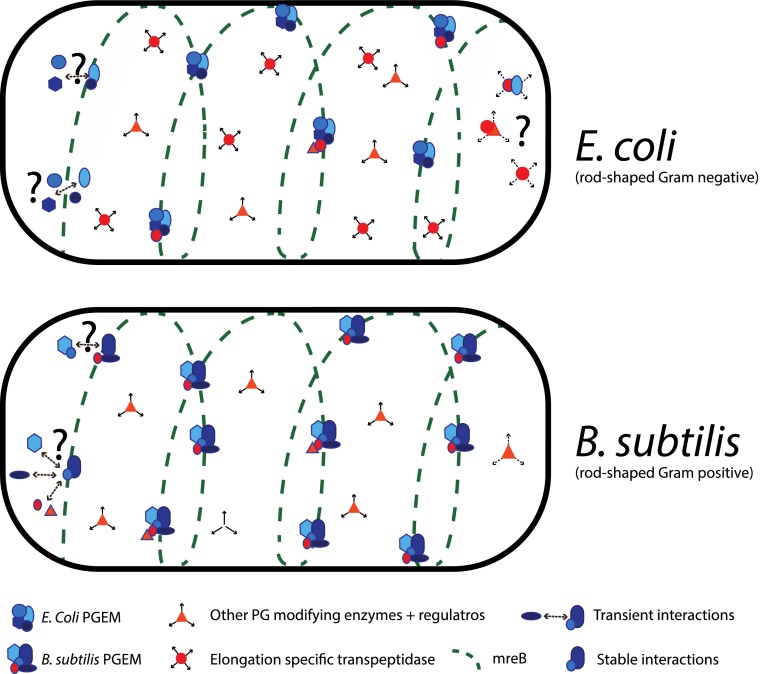
Bacterial cytoskeletal elements provide the framework for the assembly of multicomponent protein machineries controlling cell growth and division. One of the main roles of these machineries is to build the cell wall, which is instrumental for cell shape and stability. Our understanding of how these machineries are spatiotemporally controlled has been rapidly changing in the last few years, fueled mostly by single-cell or systems-based approaches. Recent studies in Bacillus subtilis described a coupled movement of the actin-like MreB and the core part of the cell wall synthesis machinery (1, 2). In PNAS, Lee et al. present evidence that the situation is different in Escherichia coli: penicillin-binding protein 2 (PBP2), a core cell wall synthesis enzyme involved in cell elongation, exhibits a diffusive motion, and dynamically associates with the rest of the elongation machinery to spatiotemporally control the insertion of new glycan strands into the cell wall (3). This new model (Fig. 1) has interesting mechanistic ramifications as to how the cell organizes cell growth. It also begs the question of whether the difference between the two model organisms is related to synthesis requirements of their architecturally distinct cell walls or is just evolution coming up with different solutions for the same problem. We anticipate this will be a recurring question in the field, as we start to accumulate knowledge in diverse organisms.
Fig. 1.
Models of PGEM assembly and movement in E. coli and B. subtilis. In E. coli, it is unknown which parts of the PGEM from stable versus transient interactions. The elongation specific TPase (PBP2) exhibits diffusive motion, uncoupled to the circumferential MreB movement. It is possible that PBP2 forms subcomplexes with other PG-related proteins or PGEM components. In B. subtilis, the elongation-specific TPases (PBP2A and PBPH) are part of a seemingly stable PGEM. Other PG-related proteins presumably associate dynamically to this complex.
The main component of the cell wall in bacteria is peptidoglycan (PG). PG is composed of long glycan chains, which run perpendicular to the long axis of the cell and are cross-linked by short peptides, forming a 3D mesh-like structure around the bacterial cytoplasmic membrane. Growth of this network requires the synchronous action of enzymes that synthesize and hydrolyze PG, as the cell needs to cleave the existing PG layer before attaching new material to it. This coordination is achieved by multicomponent protein machineries (4), which mainly consist of (i) PG synthases and hydrolases and their regulatory factors; (ii) PG precursor and/or recycling material transporters; and (iii) cytoskeletal proteins and associated factors. In terms of cell elongation, our view until recently was that MreB helical filaments, spanning the entire cell, could act as templates for these machineries to move and extend the cell wall. We know now that MreB forms small filaments [which may reach up to 1 µm in length (5)] that move around the cell circumference, in a manner that requires active PG synthesis (1, 2, 6). In addition, in B. subtilis, a large part of the PG elongation machinery (PGEM), which includes cytoskeletal proteins and associated factors and the monofunctional PG synthases, seems to move as a large stable complex (1, 2). In contrast, in E. coli, PBP2, the monofunctional PG synthase [transpeptidase (TPase)] that is required for elongation is shown to move in a diffusive manner (3), despite being required for the circumferential MreB motion (3, 6).
