Significance
The circadian clock protein KaiC cyclically phosphorylates and dephosphorylates itself with a periodicity of ∼24 h. Unlike the reactions mediated by conventional protein phosphatases, dephosphorylation of KaiC occurs via reversal of the phosphorylation reaction. This unusual mechanism suggests that the KaiC phosphorylation rhythm is sustained by periodic shifts in the equilibrium of the reversible autophosphorylation reaction, the molecular basis of which has never been elucidated. In this study, we found that KaiA promoted the forward reaction by stimulating exchange of KaiC-bound ADP for exogenous ATP. KaiB inhibited KaiA, promoting retention of KaiC-bound ADP, and thereby facilitating the reverse reaction. We propose that KaiA and KaiB sustain the circadian oscillation by regulating this reversible reaction at the level of substrate availability.
Keywords: in vitro reconstitution, self-sustained oscillation, nucleotide exchange
Abstract
The cyanobacterial circadian oscillator can be reconstituted in vitro. In the presence of KaiA and KaiB, the phosphorylation state of KaiC oscillates with a periodicity of ∼24 h. KaiC is a hexameric P-loop ATPase with autophosphorylation and autodephosphorylation activities. Recently, we found that dephosphorylation of KaiC occurs via reversal of the phosphorylation reaction: a phosphate group attached to Ser431/Thr432 is transferred to KaiC-bound ADP to generate ATP, which is subsequently hydrolyzed. This unusual reaction mechanism suggests that the KaiC phosphorylation rhythm is sustained by periodic shifts in the equilibrium of the reversible autophosphorylation reaction, the molecular basis of which has never been elucidated. Because KaiC-bound ATP and ADP serve as substrates for the forward and reverse reactions, respectively, we investigated the regulation of the nucleotide-bound state of KaiC. In the absence of KaiA, the condition in which the reverse reaction proceeds, KaiC favored the ADP-bound state. KaiA increased the ratio of ATP to total KaiC-bound nucleotides by facilitating the release of bound ADP and the incorporation of exogenous ATP, allowing the forward reaction to proceed. When both KaiA and KaiB were present, the ratio of ATP to total bound nucleotides exhibited a circadian rhythm, whose phase was advanced by several hours relative to that of the phosphorylation rhythm. Based on these findings, we propose that the direction of the reversible autophosphorylation reaction is regulated by KaiA and KaiB at the level of substrate availability and that this regulation sustains the oscillation of the phosphorylation state of KaiC.
The circadian clock is an endogenous timing mechanism that coordinates various biological functions with daily environmental changes (1). The cyanobacterial circadian clock is the sole example of such a system that can be reconstituted in vitro: when KaiA, KaiB, KaiC, and ATP are mixed in a test tube, the phosphorylation state of KaiC exhibits a robust circadian rhythm (2). Previously, it was believed that eukaryotic circadian oscillations were exclusively regulated by a transcriptional–translational feedback loop in which clock gene products repress their own transcription (3). However, recent work has revealed that protein-based circadian oscillations are ubiquitous in all domains of life (4). The cyanobacterial in vitro system provides an accessible and useful model for understanding the principles underlying such oscillations.
KaiC is a P-loop ATPase consisting of two ATPase domains: the N-terminal CI domain and the C-terminal CII domain (5). An X-ray crystallographic study revealed that KaiC assembles as a hexamer in the presence of ATP and forms CI–CII double rings with an ATP molecule bound at each subunit interface in both rings (6). ATPγS, a nonhydrolyzable ATP analog, binds to the nucleotide-free KaiC monomer with a dissociation constant of 34 μM (7). KaiC monomers associate into hexamers in the presence of nucleotide triphosphates and their analogs, whereas ADP does not induce any hexamerization, even at a concentration of 1 mM (8). Both the CI and CII domains catalyze ATP hydrolysis, which occurs at the subunit interfaces of both CI and CII (9). In addition to ATPase activity, KaiC has autophosphorylation and autodephosphorylation activity (10–12). The autophosphorylation sites, serine (Ser) 431 and threonine (Thr) 432 (13), are located at the subunit interface of CII, facing the ATP molecule on the neighboring subunit (14), indicating that autophosphorylation, autodephosphorylation, and ATP hydrolysis share the same active sites in CII. Although KaiC hydrolyzes ∼15 ATP molecules during one period of the circadian cycle, as little as 0.3–0.4 molecules of ATP are consumed for phosphorylation (9).
KaiA and KaiB control the phosphorylation state of KaiC. In the absence of KaiA, KaiC undergoes autodephosphorylation (11, 12). KaiA enhances the autophosphorylation of KaiC (15), whereas KaiB inhibits the effect of KaiA, allowing KaiC to dephosphorylate itself (11, 12). KaiB interacts with KaiC in a Ser431 phosphorylation-dependent manner (16, 17). The resultant KaiB–KaiC complex sequesters KaiA, thereby temporarily decreasing the effective concentration of KaiA, and thus sustains the circadian phosphorylation rhythm of KaiC (16, 17). In addition, KaiA is an activator, and KaiB is an inhibitor, of the ATPase activity of KaiC (9).
Recently, we and another group found that KaiC undergoes autodephosphorylation via a mechanism that is completely distinct from that of typical Ser/Thr- and tyrosine-specific protein phosphatases (18, 19): a phosphate group covalently attached to Ser/Thr is transferred to KaiC-bound ADP to generate ATP as an intermediate; thus, this reaction can be regarded as the reversal of the autophosphorylation reaction. Subsequently, the newly generated ATP molecule is hydrolyzed to form inorganic phosphate, the final product (18). This unique reaction mechanism indicates that autophosphorylation and autodephosphorylation are not two independent reactions, but are instead the forward and reverse directions of a single phospho-transfer reaction between KaiC-bound nucleotides and phosphorylation sites. On the basis of this mechanism, it is reasonable to predict that KaiC phosphorylation rhythm is sustained by periodic shifts in the equilibrium of this reaction (18). However, the underlying mechanism of these equilibrium shifts has never been elucidated.
