Motor proteins are a vital contributor to life, where they manifest themselves in the directed movement of organisms, in cell division, and in the transport of organelles, proteins, and nucleic acids. The “Big Three” superfamilies of linear motor proteins consist of myosin, kinesin, and dynein, which move along actin or microtubules. Over the past 60 y, since the discovery of the sliding filament model for muscle contraction and the identification of myosin as the active component, considerable effort has been devoted to understanding the molecular properties of the motor domains that generate directed movement. In contrast, considerably less is known about the structural interactions of these motors with their partners, which is ironic because motors only exist to move or apply force to something! Not surprisingly, this is a rich area for investigation that is now yielding an understanding of not only the structural themes but also the regulation of motors by their cargoes. An example of this is seen in PNAS, where Shi et al. (1) reveal how the budding yeast myosin Myo4p, a class V myosin, interacts with its partners She3p and She2p to facilitate transport of mRNA. The report by Shi et al. provides structural insight into how cargo binding efficiently regulates this myosin through oligomerization rather than through a disruption of a direct head-tail inhibitory interaction as often seen in many myosins (2) and members of the kinesin superfamily (3, 4).
In yeast, Myo4p transports ∼20 types of mRNA molecules along actin cables to specific locations in the cytoplasm to generate local concentrations of the corresponding gene product. The mRNAs are targeted by specific cis-acting elements, which have been called “zip codes,” that are up to 100 nucleotides long. This process facilitates transport of mRNA to the daughter cell. There are two adapter proteins involved in this transport system, She2p and She3p. She2p is a tetramer that binds the mRNA zip code in the nucleus and once in the cytoplasm forms a complex with Myo4p and Shep3 (5, 6). She3p interacts with She2p and Myo4p, but also contains an additional binding site for the cotransport of tubular endoplasmic reticulum (ER) to the yeast bud (7). Myo4p belongs to the myosin V family, the members of which take characteristically long steps along the actin filament.
Myo4p is an unusual myosin V because by itself it is not a processive motor, as are most other myosins in this class. Instead, it exists as a constitutive monomer instead of a dimer, where dimerization is needed to take long coordinated steps along actin (8, 9). Nonprocessivity in class V myosins has also been proposed for the second myosin V in yeast: Myo2p, a Drosophila myosin V, and human myosin Vc (10–12). It is well established that monomeric motors must work together to generate directed movement; thus, the initial question was whether this requires an organized oligomeric structure or whether it can be accomplished by multiple motors attached to the cargo that is being transported. Elegant work by Trybus and colleagues demonstrated that Myo4p forms a tight single-headed complex with She3p, which alone is not capable of processive movement and has no tendency to dimerize, but on addition of She2p forms a two-headed processive assembly (5, 13). Recent single-molecule in vitro reconstruction of the transport complex shows that mRNA is required to form a stable processive assembly at physiological ionic strength, and that increasing the number of zip codes on the mRNA increases the run length of the complex (14). This observation showed that motor activity is controlled by cargo binding and that large assemblies or groups of motors are not needed for transport.
Separate structures of She2p and the cargo-binding domain of Myo4p have been determined previously, but the structure of She3p was unknown (15, 16) nor how it interacts with its partners. Approximately one-third of She3p is predicted to form a stable dimeric coiled-coil, so the question arises regarding the stoichiometry of the Myo4p-She3p complex as to whether it is 1:1 or 1:2. The paper by Shi et al. resolves this issue (1).