What is the advantage of PBP2 being only transiently associated to a circumferential moving MreB and/or PGEM? The authors argue that less PBP2 molecules are required to maintain a high growth rate since a single PBP2 molecule can then contribute at multiple active sites of synthesis, making the system more robust to PBP2 fluctuations and/or defective PBP2 molecules. Indeed, theoretical calculations of PBP2 molecules required for sustaining growth better match the dynamic interaction model, allowing E. coli to retain its growth rate even when losing half of its PBP2 molecules (3). Why then in B. subtilis do PBP2A and PBP2H (B. subtilis has two PBP2 paralogues) seem to be more stably associated and cotrack with the MreB? Although this is hard to answer at this point, experiments that will test the predictions of the two models will help toward this direction. Are PBP2 (A and H) levels in B. subtilis higher and/or is elongation more sensitive to fluctuations of PBP2 levels or to inactive PBP2 versions? Interestingly, PBP2A cotracks with MreB, only in the absence of PBP2H; otherwise, its motion is diffusive (1, 2). In addition, a fraction of PBP2H is always moving diffusively (1). This could be due to excess abundance and/or redundancy of the two PBP2s, or it could also mean that both stable and dynamic associations are integral to PBP2 function in B. subtilis. In any case, cotracking of PBP2A and MreB requires active PG synthesis, as vancomycin treatment stops MreB movement and makes PBP2A diffusive (2). This implies that whether the PGEM and PBP2 form stable or dynamic associations is not a mere issue of different binding affinities between PBP2 and PGEM components in the two organisms.
Although moving diffusively, the diffusion constant of PBP2 was one to two orders of magnitude lower than that expected based on the size of the protein, implying that it either does not move on its own or interactions with other proteins slow it down (3). Consistent with both scenarios, a catalytically dead PBP2 diffused slowly, while a truncated mutant lacking the entire TPase domain moved close to the speed predicted by its size (3). It remains to be seen which protein interaction is responsible for this difference and whether subparts of the PGEM can diffuse together. Lee et al. find that neither MreB activity nor the presence of PBP1A, a bifunctional PG synthase [TPase and glycosytransferase (GTase)] involved in elongation and recently shown to bind PBP2 strongly (7), is responsible for slowing down PBP2 (3).
Similar to PBP2, quantifying the single-particle dynamics of other core PGEM members (MreC, MreD, RodA, and RodZ) and associated PG-related enzymes could help us understand the underlying design principles for coordinating the complex process of cell wall growth. In B. subtilis, core PGEM components, such as PBP2, cotrack with MreB in a circumferential motion, perpendicular to the long axis of the cell (Fig. 1). However, other PG-modifying enzymes that associate with the PGEM, but are not required for MreB movement, move in a diffusive manner, implying that their association with the PGEM is more dynamic (1). In E. coli, only the movements of MreB and PBP2 have been measured (3, 6) (Fig. 1). Following other associated PG-modifying enzymes of PGEM can be challenging, as redundancy and/or excess of the enzymes may mask the manner these proteins normally associate with the PGEM machinery. For example, although PBP1A is associated with cell elongation, a second bifunctional PG synthase, PBP1B, can cover for its absence. A similar redundancy exists for the hydrolases involved in cell elongation (8). Furthermore, in E. coli, the bifunctional PG synthases, PBP1A and PBP1B, have dedicated regulators, LpoA and LpoB, which localize independently to elongation and divisions sites, respectively (9, 10). In contrast, some PG hydrolases require their regulators for localization (11, 12), whereas others do not (13). It remains to be seen if the regulators and/or the PG enzymes associate dynamically with the PGEM (or the division machinery), but in cases of independent localization, it is unlikely that both form stable interactions with the moving macromolecular machineries. In cases of codependent localization, it is unlikely that both move independently and in a diffusive manner. More importantly, if a number of PG-modifying enzymes (synthases and hydrolases) and their regulators move diffusively on their own and only transiently assemble in a multicomponent machinery that initiates and cross-links new strands in the cell wall, then this would imply that all of them recognize the same biochemical feature/event in cell wall growth.
