Significance
Microbial natural products are a major source of new drug leads, yet discovery efforts are constrained by the lack of information describing the diversity and distributions of the associated biosynthetic pathways among bacteria. Using the marine actinomycete genus Salinispora as a model, we analyzed genome sequence data from 75 closely related strains. The results provide evidence for high levels of pathway diversity, with most being acquired relatively recently in the evolution of the genus. The distributions and evolutionary histories of these pathways provide insight into the mechanisms that generate new chemical diversity and the strategies used by bacteria to maximize their population-level capacity to produce diverse secondary metabolites.
Keywords: genome sequencing, comparative genomics
Abstract
Access to genome sequence data has challenged traditional natural product discovery paradigms by revealing that the products of most bacterial biosynthetic pathways have yet to be discovered. Despite the insight afforded by this technology, little is known about the diversity and distributions of natural product biosynthetic pathways among bacteria and how they evolve to generate structural diversity. Here we analyze genome sequence data derived from 75 strains of the marine actinomycete genus Salinispora for pathways associated with polyketide and nonribosomal peptide biosynthesis, the products of which account for some of today’s most important medicines. The results reveal high levels of diversity, with a total of 124 pathways identified and 229 predicted with continued sequencing. Recent horizontal gene transfer accounts for the majority of pathways, which occur in only one or two strains. Acquired pathways are incorporated into genomic islands and are commonly exchanged within and between species. Acquisition and transfer events largely involve complete pathways, which subsequently evolve by gene gain, loss, and duplication followed by divergence. The exchange of similar pathway types at the precise chromosomal locations in different strains suggests that the mechanisms of integration include pathway-level homologous recombination. Despite extensive horizontal gene transfer there is clear evidence of species-level vertical inheritance, supporting the concept that secondary metabolites represent functional traits that help define Salinispora species. The plasticity of the Salinispora secondary metabolome provides an effective mechanism to maximize population-level secondary metabolite diversity while limiting the number of pathways maintained within any individual genome.
Microbial secondary metabolites have long benefited human health and industry. They include important pharmaceutical agents such as the antibiotic penicillin, the anticancer agent vancomycin, and the immunosuppressant rapamycin among the more than 20 thousand biologically active microbial natural products reported as of 2002 (1). Secondary metabolites also have important ecological roles for the organisms that produce them, particularly in terms of nutrient acquisition, chemical communication, and defense (2). Many of these compounds are the products of polyketide synthase (PKS) and nonribosomal peptide synthetase (NRPS) pathways or hybrids thereof. These pathways are generally organized into gene clusters that can exceed 100 kb and include regulatory, resistance, and transport elements (3), thus making them well-suited for horizontal gene transfer (HGT) (4, 5). The architectures and functional attributes of PKS and NRPS genes have been reviewed in detail (3, 6, 7) and account for much of the structural diversity that is the hallmark of microbial natural products. Remarkably, PKS and NRPS enzymes build these complex secondary metabolites via the controlled assembly of simple biosynthetic building blocks such as acetate, propionate, and amino acids. These building blocks are incorporated in a combinatorial fashion via a series of sequential chemical condensation reactions encoded by ketosynthase (KS) and condensation (C) domains within PKS and NRPS genes, respectively (3).
The pathways responsible for secondary metabolite biosynthesis are among the most rapidly evolving genetic elements known (5). It has been shown that gene duplication, loss, and HGT have all played important roles in the distribution of PKSs among microbes (8, 9). Changes within PKS and NRPS genes also include mutation, domain rearrangement, and module duplication (5), all of which can account for the generation of new small-molecule diversity. The evolutionary histories of specific PKS and NRPS domains have proven particularly informative, with KS and C domains providing insight into enzyme architecture and function (10, 11). These studies have helped establish the extensive nature of HGT among biosynthetic genes (4, 12), which is reflected in the incongruence between PKS and NRPS gene phylogenies and those of the organisms in which they reside (13). Although resolving the evolutionary histories of entire pathways remains more challenging than individual genes or domains, comparative analyses of biosynthetic gene clusters have proven useful for the identification of pathway boundaries (14).
The exchange of PKS and NRPS pathways by HGT confounds the relationships between taxonomy and secondary metabolite production. This may in part explain the historical reliance on chance for the discovery of natural product drug leads from chemically prolific but taxonomically complex taxa such as the genus Streptomyces. Genome sequencing has changed the playing field by providing bioinformatic opportunities to “mine” the biosynthetic potential of strains before chemical analysis and to target the products of specific pathways that are predicted to yield compounds of interest (15). Sequence-based methodologies not only hold great promise for natural product discovery; they are providing a wealth of information that will ultimately improve our understanding of pathway diversity and distributions and the evolutionary events that generate new chemical diversity.
Here we report the analysis of PKS and NRPS biosynthetic gene clusters in 75 Salinispora genome sequences. This obligate marine actinomycete is composed of three closely related species (16, 17) that are clearly delineated using phylogenetic approaches (18). Salinispora spp. share 99% 16S rRNA gene sequence identity (19), thus making them more narrowly defined than many taxa, which can include up to 3% sequence divergence (20). Salinispora spp. are a rich source of secondary metabolites, including salinosporamide A (21), which has undergone a series of phase I clinical trials for the treatment of cancer (22). They devote ca. 10% of their genomic content to secondary metabolism (23, 24) and represent a tractable model with which to address correlations between fine-scale molecular systematics and secondary metabolite production (25). The results presented here describe the diversity and distributions of biosynthetic pathways among a closely related group of bacteria and reveal high levels of pathway acquisition via horizontal gene transfer, with more than half of the pathways occurring in only one or two strains. The data provide evidence of the evolutionary mechanisms that generate new pathway diversity and a striking example of the plasticity of the bacterial secondary metabolome.
Results
Pathway Identification.
Draft and complete genome sequences from 75 Salinispora strains were analyzed (Table S1). These strains encompass the major biogeographic regions from which the three currently described species have been reported (Fig. S1) and include representatives of 11 previously identified 16S rRNA gene sequence variants that differ by as little as a single nucleotide change (26). KS and C domains were extracted from the sequence data and used for the initial identification of PKS and NRPS pathways, respectively. In total, 2,079 KS and 1,693 C domains were detected. Of the KS domains, BLAST, antiSMASH (27), and manual analyses that included the gene environments in which these domains occurred linked 75 to fatty acid biosynthesis (one per strain), whereas 80 were identified as false positives (N-acetyltransferases) and the remaining 1,924 (92.5%) were associated with secondary metabolism. All of the C domains were linked to secondary metabolism.
