
- 4.Deluc LG, Grimplet J, Wheatley MD, Tillet RL, Quilici DR, Osborne C, Schooley DA, Schlauch KA, Cushman JC, Cramer GR. Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development. BMC Genomics. 2007;8:429. doi: 10.1186/1471-2164-8-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deytieux C, Geny L, Lapaillerie D, Claverol S, Bonneu M, Done Che B. Proteome analysis of grape skins during ripening. J. Exp. Bot. 2007;58(7):1851–1862. doi: 10.1093/jxb/erm049. [DOI] [PubMed] [Google Scholar]
- 6.Grimplet J, Wheatley MD, Jouira HB, Deluc LG, Cramer GR, Cushman JC. Proteomic and selected metabolite analysis of grape berry tissues under well-watered and water-deficit stress conditions. Proteomics. 2009;9(9):2503–2528. doi: 10.1002/pmic.200800158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Negri AS, Prinsi B, Rossoni M, Failla O, Scienza A, Cocucci M, Espen L. Proteome changes in the skin of the grape cultivar Barbera among different stages of ripening. BMC Genomics. 2008;9:378. doi: 10.1186/1471-2164-9-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giribaldi M, Perugini I, Sauvage FX, Shubert A. Analysis of protein changes during grape berry ripening by 2-DE and MALDI-TOF. Proteomics. 2007;7(17):3154–3170. doi: 10.1002/pmic.200600974. [DOI] [PubMed] [Google Scholar]
- 9.Lucker J, Laszczak M, Smith D, Lund ST. Generation of a predicted protein database from EST data and application to iTRAQ analyses in grape (Vitis vinifera cv. Cabernet Sauvignon) berries at ripening initiation. BMC Genomics. 2009;10:50. doi: 10.1186/1471-2164-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Esteso MJ, Casado-Vela J, Selles-Marchart S, Elortza F, Pedreno MA, Bru-Martínez R. iTRAQ-based profiling of grape berry exocarp proteins during ripening using a parallel mass spectrometric method. Mol. Biosyst. 2011;7(3):749–765. doi: 10.1039/c0mb00194e. [DOI] [PubMed] [Google Scholar]
- 11.Cramer GR, Van Sluyter SC, Hopper DW, Pascovici D, Keighley T, Haynes PA. Proteomic analysis indicates massive changes in metabolism prior to the inhibition of growth and photosynthesis of grapevine (V. vinifera L.) in response to water deficit. BMC Plant Biol. 2013;13:49. doi: 10.1186/1471-2229-13-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marsh E, Alvarez S, Hicks LM, Barbazuk WB, Qiu WB, Kovacs L, Schachtman D. Changes in protein abundance during powdery mildew infection of leaf tissues of Cabernet Sauvignon grapevine (Vitis vinifera L.) Proteomics. 2010;10(10):2057–2064. doi: 10.1002/pmic.200900712. [DOI] [PubMed] [Google Scholar]
- 13.Da Silva FG, Landolino A, Al-Kayal F, Bohlmann MC, Cushman MA, Lim H, Ergul A, Figueroa R, Kabuloglu EK, Osborne C, Rowe J, Tattersall E, Leslie A, Xu J, Baek J, Cramer GR, Cushman JC, Cook DR. Characterizing the grape transcriptome. Analysis of expressed sequence tags from multiple Vitis species and development of a compendium of gene expression during berry development. Plant Physiol. 2005;139(2):574–597. doi: 10.1104/pp.105.065748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coombe B. Growth Stages of the Grapevine: Adoption of a system for identifying grapevine growth stages. Aust. J. Grape Wine Res. 1995;1:104–110. [Google Scholar]
- 15.Lijavetzky D, Carbonell-Bejerano P, Grimplet J, Bravo G, Flores P, Fenoll J, Hellín P, Carlos Oliveros J, Martínez-Zapater JM. Berry Flesh and Skin Ripening Features in Vitis vinifera as Assessed by Transcriptional Profiling. PLoS ONE. 2012;7(6):e39547. doi: 10.1371/journal.pone.0039547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vincent D, Wheatley MD, Cramer GR. Optimization of protein extraction and solubilization for mature grape berry clusters. Electrophoresis. 2006;27(9):1853–1865. doi: 10.1002/elps.200500698. [DOI] [PubMed] [Google Scholar]