Because the forward and reverse autophosphorylation reactions require KaiC-bound ATP and ADP as substrates, respectively, we decided to investigate the effects of KaiA and KaiB on KaiC-bound nucleotides. In the absence of KaiA, KaiC favored the ADP-bound state. KaiA increased the ratio of ATP to total KaiC-bound nucleotides by facilitating the release of bound ADP, followed by the incorporation of exogenous ATP. When both KaiA and KaiB were present, the nucleotide-bound state of KaiC and the rate of exogenous ATP incorporation into KaiC exhibited circadian oscillation, whose phases were advanced by several hours relative to that of the phosphorylation rhythm. Based on these results, we propose that the direction of the reversible phospho-transfer reaction is regulated by KaiA and KaiB at the level of substrate availability, which sustains the oscillation of the phosphorylation state of KaiC.
Results
KaiA Regulates the Nucleotide-Bound State of KaiC.
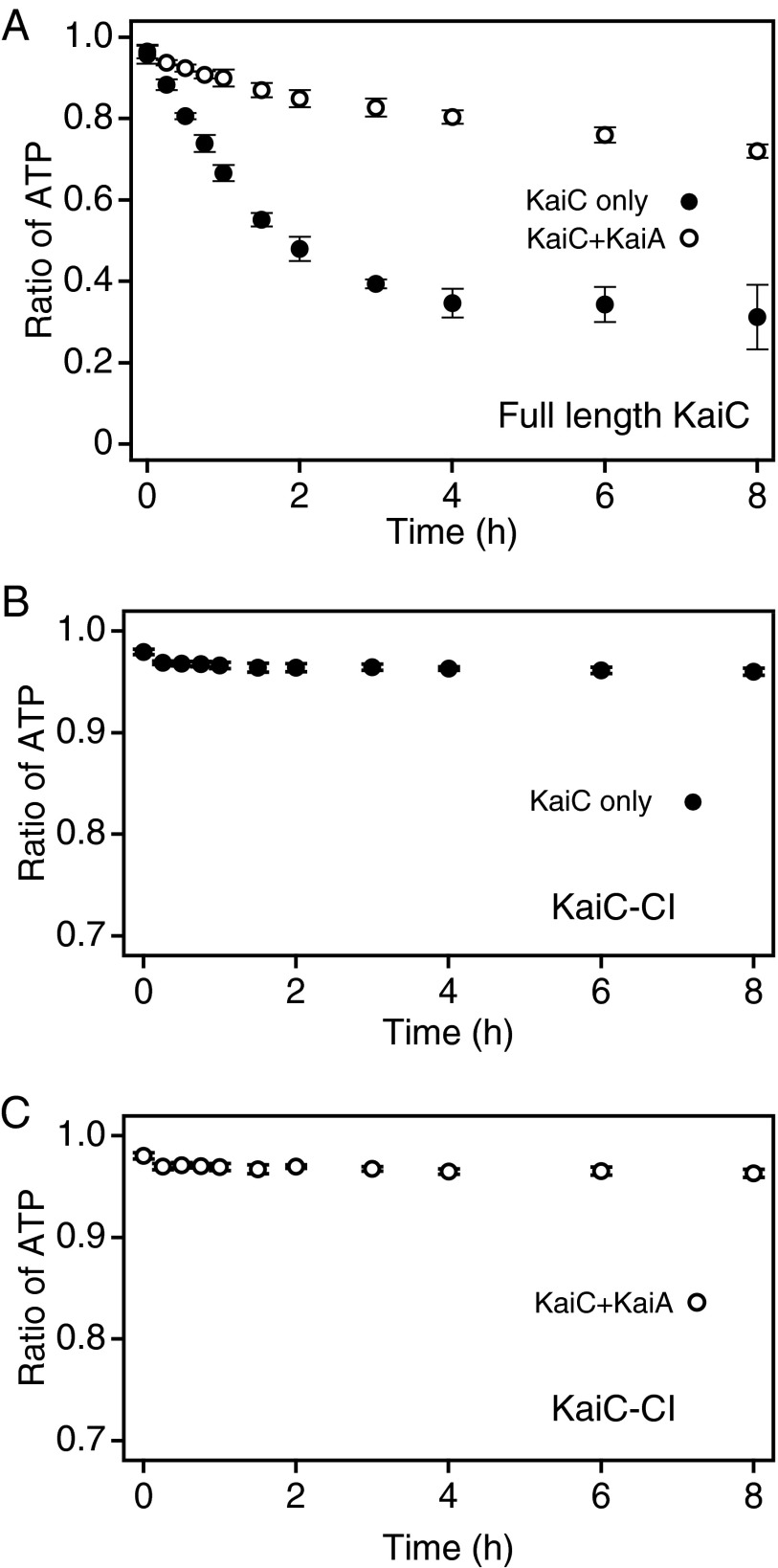
The phosphorylation level of KaiC increases in the presence of KaiA (15) but decreases in its absence (11, 12). To investigate the molecular basis of these phenomena, we first examined the effect of KaiA on the KaiC-bound nucleotides that serve as substrates in the reversible phospho-transfer reaction. We reconstituted KaiC hexamers from monomers in the presence of 2 mM [α-32P]ATP; this concentration of ATP is ∼60-fold higher than the dissociation constant of ATPγS (34 µM), a nonhydrolyzable analog of ATP (7). We incubated these hexamers at 30 °C, with or without KaiA, and plotted the ratio of KaiC-bound ATP to total KaiC-bound nucleotides as a function of time over an 8-h reaction. KaiC has two ATP-binding sites in each protomer, both of which are constantly occupied by either ATP or ADP (18). Before incubation, ATP accounted for more than 90% of the KaiC-bound nucleotides. In the absence of KaiA, the ratio of ATP to total bound nucleotide decreased, reaching ∼0.3 after 4 h of incubation (Fig. 1A, ●), consistent with our previous results (18). In the presence of KaiA, the decrease in the ATP ratio was slower than in its absence (Fig. 1A, ○): during 8 h of incubation, the ratio did not drop below 0.7. These results suggest that KaiC favors the ATP-bound state more strongly in the presence of KaiA than in its absence. In the case of KaiC-CI, a truncated protein that lacks the CII domain, ∼95% of the KaiC-bound nucleotides were ATP, regardless of whether KaiA was present (Fig. 1 B and C), suggesting that the CII domain is responsible for retaining ADP on KaiC.