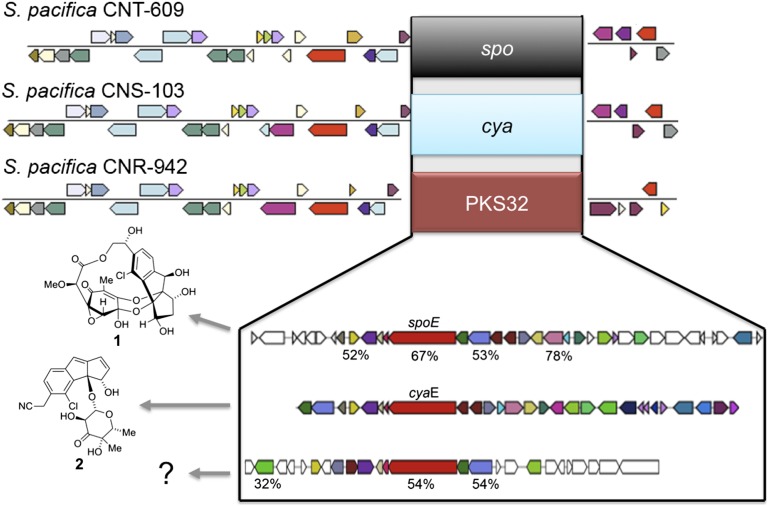
The next step was to assemble pathways that appeared to be split among different contigs, which was generally the case for highly repetitive modular type I PKSs and some NRPSs. This was accomplished using reference pathways from prior studies (23, 24) and better-assembled Salinispora genomes that included seven strains that were assembled into single contigs that exceeded 5 Mb. In the absence of a reference pathway, contigs were assembled when the KS- or C-domain phylogenies indicated close evolutionary relationships. Pathways that contained similar gene content and organization were grouped into “operational biosynthetic units” (OBUs) based on predictions they produced related secondary metabolites. These groups were defined based on sequence identity (SI) values of 90% and 85%, respectively, among homologous KS and C domains (10). The stringency of these cutoff values is supported by the cya and spo enediyne KSs, which share ca. 88% SI yet yield compounds that possess fundamentally different carbon skeletons (Fig. 1) (28, 29). Likewise, homologous C domains associated with NRPS4 and 19 share ca. 80% SI, yet they occur in pathways that differ not only in gene content but also in the composition of the NRPS genes (Fig. S2). In all cases where the secondary metabolic products of the pathways were known, fundamentally different carbon skeletons were observed when the KS- and C-domain SIs fell below these cutoff values. MultiGeneBlast analyses were performed on pathways that occurred in at least five strains to better assess the OBU assignments. These analyses revealed high levels of synteny and SI among the shared genes within each OBU and sharply lower cumulative BLAST bit scores for strains that lacked the pathway. Intra-OBU differences occurred largely among genes predicted to encode tailoring enzymes, as observed in the cya pathway (Fig. 2). Nonetheless, given that minor structural differences can have a major impact on secondary metabolite biological activity, the products of different versions of a pathway that have been grouped into a single OBU may have different ecological functions. Although the KS- and C-domain clustering values used here appear appropriate for Salinispora species, it remains to be seen how well they will apply to other taxonomic groups.
Fig. 1.
Enediyne pathway exchange. Three different OBUs were detected in the same chromosomal position in three different S. pacifica strains. These OBUs were classified as enediyne PKSs based on a NaPDoS analysis of the KS domains derived from the type I PKS genes (in red) in each pathway. The different OBU assignments are supported by the products of the sporolide (spo) and cyanosporaside (cya) gene clusters, which include sporolide A (1) and cyanosporaside A (2), respectively. These compounds, which are shown to the left of the pathways responsible for their production, possess fundamentally different carbon skeletons and are predicted to originate from enediyne precursors (28, 65, 66). Amino acid sequence identities relative to orthologs in the cya pathway are shown for representative genes. Products have yet to be identified from PKS32, which appears at the bottom.
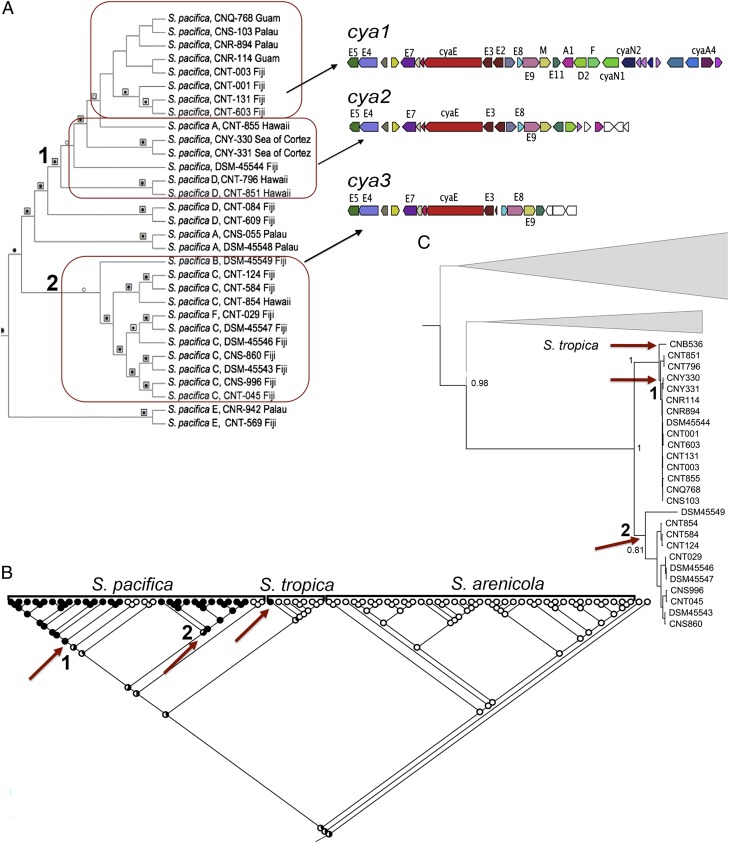
Fig. 2.
Evolution of the cya pathway in S. pacifica. (A) The cya OBU contains three different versions of the pathway. The cya1 version is responsible for the biosynthesis of cyanosporaside A, which is derived from an enediyne precursor (28). It was observed in all strains in the uppermost clade of the Salinispora species tree (boxed in red). Two truncated versions of the pathway were observed, with cya2 appearing ancestral to cya1 in the species tree. Genes missing in cya2 and 3 include cyaA4 (O-acyltransferase), cyaN1 (epoxide hydrolase), and cyaN2 (oxidoreductase), which are predicted to encode tailoring enzymes. Together, the cya1- and 2-containing strains form a single clade (clade 1) in this region of the S. pacifica species tree. The cya3 pathway occurs in a separate S. pacifica lineage (clade 2). Products have yet to be identified from cya2 and 3. (B) A likelihood analysis predicts three independent acquisition events for the cya pathway, one in S. tropica and two in S. pacifica (red arrows). The S. pacifica acquisition events correspond to clades 1 and 2 in the species tree. (C) Maximum-likelihood phylogeny of the cyaE enediyne PKS gene including the top 10 BLASTp matches (bootstrap values for 100 replicates are shown at major nodes) reveals two major lineages (red arrows, numbers 1 and 2) that correspond to the strains in clades 1 and 2 of the species tree. This supports the vertical inheritance of this gene subsequent to acquisition. The position of the S. tropica (CNB-536) cyaE homolog within S. pacifica clade 1 suggests that the acquisition event in S. tropica is the result of horizontal gene transfer with S. pacifica clade 1. Gene gain and loss are assumed to account for the variations in the cya1–3 pathways.
