Significance
The pathogen Legionella pneumophila replicates within human alveolar macrophages, causing a potentially fatal pneumonia known as Legionnaires’ disease. We identified that the effector protein VipD, which is injected into infected cells by L. pneumophila, localizes to degradative host organelles called endosomes. There, VipD binds to the endosomal regulator Rab5, an event that triggers the hydrolytic phospholipase A1 activity in VipD, which causes the removal of the lipid phosphatidylinositol 3-phosphate. Without this key lipid, endosomes can no longer execute their function and L. pneumophila is protected from their harmful effect. Our finding opens new avenues for the development of therapeutics that interfere with the activation of VipD and with endosomal avoidance by L. pneumophila.
Abstract
A crucial step in the elimination of invading microbes by macrophages is phagosomal maturation through heterotypic endosomal fusion. This process is controlled by the guanine nucleotide binding protein Rab5, which assembles protein microdomains that include the tethering protein early endosomal antigen (EEA) 1 and the phosphatidylinositol (PI) 3-kinase hVps34, which generates PI(3)P, a phospholipid required for membrane association of EEA1 and other fusion factors. During infection of macrophages, the pathogen Legionella pneumophila bypasses the microbicidal endosomal compartment by an unknown mechanism. Here, we show that the effector protein VipD from L. pneumophila exhibits phospholipase A1 activity that is activated only upon binding to endosomal Rab5 or Rab22. Within mammalian cells, VipD localizes to endosomes and catalyzes the removal of PI(3)P from endosomal membranes. EEA1 and other transport and fusion factors are consequently depleted from endosomes, rendering them fusion-incompetent. During host cell infection, VipD reduces exposure of L. pneumophila to the endosomal compartment and protects their surrounding vacuoles from acquiring Rab5. Thus, by catalyzing PI(3)P depletion in a Rab5-dependent manner, VipD alters the protein composition of endosomes thereby blocking fusion with Legionella-containing vacuoles.
Phagosomal maturation is a fundamental innate immune mechanism of eukaryotic cells that facilitates the killing and degradation of ingested microbes. After separation from the plasma membrane, the nascent phagosome rapidly undergoes a series of fusion events with early endosomes, late endosomes, and eventually lysosomes, resulting in the gradual acidification of the phagosome’s lumen and the destruction of its contents (1). The various stages of phagosome maturation are interconnected through, and mainly controlled by, small guanine nucleotide binding proteins (GTPases) of the Rab (ras genes from rat brain) family. Rab5 is a signature protein of early endosomes and crucial for the initial fusion with nascent phagosomes. Once activated by the guanine nucleotide-exchange factor Rabex-5, GTP-loaded Rab5 orchestrates the recruitment of several downstream ligands, such as the tethering protein early endosomal antigen (EEA) 1 and the type III phosphatidylinositol (PI) 3-kinase vacuolar protein sorting 34 (hVps34) and their assembly into membrane microdomains capable of instigating lipid bilayer fusion (2). hVps34 plays a supporting yet critical role in the assembly of these multimeric fusion protein complexes by catalyzing the phosphorylation of PI at the 3′ hydroxyl position, thereby generating PI 3-phosphate (PI(3)P), which in turn enhances association of EEA1, rabenosyn-5, sorting nexins (SNX), and other proteins that possess PI(3)P-specific binding modules with the endosomal surface (3). Failure of cells to generate endosomal PI(3)P, for example after chemical inhibition of the PI 3-kinase activity of hVps34, results in the reduced accumulation of EEA1 on early endosomes and an attenuated maturation of phagosomes into phagolysosomes (4, 5).
The ability to delay or block a particular step during phagosomal maturation is a key virulence trait of a variety of intravacuolar pathogens. The Gram-negative bacterium Legionella pneumophila, the causative agent of Legionnaires’ pneumonia, possesses the remarkable ability to block phagosomal maturation at the earliest stage in the endocytic pathway. Upon uptake by macrophages, the Legionella-containing vacuole (LCV) does not acquire any early or late endosomal markers (6). Instead, the pathogen attracts proteins and vesicles from the early secretory pathway and transforms its surrounding phagosome into a specialized replication compartment that morphologically resembles host-cell rough endoplasmic reticulum (7).
Endolysosomal avoidance and intracellular proliferation of L. pneumophila relies on more than 250 bacterial proteins, so called “effectors,” which are delivered through the Dot/Icm type IV secretion system (T4SS) into the infected host cell (8). Although several L. pneumophila effector proteins have been functionally characterized over the past decade, surprisingly few have been ascribed a function in manipulating endolysosomal maturation.
One effector known to target host cell endolysosomal trafficking, and the focus of this study, is L. pneumophila VPS inhibitor protein D (VipD) (9, 10). The C-terminal part of VipD lacks sequence homology to other proteins but, when overproduced in the surrogate host Saccharomyces cerevisiae, interferes with protein sorting to the vacuole (9), the yeast equivalent of mammalian lysosomes. A possible explanation for this phenomenon came from a recent study showing that the C-terminal domain of VipD (residues 316–621) binds endosomal Rab5 and Rab22 (11), another GTPase regulating endosomal trafficking. Ku et al. (11) consequently proposed that VipD overproduction interferes with endosomal maturation in transiently transfected mammalian cells by competing with EEA1 and other cellular ligands for Rab5 binding. However, it was inconclusive from their studies whether VipD is in fact involved in endosomal avoidance by L. pneumophila.
The N-terminal region of VipD shows significant sequence homology to the patatin-like phospholipase domain of Pseudomonas aeruginosa ExoU (Fig. 1A), a type III-translocated toxin with phospholipase A2 (PLA2) activity (12). Despite the obvious homology to ExoU, several laboratories failed over the past decade to detect robust PLA2 activity in VipD (10, 11), raising the question about the role of the catalytic domain for the biological function of this effector.