Fig. 1.
Effect of KaiA on the nucleotide-bound state of KaiC. (A) Full-length KaiC hexamers were reconstituted from monomers in the presence of 2 mM [α-32P]ATP and incubated at 30 °C in the presence (○) or absence (●) of KaiA. At the indicated time points, aliquots of the reaction mixtures were collected, unbound nucleotides were removed, and the samples were analyzed. The ratio of ATP bound to KaiC to total bound nucleotides was plotted against time. The data represent the means ± SD from three independent experiments. (B and C) The nucleotide-bound state of KaiC-CI (0.5 mg/mL) was monitored in the presence of 1 mM [α-32P]ATP with (C) or without (B) KaiA. The data were plotted as the ratio of ATP to total bound nucleotides. ATP accounted for over 95% of nucleotides bound to KaiC-CI, regardless of whether KaiA was present.
We suspect that KaiC-bound ADP is the hydrolysis product of KaiC-bound ATP for the following reasons: (i) we did not add ADP to the reaction mixture, and (ii) kinetic analysis of the KaiC autodephosphorylation process, based on the assumption that KaiC-bound ADP was generated by hydrolysis of bound ATP, gave a good fit to the data (18). Therefore, we hypothesized that KaiA increases the bound ATP percentage by facilitating the release of bound ADP and the incorporation of external ATP.
KaiA Facilitates the Release of ADP from KaiC Hexamers.
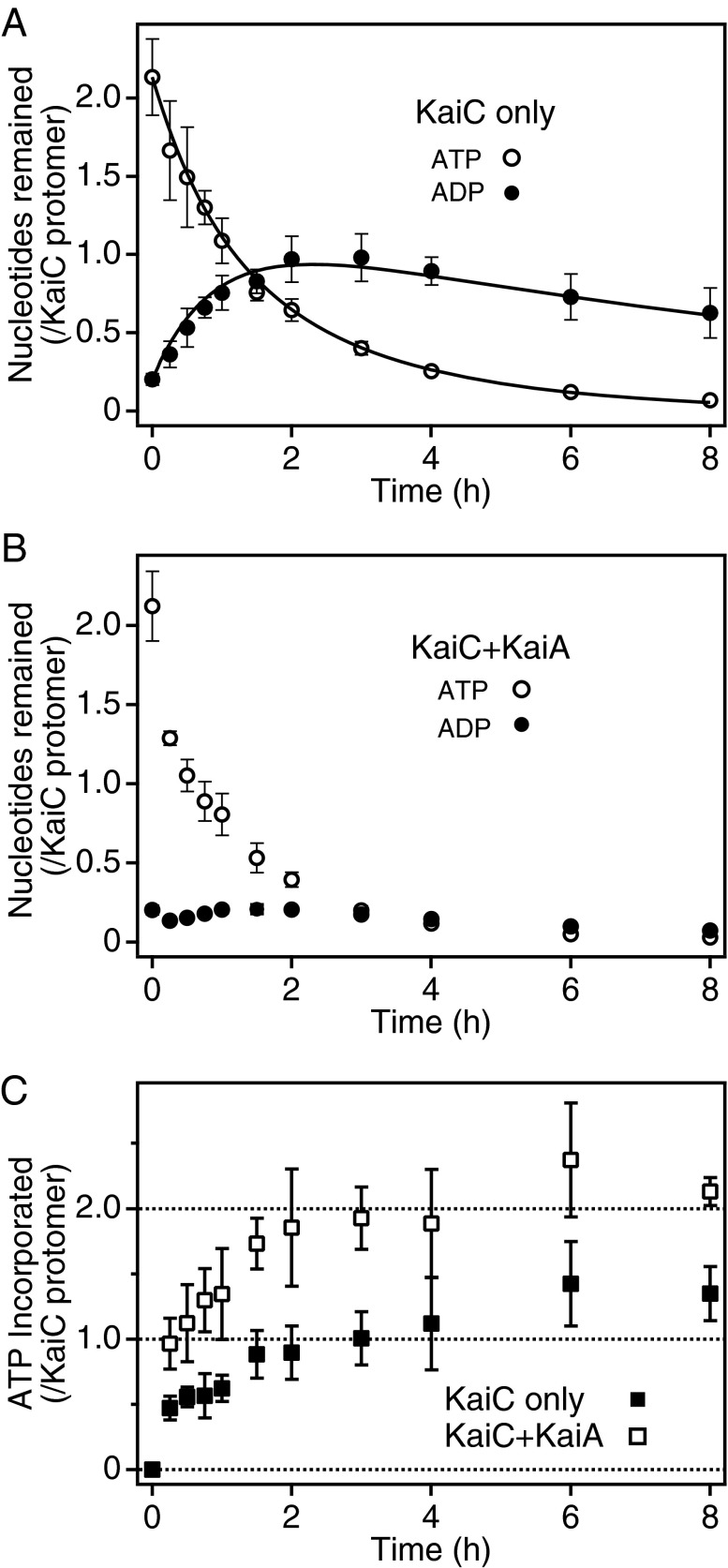
To test the possibility described above, we first performed experiments designed to trace the fate of ATP prebound to KaiC (Fig. 2 A and B). KaiC hexamers were generated by mixing KaiC monomers with 2 mM [α-32P]ATP, unbound radioactive ATP was removed, and the KaiC-bound radioactive nucleotides were chased for 8 h with 2 mM nonradioactive ATP. Approximately 2.1 ATP and 0.2 ADP were initially bound per subunit (Fig. 2 A and B), consistent with our previous observation that the nucleotide-binding sites of both CI and CII are occupied (18). In the absence of KaiA, about half of the prebound ATP disappeared within 1 h, and little ATP could be detected after 8 h (Fig. 2A, ○). On the other hand, the amount of the KaiC-bound radioactive ADP, which can be interpreted as the hydrolysis product of KaiC-bound ATP, increased to approach one molecule per KaiC protomer during the initial 2 h; ∼0.6 ADP per subunit remained even after 8 h of reaction (Fig. 2A, ●). In the presence of KaiA, no more than 0.2 ADP per subunit was retained on KaiC throughout the reaction (Fig. 2B, ●), suggesting that KaiA facilitates the release of bound ADP after the hydrolysis of KaiC-bound ATP. In the presence of KaiA, the level of prebound ATP decreased to about half of its initial value within 0.5 h, slightly faster than in the absence of KaiA (Fig. 2B, ○).