- 17.Bradford MM. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 1976;7(72):248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 18.Zhu M, Simons B, Zhu N, Oppenheimer DG, Chen S. Analysis of abscisic acid responsive proteins in Brassica napus guard cells by multiplexed isobaric tagging. J. Proteomics. 2010;73(4):790–805. doi: 10.1016/j.jprot.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Koh J, Chen S, Zhu N, Yu F, Soltis PS, Soltis DE. Comparative proteomics of the recently and recurrently formed natural allopolyploid Tragopogon mirus (Asteraceae) and its parents. New Phytol. 2012;196(1):292–305. doi: 10.1111/j.1469-8137.2012.04251.x. [DOI] [PubMed] [Google Scholar]
- 20.Glen A, Gan CS, Hamdy FC, Eaton CL, Cross SS, Catto WF, Wright PC, Rehman I. iTRAQ-Facilitated Proteomic Analysis of Human Prostate Cancer Cells Identifies Proteins Associated with Progression. J. Proteome Res. 2008;7(3):897–907. doi: 10.1021/pr070378x. [DOI] [PubMed] [Google Scholar]
- 21.Conesa S, Götz JM, Garcia-Gomez J, Terol M, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 22.Kristoffer TG, Jens TR, Blagoev VB. GProX, a User-Friendly Platform for Bioinformatics Analysis and Visualization of Quantitative Proteomics Data. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.O110.007450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fortes AM, Agudelo-Romero P, Silva MS, Ali K, Sousa L, Maltese F, Choi YH, Grimplet J, Martinez-Zapater JM, Verpoorte R, Pais MS. Transcript and metabolite analysis in Trincadeira cultivar reveals novel information regarding the dynamics of grape ripening. BMC Plant Biol. 2011;11:149. doi: 10.1186/1471-2229-11-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pastore C, Zenoni S, Tornielli GB, Allegroqq G, Tornielli GB, Allegro G, Dal Santo S, Valentini G, Intrieri C, Pezzotti M, Filippetti I. Increasing the source/sink ratio in Vitis vinifera (cv Sangiovese) induces extensive transcriptome reprogramming and modifies berry ripening. BMC Genomics. 2011;12:63. doi: 10.1186/1471-2164-12-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giribaldi M, Perugini I, Sauvage FX, Schubert A. Analysis of protein changes during grape berry ripening by 2-DE and MALDI-TOF. Proteomics. 2007;7(17):3154–3170. doi: 10.1002/pmic.200600974. [DOI] [PubMed] [Google Scholar]
- 26.Famiani F, Walker RP, Tecsi L, Chen ZH, Proietti P, Leegood RC. An immune histochemical study of the compartmentation of metabolism during the development of grape (Vitis vinifera L.) berries. J. Exp. Bot. 2000;51(345):675–683. [PubMed] [Google Scholar]
- 27.Carli MD, Zamboni A, Pezzotti ME, Lilley KS, Pezzotti M, Lilley KS, Benvenuto E, Desiderio A. Two-Dimensional Differential in Gel Electrophoresis (2D-DIGE) Analysis of Grape Berry Proteome during Postharvest Withering. J. Proteome Res. 2011;10(2):429–446. doi: 10.1021/pr1005313. [DOI] [PubMed] [Google Scholar]
- 28.Fowler JH, Narvá ez-Vá squez J, Aromdee DN, Pautot V, Holzer FM, Walling LL. Leucine Aminopeptidase Regulates Defense and Wound Signaling in Tomato Downstream of Jasmonic Acid. Plant Cell. 2009;21:1239–1251. doi: 10.1105/tpc.108.065029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niu N, Cao Y, Duan W, Wu B, Li S. Proteomic analysis of grape berry skin responding to sunlight exclusion. J. Plant Physiol. 2013;170(8):748–757. doi: 10.1016/j.jplph.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Luca VD. The biosynthesis and regulation of biosynthesis of Concord grape fruit esters, including ‘foxy’ methylanthranilate. Plant J. 2005;44(4):606–619. doi: 10.1111/j.1365-313X.2005.02552.x. [DOI] [PubMed] [Google Scholar]