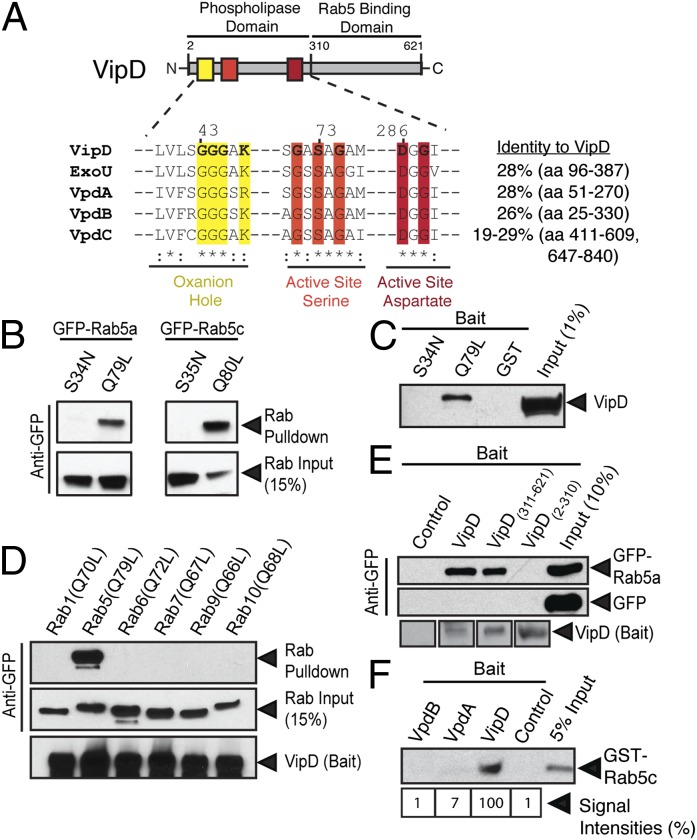
Fig. 1.
VipD interacts with active Rab5. (A) Domain organization of VipD. Amino acid (aa) residues predicted to be required for phospholipase activity are colored. The equivalent sequences present in VpdA-C and ExoU are aligned, with highly (*) or moderately (:) conserved side chains being marked. The sequence identity (in percent) is shown for the maximum region of homology (in parentheses) of each protein. (B) VipD preferentially binds active Rab5. VipD2–621-coated beads precipitate the active (Q→L) but not inactive (S→N) mutant forms of GFP-Rab5a or Rab5c from lysate of transiently transfected 293T cells. Proteins were detected by immunoblot using anti-GFP antibody. (C) Pull-down of endogenous VipD from L. pneumophila lysate by active (Q79L) but not inactive (S34N) GST-Rab5 or by GST (control). (D) VipD-coated beads precipitate Rab5a(Q79L) but none of the other constitutively active Rab proteins tested. (E) The C-terminal domain of VipD mediates GFP-Rab5 binding. Beads coated with the indicated VipD fragments or uncoated beats (control) were incubated with 293T cell lysate containing either GFP-Rab5a or GFP, and proteins retained by the beads were detected using anti-GFP antibody. (F) Rab5 interacts with VipD but not its paralogs. Uncoated beads (control) or equal amounts of bead-immobilized VipD, VpdA, or VpdB were incubated with GST-Rab5c, and GST-Rab5c binding was detected by immunoblot. The immunoblot was quantified using densitometry and signal intensities are listed in percent and normalized to VipD, which was arbitrarily set to 100%.
Despite the aforementioned efforts, the precise mechanisms underlying endosomal avoidance by L. pneumophila remained elusive. Our study now provides detailed insight into this key virulence process of L. pneumophila by revealing that a previously unrecognized phospholipase activity in VipD plays a major role in altering the lipid composition of endosomes, thereby interfering with their ability to target LCVs during infection. Our findings also provide a remarkable example for the emerging concept of spatiotemporal regulation of microbial effectors by coupling their catalytic activity to the arrival in a certain subcellular location.
Results
VipD Interacts with Active Rab5 via its C-Terminal Domain.
The finding that L. pneumophila VipD binds mammalian Rab5 (11) led the authors of a recent study to suggest that competitive binding between VipD and cellular Rab5 ligands was the reason for the block in endosomal maturation observed in transiently transfected mammalian cells overproducing VipD. Given that most microbial pathogens translocate only trace amounts of their effector proteins into infected host cells, and that stoichiometric binding, as opposed to catalysis, is a very inefficient mechanism of manipulating host proteins, it seemed unlikely that such a strategy would allow L. pneumophila VipD to successfully interfere with endosomal fusion during host cell infection. In fact, we determined that on average, each L. pneumophila bacterium delivers no more than 3.85 × 10−7 fmol (or 232 molecules) of VipD into an infected mammalian cell, but that each host cell contains on average 5.31 × 10−4 fmol (or 319,771 molecules) of active Rab5. Thus, assuming the most likely scenario of a single bacterium infecting a single mammalian macrophage, active Rab5 outnumbers VipD by a factor 1,379 (Fig. S1). This large stoichiometric difference between VipD and Rab5 strongly argues against VipD manipulating the early endosomal pathway solely by competitive binding to Rab5. As a result, we reevaluated the functional relation between L. pneumophila VipD and host-cell Rab5.
Given that the VipD fragments used in our study differed in length and composition from those used by Ku et al. (11) (Fig. 1A and Fig. S2A), we first confirmed the interaction between Rab5 and our VipD and its truncated variants in protein-protein binding studies. As expected, we found that binding of Rab5 to full-length VipD (residues 2–621; VipD2–621) was nucleotide-dependent, with a strong preference of VipD for the constitutively active conformation of Rab5 [Rab5a(Q79L) or Rab5c(Q80L)] compared with the constitutively inactive form [Rab5a(S34N) or Rab5c(S35N)] (Fig. 1 B and C). VipD2–621 binding was specific for Rab5 but active variants of other endosomal Rab GTPases, such as Rab7, Rab9, or Rab10, were not precipitated from 293T cell lysate by bead-immobilized VipD2–621 (Fig. 1D). Moreover, using our truncated VipD variants, we confirmed that Rab5 binding was mediated by the C-terminal domain of VipD (VipD311–621) (Fig. 1E), with the shortest of our fragments capable of Rab5 binding being VipD311–544 (Fig. S2 B and C). Thus, the VipD proteins used in this study exhibited a behavior similar to those described previously (11).