Fig. 2.
KaiA-stimulated nucleotide exchange on KaiC. (A and B) KaiC-bound radioactive nucleotides were chased with exogenous nonradioactive ATP. KaiC hexamers were prepared from monomers in the presence of 2 mM [α-32P]ATP, unbound nucleotides were removed, 2 mM nonradioactive ATP was added, and the samples were incubated at 30 °C in the presence (B) or absence (A) of KaiA. Aliquots were collected at the indicated time points, unbound nucleotides were removed, and the KaiC-bound radioactive signals were quantitated. The amounts of KaiC-bound ATP (○) and ADP (●) were plotted against time. The data represent the means ± SD from three independent experiments. (C) Incorporation of exogenous ATP into KaiC hexamers. KaiC hexamers were reconstituted from monomers with nonradioactive ATP and incubated in the presence of 2 mM [α-32P]ATP, either with (□) or without (■) KaiA. Aliquots were collected at predetermined time points, unbound nucleotides were removed, and KaiC-bound radioactivity was quantitated. The amount of ATP incorporated into KaiC was plotted against time. The data represent the means ± SD from three independent experiments. Solid lines in A indicate fits of the model as shown in Eqs. S1–S3 in the SI Materials and Methods section. The rate constants were estimated to be 0.383 ± 0.013 for k1, 1.11 ± 0.19 for k2, and 0.090 ± 0.047 for k3, where k1 is the overall rate constant for the consecutive processes, including ATP hydrolysis and ADP release, from Site 1; k2 is the rate constant for ATP hydrolysis at Site 2; and k3 is the rate constant of ADP release from Site 2. These parameters are presented as the best fit values ± SE. It is likely that Sites 1 and 2 correspond to the ATPase active sites in domains CI and CII, respectively (see Discussion).
Effects of KaiA on ATP Incorporation into KaiC Hexamers.
Next, we tested the ability of KaiA to stimulate the incorporation of exogenous ATP into KaiC. We prepared KaiC hexamers from monomers in the presence of 2 mM unlabeled ATP, added [α-32P]ATP at a final specific activity of 3.0 GBq/mmol, and then monitored the accumulation of radioactivity in the KaiC hexamers (Fig. 2C). In the absence of KaiA, radioactivity corresponding to approximately one molecule of ATP was incorporated per KaiC protomer by 2 h, and the signals increased slightly until 8 h (Fig. 2C, ■), suggesting that one prebound nucleotide was exchanged for exogenous ATP within the initial 2 h. KaiA increased ATP uptake by KaiC, which reached approximately two molecules per subunit by 2 h (Fig. 2C, □). These results suggest that KaiC has two ATPase active sites with different turnover rates and that KaiA acts on the site with the slower rate to facilitate nucleotide exchange (Discussion).
Nucleotide-Bound State of KaiC and the Rate of ATP Incorporation into KaiC Exhibited a Circadian Rhythm in the Presence of KaiA and KaiB.
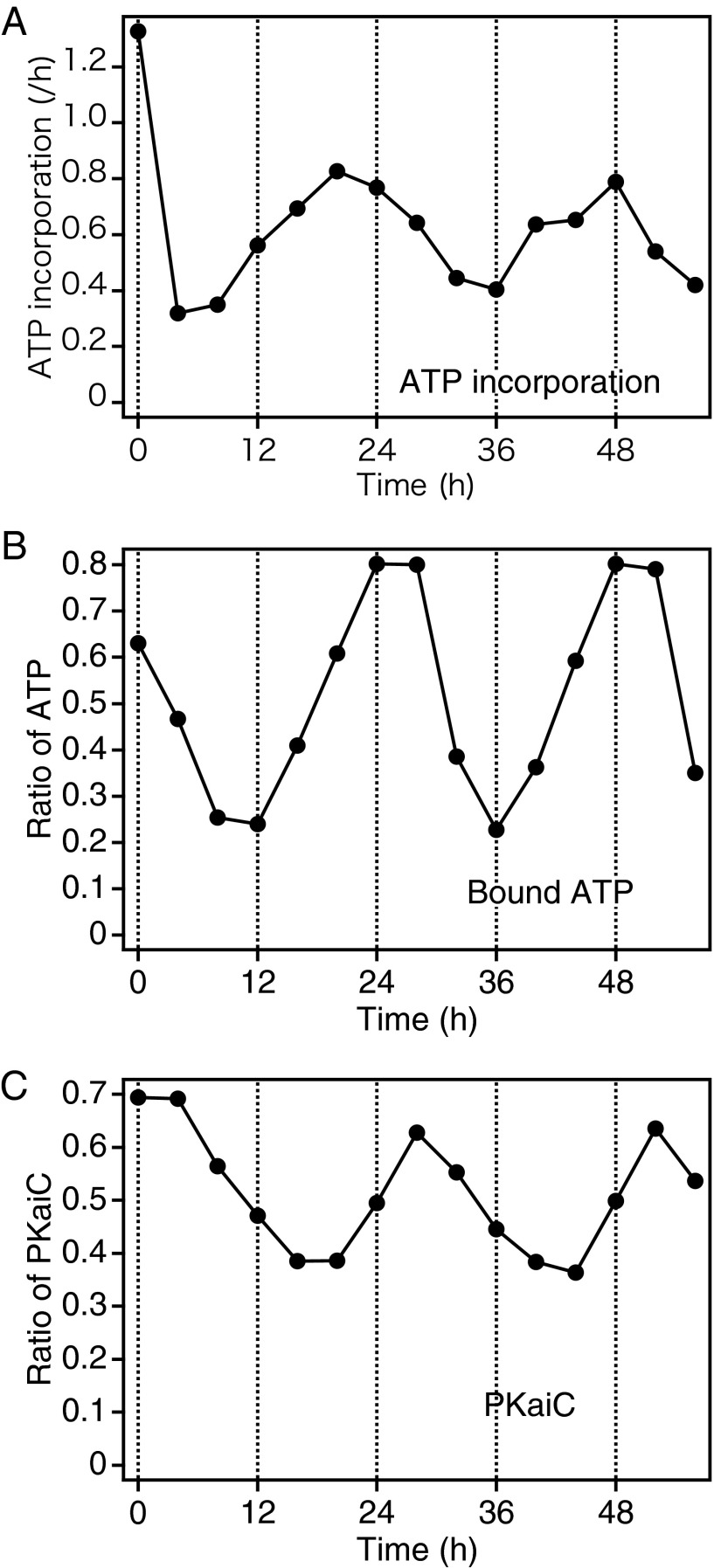
KaiB inhibits KaiA, allowing autodephosphorylation of KaiC (11, 12). We also investigated whether KaiB is involved in the regulation of KaiC-bound nucleotide in the presence of KaiA. To this end, we mixed KaiC (0.5 mg/mL) with KaiA (0.1 mg/mL), KaiB (0.1 mg/mL), and 2 mM [α-32P]ATP, and incubated this mixture at 30 °C. The phosphorylation level of KaiC exhibited a clear circadian rhythm (Fig. 3C). In addition, we monitored the nucleotide-bound state of KaiC at 4-h intervals for 56 h and found that the ratio of ATP to total KaiC-bound nucleotides also exhibited a robust circadian rhythm (Fig. 3B), whose phase was advanced by several hours relative to the phosphorylation rhythm.