Pathway Diversity.
Comparable to prior studies (23, 24), the Salinispora genomes were enriched in PKS and NRPS biosynthetic pathways. On average, S. arenicola genomes were 300 kb larger and contained four more OBUs per genome than S. pacifica or S. tropica (Table 1). Although more OBUs were detected per S. arenicola genome, considerably more OBU diversity was observed in S. pacifica, which contained a total of 88 different OBUs compared with 47 and 19 for S. arenicola and S. tropica, respectively. In total, 124 distinct OBUs were identified, including representatives of diverse PKS types (Fig. S3). Only nine of these OBUs have been formally linked to the production of specific secondary metabolites. These are sal (salinosporamides) (30), slm (salinilactam) (23), cyl (cyclomarins) (31), cya (cyanosporasides) (28), spo (sporolides) (29), arn (arenimycin) (32), rif (rifamycins) (33), lym (lymphostin) (34), and lom (lomaiviticin) (35), whereas two others are predicted to yield enterocin (36) (PKS31) and arenicolide (37) (PKS28) based on bioinformatic analyses. Although there is no evidence that all pathways are functional, the 113 remaining OBUs far exceed the four Salinispora secondary metabolites (arenamides, pacificanones, salinipyrones, and saliniquinones) that have yet to be linked to specific pathways, suggesting that considerable chemical diversity remains to be discovered from this genus.
Table 1.
Salinispora genome composition and pathway (OBU) statistics
| Species | No. genomes analyzed | Avg. genome size, Mb | Avg. no. contigs* | Avg. no. OBUs per genome | OBU richness† | Avg. no. singletons‡ per genome |
| S. arenicola | 37 | 5.7 ± 0.14 | 78 ± 19 | 17.5 ± 1.9 | 47 | 0.49 |
| S. pacifica | 31 | 5.4 ± 0.19 | 93 ± 33 | 14.1 ± 2.7 | 88 | 1.00 |
| S. tropica | 7 | 5.4 ± 0.19 | 90 ± 19 | 13.6 ± 1.8 | 19 | 0.57 |
Averages reported are ±1 SD.
Does not include the closed genomes of CNB-440 and CNS-205.
Number of different OBUs observed.
OBUs observed in only one strain.
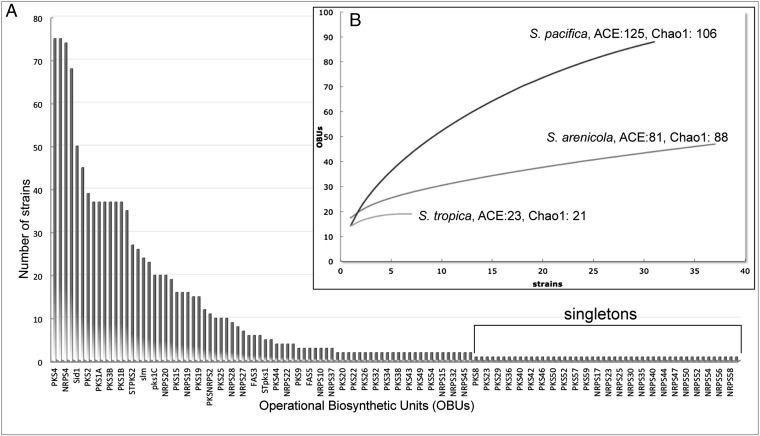
A rank-abundance curve describing the distribution of the OBUs among the 75 strains reveals a long right-hand tail, as is characteristic of a highly diverse community (Fig. 3). Remarkably, 48 of the OBUs were only observed in one strain (singletons), with an additional 24 occurring in two strains. These 72 OBUs account for 58% of the total number observed in the 75 genomes and illustrate extensive acquisition via horizontal gene transfer. In the case of S. pacifica, the most phylogenetically diverse of the three species (26), an average of one singleton was detected per genome sequenced (Table 1). Rarefaction curves, used primarily in community ecology to assess species richness (38), provide an assessment of OBU richness for the given sequencing effort and reveal that considerable diversity has yet to be detected (Fig. 3). This is particularly evident for S. pacifica, which shows little evidence of saturation, and is further supported by ACE and Chao1 diversity estimators, which predict as many as 229 distinct OBUs with continued sequencing of the three species (Fig. 3). This represents an extraordinary level of biosynthetic diversity for three bacterial species that share 99% 16S rRNA sequence identity (19).
Fig. 3.
Distribution and diversity of PKS and NRPS OBUs. (A) Rank-abundance curve showing the abundance of each OBU among the 75 strains analyzed (representative OBU names are shown). (B) Rarefaction curves with diversity estimators for each species.
Pathway Distributions.
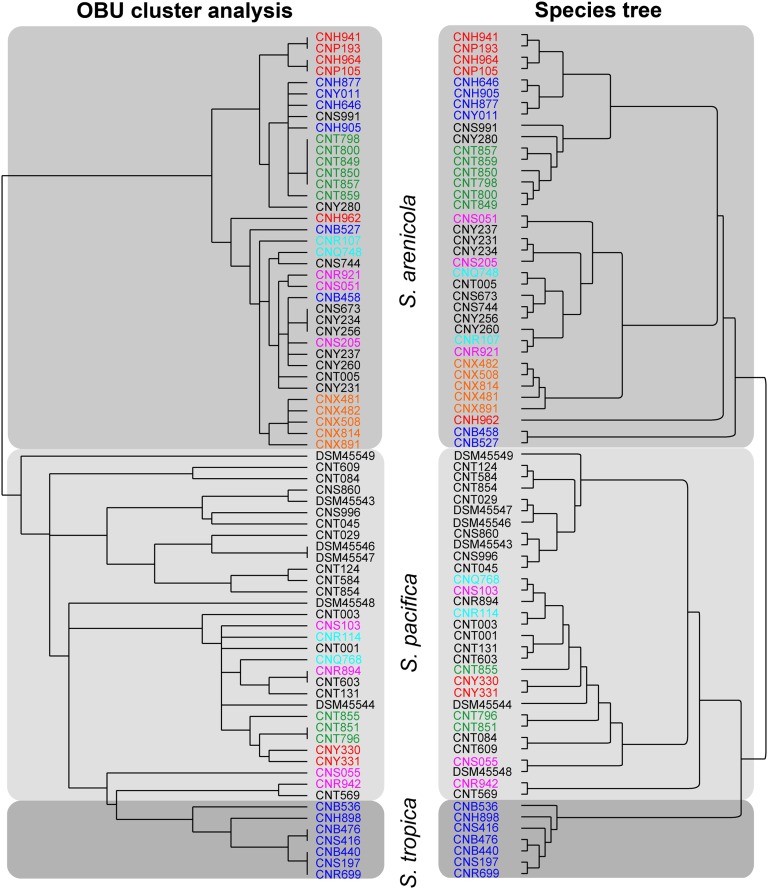
We next generated a well-supported Salinispora species phylogeny (Fig. S4) and a hierarchical cluster analysis based on pathway presence or absence. Despite the large number of OBUs that occur in only one or two strains, these two dendrograms are highly congruent, with the exception that S. pacifica is paraphyletic with respect to S. tropica in the OBU cluster analysis (Fig. 4). Contributing to this congruence are species-specific OBUs (i.e., pathways commonly observed in one species but generally not in others). In S. arenicola, these include rif, PKS1A/B, PKS2, PKS3A/B, PKS5, NRPS1, and NRPS2 (Table S2). In S. tropica, these include spo, slm, sal, Sid3, Sid4, NRPS3, and STPKS1 (Table S3). Interestingly, only one OBU (NRPS20) appears commonly in S. pacifica and not in others (Table S4). These results support previous culture-based studies and KS fingerprinting analyses that revealed species-specific patterns of secondary metabolite production and gene distributions in S. arenicola and S. tropica (25, 39). There is some evidence of OBU clustering based on the location from which the strains originate (Fig. 4); however, a permutational multivariate analysis of variance (PERMANOVA) revealed a significant correlation between OBU and species (R2 = 0.54, P = 0.001) and not location (P = 0.075), indicating the importance of taxonomy over biogeographic origin in terms of OBU distributions.