L. pneumophila, in addition to VipD, encodes three paralogs named VpdA, VpdB, and VpdC. These paralogs all possess sequence homology to the N-terminal phospholipase domain of VipD but show little homology to its C-terminal region (Fig. 1A). Upon closer examination, we found that, unlike VipD, neither VpdA nor VpdB interacted with recombinant Rab5 (Fig. 1F and Fig. S2D). Taken together, these findings illustrated that the L. pneumophila effector VipD specifically targets active Rab5 through its C-terminal region, a feature that is not shared by its paralogs.
Inhibitory Effect of VipD on Yeast Growth Is Exacerbated by Rab5.
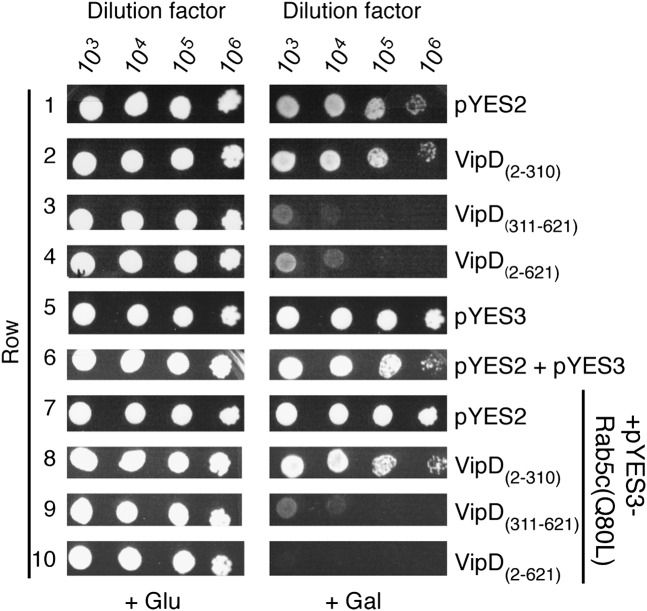
Translocated bacterial effectors often manipulate conserved host-cell pathways that are essential for eukaryotic cell growth and survival. To decipher the functional relationship between VipD and Rab5 we studied the two proteins in a yeast sensitivity assay. When overproduced in S. cerevisiae INVSc1, VipD2–621 caused a slow-growth phenotype both on media plates (Fig. 2, row 4) and in liquid media (Fig. S3), a phenomenon not observed in cells containing the empty vector (Fig. 2, row 1). Synthesis of VipD311–621 in yeast also resulted in a slow-growth phenotype, most likely by binding to and interfering with the function of yeast Rab (Ypt) GTPases, whereas VipD2–310 did not have an impact on yeast growth (Fig. 2, rows 2 and 3, and Fig. S3). Given that VipD targets active Rab5 (Fig. 1 B and C), we hypothesized that overproduction of this GTPase might rescue the growth defect caused by VipD2–621. Unexpectedly, we discovered that the simultaneous presence of both VipD2–621 and constitutively active Rab5c(Q80L) in yeast exacerbated the growth defect relative to that of cells producing only VipD2–621 (Fig. 2, row 10 vs. 4). Because Rab5c(Q80L) alone did not negatively impact yeast growth (Fig. 2, row 7), it was possible that the presence of active Rab5 had triggered an otherwise quiescent activity in VipD that was harmful to yeast.
Fig. 2.
Rab5 enhances the growth defect of yeast cells producing VipD. Yeast sensitivity plating assay showing serial (103- to 106-fold) dilutions of S. cerevisiae INVSc1 cells grown under inducing (+Gal) or noninducing (+Glu) conditions that contained plasmids encoding the indicated proteins. pYES2 and pYES3 are the empty control plasmids.
VipD Is a Rab5-Activated Phospholipase A1.
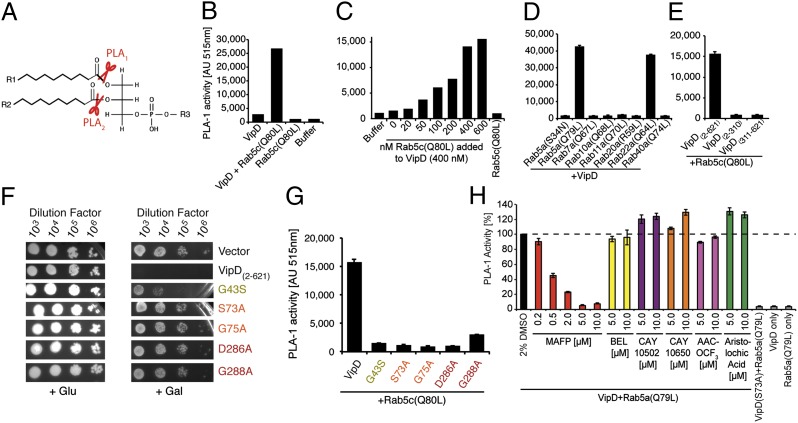
We consequently tested if VipD possessed Rab5-dependent phospholipase activity using a fluorometric in vitro assay. PLA1 and PLA2 enzymes hydrolyze the carboxylester bond of phospholipids at the sn-1 or sn-2 position, respectively, releasing lysophospholipids and free fatty acids that can either be further degraded or function as signaling molecules (Fig. 3A). Although VipD2–621 alone exhibited neither PLA1 (Fig. 3B) nor PLA2 activity (Fig. S4A), we detected robust PLA1 activity in VipD2–621 upon addition of active Rab5c(Q80L) (Fig. 3B). The stimulatory effect of active Rab5 was concentration-dependent, with the strongest PLA1 activity at an equimolar or higher ratio of Rab5c(Q80L) to VipD2–621 (Fig. 3C), suggesting a 1:1 stoichiometry for the VipD–Rab5 complex. Addition of constitutively inactive Rab5a(S34N) to VipD2–621 failed to stimulate PLA1 activity (Fig. 3D), consistent with the lack of interaction between both proteins (Fig. 1 B and C). Similarly, no other constitutively active Rab protein tested here (Rab7a, -10a, -11a, -20a, -40a) had a stimulatory effect on the PLA1 activity of VipD2–621, with the exception of Rab22a(Q64L) (Fig. 3D), the only other GTPase shown to interact with VipD (11). Consistent with its inability to bind Rab5 (Fig. 1F), L. pneumophila VpdB did not exhibit PLA1 activity, even in the presence of Rab5c(Q80L) (Fig. S4B), suggesting that this paralog requires a different host factor for activation. Recent reports (11, 13) that VipD exhibited PLA2 activity were not supported by our study (Fig. S4A).