Fig. 3.
Effect of the presence of both KaiA and KaiB on the nucleotide-bound state and ATP incorporation rate of KaiC. ATP incorporation rate (A) and the nucleotide-bound state (B) were monitored at 4-h intervals for 56 h in the presence of KaiA and KaiB. The nucleotide-bound state of KaiC was examined by the same procedure as described in Fig. 1, except that both KaiA and KaiB were present (B). In this condition, the phosphorylation state exhibits a clear circadian rhythm (C). To determine the rate of ATP incorporation, we reconstituted the KaiC phosphorylation rhythm in the presence of 2 mM nonradioactive ATP. Aliquots were removed at the indicated time points, supplemented with [α-32P]ATP, and incubated for another 20 min, followed by removal of the unbound nucleotides. The rate of ATP incorporation was calculated from the KaiC-bound radioactivity and plotted against time (A). The data are representative of three experiments.
To determine whether the uptake of exogenous ATP also exhibits a circadian rhythm, we set up another reaction in the presence of 2 mM nonradioactive ATP. The phosphorylation rhythms observed in these two sets of experiments were almost identical (Fig. S1). At the indicated time points, the aliquots were supplemented with [α-32P]ATP to a final specific activity of 3.0 GBq/mmol and incubated for another 20 min. As expected, the rate of ATP incorporation exhibited a circadian rhythm (Fig. 3A), the phase of which was further advanced relative to the oscillation in the ratio of bound ATP to total KaiC-bound nucleotides. We also performed experiments in which we chased the KaiC-bound radioactive nucleotides with nonradioactive ATP starting from the reaction times of 4 and 20 h, corresponding to the first trough and the first peak of the ATP incorporation rhythm, respectively (Fig. S2). The decrease in bound ADP was accelerated at 20 h relative to 4 h, whereas the change in the bound ATP exhibited little difference between these time points. It is possible that ADP release also exhibits a daily fluctuation that is in phase with ATP incorporation.
The interaction among Kai proteins exhibits a circadian rhythm that is in phase with the rhythm of accumulation of S431-phosphorylated KaiC (16, 17). We performed long-distance SDS/PAGE (16) to separate bands of KaiC proteins with different phosphorylation states (Fig. S3). The rhythm of exogenous ATP incorporation was in antiphase with that of the accumulation of S431-phosphorylated KaiC, suggesting that KaiABC complex inhibits nucleotide exchange on KaiC.
Based on the phase relationships among these oscillations, we propose that the KaiC phosphorylation rhythm is driven by the oscillation of the nucleotide-bound state of KaiC, which is in turn determined by the rate of ADP/ATP exchange, and that this oscillation is regulated by the antagonistic effects of KaiA and KaiB (Fig. 4).
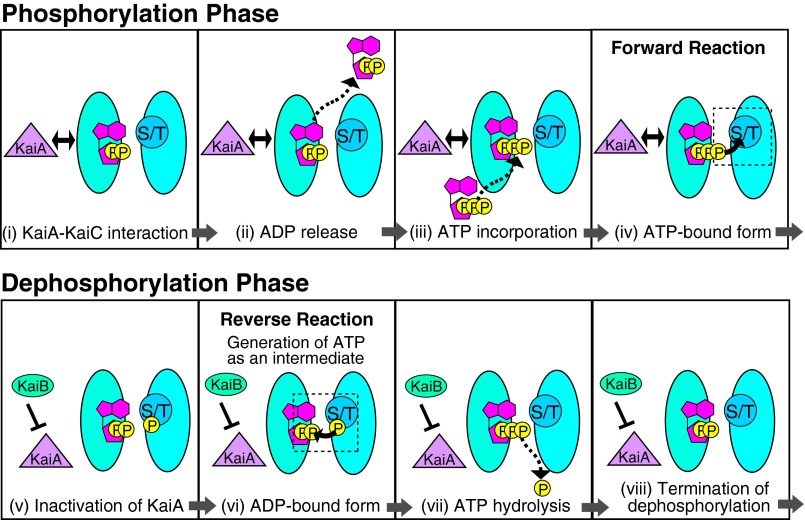
Fig. 4.