Fig. 4.
OBU hierarchical cluster analysis and Salinispora species tree. Hierarchical cluster analysis based on OBU presence or absence. Maximum-likelihood species phylogeny generated from 10 housekeeping genes. Colors indicate the collection site: blue, Bahamas; pink, Palau; black, Fiji; red, Sea of Cortez; orange, Palmyra; turquoise, Guam; green, Hawaii.
Pathway Evolution.
To explore the evolutionary history of the pathways in relation to the strains in which they reside, likelihood analyses were performed on the KS- and C-domain sequences to assign the ancestral node(s) for each OBU in the species tree. The results for the 124 OBUs were overlaid onto a simplified Salinispora phylogeny (Fig. 5) generated by collapsing the species tree (Fig. S4) into 12 lineages. The analysis reveals that only five OBUs were present in the common ancestor of the genus and only two of these (FAS1 and PKS4) were shared with the closely related genus Micromonospora. It can thus be inferred that the remaining pathways (96% of the total) were acquired by HGT at various points during the evolution of the genus. Phylogenetic analyses of key biosynthetic genes from each OBU confirm these evolutionary histories and indicate, based on congruence with the species tree, vertical inheritance for 65 of the OBUs subsequent to acquisition. Seven OBUs appear to have been acquired early in the evolution of S. arenicola. These include rif, which supports the consistent production of rifamycins by S. arenicola (25, 40). Likewise, six OBUs appear early in the evolutionary history of S. tropica and one in S. pacifica (Fig. 5). Most of the OBUs, however, were acquired relatively recently in the evolution of the genus, appearing toward the branch terminals in the tree. Based on BLAST analyses of the singleton PKS and NRPS genes, it appears that most of these pathways were acquired from other high-G+C bacteria such as Streptomyces spp. (Fig. S5), which also occur in marine sediments (41). The results for PKS17 suggest the independent acquisition of this pathway by four S. arenicola strains from Fiji and one S. pacifica strain from the Sea of Cortez (Fig. S6). Although these results may reflect sampling effort, they suggest that location-dependent pathway acquisition warrants future study.
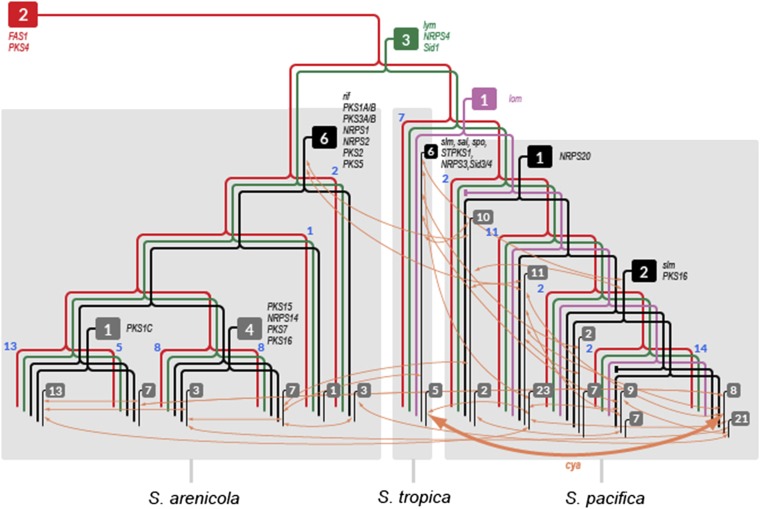
Fig. 5.
Salinispora phylogeny depicting OBU inferred ancestry. A simplified species tree generated from 10 housekeeping genes (Fig. S4) shows 12 major Salinispora lineages with the number of strains in each indicated in blue adjacent to the node branch points. Boxes indicate the number of OBUs originating at various points in the species tree. Red, shared with a common ancestor of the genus; green, genus-specific; purple, shared with S. tropica and S. pacifica; black, species-specific; gray, clade-specific. Representative OBU names are indicated next to the point of acquisition. Orange arrows describe inter- and intraspecies OBU exchange events (cya exchange between S. tropica and S. pacifica is indicated in bold).
Phylogenetic analyses of key biosynthetic genes were also used to infer that 36 of the 124 OBUs identified (29%) were exchanged within or between species. One example is the cya pathway, which was exchanged between S. pacifica and S. tropica (Fig. 2). These transfer events were added to the simplified species tree to depict the complexity of pathway movement within the genus (Fig. 5). In total, it could be inferred that 23 OBUs moved once, 9 moved twice, and 4 (PKS17, sal, Sid1, and PKSNRPS2) moved three times. There was no evidence for KS- or C-domain exchange among OBUs or the formation of chimeric pathways, although events of these types may have been missed with the assembly methods used. Instead, OBUs evolved largely by gene gain, gene loss, and duplication followed by divergence. In the last case, NRPS4 is a genus-specific pathway observed in 72 of the 75 strains. A subset of S. arenicola (clade 6) and S. pacifica (clade 12) contains a second copy of this pathway (NRPS19) that is sufficiently diverged (i.e., shares <85% C-domain SI) to be considered a new OBU (Fig. S2). Thus, pathway duplication followed by divergence appears to be another mechanism by which OBU diversity is created in Salinispora spp.
Genomic Islands as Hot Spots for Secondary Metabolism.
Pseudochromosomes were generated by mapping sequence contigs onto the closed genomes of S. tropica (CNB-440), S. arenicola (CNS-205), and a number of high-quality S. pacifica draft genomes that were generated as part of this study. The results show that the OBUs are clustered in genomic islands (GIs) (Fig. 6), regions of bacterial chromosomes known to encode acquired, adaptive traits (42). Salinispora GIs were also enriched in mobile genetic elements, which may play a role in OBU acquisition and transfer, relative to other regions of the genome (Wilcoxon rank-sum test, P < 0.05). Remarkably, the flanking regions of 21 previously identified Salinispora GIs (24) are conserved across all 75 genome sequences, suggesting that island boundaries can be used as queries to identify similar regions in other strains. In some cases, OBUs that encode the biosynthesis of similar classes of compounds were exchanged at precisely the same island location. This type of pathway “swapping” was observed with three enediyne OBUs in S. pacifica (Fig. 1), and may represent an example of pathway-level homologous recombination that is yet to be described.