Fig. 3.
VipD exhibits PLA1 activity upon binding to active Rab5. (A) Schematic representation of PLA1 and PLA2 cleavage sites on phospholipids. (B–E) Fluorometric PLA1 assays. The substrate PED-A1 is a BODIPY FL dye-labeled glycerophosphoethanolamine, the fluorescence emission of which is dequenched upon PLA1 hydrolysis. (B) VipD exhibits PLA1 activity in the presence of Rab5c(Q80L). (C) The stimulatory effect of Rab5 on the PLA1 activity of VipD is concentration-dependent. VipD (400 nM) was incubated with increasing amounts of Rab5c(Q80L), and substrate hydrolysis was determined 40 min after incubation. (D) Active Rab22a, but none of the other Rabs tested, triggered the PLA1 activity of VipD equally efficient as Rab5a(Q79L). (E) Neither the N- nor C-terminal domain of VipD exhibited PLA1 activity in the presence of Rab5c(Q80L). (F) Substitution of catalytically relevant residues in VipD attenuated their ability to interfere with yeast growth. (G) VipD point mutants are catalytically inactive. Rab5c(Q80L) was incubated with an equimolar amount of VipD or the indicated point mutants, and substrate (PED-1A) hydrolysis was determined fluorometrically. (H) Screen for PLA2 inhibitors capable of blocking the PLA1 activity of VipD. VipD was incubated with Rab5a(Q79L) in the presence of increasing concentrations of the indicated PLA2 inhibitors. The level of uncleaved substrate 40 min after the PLA1 reaction is shown (normalized to the VipD+DMSO control).
Unlike full-length VipD2–621, neither the N- nor the C-terminal domain on its own exhibited notable PLA1 activity in vitro when incubated with Rab5c(Q80L) (Fig. 3E). Similar results were obtained in a yeast sensitivity assay where overproduction of VipD2–310 alone or together with Rab5c(Q80L) did not reduce cell growth (Fig. 2, rows 2 and 8, and Fig. S3). Similarly, the intermediate growth reduction caused by VipD311–621 in yeast was not exacerbated by Rab5c(Q80L) (Fig. 2, rows 3 and 9) showing that active Rab5 stimulates phospholipase activity only in full-length VipD.
The ability of VipD2–621 to efficiently attenuate yeast growth depended on its PLA1 activity because substitution of individual residues within the consensus motifs predicted to be essential for substrate hydrolysis (Fig. 1A) rendered VipD mutant proteins less toxic to yeast (Fig. 3F and Fig. S3). Similarly, we found that the in vitro PLA1 activity of purified VipD2–621 mutant proteins was strongly attenuated (Fig. 3G), confirming the importance of the active site residues for the catalytic activity of VipD. The loss of PLA1 activity was likely not because of a folding defect of the VipD2–621 point mutants because they interacted with Rab5c(Q80L) to an extent comparable to wild-type VipD2–621 (Fig. S4C).
Enzymes with PLA1 activity, although present in many organisms and cell types, are not studied and understood as well as PLA2 proteins, and the biological function for most—if not all—PLA1 enzymes has yet to be defined. To characterize VipD in greater detail, we determined its sensitivity toward phospholipase inhibitors. Because inhibitors specific for PLA1 enzymes have not been identified, we analyzed a variety of PLA2 inhibitors for their ability to interfere with VipD activity (Fig. 3H). Bromoenol lactone (BEL), Cay10502, Cay10650, AACOCF3, or aristolochic acid showed no inhibitory effect on VipD2–621 even at high concentrations (10 μM), as was expected for an enzyme that does not possess PLA2 activity. Interestingly, MAFP (methyl arachidonyl fluorophosphonate), a PLA2 inhibitor that had earlier been shown to also inhibit enzymes with PLA1 activity (14, 15), potently blocked substrate hydrolysis by VipD, even at low concentrations (0.5 μM) (Fig. 3H).
Taken together, these findings demonstrate that L. pneumophila VipD possesses PLA1 but not PLA2 activity, and that this activity is triggered upon binding of the C-terminal domain of VipD to either active Rab5 or Rab22. Our findings also explain why earlier studies that did not include Rab5 in their assays were unable to detect any notable phospholipase activity in VipD (10, 11, 13).
VipD Alters the Endosomal Protein and Lipid Composition.