Schematic model for the regulation of the direction of reversible autophosphorylation of KaiC. Autophosphorylation, autodephosphorylation, and ATP hydrolysis take place at the subunit interface of the CII domain in KaiC hexamers. The autodephosphorylation of KaiC occurs via reversal of the autophosphorylation reaction, indicating that the KaiC phosphorylation rhythm is sustained by periodic inversion of the direction in which the reaction proceeds. We propose that the reaction direction is regulated at the level of substrate availability, as follows: In the phosphorylation phase, KaiA interacts with KaiC (i) to facilitate the release of bound ADP (ii) and incorporation of exogenous ATP (iii), thereby supplying the active sites with the substrate for the forward reaction (iv). In the dephosphorylation phase, KaiB sequesters KaiA by S431 phosphorylated KaiC-dependent manner (v) to inhibit ADP–ATP exchange (vi); this results in retention of ADP at the active sites, allowing the reaction to proceed in the reverse direction to generate ATP as an intermediate of autodephosphorylation. This ATP is subjected to hydrolysis (vii), resulting in termination of autodephosphorylation reaction (viii).
Discussion
We previously reported that the autodephosphorylation of KaiC occurs via the reversal of the autophosphorylation reaction, in which a phosphate group covalently attached to a Ser or Thr residue is transferred to KaiC-bound ADP to generate ATP (18). This reaction mechanism leads to the prediction that the KaiC phosphorylation rhythm is generated by periodic shifts in the equilibrium of reversible phospho-transfer between phosphorylation sites and KaiC-bound nucleotides. Here, we propose that KaiA and KaiB regulate the direction of this reaction at the level of substrate availability.
KaiC protomer consists of the N-terminal CI and C-terminal CII domains (5), each of which contains an ATP-binding site constantly occupied by either ATP or ADP (18). In the KaiA-free condition, during the initial 2 h, KaiC released approximately one prebound nucleotide per protomer and retained approximately one ADP (Fig. 2A). The incorporation of exogenous ATP into KaiC protomer seemed to be biphasic (Fig. 2C). In the absence of KaiA, approximately one ATP per protomer was rapidly incorporated (within 2 h), followed by a slower incorporation phase (Fig. 2C, ■). These results suggest that ATPase active sites in CI and CII have different rates of nucleotide exchange, i.e., dissociation of bound ADP is followed by incorporation of exogenous ATP. In the presence of KaiA, the amount of incorporated ATP reached approximately two molecules per protomer by 2 h (Fig. 2C, □), and little ADP was retained on KaiC throughout the reaction (Fig. 2B). Therefore, it is likely that KaiA promotes the nucleotide exchange at the site with the lower exchange rate.
To confirm our hypothesis, we performed kinetic analysis of ADP release in the absence of KaiA (Fig. 2A). We fit the equations derived from our model to the data in Fig. 2A, which assumes that the KaiC protomer has two nucleotide-binding sites with different properties (SI Materials and Methods). We assumed that ADP bound to site 1 is released immediately after it is generated, whereas the ATPase cycle at site 2 pauses before the release of ADP. As shown in Fig. 2A, this model adequately explained the experimental data. However, our model did not give a good fit when KaiA was present together with KaiC. This discrepancy is most likely due to the insufficient time resolution of this experiment, which limited our ability to monitor the initial rapid decrease in the level of ATP (Fig. 2B).
We propose that site 1 and site 2 correspond to the ATPase active sites in CI and CII, respectively, for the following reasons. (i) The autophosphorylation sites are located at subunit interfaces of CII, and each site faces the ATP molecule on the neighboring subunit (14). (ii) KaiA acts on the CII domain to promote autophosphorylation and ATP hydrolysis (20–22), whereas KaiA has little effect on ATPase activity in the CI domain (22). (iii) ATP accounted for more than 95% of nucleotides bound to KaiC-CI, regardless of the presence or absence of KaiA (Fig. 1 B and C). It is likely that ADP release is the rate-limiting step of the ATPase reaction in the CII domain and that KaiA facilitates ADP/ATP exchange at the active site of CII. By contrast, CI-bound ADP seems to be exchanged constantly with exogenous ATP, irrespective of the existence of KaiA; this might allow CI to act as a circadian pacemaker, as we previously discussed (9).
Fig. 4 summarizes our model of how KaiA and KaiB stimulate the forward and the reverse reactions of autophosphorylation, respectively, to sustain the phosphorylation rhythm of KaiC. Reversible autophosphorylation and ATP hydrolysis take place at the active site in CII. In the phosphorylation phase, KaiA interacts with KaiC (i) to release ADP bound to the subunit interfaces of KaiC (ii), followed by incorporation of ATP from the outside (iii), which converts KaiC into the ATP-bound form (iv). This ATP serves as the substrate for the forward reaction. In the dephosphorylation phase, KaiB associates with S431-phosphorylated KaiC (v) to form the KaiB–KaiC complex that sequesters KaiA, resulting in inhibition of KaiA (16, 17), which causes ADP to be retained on KaiC (vi). This ADP serves as the substrate for the reversal of autophosphorylation to generate ATP as an intermediate of autodephosphorylation. Subsequently, this ATP is hydrolyzed (vii) to terminate the autodephosphorylation (viii).
Kim et al. demonstrated that KaiA interacts with the CII domain via a segment called the A-loop, which located near the enzymatic active site of CII (20). They proposed that the structure of the A-loop changes dynamically between the buried and exposed states and that KaiA stabilizes the A-loop in the exposed state. It is possible that the structural equilibrium of the A-loop determines the rate of ADP–ATP exchange in the ATPase active sites of CII, thereby shifting the equilibrium of the reversible autophosphorylation reaction.
Phosphorylation of S431, which allows KaiC to enter the dephosphorylation phase (16, 17), rigidifies the CII ring of KaiC (21), and enhances the stacking between the CI and CII rings (22). However, it is not fully understood how these conformational changes enhance the reversal of autophosphorylation. Recently, we revealed that substitution of the catalytic carboxylates E77 and E78 to glutamine residues, which decreases the ATPase activity of CI, does not affect the profile of autodephosphorylation (23). Therefore, it is unlikely that these conformational changes facilitate the coupling of ATP hydrolysis in CI to formation of ATP by reversing autodephosphorylation in CII. It is possible that the rigid conformation of the CII ring causes the distance between ADP and phosphorylated Ser/Thr residues to be appropriate for ATP generation.