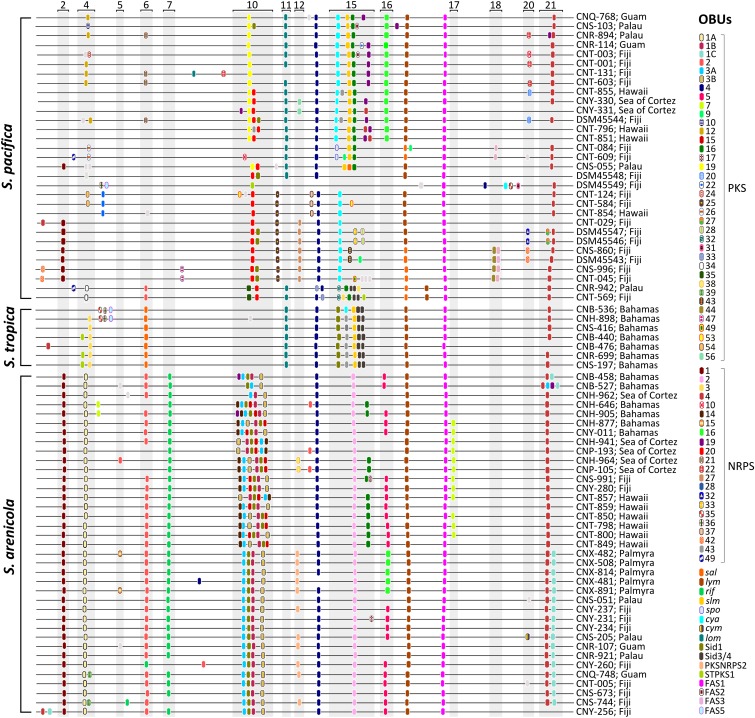
Fig. 6.
Linear pseudochromosomes reveal the positioning of Salinispora OBUs within genomic islands (numbered and shaded) as identified based on previously defined boundaries (24). Pathway positions were mapped using PKS and NRPS genes as reference. Only the pathways that could be mapped onto the pseudochromosomes are depicted. Pathways are color-coded and listed on the right next to the strain name and geographic origin. Singletons are depicted in light gray.
Discussion
Major advances in our understanding of the molecular genetics of natural product biosynthesis have created unprecedented opportunities for pathway engineering (43) and the generation of new chemical diversity in high-priority scaffolds (44). Coupled with increased access to genome sequence data and the revelation that even well-studied taxa can harbor a wealth of biosynthetic pathways for which the products have yet to be discovered (45, 46), natural product research is undergoing a renaissance driven by the development of new discovery methods (47). Despite these advances, we have yet to gain perspective on the diversity and distributions of the pathways responsible for secondary metabolism among groups of related bacteria and how these pathways evolve to generate new chemical diversity.
The 75 genome sequences analyzed here provide insight into the remarkable levels of pathway diversity that can be maintained among a group of bacteria that share 99% 16S rRNA gene sequence identity. This diversity can largely be attributed to the many pathways that were observed in only one or two strains and that are inferred to be the result of HGT events that occurred relatively recently in the evolutionary history of the genus. Although the effects of geographic origin on the OBUs maintained by individual strains warrant further study, the potential for location-specific acquisition suggests that differences in the local gene pool may account for some of the diversity reported here. Although the total number of OBUs maintained by these three closely related species remains unknown, it is extraordinarily high relative to the numbers observed in the individual strains (Table 1), with a total of 229 distinct PKS and NRPS OBUs predicted with continued sequencing.
Mapping the inferred ancestral nodes of the individual OBUs onto the Salinispora species phylogeny made it possible to trace pathway evolutionary histories relative to the strains in which they reside (Fig. 5). These analyses reveal that 105 of the 124 OBUs (85%) were acquired subsequent to the speciation events within the genus, which suggests that the ecological functions of secondary metabolites act largely at the subspecies level. However, the congruence observed between the species tree and the OBU cluster analysis (Fig. 4) suggests that secondary metabolites nonetheless represent functional traits that help define Salinispora spp. (25). The fixation of certain pathways within S. arenicola and S. tropica could be the result of periodic selection (48), which if driven by the products of these OBUs would indicate that they provide a strong selective advantage. Species-specific OBUs include rif and sal, which encode the production of the potent antibiotic rifamycin and the proteasome inhibitor salinosporamide A in S. arenicola and S. tropica, respectively. In S. pacifica, the most diverse of the three species (26), similar levels of fixation are not observed, yet many OBUs appear fixed among major clades within the species. Based on this, it could be speculated that S. pacifica is undergoing a series of nascent speciation events, with ecological divergence preventing periodic selection from fixing pathways at the currently defined species level.
The OBUs were concentrated in GIs whose boundaries were highly conserved among all strains. These GIs were enriched in mobile genetic elements, suggesting they are hot spots for pathway acquisition and evolution. The observed swapping of enediyne OBUs at the precise chromosomal locations in different strains (Fig. 1) suggests that recombination may function at the pathway level in a manner comparable to the domain-level homologous recombination observed in PKS and NRPS analyses (49). The absence of KS- or C-domain exchange among OBUs, a process that is generally considered important in PKS and NRPS evolution (5, 50), suggests that pathway HGT followed by gene gain or loss events is the major force driving the creation of OBU diversity in Salinispora spp. Although it is unclear how these results apply to other bacteria, the continued sequencing of large numbers of closely related strains will provide additional insight into the evolutionary processes by which bacteria generate new secondary metabolite diversity.
A better understanding of the taxonomic distributions and evolutionary histories of the pathways responsible for secondary metabolite biosynthesis will provide opportunities for the development of theory-based sampling strategies that capitalize on the genetic potential of individual strains to produce new chemical scaffolds or compounds within a privileged chemical class. Recognition that some pathways diverge in lineage-specific patterns indicates that related strains within the same species can be the source of related compounds within the same chemical class (39), thus providing an alternative to synthetic chemistry as an approach to generating structural diversity. The plasticity of secondary metabolism in Salinispora spp. provides a glimpse into the evolutionary strategies by which bacteria capitalize on the benefits afforded by these compounds. Despite not knowing the ecological functions of most Salinispora secondary metabolites, extensive pathway sampling provides a mechanism to maximize the population-level secondary metabolome while limiting the number of pathways maintained within any individual genome. The potentially vast array of molecules produced at the population level would increase the likelihood of an effective response to new selective pressures and thus provide an ecological rationale for the extensive pathway diversity observed in this study.
Materials and Methods
Genome Sequencing and Assembly.