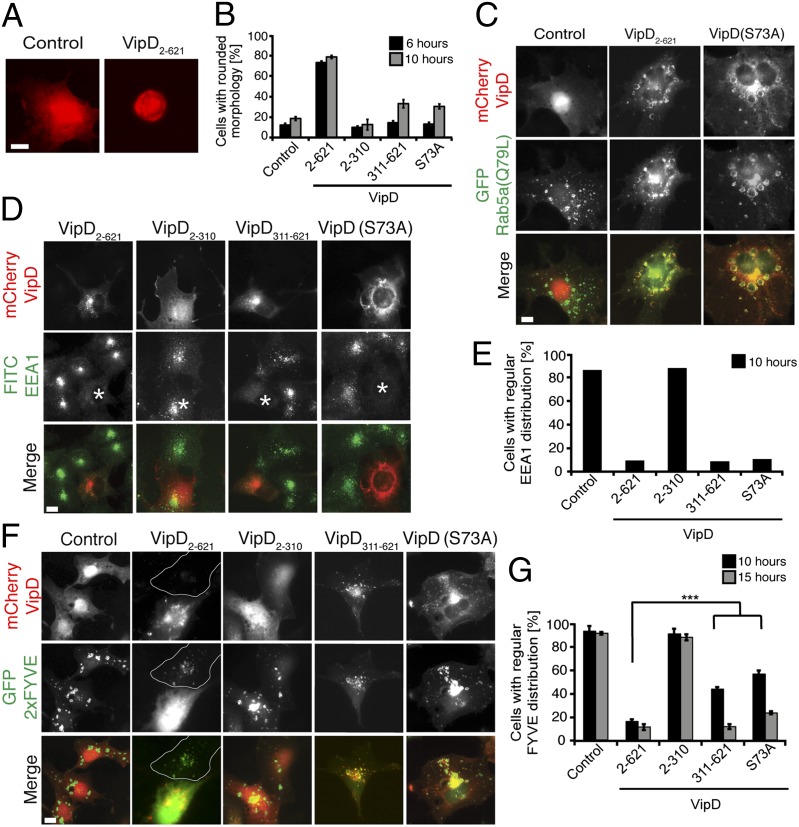
Having deciphered the molecular mechanism that activates the PLA1 domain of VipD, we subsequently analyzed the effect of its catalytic activity on endosomes, the compartment where active Rab5 is predominantly localized within eukaryotic cells. Similar to our observations in the yeast sensitivity assay (Fig. 2), we found that overproduction of VipD2–621 in transiently transfected COS-1 cells severely affected cell viability, with 79 ± 3% of VipD2–621-producing cells showing signs of cell rounding and death, as opposed to 18 ± 4% for control cells (Fig. 4 A and B). Notably, VipD311–621 and catalytically inactive VipD(S73A) caused only an intermediate phenotype (33% and 31% cytotoxicity, respectively), indicating that the PLA1 activity of VipD contributed to the cytotoxic effect of VipD. VipD2–310 caused no noticeable change in COS-1 cell morphology (Fig. 4B), consistent with the idea that PLA1 activity requires activation through the C-terminal Rab5-binding domain. To reduce any secondary effects attributable to cytotoxicity caused by overproduction of VipD or its variants, all subsequent analyses were limited to transfected cells with intermediate or low protein production levels.
Fig. 4.
VipD alters the protein and lipid composition of endosomes. All experiments were performed in transiently transfected COS-1 cells producing the indicated proteins. (A and B) High levels of VipD affect cell morphology and viability. (A) Representative images showing COS-1 cells producing either mCherry (control) or mCherry-VipD2–621. (Scale bar, 2 μm.) (B) Quantification of the amount of rounded cells 6 h and 10 h after transfection (black and gray bars, respectively) from the experiment shown in A. Control, mCherry. (C) VipD localization to endosomes does not require PLA1 activity. COS-1 cells producing Rab5a(Q79L) were transfected with plasmids producing mCherry-tagged VipD or catalytically inactive VipD(S73A), and protein localization was determined by fluorescence microscopy. The merged images (Bottom row) show Rab5a(Q79L) in green and VipD variants in red. Control, mCherry. (Scale bar, 2 μm.) (D and E) VipD displaces endogenous EEA1 form endosomes. (D) Representative fluorescence microscopy images showing the intracellular localization of endogenous EEA1 (green) in COS-1 cells producing the indicated mCherry-tagged VipD variants (red). An asterisk (*) represents transfected cell. The quantification of the experiment is shown in E. (Scale bar, 2 μm.) (F and G) Similar to D and E but with GFP-2 × FYVE as a PI(3)P marker. Control, mCherry. Outlined, cell not transfected with mCherry-VipD. The graphs are a summary of three independent experiments. ***P ≤ 0.005 (Student t test).
It has been reported that VipD heterologously produced in HeLa cells colocalizes with Rab5 and Rab22 on early endosomes, and that this colocalization required its C-terminal domain (11). We analyzed the intracellular distribution pattern of our VipD variants and obtained similar results (Fig. 4C). Although VipD2–310 showed a primarily cytosolic localization in COS-1 cells (Fig. S5), VipD2–621 and VipD311–621 colocalized with enlarged endosomal compartments enriched in Rab5a(Q79L) (Fig. 4C and Fig. S5), consistent with these fragments binding active Rab5 (Fig. 1E). A similar distribution was observed for catalytically inactive VipD(S73A), showing that PLA1 activity was dispensable for endosomal localization.
Concomitant with these findings, we also noticed that endogenous EEA1, unlike Rab5a(Q79L), assumed a primarily cytosolic localization in VipD2–621-producing cells, whereas in untransfected cells or control cells producing mCherry, EEA1 displayed a vesicular distribution in accordance with its accumulation on endosomes (Fig. 4 D and E). This finding was in stark contrast to earlier reports (11), which claimed that EEA1 was still associated with endosomes in VipD-producing cells. We found that EEA1 displacement from endosomes was also observable in cells producing either VipD311–621 or VipD(S73A) but not in cells producing only VipD2–310 (Fig. 4 D and E), indicating that the phenomenon of EEA1 displacement was, at least in part, mediated by the C-terminal domain of VipD, most likely a consequence of its ability to compete with EEA1 and other cellular ligands for Rab5 binding (Fig. S6A).