- 31.Boss PK, Davies C, Robinson SP. Analysis of the Expression of Anthocyanin Pathway Genes in Developing Vitis vinifera L. cv Shiraz Grape Berries and the Implications for Pathway Regulation. Plant Physiol. 1996;111(4):1059–1066. doi: 10.1104/pp.111.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feussner I, Wasternack C. The Lipoxygenase pathway. Ann. Rev. Plant Biol. 2002;53:275–297. doi: 10.1146/annurev.arplant.53.100301.135248. [DOI] [PubMed] [Google Scholar]
- 33.Zhu BQ, Xu XQ, Wu YW, Duan CQ, Pan QH. Isolation and characterization of two hydroperoxide lyase genes from grape berries: HPL isogenes in Vitis vinifera grapes. Mol. Biol. Rep. 2012;39(7):7443–7455. doi: 10.1007/s11033-012-1577-0. [DOI] [PubMed] [Google Scholar]
- 34.Tesniere C, Davies C, Sreekantan L, Bogs J, Thomas M, Torregrosa L. Analysis of the transcript levels of VvAdh1, VvAdh2 and VvGrip4, three genes highly expressed during Vitis vinifera L. berry development. Vitis. 2006;45(2):75–79. [Google Scholar]
- 35.Pilati S, Perazzolli M, Malossini A, Cestaro A, Dematté L, Fontana P, Dal Ri A, Viola R, Velasco R, Moser C. Genome-wide transcriptional analysis of grapevine berry ripening reveals a set of genes similarly modulated during three seasons and the occurrence of an oxidative burst at véraison. BMC Genomics. 2007;22(8):428. doi: 10.1186/1471-2164-8-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colas S, Afoufa-Bastien D, Jacquens L, Clément C, Baillieul F, Mazeyrat-Gourbeyre F, Monti-Dedieu L. Expression and in situ localization of two major PR proteins of grapevine berries during development and after UV-C exposition. PLoS One. 2012;7(8):e43681. doi: 10.1371/journal.pone.0043681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caporale C, Facchiano A, Bertini L, Leonardi L, Chilosi G, Buonocore V, Caruso C. Comparing the modeled structures of PR-4 proteins from wheat. J. Mol. Model. 2003;9(1):9–15. doi: 10.1007/s00894-002-0103-z. [DOI] [PubMed] [Google Scholar]
- 38.Robert N, Ferran J, Breda C, Coutos-Thévenot P, Boulay M, Buffard D, Esnault R. Molecular characterization of the incompatible interaction of Vitis vinifera leaves with Pseudomonas syringae pv. pisi: Expression of genes coding for stilbene synthase and class 10 PR protein. Eur. J. Plant Pathol. 2001;107(2):249–261. [Google Scholar]
- 39.Lebel S, Schellenbaum P, Walter B, Maillot P. Characterisation of the Vitis vinifera PR10 multigene family. BMC Plant Biol. 2010;20(10):184. doi: 10.1186/1471-2229-10-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osmark P, Boyle B, Brisson N. Sequential and structural homology between intracellular pathogenesis-related proteins and a group of latex proteins. Plant Mol. Biol. 1998;38(6):1243–1246. doi: 10.1023/a:1006060224012. [DOI] [PubMed] [Google Scholar]
- 41.Lytle BL, Song J, de la Cruz NB, Peterson FC, Johnson KA, Bingman CA, Phillips GN, Volkman BF. Structures of two Arabidopsis thaliana major latex proteins represent novel helix-grip folds. Proteins. 2009;2:237–243. doi: 10.1002/prot.22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoekstra FA, Golovina EA, Tetteroo FA, Wolkers WF. Induction of desiccation tolerance in plant somatic embryos: how exclusive is the protective role of sugars? Cryobiology. 2001;43:140–150. doi: 10.1006/cryo.2001.2358. [DOI] [PubMed] [Google Scholar]