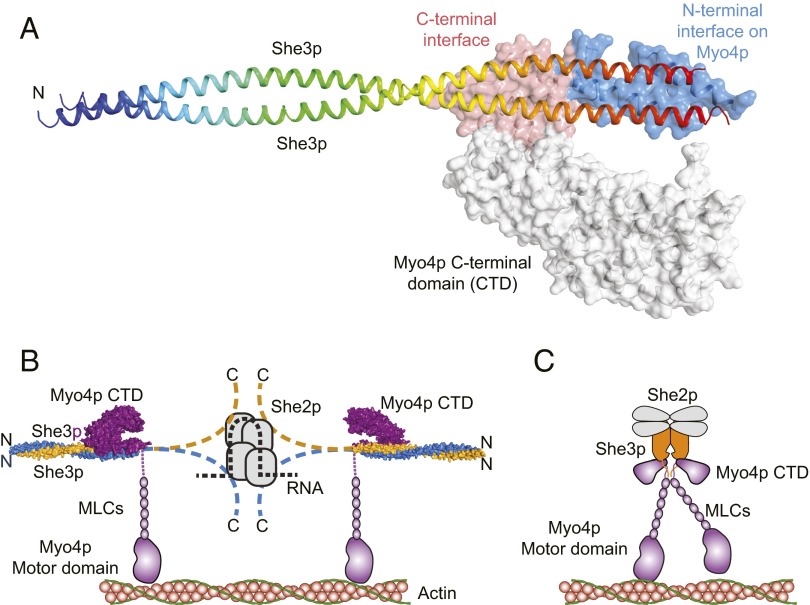
Shi et al. (1) show that the cargo-binding domain of Myo4p binds to a truncated dimeric fragment of She3p to form a 1:2 complex. This process was demonstrated though an X-ray structure determined to 3.6 Å resolution (Fig. 1A), and also by multiangle light scattering in solution. The stoichiometry was an unanticipated finding. The protein utilized for crystallization included residues 81–311 of She3p; however, electron density was only observed for residues 81–196, where presumably the latter 115 residues were disordered in the lattice. The C-terminal region contains the binding site for She2p (17). The observed polypeptide for She3p forms an extended coiled-coil that deviates from a symmetrical helical structure in the region that binds Myo4p. Examination of the coordinates (PDB ID code 4LL8) reveals that the asymmetric distortion of the She3p coiled-coil precludes the binding of a second molecule of the motor cargo domain. The distortion from a canonical coiled-coil exposes hydrophobic side chains that form the interface with Myo4p, which contains many conserved residues for both proteins. Unexpectedly, there is an area toward the N terminus of She3p that is also conserved, which is proposed to contain the binding site for tubular ER.
Fig. 1.
(A) The structure of Myo4p918-1417·She3p81-311. In She3p only residues 81–195 were visible in the electron density for She3p where the C-terminal segment (96–311) is assumed to be disordered. She3p81-195 adopts a regular coiled-coil at its N-terminal region but is distorted where it binds to Myo4p. (B and C) Potential models for the active transport complex. B was adapted from Shi et al. (1), whereas C was proposed by Trybus and colleagues (13, 14). The latter is in agreement with the step size and would provide tight coordination between the motor domain, but needs to be modified to accommodate the structure of Myo4p·She3p described in Shi et al. (1). The structure in A was generated from PDB coordinates 4LL8 (1). MLCs, myosin light chains.
The structure of the cargo binding region of Myo4p itself is essentially identical to earlier structures with small changes in the domain organization with the exception of the extra ∼100 amino acid residues at the N terminus (16). This section, from 1,022–1,061, forms a helical hairpin that constitutes a substantial fraction of the binding interface with She3p. A binding surface on Myo4p is provided by the segment extending from 1,325–1,402, where this is structurally adjacent to the N-terminal binding surface, even though is it separated by 264 residues in the sequence. As noted before, the structure of the cargo-binding domain of Myo4p is very different from that of mammalian class V myosins where the latter exist as constitutive dimers and are regulated via a head–tail interaction (18).
The structure of the complex is highly asymmetric and forms a “claw-like” assembly where the cargo domain extends away from the extended coiled-coil of She3p (Fig. 1A).
The report by Shi et al. establishes important constraints on the regulation and assembly of Myo4p as a functional motor.
Shi et al. (1) also confirm that mRNA is needed to form a stable complex with She2p, where only one zip code (∼44 nucleotides) is needed per tetramer to form a complex. Based on this information a new model for the complete Myo4p·She3p·She2p·mRNA complex was proposed (Fig. 1B). This model differs from that proposed by Trybus and colleagues (13, 14) (Fig. 1C) because it does not include a close interaction between the two motors; this is almost certainly needed because the step size of the Myo4p·She3p·She2p·mRNA complex is almost identical to that seen for conventional class V myosins (13). It is well established in these myosins that the step size is proportional to the length of the lever arm, hence the distance from the motor domain to the hinge region must be similar. The model depicted in Fig 1B would increase the distance between the motors and would not provide tight coupling. A larger step size would be expected for such an arrangement. The earlier model proposed by Trybus et al. would result in the same arrangement of motors as seen in dimeric class V myosins and hence should have the same step size (14). This latter model is consistent with images of the complex seen by electron microscopy, but now must be modified to reflect the stoichiometry of the Myo4p·She3p complex. Dimerization might occur through the predicted coiled-coil that occurs following the six myosin light-chain binding motifs.
The report by Shi et al. (1) establishes important constraints on the regulation and assembly of Myo4p as a functional motor. The report extends our understanding of how class V myosins, which transport an exceedingly diverse range of cargoes, recognize and bind their targets. Structural questions remain, though, concerning the manner in which the Myo4p·She3p heterotrimers interact with each other and how She3p mediates this process. This will be an exciting area of investigation in years to come.