We also found that SNX2, a component of the retromer and marker of recycling endosomes, was redistributed to the cytosol in COS-1 cells producing either VipD2–621 or VipD311–621 (Fig. S6B). Because recruitment of SNX2 to endosomes does not directly depend on Rab5 (16), it seemed unlikely that competitive binding of VipD to Rab5 caused the displacement of SNX2 from endosomes. Instead, given that both EEA1 and SNX2 require PI(3)P to associate with endosomal membranes, we hypothesized that VipD affected the pool of endosomal PI(3)P, thereby causing the loss of PI(3)P ligands from membranes.
Using the FYVE domain of EEA1 (GFP-2×FYVE) as a PI(3)P sensor, we found that PI(3)P was highly enriched on endosomal structures both in untransfected COS-1 cells or in control cells producing mCherry. In contrast, production of even low levels of VipD2–621 resulted in the redistribution of GFP-2×FYVE to the cytosol (Fig. 4 F and G), indicating that PI(3)P had been efficiently removed from endosomal membranes by VipD2–621. PI(3)P displacement from endosomes was less pronounced in cells producing either VipD311–621 or the catalytically inactive mutant VipD(S73A), demonstrating that the PLA1 activity of VipD2–621 was primarily responsible for PI(3)P removal. VipD311–621 depleted PI(3)P more slowly from endosomes, most likely by blocking Rab5 from recruiting PI (3) kinases like hVps34, thus preventing the de novo synthesis of PI(3)P (Fig. 4 F and G and Fig. S6A). Notably, VipD2–621 did not alter the distribution of other organelle markers such as Giantin (Golgi compartment), Sec61b (endoplasmic reticulum), Hoechst (DNA), TOM70 (mitochondria), or lysosomal-associated membrane protein-2 (LAMP-2, lysosomes) (Fig. S7A). Similarly, the localization of GFP-SidM451–647, a PI(4)P probe (17), was not affected by VipD (Fig. S7 B and C), demonstrating that its PLA1 activity was directed only against Rab5-containing endosomes. Taken together, these findings define a unique role for the PLA1 domain of VipD in manipulating the lipid and, consequently, protein composition of endosomal membranes.
VipD Is Required for Efficient Endosomal Avoidance by L. pneumophila.
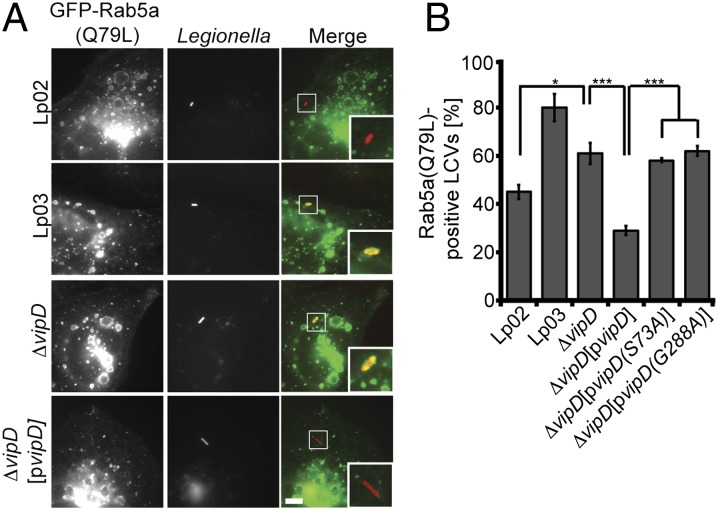
Given the striking effect of exogenous VipD on the molecular assembly of endosomes (Fig. 4), we subsequently examined the involvement of this effector protein in early endosomal avoidance by L. pneumophila. We used CHO-FcγRII cells challenged with L. pneumophila to monitor, by fluorescence microscopy, the acquisition of endosomal markers on LCVs shortly after bacterial uptake. The parental strain L. pneumophila Lp02 avoided endosomal fusion with much greater efficiency than the avirulent strain Lp03, with only 45 ± 3% Rab5a(Q79L)-positive Lp02-containing vacuoles as opposed to 80 ± 6% Rab5a(Q79L)-positive LCVs observed for Lp03 (Fig. 5). A L. pneumophila mutant lacking VipD (Lp02ΔvipD) showed a significant increase (61 ± 4%) in the colocalization with Rab5a(Q79L) compared with Lp02, and this phenotype was efficiently rescued (29 ± 2%) by complementing Lp02ΔvipD with a plasmid encoding VipD (Fig. 5). On the other hand, catalytically inactive VipD(S73A) and VipD(G288A) were unable to protect Lp02ΔvipD from Rab5a(Q79L)-positive compartments, confirming the biological significance of the PLA1 activity of VipD for endosomal avoidance by L. pneumophila.
Fig. 5.
VipD is required for endosomal avoidance by L. pneumophila. (A and B) CHO-FcγRII cells producing GFP-tagged Rab5a(Q79L) were challenged with the indicated L. pneumophila strains. Colocalization of LCVs with GFP-Rab5a(Q79L) 10 min after uptake was determined by immunofluorescence microscopy. (Scale bar, 1 μm.) *P < 0.05; **P < 0.01; ***P < 0.005 (Student t test).
Discussion
The ability of L. pneumophila to avoid endolysosomal trafficking is a key virulence feature. Despite recent observations pointing toward a role of the T4SS-translocated effector protein VipD in this process, the true biological function of VipD and the molecular mechanisms underlying endosomal avoidance by L. pneumophila remained unclear. Our study now shows that VipD possesses robust PLA1 (but not PLA2) activity, which alters the lipid and, consequently, protein composition of endosomal membranes, thereby preventing them from fusing with LCVs. The truly unexpected finding, however, was that VipD, to exhibit PLA1 activity, has to be bound and activated by host-cell endosomal Rab GTPases, namely Rab5 and Rab22.