- 43.Dure L. Structural motifs in LEA proteins. In: Close TJ, Bray EA, editors. Plant Responses to Cellular Dehydration during Environmental Stress. Am. Soc. Plant Physiol. Vol. 91. 1993. p. 103. [Google Scholar]
- 44.Buchanan CD, Lim SY, Salzman RA, Kagiampakis L. Sorghum bicolor’s transcriptome response to dehydration, high salinity and ABA. Plant Mol. Biol. 2005;58:699–720. doi: 10.1007/s11103-005-7876-2. [DOI] [PubMed] [Google Scholar]
- 45.Rampino P, Pataleo S, Gerardi C, Mita G, Perrotta C. Drought stress response in wheat: physiological and molecular analysis of resistant and sensitive genotypes. Plant Cell Environ. 2006;29:2143–2152. doi: 10.1111/j.1365-3040.2006.01588.x. [DOI] [PubMed] [Google Scholar]
- 46.Funato Y, Miki H. Redox regulation of Wnt signaling via nucleoredoxin. Free Radical Res. 2010;44(4):379–388. doi: 10.3109/10715761003610745. [DOI] [PubMed] [Google Scholar]
- 47.Bridges D, Moorhead GB. 14-3-3 proteins: a number of functions for a numbered protein. Sci. STKE. 2005;296:re10. doi: 10.1126/stke.2962005re10. [DOI] [PubMed] [Google Scholar]
- 48.Duby G, Poreba W, Piotrowiak D, Bobik K, Derua R, Waelkens E, Boutry M. Activation of plant plasma membrane H+-ATPase by 14-3-3 proteins is negatively controlled by two phosphorylation sites within the H+-ATPase C-terminal region. J. Biol. Chem. 2009;284(7):4213–4221. doi: 10.1074/jbc.M807311200. [DOI] [PubMed] [Google Scholar]
- 49.Eduardo L, Bem VD. The evolutionary history of calreticulin and calnexin genes in green plants. Genetica. 2011;139(2):255–259. doi: 10.1007/s10709-010-9544-y. [DOI] [PubMed] [Google Scholar]
- 50.País SM, Téllez-Iñón MT, Capiat DA. Serine/threonine protein phosphatases type 2A and their roles in stress signaling. Plant Signal. Behav. 2009;4(11):1013–1015. doi: 10.4161/psb.4.11.9783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marmagne A, Rouet MA, Ferro M, Rolland N, Alcon C, Joyard J, Garin J, Barbier-Brygoo H, Ephritikhine G. Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol. Cell. Proteomics. 2004;3:675–691. doi: 10.1074/mcp.M400001-MCP200. [DOI] [PubMed] [Google Scholar]
- 52.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 2002;82(2):373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 53.Chrispeels MJ, Maurel C. Aquaporins: the molecular basis of facilitated water movement through living plant cells? Plant Physiol. 1994;105(1):9–13. doi: 10.1104/pp.105.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fouquet R, Leon C, Ollat N, Barrieu F. Identification of grapevine aquaporins and expression analysis in developing berries. Plant Cell Rep. 2008;27(9):1541–1550. doi: 10.1007/s00299-008-0566-1. [DOI] [PubMed] [Google Scholar]
- 55.Taiz L. The plant vacuole. J. Exp. Biol. 1992;172:113–122. doi: 10.1242/jeb.172.1.113. [DOI] [PubMed] [Google Scholar]
- 56.Bianco L, Alagna F, Baldoni L, Finnie C, Svensson B. Proteome Regulation during Olea europaea. Fruit Development. PLoS ONE. 2013;8(1):e53563. doi: 10.1371/journal.pone.0053563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Terrier N, Francois-Xavier S, Ageorges A, Romieu C. Changes in acidity and in proton transport at the tonoplast of grape berries during development. Planta. 2001;213:20–28. doi: 10.1007/s004250000472. [DOI] [PubMed] [Google Scholar]