Salinispora strains were obtained in culture as previously described (41, 51). DNA was extracted following US Department of Energy Joint Genome Institute (JGI) protocols (http://my.jgi.doe.gov/general/protocols.html) and submitted to the JGI for sequencing, assembly, and annotation. The sequencing and annotation of S. arenicola CNS-205 and S. tropica CNB-440 were as previously described (23, 24). For the remaining 73 strains, short- and long-insert paired-end libraries were constructed and sequenced by the JGI using the Illumina HiSeq 2000 system. Filtered reads were assembled using Velvet (52) and ALLPATHS-LG (53), and possible misassemblies were corrected with manual editing in Consed (54). Gap closure was accomplished using repeat resolution software and sequencing of bridging PCR fragments with Sanger and/or PacBio technologies. Genes were identified using Prodigal (55), followed by a round of manual curation using GenePRIMP (56). Predicted coding DNA sequences were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant (nr) database, UniProt, TIGRFam, Pfam, Kyoto Encyclopedia of Genes and Genomes, Clusters of Orthologous Groups, and InterPro databases. Strains and accession numbers are provided in Table S1.
Pathway and OBU Identification.
Genome sequences in FASTA format were screened for PKS and NRPS genes by searching for KS and C domains, respectively, using NaPDoS (http://napdos.ucsd.edu) with default settings (57). The associated genes and gene environments were then analyzed using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi), IMG/ER (https://img.jgi.doe.gov/cgi-bin/er/main.cgi), and antiSMASH (27) to confirm association with secondary metabolism, assess the similarities among pathways, and create links to known secondary metabolites based on homology to experimentally characterized pathways. Pathways split onto different contigs were assembled with the aid of complete pathways or when KS- and C-domain phylogenies revealed that the sequences claded together. Pathways were grouped into OBUs when BLAST analyses revealed that homologous KS and C domains shared ≥90% and ≥85% amino acid sequence identity, respectively. OBUs were assigned a unique identifier (e.g., PKS1, NRPS1) or a formal name if linked to an experimentally characterized pathway. A database of all genomes was created and OBU assignments were verified for pathways that occurred in five or more strains using MultiGeneBlast (58) based on the synteny and SI of conserved genes in each pathway and cumulative BLAST bit scores, which dropped precipitously in strains that did not possess the pathway (10).
Salinispora Species Phylogeny.
Nucleotide sequences for 10 unlinked, single-copy genes (dnaA, gyrB, pyrH, recA, pgi, trpB, atpD, sucC, rpoB, topA) were extracted, aligned using Muscle in Geneious Pro v5.5 (Biomatters; www.geneious.com), and concatenated using Mesquite v2.75 (59). MODELTEST (60) was run and the best model [generalized time reversible (GTR)+G] was used to create a maximum-likelihood (ML) tree using PhyML 3.0 (61) and a neighbor-joining tree using MEGA5 (62). Nodal support values were obtained using 1,000 bootstrap replicates. Concatenated Bayesian tree and posterior probabilities were created using MRBAYES (63) with 1 million generations.
OBU Phylogeny.
Nucleotide sequences from at least two conserved genes from each OBU observed in two or more of the 12 major Salinispora clades presented in Fig. 5 were aligned in Muscle and manually curated. ML phylogenies were created using PhyML 3.0 under the GTR model of nucleotide substitution with 100 bootstrap replicates or a fast approximate likelihood-ratio test performed as a measure of branch support. Other common models of nucleotide substitution were used with no significant changes in the results. If the phylogenies for the genes within an OBU were congruent, this phylogeny was assumed for the whole pathway.
Statistics.
Hierarchical cluster analyses were performed using Cluster 3.0 (http://bonsai.hgc.jp/∼mdehoon/software/cluster/software) with presence/absence OBU matrices as the input files (Tables S2–S4) using a correlation-centered similarity metric with the complete linkage clustering method. A PERMANOVA was implemented with the vegan package in R (www.r-project.org). EstimateS (http://viceroy.eeb.uconn.edu/estimates) was used to generate rarefaction curves and diversity estimates. The Wilcoxon rank-sum test implemented in R was used to compare the fraction of mobile genetic elements inside and outside of GIs.
Ancestral State Reconstruction.
The ancestral node for each OBU was inferred in the species tree using the trace character history function implemented in Mesquite v2.75 (59). A categorical character matrix was created for all OBUs, and likelihood calculations were performed using the Mk1 model. Likelihood scores >50% were used to infer the points of OBU acquisition (ancestral nodes) in the species tree. OBU ML phylogenies were used to corroborate points of acquisition based on congruence with the species tree and to infer inter- and intraspecies exchange events as shown in Fig. 2.
Pseudochromosome Assembly, OBU Localization, and Genomic Island Analysis.
Draft genomes were assembled into linear “pseudochromosomes” using the CONTIGuator 2 web application (64) and oriented with dnaA as the first gene. The closed genomes S. arenicola CNS-205 (24) and S. tropica CNB-440 (23) were used as templates for the assembly of these species. High-quality draft S. pacifica genomes (one 5-Mb scaffold and one to three contigs of 10–100 kb) from strains DSM-45544, DSM-45548, and DSM-45543 were used as reference templates for the assembly of S. pacifica phylotypes ST, A, and C. For other phylotypes, the template that gave the best assembly was used. The chromosomal position of the OBUs present in ≥3 strains was determined using the Assembly function in Geneious Pro v5.5 and a PKS or NRPS gene from the predicted OBU as reference. All remaining OBUs were mapped by searching for KS- and C-domain amino acid sequences using Custom-BLAST in Geneious Pro v5.5. In a previous study of S. arenicola CNS-205 and S. tropica CNB-440, 21 GIs were identified based on regions of conservation flanking regions >20 kb that shared <40% gene orthology (24). Conserved regions 5 kb up- and downstream of genomic islands were extracted from CNS-205 and located in the pseudochromosomes by BLAST in Geneious Pro v5.5. Mobile genetic elements were quantified in closed and high-quality draft genomes (S. arenicola CNS-205 and CNS-991, S. tropica CNB-440, and S. pacifica DSM-45543, DSM-45544, DSM-45546, DSM-45547, DSM-45548, and DSM-45549) by counting annotated recombinase, transposase, phage, integrase, and tRNA genes inside and outside of GIs.
Source of Singleton OBUs.
All KS and C domains that occurred in one Salinispora strain (singletons) were subjected to BLAST analyses using the NCBI/nr protein database to assess the taxonomic distribution of homologous domains in other microorganisms. A total of 330 KS domains (from 16 pathways) and 1,100 C domains (from 26 pathways) was analyzed. The top 10 BLAST hits of every query were sorted by taxonomy in Geneious Pro v5.5 to calculate the distribution per taxonomic group.