A challenging open question is what drives MreB motion. Both PG synthesis enzymatic activities, transpeptidation and transglycosylation, are necessary (6). Whether they drive the motion (motor) or provide feedback to it is still unknown. Nonetheless, it has been established that MreB polymerization does not self-promote this motion (2, 6). As Lee et al. provide evidence that PBP2 only dynamically associates with the PGEM in E. coli, yet its presence and activity are absolutely required for MreB motion (3), it seems more likely that the activity of PBP2 on the cell wall drives/feeds back to MreB motion rather than PBP2 per se. Nevertheless, the activity of PBP2 on the cell wall is not the only thing required, as blocking transglycosylation with moenomycin can also stop MreB movement (6). The molecular feature of cell wall synthesis that drives MreB motion is the focus of extensive ongoing research. Notably, a recent model proposes multiple unidirectionally moving motors that are equally distributed in the cell periphery and dynamically associate with the PGEM (5).
In summary, disentangling how large protein machineries assemble and orchestrate cell growth is of broad biological interest. It is expected that the process will be significantly different among microbes: many subunits are not ubiquitously conserved, cytoskeletal elements can vary in structure and function (14), recruitment hierarchy is different even in organisms with conserved subunits (15), and niche-specific regulators of PG growth and interchangeability of enzymatic components seem to be prevalent (4, 10). Mapping the differences and also linking them to the physiology of diverse organisms will be in the core of future studies in the field and will help us dissect the general underlying principles of these machineries from the niche-specific ones. This will only be achieved by an interdisciplinary approach, combining biochemistry (to map interactions, protein levels, binding affinities, and to reconstitute complexes in vitro), cell biology (single-particle tracking, and also methods for following protein–protein interactions in vivo, such as FRET), modeling, and systems-based approaches (to identify new players of these machineries across organisms). Ultimately, how cells are built is a complex question fundamental to our understanding of biology, and the application of new technologies will surely bring many surprising revelations.
Acknowledgments
We thank Waldemar Vollmer (Newcastle, UK) for critically reading the manuscript and providing feedback.
Footnotes
The authors declare no conflict of interest.
See companion article on page 4554.
References
- 1.Domínguez-Escobar J, et al. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333(6039):225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 2.Garner EC, et al. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333(6039):222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee TK, et al. A dynamically assembled cell wall synthesis machinery buffers cell growth. Proc Natl Acad Sci USA. 2014;111:4554–4559. doi: 10.1073/pnas.1313826111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10(2):123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olshausen PV, et al. Superresolution imaging of dynamic MreB filaments in B. subtilis-a multiple-motor-driven transport? Biophys J. 2013;105(5):1171–1181. doi: 10.1016/j.bpj.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Teeffelen S, et al. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc Natl Acad Sci USA. 2011;108(38):15822–15827. doi: 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banzhaf M, et al. Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Mol Microbiol. 2012;85(1):179–194. doi: 10.1111/j.1365-2958.2012.08103.x. [DOI] [PubMed] [Google Scholar]
- 8.Singh SK, SaiSree L, Amrutha RN, Reddy M. Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Mol Microbiol. 2012;86(5):1036–1051. doi: 10.1111/mmi.12058. [DOI] [PubMed] [Google Scholar]
- 9.Paradis-Bleau C, et al. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell. 2010;143(7):1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Typas A, et al. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell. 2010;143(7):1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domínguez-Cuevas P, Porcelli I, Daniel RA, Errington J. Differentiated roles for MreB-actin isologues and autolytic enzymes in Bacillus subtilis morphogenesis. Mol Microbiol. 2013;89(6):1084–1098. doi: 10.1111/mmi.12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meisner J, et al. FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Mol Microbiol. 2013;89(6):1069–1083. doi: 10.1111/mmi.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters NT, Dinh T, Bernhardt TG. A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators. J Bacteriol. 2011;193(18):4973–4983. doi: 10.1128/JB.00316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cabeen MT, Jacobs-Wagner C. The bacterial cytoskeleton. Annu Rev Genet. 2010;44:365–392. doi: 10.1146/annurev-genet-102108-134845. [DOI] [PubMed] [Google Scholar]
- 15.Goley ED, et al. Assembly of the Caulobacter cell division machine. Mol Microbiol. 2011;80(6):1680–1698. doi: 10.1111/j.1365-2958.2011.07677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]