We examined the effect of phosphorylation state on the nucleotide specificity using KaiC proteins containing mutations in the phosphorylation sites (Fig. S4). KaiC-DE, in which S431 is mutated to aspartate and T432 is mutated to glutamate, mimics the fully phosphorylated state (16). We found that KaiC-DE favored ADP to a greater degree than the WT protein (Fig. S4A). Rigidification of the CII ring and enhanced stacking between the CI and CII rings (21, 22) may inhibit exchange of bound ADP to external ATP. On the other hand, ATP consistently accounted for ∼60% of KaiC-AA–bound nucleotide, regardless of the presence or absence of KaiA (Fig. S4B). In the KaiA-free condition, the ratio of ATP to total bound nucleotides was higher in KaiC-AA than in KaiC-DE as expected. However, it is unclear why KaiA had no effect on the nucleotide-bound state of KaiC-AA. We previously reported that ATP hydrolysis on KaiC-AA is accelerated for an unknown reason (9); this acceleration might influence the effectiveness of KaiA in regulating KaiC-AA.
In the absence of KaiA, the steady-state ratio of bound ADP to total KaiC-bound nucleotides was ∼0.7 (Fig. 1). Because the KaiC protomer contains two nucleotide-binding sites, a ratio greater than 0.5 indicates that ADP is present at both the CI and CII domains. In KaiC-CI, ∼95% of the bound nucleotides were ATP, regardless of the presence or absence of KaiA (Fig. 1 B and C), suggesting that CII is necessary not only for retaining ADP on itself but also for regulating the nucleotide-bound state of CI. A recent study showed that the KaiB–KaiC interaction is mediated by the CI domain and that the binding of ADP to CI is necessary for KaiC’s association with KaiB (22). Previously, we and another group reported that the phosphorylation of S431 promotes the KaiB–KaiC interaction (16, 17). It is possible that the CII domain affects the nucleotide-bound state of the CI domain through the phosphorylation of S431 and the resultant enhanced stacking between the two rings, which might regulate the KaiB–KaiC interaction. However, the identity of the KaiC domain that interacts with KaiB remains controversial (24).
Changes in environmental illumination induce phase shifts in circadian clocks (1). The magnitude and the direction of these shifts are dependent on the initial phase of the oscillation to which the stimulation is applied (1). In cyanobacterial cells, a light/dark signal from the environment is transduced to the circadian clock as a change in the intracellular ATP/ADP ratio (25). In the reconstituted circadian system, transient changes in the ATP/ADP ratio entrain the KaiC phosphorylation rhythm in a phase-dependent manner (25). This rhythm is sensitive to the change in the ATP/ADP ratio when the phosphorylation level of KaiC is increasing, but insensitive when the phosphorylation level is decreasing (25). Our current model for the KaiC phosphorylation rhythm can explain the phase dependency of entrainment. In the phosphorylation phase, during which the forward reaction of autophosphorylation is favored, adenine nucleotides are actively exchanged between the protein and solution, and the exchange rate should be influenced by the ATP/ADP ratio in solution. In the dephosphorylation phase, when the reversal of the autophosphorylation reaction proceeds, bound ADP is not efficiently exchanged with exogenous ATP and is instead retained on KaiC. This behavior may explain why changes in the external ATP/ADP ratio do not affect the dephosphorylation phase.
Previously, we proposed that the physical state of KaiC maintained by ATP hydrolysis at the active site in CI is the pacemaker of the cyanobacterial circadian clock, i.e., that it defines the period of the oscillation (9). In future work we will dissect the cross-talk between CI and CII domains to clarify how the phosphorylation cycle is sustained with a temperature-compensated period of 24 h.
Materials and Methods
Detection and Quantification of KaiC-Bound Nucleotides.
Monomeric KaiC was prepared as previously described (18). KaiC-bound nucleotides were analyzed as described previously, with some modifications (18). Briefly, KaiC hexamers (0.5 mg/mL) were reconstituted from KaiC monomers in the presence of 2 mM [α-32P]ATP (NEG003H, 370 MBq/mL, 111 TBq/mmol; Perkin Elmer) in MD-2 buffer, followed by incubation at 30 °C in the presence or absence of KaiA (0.1 mg/mL) and KaiB (0.1 mg/mL). The specific radioactivity of ATP in the reaction mixture was 3.0 GBq/mmol. Aliquots were collected at predetermined time points and applied twice to Micro Bio-Spin P-30 columns (BioRad) equilibrated with MD-2 buffer to remove unbound nucleotides. The eluate was mixed with an equal volume of Laemmli sample buffer (BioRad) containing 5% (vol/vol) 2-mercaptoethanol to release the KaiC-bound nucleotides from the proteins. The samples were subjected to thin-layer chromatography, and the signals for 32P-labeled ATP and ADP were quantitated as described previously (18), except that the standard curves were obtained with serial dilutions of the 2 mM [α-32P]ATP solution.
Chase of Prebound Nucleotides on KaiC Hexamers.
KaiC hexamers were prepared from monomers in the presence of 2 mM [α-32P]ATP (3.0 GBq/mmol), and unbound nucleotides were removed using a HiTrap Desalting column (GE Healthcare) equilibrated with MD-2 buffer. The KaiC hexamer concentration in the eluate was adjusted to 0.5 mg/mL; 2 mM nonradioactive ATP was added; and the reaction was started at 30 °C in the presence or absence of KaiA. Aliquots were collected at the indicated time points and passed twice through Bio-Spin P-30 columns (BioRad) to remove unbound nucleotides. The signals from the KaiC-bound 32P-labeled nucleotides were quantitated as described above.
Assay for ATP Incorporation into KaiC Hexamers.