Acknowledgments
This work was supported by National Institutes of Health Grant GM054141 to Susan Gilbert (Rensselaer Polytechnic Institute) and I.R.
Footnotes
The author declares no conflict of interest.
See companion article on page E1082.
References
- 1.Shi H, Singh N, Esselborn F, Blobel G. Structure of a myosin·adaptor complex and pairing by cargo. Proc Natl Acad Sci USA. 2014;111:E1082–E1090. doi: 10.1073/pnas.1401428111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sellers JR, Knight PJ. Folding and regulation in myosins II and V. J Muscle Res Cell Motil. 2007;28(7-8):363–370. doi: 10.1007/s10974-008-9134-0. [DOI] [PubMed] [Google Scholar]
- 3.Kaan HY, Hackney DD, Kozielski F. The structure of the kinesin-1 motor-tail complex reveals the mechanism of autoinhibition. Science. 2011;333(6044):883–885. doi: 10.1126/science.1204824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verhey KJ, Hammond JW. Traffic control: Regulation of kinesin motors. Nat Rev Mol Cell Biol. 2009;10(11):765–777. doi: 10.1038/nrm2782. [DOI] [PubMed] [Google Scholar]
- 5.Hodges AR, Krementsova EB, Trybus KM. She3p binds to the rod of yeast myosin V and prevents it from dimerizing, forming a single-headed motor complex. J Biol Chem. 2008;283(11):6906–6914. doi: 10.1074/jbc.M708865200. [DOI] [PubMed] [Google Scholar]
- 6.Müller M, et al. Formation of She2p tetramers is required for mRNA binding, mRNP assembly, and localization. RNA. 2009;15(11):2002–2012. doi: 10.1261/rna.1753309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmid M, Jaedicke A, Du TG, Jansen RP. Coordination of endoplasmic reticulum and mRNA localization to the yeast bud. Curr Biol. 2006;16(15):1538–1543. doi: 10.1016/j.cub.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 8.Reck-Peterson SL, Tyska MJ, Novick PJ, Mooseker MS. The yeast class V myosins, Myo2p and Myo4p, are nonprocessive actin-based motors. J Cell Biol. 2001;153(5):1121–1126. doi: 10.1083/jcb.153.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purcell TJ, Morris C, Spudich JA, Sweeney HL. Role of the lever arm in the processive stepping of myosin V. Proc Natl Acad Sci USA. 2002;99(22):14159–14164. doi: 10.1073/pnas.182539599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tóth J, Kovács M, Wang F, Nyitray L, Sellers JR. Myosin V from Drosophila reveals diversity of motor mechanisms within the myosin V family. J Biol Chem. 2005;280(34):30594–30603. doi: 10.1074/jbc.M505209200. [DOI] [PubMed] [Google Scholar]
- 11.Takagi Y, et al. Human myosin Vc is a low duty ratio, nonprocessive molecular motor. J Biol Chem. 2008;283(13):8527–8537. doi: 10.1074/jbc.M709150200. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe S, et al. Human myosin Vc is a low duty ratio nonprocessive motor. J Biol Chem. 2008;283(16):10581–10592. doi: 10.1074/jbc.M707657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krementsova EB, et al. Two single-headed myosin V motors bound to a tetrameric adapter protein form a processive complex. J Cell Biol. 2011;195(4):631–641. doi: 10.1083/jcb.201106146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sladewski TE, Bookwalter CS, Hong MS, Trybus KM. Single-molecule reconstitution of mRNA transport by a class V myosin. Nat Struct Mol Biol. 2013;20(8):952–957. doi: 10.1038/nsmb.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niessing D, Hüttelmaier S, Zenklusen D, Singer RH, Burley SK. She2p is a novel RNA binding protein with a basic helical hairpin motif. Cell. 2004;119(4):491–502. doi: 10.1016/j.cell.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 16.Heuck A, et al. The structure of the Myo4p globular tail and its function in ASH1 mRNA localization. J Cell Biol. 2010;189(3):497–510. doi: 10.1083/jcb.201002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Böhl F, Kruse C, Frank A, Ferring D, Jansen RP. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 2000;19(20):5514–5524. doi: 10.1093/emboj/19.20.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nascimento AF, et al. Structural insights into functional overlapping and differentiation among myosin V motors. J Biol Chem. 2013;288(47):34131–34145. doi: 10.1074/jbc.M113.507202. [DOI] [PMC free article] [PubMed] [Google Scholar]