To our knowledge, VipD is the first T4SS-translocated effector with phospholipase activity. Unlike ExoU and most secreted bacterial phospholipases, VipD alters the composition of endosomes in a bimodal manner (Fig. S8). The PLA1 domain catalyzes the removal of PI(3)P from the endosomal compartment by either directly hydrolyzing this phospholipid or by targeting its precursor molecules (Fig. 4 F and G), the C-terminal domain competes with cellular ligands like EEA1 or hVps34 for Rab5 binding (Fig. S6A), thereby preventing their de novo recruitment to endosomes. Simultaneously depleting the existing pool of PI(3)P and interfering with its replenishment through PI 3-kinase exclusion may explain how VipD can efficiently transform endosomes into membrane structures that lack many of their endosome-defining features, most likely rendering them fusion-incompetent. Given that the amount of VipD translocated during infection is very low (Fig. S1), we do not expect VipD to target the entire cellular pool of endosomes but rather only those organelles in the immediate vicinity of the LCV.
The finding that L. pneumophila VipD does not simply deactivate Rab5, for instance by functioning as a GTPase-activating protein, but rather exploits Rab5 for the activation of its PLA1 activity may seem extraneous at first. However, given that endosomal function is controlled by several Rab GTPases (18), their collective deactivation by L. pneumophila would require numerous effectors with GTPase-activating protein activity. PI(3)P, on the other hand, is the only phospholipid known to be essential for the assembly of tethering and fusion protein complexes into endosomal microdomains, and its removal can be efficiently accomplished by a single bacterial phospholipase, as shown here.
The strategy of removing endosomal PI(3)P is an efficient tactic used by other intracellular pathogens to combat the host's bactericidal defenses. For example, Mycobacterium tuberculosis (Mtb) secretes SapM, a phosphatase that removes PI(3)P from Mtb-containing vacuoles by converting it to PI, thereby arresting endosomal maturation (19). VipD differs from SapM because it targets PI(3)P within the endosomal membrane rather than the pathogen-containing vacuole, which may explain why earlier attempts to detect VipD on the surface of LCVs were unsuccessful (10). Despite the importance of VipD for endosomal avoidance, L. pneumophila mutants lacking vipD, either individually or in combination with its three paralogs, were not attenuated for growth in mammalian or amoebean cells (10), arguing for the existence of additional effector proteins that interfere with endosomal maturation by a yet unknown mechanism. This hypothesis may also explain why the defect in endosomal avoidance observed for L. pneumophila mutants lacking VipD, although statistically significant, was subtle (Fig. 5).
The involvement of VipD in endosomal avoidance has recently been challenged by a study proposing that VipD stimulates caspase-3 activation by targeting the mitochondrial membrane and causing cytochrome c release (13). Several points of evidence argue against such a role of VipD. First, VipD does not colocalize with mitochondria (or the plasma membrane) but is highly enriched on endosomal membranes of transfected mammalian cells (Fig. 4C) (11). Second, VipD requires endosomal Rab GTPases for the activation of its PLA1 domain (Fig. 3D), and because these GTPases are absent from mitochondrial membranes, it seems difficult to envision how they could logistically direct the PLA1 activity of VipD toward the mitochondrial membrane. Finally, caspase-3 activation was blocked by the PLA2 inhibitor BEL (13), yet our study clearly demonstrates that the PLA1 activity of VipD is insensitive even to high concentrations of BEL (Fig. 3H), suggesting that other contaminating phospholipases caused the observed effects on caspase-3. This convincing evidence against an involvement of VipD in altering mitochondrial membranes and causing caspase-3 activation combined with our own data prompt us to favor a model in which the primary role of this L. pneumophila effector is to attenuate endosome function by altering their protein and lipid composition (Fig. S8).
The finding that the PLA1 activity of VipD is entirely dependent on the binding of active endosomal Rab GTPases adds an intriguing case to a very short list of related examples in which microbial proteins require a host-cell GTPase as a trigger for their catalytic activity (20–22). Linking catalytic activity to subcellular localization most likely allows L. pneumophila and other microbial pathogens to remotely control the activity of their effectors and toxins after their translocation into the host cell and to prevent them from indiscriminately targeting other host compartments or even the bacterium’s own physiology (Fig. S8). We hypothesize that the VipD paralogs and probably a variety of other effectors from L. pneumophila and related pathogens possess safety mechanisms similar to that described here for VipD. Understanding their mechanism of activation not only provides valuable insight into microbial virulence strategies, but also opens the way for the discovery of novel therapeutics designed to interfere with the activation processes.
Experimental Procedures
Phospholipase Assays.
PLA1 or PLA2 activities were monitored using EnzChek Phospholipase Assay Kits (Invitrogen) according to the manufacturer's specifications. For the assays, 400 nM of each recombinant protein was used in 100-µL reactions in black 96-well microtiter plates for 40 min at room temperature. In protein complex reactions involving His6-VipD and GST-Rabs, 400 nM of each protein (unless otherwise indicated) was preincubated at 4 °C for 2 h to allow protein complex formation followed by the addition of fluorescent substrates. Phospholipase inhibitors were added after protein complex formation at room temperature for 1 h before the addition of fluorescent substrates. Fluorescence intensities, representing substrate cleavage, were detected after 40 min at an emission wavelength of 515 nm using a microplate reader (BioTek Synergy 4).
Immunofluorescence Microscopy.
VipD localization, cell morphology, and endosomal modification was analyzed in COS-1 cells transiently transfected using Lipofectamine 2000 (Invitrogen). Cells were fixed at the indicated time points, and endogenous proteins were detected with antibodies at the following concentrations: EEA1 (1:200), SNX2 (1:1,250), LAMP-2 (1:2,000), Sec61b (1:200), Giantin (1:3,000), and anti-rabbit FITC (1:1,000). All images were analyzed on a Zeiss Axio Observer.Z1 inverted light microscope using a Zeiss Plan-Apochromat 63×/oil M27 objective (COS-1 imaging) or a Zeiss Plan-Apochromat 100×/1.4 oil DIC M27 objective (CHO infections) and processed with Zeiss AxioVision 4.7.2 software.