KaiC monomers (0.5 mg/mL) were prepared and added to 2 mM nonradioactive ATP. The resultant KaiC hexamers were supplemented with [α-32P]ATP to a final specific activity of 3.0 GBq/mmol, and then incubated at 30 °C in the presence or absence of KaiA (0.1 mg/mL) for 8 h. Aliquots of the reaction mixture were collected at predetermined time points, unbound nucleotides were removed, and the amount of ATP incorporated was calculated using serial dilutions of the 2 mM [α-32P]ATP solution as standards. To measure the rate of ATP incorporation into KaiC, we reconstituted the KaiC phosphorylation rhythm in the presence of 2 mM nonradioactive ATP. Aliquots of the reaction mixture were removed at 4-h intervals for 56 h. Samples were then supplemented with [α-32P]ATP to a final specific activity of 3.0 GBq/mmol and incubated for another 20 min. Aliquots of the reaction mixture were collected at predetermined time points, and unbound nucleotides were removed. The rate of ATP incorporation was calculated from the level of KaiC-bound radioactivity.
Supplementary Material
Acknowledgments
We thank N. Kawakami, M. Ikeda, K. Miwa, and Y. Onoue for helpful discussion and advice, and K. Nagiri, J. Furuta, H. Yoshimura, S. Yi, T. Nishikawa, and Y. Ide for technical support. This work was supported in part by Ministry of Education, Culture, Sports, Science and Technology of Japan Grants-in-Aid 23118708 (to T.N.-O.), 24770043 (to Y.K.), and 24000016 (to T.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319353111/-/DCSupplemental.
References
- 1.Pittendrigh CS. Temporal organization: Reflections of a Darwinian clock-watcher. Annu Rev Physiol. 1993;55:16–54. doi: 10.1146/annurev.ph.55.030193.000313. [DOI] [PubMed] [Google Scholar]
- 2.Nakajima M, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308(5720):414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 3.Lakin-Thomas PL. Transcriptional feedback oscillators: Maybe, maybe not…. J Biol Rhythms. 2006;21(2):83–92. doi: 10.1177/0748730405286102. [DOI] [PubMed] [Google Scholar]
- 4.Edgar RS, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485(7399):459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iwasaki H, Taniguchi Y, Ishiura M, Kondo T. Physical interactions among circadian clock proteins KaiA, KaiB and KaiC in cyanobacteria. EMBO J. 1999;18(5):1137–1145. doi: 10.1093/emboj/18.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pattanayek R, et al. Visualizing a circadian clock protein: Crystal structure of KaiC and functional insights. Mol Cell. 2004;15(3):375–388. doi: 10.1016/j.molcel.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi F, et al. Roles of two ATPase-motif-containing domains in cyanobacterial circadian clock protein KaiC. J Biol Chem. 2004;279(50):52331–52337. doi: 10.1074/jbc.M406604200. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi F, et al. ATP-induced hexameric ring structure of the cyanobacterial circadian clock protein KaiC. Genes Cells. 2003;8(3):287–296. doi: 10.1046/j.1365-2443.2003.00633.x. [DOI] [PubMed] [Google Scholar]
- 9.Terauchi K, et al. ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2007;104(41):16377–16381. doi: 10.1073/pnas.0706292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishiwaki T, Iwasaki H, Ishiura M, Kondo T. Nucleotide binding and autophosphorylation of the clock protein KaiC as a circadian timing process of cyanobacteria. Proc Natl Acad Sci USA. 2000;97(1):495–499. doi: 10.1073/pnas.97.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 2003;22(9):2127–2134. doi: 10.1093/emboj/cdg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y, Mori T, Johnson CH. Cyanobacterial circadian clockwork: Roles of KaiA, KaiB and the kaiBC promoter in regulating KaiC. EMBO J. 2003;22(9):2117–2126. doi: 10.1093/emboj/cdg168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishiwaki T, et al. Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC 7942. Proc Natl Acad Sci USA. 2004;101(38):13927–13932. doi: 10.1073/pnas.0403906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y, et al. Identification of key phosphorylation sites in the circadian clock protein KaiC by crystallographic and mutagenetic analyses. Proc Natl Acad Sci USA. 2004;101(38):13933–13938. doi: 10.1073/pnas.0404768101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc Natl Acad Sci USA. 2002;99(24):15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishiwaki T, et al. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 2007;26(17):4029–4037. doi: 10.1038/sj.emboj.7601832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rust MJ, Markson JS, Lane WS, Fisher DS, O’Shea EK. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science. 2007;318(5851):809–812. doi: 10.1126/science.1148596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishiwaki T, Kondo T. Circadian autodephosphorylation of cyanobacterial clock protein KaiC occurs via formation of ATP as intermediate. J Biol Chem. 2012;287(22):18030–18035. doi: 10.1074/jbc.M112.350660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egli M, et al. Dephosphorylation of the core clock protein KaiC in the cyanobacterial KaiABC circadian oscillator proceeds via an ATP synthase mechanism. Biochemistry. 2012;51(8):1547–1558. doi: 10.1021/bi201525n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim YI, Dong G, Carruthers CW, Jr, Golden SS, LiWang A. The day/night switch in KaiC, a central oscillator component of the circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2008;105(35):12825–12830. doi: 10.1073/pnas.0800526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang YG, Kuo NW, Tseng R, LiWang A. Flexibility of the C-terminal, or CII, ring of KaiC governs the rhythm of the circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2011;108(35):14431–14436. doi: 10.1073/pnas.1104221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang YG, Tseng R, Kuo NW, LiWang A. Rhythmic ring-ring stacking drives the circadian oscillator clockwise. Proc Natl Acad Sci USA. 2012;109(42):16847–16851. doi: 10.1073/pnas.1211508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitayama Y, Nishiwaki-Ohkawa T, Sugisawa Y, Kondo T. KaiC intersubunit communication facilitates robustness of circadian rhythm in cyanobacteria. Nat Commun. 2013 doi: 10.1038/ncomms3897. 10.1038/ncomms3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villarreal SA, et al. CryoEM and molecular dynamics of the circadian KaiB-KaiC complex indicates that KaiB monomers interact with KaiC and block ATP binding clefts. J Mol Biol. 2013;425(18):3311–3324. doi: 10.1016/j.jmb.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rust MJ, Golden SS, O’Shea EK. Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science. 2011;331(6014):220–223. doi: 10.1126/science.1197243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.