Supplementary Material
Acknowledgments
We acknowledge the Joint Genome Institute/Community Sequencing Program for providing sequence data, assembly, and annotation and for helpful advice on sample preparation. Greg Rouse and Nastassia Patin are acknowledged for assistance with the phylogenetic and statistical analyses, respectively. Brad Moore is acknowledged for helpful discussions about the data. Kelley Gallagher, Anindita Sarkar, Eun Ju Choi, Kevin Penn, and Nastassia Patin assisted with DNA extractions. Financial support was provided by the National Institutes of Health under Grants U01-TW0007401, GM085770, and GM086261 (to P.R.J.); the National Science Foundation Graduate Research Fellowship under Grant DGE-1144086 (to K.L.C.); and the German Academic Exchange Service [Deutscher Akademischer Austauschdienst (DAAD)] (to M.W.). The work conducted by the US Department of Energy Joint Genome Institute is supported by the Office of Science of the US Department of Energy under Contract no. DE-AC02-05CH11231.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: Genome sequences reported in this paper have been deposited in the Joint Genome Institute’s Integrated Microbial Genomes (IMG) database, http://img.jgi.doe.gov/cgi-bin/w/main.cgi (accession nos. 2517572210, 2515154093, 2515154181, 2519103192, 2515154183, 2515154193, 2519103193, 2515154125, 2518285551, 2518285552, 2515154180, 2519103194, 2515154203, 641228504, 2516143022, 2518285553, 2519103185, 2518285554, 2517572137, 2515154186, 2515154088, 2515154135, 2515154127, 2517572233, 2518285555, 2515154137, 2515154188, 2517572152, 2515154187, 2517572153, 2518285558, 2519103195, 2518285559, 2518285560, 2517572154, 2517572155, 2515154178, 2515154194, 2518285561, 2518285562, 2515154129, 2518285563, 2517572157, 2515154184, 2515154126, 2515154177, 2517572158, 2515154202, 2517572159, 2515154200, 2515154124, 2517572160, 2515154185, 2517572161, 2515154182, 2518285550, 2517572162, 2515154170, 2515154128, 2517572163, 2518645626, 2518645627, 2517572194, 2517287019, 2516653042, 2516493032, 2517287023, 2517434008, 640427140, 2517572211, 2517572212, 2515154094, 2518645624, 2515154163, and 2517572164).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1324161111/-/DCSupplemental.
References
- 1.Bérdy J. Bioactive microbial metabolites. J Antibiot (Tokyo) 2005;58(1):1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 2.Wietz M, Duncan K, Patin NV, Jensen PR. Antagonistic interactions mediated by marine bacteria: The role of small molecules. J Chem Ecol. 2013;39(7):879–891. doi: 10.1007/s10886-013-0316-x. [DOI] [PubMed] [Google Scholar]
- 3.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem Rev. 2006;106(8):3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 4.Jenke-Kodama H, Dittmann E. Evolution of metabolic diversity: Insights from microbial polyketide synthases. Phytochemistry. 2009;70(15-16):1858–1866. doi: 10.1016/j.phytochem.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 5.Fischbach MA, Walsh CT, Clardy J. The evolution of gene collectives: How natural selection drives chemical innovation. Proc Natl Acad Sci USA. 2008;105(12):4601–4608. doi: 10.1073/pnas.0709132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hertweck C. The biosynthetic logic of polyketide diversity. Angew Chem Int Ed Engl. 2009;48(26):4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 7.Mootz HD, Schwarzer D, Marahiel MA. Ways of assembling complex natural products on modular nonribosomal peptide synthetases. ChemBioChem. 2002;3(6):490–504. doi: 10.1002/1439-7633(20020603)3:6<490::AID-CBIC490>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 8.Jenke-Kodama H, Sandmann A, Müller R, Dittmann E. Evolutionary implications of bacterial polyketide synthases. Mol Biol Evol. 2005;22(10):2027–2039. doi: 10.1093/molbev/msi193. [DOI] [PubMed] [Google Scholar]
- 9.Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc Natl Acad Sci USA. 2003;100(26):15670–15675. doi: 10.1073/pnas.2532165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ziemert N, Jensen PR. Phylogenetic approaches to natural product structure prediction. Methods Enzymol. 2012;517:161–182. doi: 10.1016/B978-0-12-404634-4.00008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rausch C, Hoof I, Weber T, Wohlleben W, Huson DH. Phylogenetic analysis of condensation domains in NRPS sheds light on their functional evolution. BMC Evol Biol. 2007;7:78. doi: 10.1186/1471-2148-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ginolhac A, et al. Type I polyketide synthases may have evolved through horizontal gene transfer. J Mol Evol. 2005;60(6):716–725. doi: 10.1007/s00239-004-0161-1. [DOI] [PubMed] [Google Scholar]
- 13.Metsä-Ketelä M, et al. Molecular evolution of aromatic polyketides and comparative sequence analysis of polyketide ketosynthase and 16S ribosomal DNA genes from various Streptomyces species. Appl Environ Microbiol. 2002;68(9):4472–4479. doi: 10.1128/AEM.68.9.4472-4479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doroghazi JR, Metcalf WW. Comparative genomics of actinomycetes with a focus on natural product biosynthetic genes. BMC Genomics. 2013;14:611. doi: 10.1186/1471-2164-14-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corre C, Challis GL. New natural product biosynthetic chemistry discovered by genome mining. Nat Prod Rep. 2009;26(8):977–986. doi: 10.1039/b713024b. [DOI] [PubMed] [Google Scholar]
- 16.Ahmed L, et al. Salinispora pacifica sp. nov., an actinomycete from marine sediments. Antonie van Leeuwenhoek. 2013;103(5):1069–1078. doi: 10.1007/s10482-013-9886-4. [DOI] [PubMed] [Google Scholar]
- 17.Maldonado LA, et al. Salinispora arenicola gen. nov., sp. nov. and Salinispora tropica sp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int J Syst Evol Microbiol. 2005;55(Pt 5):1759–1766. doi: 10.1099/ijs.0.63625-0. [DOI] [PubMed] [Google Scholar]
- 18.Freel KC, Millán-Aguiñaga N, Jensen PR. Multilocus sequence typing reveals evidence of homologous recombination linked to antibiotic resistance in the genus Salinispora. Appl Environ Microbiol. 2013;79(19):5997–6005. doi: 10.1128/AEM.00880-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen PR, Mafnas C. Biogeography of the marine actinomycete Salinispora. Environ Microbiol. 2006;8(11):1881–1888. doi: 10.1111/j.1462-2920.2006.01093.x. [DOI] [PubMed] [Google Scholar]
- 20.Gevers D, et al. Re-evaluating prokaryotic species. Nat Rev Microbiol. 2005;3(9):733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- 21.Feling RH, et al. Salinosporamide A: A highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew Chem Int Ed Engl. 2003;42(3):355–357. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- 22.Fenical W, et al. Discovery and development of the anticancer agent salinosporamide A (NPI-0052) Bioorg Med Chem. 2009;17(6):2175–2180. doi: 10.1016/j.bmc.2008.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Udwary DW, et al. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc Natl Acad Sci USA. 2007;104(25):10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Penn K, et al. Genomic islands link secondary metabolism to functional adaptation in marine Actinobacteria. ISME J. 2009;3(10):1193–1203. doi: 10.1038/ismej.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen PR, Williams PG, Oh DC, Zeigler L, Fenical W. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl Environ Microbiol. 2007;73(4):1146–1152. doi: 10.1128/AEM.01891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freel KC, Edlund A, Jensen PR. Microdiversity and evidence for high dispersal rates in the marine actinomycete ‘Salinispora pacifica’. Environ Microbiol Rep. 2012;14(2):480–493. doi: 10.1111/j.1462-2920.2011.02641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Medema MH, et al. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39(Web Server issue) Suppl 2:W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lane AL, et al. Structures and comparative characterization of biosynthetic gene clusters for cyanosporasides, enediyne-derived natural products from marine actinomycetes. J Am Chem Soc. 2013;135(11):4171–4174. doi: 10.1021/ja311065v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGlinchey RP, Nett M, Moore BS. Unraveling the biosynthesis of the sporolide cyclohexenone building block. J Am Chem Soc. 2008;130(8):2406–2407. doi: 10.1021/ja710488m. [DOI] [PubMed] [Google Scholar]