Legionella Infections.
CHO-FcgRII cells producing GFP-Rab5a(Q79L) were challenged with opsonized L. pneumophila at a multiplicity of infection of 15 for 10 min at 37 °C. Cells were chemically fixed, and intracellular and extracellular bacteria were differentially stained (23). Statistical representation of GFP-endosomal markers localizing to LCVs is derived from the means of four independent experiments (n = 25).
Supplementary Material
Acknowledgments
We thank all members of the M.P.M. laboratory, and Drs. G. Storz, J. Donaldson, T. Updegrove, M. Barzik, and J. Bonifacino for critical reading of the manuscript and insightful discussion. This work was supported by the Intramural Research Program of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316376111/-/DCSupplemental.
References
- 1.Fairn GD, Grinstein S. How nascent phagosomes mature to become phagolysosomes. Trends Immunol. 2012;33(8):397–405. doi: 10.1016/j.it.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: Achieving specificity in membrane traffic. Proc Natl Acad Sci USA. 2006;103(32):11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kutateladze TG. Mechanistic similarities in docking of the FYVE and PX domains to phosphatidylinositol 3-phosphate containing membranes. Prog Lipid Res. 2007;46(6):315–327. doi: 10.1016/j.plipres.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vieira OV, et al. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J Cell Biol. 2001;155(1):19–25. doi: 10.1083/jcb.200107069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fratti RA, Backer JM, Gruenberg J, Corvera S, Deretic V. Role of phosphatidylinositol 3-kinase and Rab5 effectors in phagosomal biogenesis and mycobacterial phagosome maturation arrest. J Cell Biol. 2001;154(3):631–644. doi: 10.1083/jcb.200106049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clemens DL, Lee BY, Horwitz MA. Deviant expression of Rab5 on phagosomes containing the intracellular pathogens Mycobacterium tuberculosis and Legionella pneumophila is associated with altered phagosomal fate. Infect Immun. 2000;68(5):2671–2684. doi: 10.1128/iai.68.5.2671-2684.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horwitz MA. Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med. 1983;158(4):1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ensminger AW, Isberg RR. Legionella pneumophila Dot/Icm translocated substrates: A sum of parts. Curr Opin Microbiol. 2009;12(1):67–73. doi: 10.1016/j.mib.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shohdy N, Efe JA, Emr SD, Shuman HA. Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc Natl Acad Sci USA. 2005;102(13):4866–4871. doi: 10.1073/pnas.0501315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.VanRheenen SM, Luo ZQ, O’Connor T, Isberg RR. Members of a Legionella pneumophila family of proteins with ExoU (phospholipase A) active sites are translocated to target cells. Infect Immun. 2006;74(6):3597–3606. doi: 10.1128/IAI.02060-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ku B, et al. VipD of Legionella pneumophila targets activated Rab5 and Rab22 to interfere with endosomal trafficking in macrophages. PLoS Pathog. 2012;8(12):e1003082. doi: 10.1371/journal.ppat.1003082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson DM, et al. Ubiquitin and ubiquitin-modified proteins activate the Pseudomonas aeruginosa T3SS cytotoxin, ExoU. Mol Microbiol. 2011;82(6):1454–1467. doi: 10.1111/j.1365-2958.2011.07904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu W, Hammad LA, Hsu F, Mao Y, Luo ZQ. Induction of caspase 3 activation by multiple Legionella pneumophila Dot/Icm substrates. Cell Microbiol. 2013;15(11):1783–1795. doi: 10.1111/cmi.12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgs HN, Glomset JA. Purification and properties of a phosphatidic acid-preferring phospholipase A1 from bovine testis. Examination of the molecular basis of its activation. J Biol Chem. 1996;271(18):10874–10883. doi: 10.1074/jbc.271.18.10874. [DOI] [PubMed] [Google Scholar]
- 15.Amara S, Delorme V, Record M, Carrière F. Inhibition of phospholipase A1, lipase and galactolipase activities of pancreatic lipase-related protein 2 by methyl arachidonyl fluorophosphonate (MAFP) Biochim Biophys Acta. 2012;1821(11):1379–1385. doi: 10.1016/j.bbalip.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 16.Rojas R, et al. Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J Cell Biol. 2008;183(3):513–526. doi: 10.1083/jcb.200804048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brombacher E, et al. Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J Biol Chem. 2009;284(8):4846–4856. doi: 10.1074/jbc.M807505200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Somsel Rodman J, Wandinger-Ness A. Rab GTPases coordinate endocytosis. J Cell Sci. 2000;113(Pt 2):183–192. doi: 10.1242/jcs.113.2.183. [DOI] [PubMed] [Google Scholar]
- 19.Vergne I, et al. Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2005;102(11):4033–4038. doi: 10.1073/pnas.0409716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nawabi P, Catron DM, Haldar K. Esterification of cholesterol by a type III secretion effector during intracellular Salmonella infection. Mol Microbiol. 2008;68(1):173–185. doi: 10.1111/j.1365-2958.2008.06142.x. [DOI] [PubMed] [Google Scholar]
- 21.Christen M, et al. Activation of a bacterial virulence protein by the GTPase RhoA. Sci Signal. 2009;2(95):ra71. doi: 10.1126/scisignal.2000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lossi NS, Rolhion N, Magee AI, Boyle C, Holden DW. The Salmonella SPI-2 effector SseJ exhibits eukaryotic activator-dependent phospholipase A and glycerophospholipid:cholesterol acyltransferase activity. Microbiology. 2008;154(Pt 9):2680–2688. doi: 10.1099/mic.0.2008/019075-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swanson MS, Isberg RR. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun. 1995;63(9):3609–3620. doi: 10.1128/iai.63.9.3609-3620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.