- 58.Golla R, Philp N, Safer D, Chintapalli J, Collins L, Nachmias VT. Co-ordinate regulation of the cytoskeleton in 3T3 cells over-expressing thymosin-β4. Cell Motil. Cytoskeleton. 1997;38(2):187–200. doi: 10.1002/(SICI)1097-0169(1997)38:2<187::AID-CM7>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 59.Feiler HS, Desprez T, Santoni V, Kronenberger J, Caboche M, Traas J. The higher plant Arabidopsis thaliana encodes a functional cdc48 homologue which is highly expressed in dividing and expanding cells. EMBO J. 1995;14:5626–5637. doi: 10.1002/j.1460-2075.1995.tb00250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwender J, Goffman F, Ohlrogge JB, Shachar-Hill Y. Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature. 2004;432:779–782. doi: 10.1038/nature03145. [DOI] [PubMed] [Google Scholar]
- 61.Blanke MM, Lenz F. Fruit photosynthesis. Plant Cell Environ. 1999;12:31–46. [Google Scholar]
- 62.Aschan G, Pfanz H. Non-foliar photosynthesis - A strategy of additional carbon acquisition. Flora. 2003;198:81–97. [Google Scholar]
- 63.Li J, Dickerson TJ, Benning SH. Contribution of proteomics in the identification of novel proteins associated with plant growth. J. Proteome Res. 2013;12:4882–4891. doi: 10.1021/pr400608d. [DOI] [PubMed] [Google Scholar]
- 64.Sweetman C, Deluc LG, Cramer GR, Ford CM, Soole KL. Regulation of malate metabolism in grape berry and other developing fruits. Phytochemistry. 2009;70(11):1329–1344. doi: 10.1016/j.phytochem.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 65.Grimplet J, Deluc LG, Tillett RL, Wheatley MD, Schlauch KA, Cramer GR, Cushman JC. Tissue-specific mRNA expression profiling in grape berry tissues. BMC Genomics. 2007;8:187. doi: 10.1186/1471-2164-8-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ali MB, Howard S, Chen S, Wang Y, Yu O, Kovacs LG, Qiu W. Berry skin development in Norton grape: distinct patterns of transcriptional regulation and flavonoid biosynthesis. BMC Plant Biol. 2011;11:7. doi: 10.1186/1471-2229-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sarry JE, Gunata Z. Plant and microbial glycoside hydrolases: volatile release from glycosidic aroma precursors. Food Chem. 2004;87:509–521. [Google Scholar]
- 68.Lackner DH, Schmidt MW, Wu S, Wolf DA, Bahler J. Regulation of transcriptome, translation, and proteome in response to environmental stress in fission yeast. Genome Biol. 2012;13:R25. doi: 10.1186/gb-2012-13-4-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lan P, Li W, Schmidit W. Complementary proteome and transcriptome profiling in phosphate-deficient Arabidopsis roots reveals multiple levels of gene regulation. Mol. Cell. Proteomics. 2012;11:1156–1166. doi: 10.1074/mcp.M112.020461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu GT, Wang JF, Cramer G, Dai ZW, Duan W, Xu HG, Wu BH, Fan PG, Wang LJ, Li SH. Transcriptomic analysis of grape (Vitis vinifera L.) leaves during and after recovery from heat stress. BMC Plant Biol. 2012;12:174. doi: 10.1186/1471-2229-12-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alexandersson E, Becker JVW, Jacobson D, Ona EM, Steyn C, Denby KJ, Vivier MA. Constitutive expression of a grapevine polygalacturonase-inhibiting protein affects gene expression and cell wall properties in uninfected tobacco. BMC Res. Notes. 2011;4:493. doi: 10.1186/1756-0500-4-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marcus JP, Goulter KC, Green JL, Harrison SJ, Manners JM. Purification, characterization and cDNA cloning of an antimicrobial peptide from Macadamia integrifolia. Eur. J. Biochem. 1997;244(3):743–749. doi: 10.1111/j.1432-1033.1997.00743.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.