- 30.Eustáquio AS, et al. Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-coenzyme A from S-adenosyl-L-methionine. Proc Natl Acad Sci USA. 2009;106(30):12295–12300. doi: 10.1073/pnas.0901237106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schultz AW, et al. Biosynthesis and structures of cyclomarins and cyclomarazines, prenylated cyclic peptides of marine actinobacterial origin. J Am Chem Soc. 2008;130(13):4507–4516. doi: 10.1021/ja711188x. [DOI] [PubMed] [Google Scholar]
- 32.Kersten RD, et al. Glycogenomics as a mass spectrometry-guided genome-mining method for microbial glycosylated molecules. Proc Natl Acad Sci USA. 2013;110(47):E4407–E4416. doi: 10.1073/pnas.1315492110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson MC, Gulder TAM, Mahmud T, Moore BS. Shared biosynthesis of the saliniketals and rifamycins in Salinispora arenicola is controlled by the sare1259-encoded cytochrome P450. J Am Chem Soc. 2010;132(36):12757–12765. doi: 10.1021/ja105891a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyanaga A, et al. Discovery and assembly-line biosynthesis of the lymphostin pyrroloquinoline alkaloid family of mTOR inhibitors in Salinispora bacteria. J Am Chem Soc. 2011;133(34):13311–13313. doi: 10.1021/ja205655w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kersten RD, et al. Bioactivity-guided genome mining reveals the lomaiviticin biosynthetic gene cluster in Salinispora tropica. ChemBioChem. 2013;14(8):955–962. doi: 10.1002/cbic.201300147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piel J, et al. Cloning, sequencing and analysis of the enterocin biosynthesis gene cluster from the marine isolate ‘Streptomyces maritimus’: Evidence for the derailment of an aromatic polyketide synthase. Chem Biol. 2000;7(12):943–955. doi: 10.1016/s1074-5521(00)00044-2. [DOI] [PubMed] [Google Scholar]
- 37.Williams PG, Miller ED, Asolkar RN, Jensen PR, Fenical W. Arenicolides A-C, 26-membered ring macrolides from the marine actinomycete Salinispora arenicola. J Org Chem. 2007;72(14):5025–5034. doi: 10.1021/jo061878x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gotelli NJ, Colwell RK. Quantifying biodiversity: Procedures and pitfalls in the measurement and comparison of species richness. Ecol Lett. 2001;4(4):379–391. [Google Scholar]
- 39.Freel KC, Nam S-J, Fenical W, Jensen PR. Evolution of secondary metabolite genes in three closely related marine actinomycete species. Appl Environ Microbiol. 2011;77(20):7261–7270. doi: 10.1128/AEM.05943-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim TK, Hewavitharana AK, Shaw PN, Fuerst JA. Discovery of a new source of rifamycin antibiotics in marine sponge actinobacteria by phylogenetic prediction. Appl Environ Microbiol. 2006;72(3):2118–2125. doi: 10.1128/AEM.72.3.2118-2125.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jensen PR, Gontang E, Mafnas C, Mincer TJ, Fenical W. Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environ Microbiol. 2005;7(7):1039–1048. doi: 10.1111/j.1462-2920.2005.00785.x. [DOI] [PubMed] [Google Scholar]
- 42.Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2(5):414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 43.Walsh CT. Combinatorial biosynthesis of antibiotics: Challenges and opportunities. ChemBioChem. 2002;3(2-3):125–134. doi: 10.1002/1439-7633(20020301)3:2/3<124::AID-CBIC124>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 44.Weissman KJ, Leadlay PF. Combinatorial biosynthesis of reduced polyketides. Nat Rev Microbiol. 2005;3(12):925–936. doi: 10.1038/nrmicro1287. [DOI] [PubMed] [Google Scholar]
- 45.Bentley SD, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417(6885):141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 46.Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26(11):1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koehn FE, Carter GT. The evolving role of natural products in drug discovery. Nat Rev Drug Discov. 2005;4(3):206–220. doi: 10.1038/nrd1657. [DOI] [PubMed] [Google Scholar]
- 48.Cohan FM. What are bacterial species? Annu Rev Microbiol. 2002;56:457–487. doi: 10.1146/annurev.micro.56.012302.160634. [DOI] [PubMed] [Google Scholar]
- 49.Jenke-Kodama H, Dittmann E. Bioinformatic perspectives on NRPS/PKS megasynthases: Advances and challenges. Nat Prod Rep. 2009;26(7):874–883. doi: 10.1039/b810283j. [DOI] [PubMed] [Google Scholar]
- 50.Hopwood DA. Genetic contributions to understanding polyketide synthases. Chem Rev. 1997;97(7):2465–2498. doi: 10.1021/cr960034i. [DOI] [PubMed] [Google Scholar]
- 51.Gontang EA, Fenical W, Jensen PR. Phylogenetic diversity of Gram-positive bacteria cultured from marine sediments. Appl Environ Microbiol. 2007;73(10):3272–3282. doi: 10.1128/AEM.02811-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gnerre S, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci USA. 2011;108(4):1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gordon D, Abajian C, Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8(3):195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 55.Hyatt D, et al. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pati A, et al. GenePRIMP: A gene prediction improvement pipeline for prokaryotic genomes. Nat Methods. 2010;7(6):455–457. doi: 10.1038/nmeth.1457. [DOI] [PubMed] [Google Scholar]
- 57.Ziemert N, et al. The natural product domain seeker NaPDoS: A phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS ONE. 2012;7(3):e34064. doi: 10.1371/journal.pone.0034064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Medema MH, Takano E, Breitling R. Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol Biol Evol. 2013;30(5):1218–1223. doi: 10.1093/molbev/mst025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Maddison WP, Maddison DR (2011) Mesquite: A Modular System for Evolutionary Analysis, Version 2.75. Available at http://mesquiteproject.org. Accessed February 17, 2014.
- 60.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 61.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 62.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 64.Galardini M, Biondi EG, Bazzicalupo M, Mengoni A. CONTIGuator: A bacterial genomes finishing tool for structural insights on draft genomes. Source Code Biol Med. 2011;6:11. doi: 10.1186/1751-0473-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oh D-C, Williams PG, Kauffman CA, Jensen PR, Fenical W. Cyanosporasides A and B, chloro- and cyano-cyclopenta[a]indene glycosides from the marine actinomycete “Salinispora pacifica”. Org Lett. 2006;8(6):1021–1024. doi: 10.1021/ol052686b. [DOI] [PubMed] [Google Scholar]
- 66.Fenical W, Jensen PR. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat Chem Biol. 2006;2(12):666–673. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.